Geographical and Ecological Analysis of Resistance, Coresistance, and Coupled Resistance to Antimicrobials in Respiratory Pathogenic Bacteria in Spain (original) (raw)
Abstract
A multicenter susceptibility surveillance (the S.A.U.C.E. project) including 2,721 Streptococcus pneumoniae, 3,174 Streptococcus pyogenes, and 2,645 Haemophilus influenzae consecutive isolates was carried out in 25 hospitals all over Spain from November 2001 to October 2002 to evaluate the current epidemiology of resistance of the main bacteria involved in community-acquired respiratory tract infections. Susceptibility testing was performed in a single centralized laboratory by a broth microdilution method. The prevalence of resistant S. pneumoniae strains was 0.4% for cefotaxime, 4.4% for amoxicillin and amoxicillin-clavulanic acid, 25.6% for cefuroxime-axetil, 34.5% for erythromycin, clarithromycin, and azithromycin, and 36.0% for cefaclor. Phenotypes of resistance to erythromycin were MLSB (macrolide-lincosamide-streptogramin B) in 89.9% (gene ermB) and M (macrolide) in 9.7% of cases (gene mefA). No strain harbored both genes simultaneously. Serotypes 19, 6, 23, 14, and 3 were the most prevalent, accounting for 54.6% of the total isolates. Resistance to macrolides seems to be the most alarming point, since among penicillin-susceptible isolates it reached 15.1% compared to 55.8% among penicillin-resistant strains. Geographically, a number of regions had rates of erythromycin resistance above 40% (even higher in children). Resistance to erythromycin was also high in S. pyogenes isolates: mean regional 33.2%, beta-lactamase-producing H. influenzae were 20%, whereas 4.4% had a beta-lactamase-negative, ampicillin-resistant phenotype. We highlight the importance of different geographical frequencies of coresistance (associations of resistance to different drugs within the same species) and coupled resistance (association of resistance between different species) probably resulting from different local coselective events.
Surveillance of antimicrobial resistance should be considered an important tool helping clinicians tailor empirical prescriptions for common infectious diseases. Surveillance data also serve as sentinel checkpoints for emerging phenotypes of resistance and enable performance of studies aiming at characterizing molecular resistance determinants alongside the possibility of tracking the genotypic or clonal relatedness of the isolates harboring the resistance traits detected (4). Besides, when coupled with appropriate data of antimicrobial consumption, they contribute to the understanding of the complex dynamics of the pharmacoepidemiology of resistance. Ideally, they should also serve to assess the impact of measures implemented in response to resistance warnings.
The most prevalent bacteria causing community-acquired respiratory tract infections are Streptococcus pneumoniae, Streptococcus pyogenes, and Haemophilus influenzae. In all of them an increase of resistance to several first- or second-line antibiotics has been observed in recent decades.
In S. pneumoniae, the high prevalence of resistance to penicillin (1, 9), and the constant rise of resistance to macrolides (9) considerably limit the therapeutic options for the different conditions. Although amoxicillin has usually displayed an optimal activity against S. pneumoniae, with MICs one or two dilutions lower than that of penicillin, in recent years a worrisome number of articles have reported the spread of clones with increased MICs for amoxicillin and/or presenting MICs higher for amoxicillin than for penicillin (22).
Empirical prescription of macrolides cannot be considered a justifiable therapeutic option for S. pyogenes any longer, given the impressive increase of resistance to this family of antibiotics, particularly in countries with a high prevalence of resistance, like Spain (21).
A substantial proportion of Haemophilus influenzae strains are resistant to aminopenicillins due to the production of TEM-1 and ROB-1 beta-lactamases (11). However, the widespread use of oral cephalosporins and amoxicillin-clavulanate associations may have contributed to the emergence of strains with PBP3 alterations leading to loss of susceptibility to aminopenicillins in the absence of beta-lactamase production, the so-called beta-lactamase-negative, ampicillin-resistant phenotype. The combination of altered PBP3 and beta-lactamase production may also give rise to a beta-lactamase-positive, amoxicillin-clavulanate-resistant phenotype (17). On the other hand, the existence of efflux pumps leads to loss of susceptibility to macrolides in more than 98% of H. influenzae strains (23).
This article presents a detailed perspective of the current situation of antimicrobial resistance of clinical respiratory isolates of S. pneumoniae, S. pyogenes, and H. influenzae in Spain. We also explore differences among regions and compare resistance results when in vitro National Committee for Clinical Laboratory Standards and pharmacodynamic breakpoints are used. On the other hand, we investigate the importance of coresistance (association of resistance to different drugs within the same species) and coupled resistance (association of resistances between different species, which could also be named as coselection of resistance) (8).
Established in 1996, the S.A.U.C.E. surveillance (the acronym stands for Susceptibility to the Antimicrobials Used in the Community in España) is an ongoing study run on average every 2 years and aiming at assessing the antimicrobial susceptibility of respiratory bacterial pathogens to the antibiotics most commonly used in the community.
MATERIALS AND METHODS
The distribution of hospitals across the country was done bearing in mind demographical considerations to include the regional population distribution of Spain. Overall data were broken into smaller regions called Autonomous Communities.
Centers.
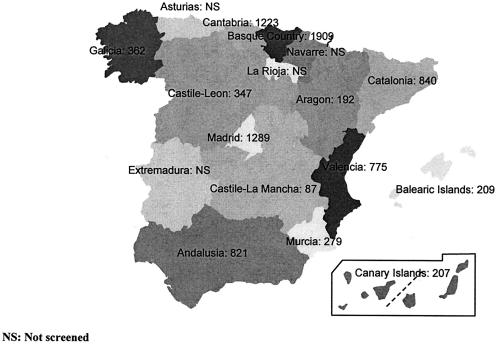
Twenty-five microbiology laboratories (two centers in Galicia: Pontevedra and La Coruña; three in Catalonia: Barcelona; two in Valencia; three in Madrid; five in Andalusia: Seville, Malaga, Jerez de la Frontera, Granada, and Cordoba; 1 in Cantabria: Santander; two centers in Castile-Leon: Salamanca and Valladolid; one in Castile-La Mancha: Ciudad Real; one in Aragon: Saragossa; one in the Canary Islands: Las Palmas de Gran Canaria; one in the Balearic Islands: Palma de Mallorca; two centers in the Basque Country: Baracaldo and San Sebastian; and one center in Murcia) corresponding to regional and university hospitals selected along Spain according to population and geographical distribution took part in this study (Fig. 1).
FIG. 1.
Map of Spain and its autonomous communities. The number of isolates of the three species tested is also given.
Isolates, susceptibility testing, and phenotypes of resistance.
The centers collected all unique consecutive S. pneumoniae, beta-hemolytic streptococci and H. influenzae from clinically significant specimens corresponding to community-acquired infections (acute pharyngitis for beta-hemolytic streptococci, acute otitis media, acute exacerbations of chronic bronchitis and pneumonia for S. pneumoniae or H. influenzae) during a one-year period (November 2001 to October 2002) and stored them at −70°C in a frozen medium containing 1% vegetable peptone and 8% glycerine. A total of 8,540 valid isolates were recovered, 2,721 S. pneumoniae, 3,174 S. pyogenes, and 2,645 H. influenzae. Demographic information on specimen collection data, specimen source, and hospital unit was also collected. Isolates were shipped once a month to a single central laboratory ISO 9001-2000 certified no. ER-0194/1998 (Instituto Valenciano de Microbiología, Valencia, Spain) where isolates were confirmed and serotyped by accepted conventional laboratory methods: optochin, bile solubility test, and the Quellung reaction for the main prevalent pneumococcal serotypes; bacitracin, PYR (pyrrolidonyl-arylamidase) test, and serogrouping for beta-hemolytic streptococci; catalase, X- and V-factor requirements, hemolysis on horse blood, and serogrouping for H. influenzae.
Isolates were stored in duplicate at −70°C for further testing, and recovered by the hot-loop touching method to avoid repeated thawing and freezing.
Antimicrobial susceptibility tests to penicillin, ampicillin, amoxicillin-clavulanate (2:1), cefaclor, axetil-cefuroxime, cefonicid, cefixime, cefotaxime, erythromycin, clarithromycin, azithromycin, and ciprofloxacin were performed by microdilution using custom-designed 96-well trays (Sensititre, Trek Diagnostics Inc. Westlake, Ohio) with a range of concentrations of each antimicrobial, including at least one dilution higher and lower of those required to detect the susceptible and resistance breakpoints for all the control strains recommended by the National Committee for Clinical Laboratory Standards (S. pneumoniae ATCC 49619; H. influenzae ATCC 49247; Staphylococcus aureus ATCC 29213; and Escherichia coli ATCC 25922 and ATCC 35218). The broth microdilution test conditions and breakpoints recommended and accepted by the National Committee for Clinical Laboratory Standards (19) were followed, except for the ciprofloxacin breakpoint, for which an arbitrary 4 μg/ml MIC was considered for resistance. Wherever they appear, National Committee for Clinical Laboratory Standards resistant breakpoints are meant for high resistance. Pharmacokinetic/pharmacodynamic breakpoints used for interpretation of MICs have already appeared in other similar surveillance studies (11).
Isolates with an MIC of erythromycin ≥0.5 μg/ml were tested by the double disk method to detect the constitutive, inducible, or efflux phenotype in S. pneumoniae and S. pyogenes (24). The beta-lactamase test using the chromogenic cephalosporin method (Nitrocefin, Becton-Dickinson) was performed on all H. influenzae isolates and an ampicillin MIC of ≥2 μg/ml was used to define beta-lactamase-negative, ampicillin-resistant isolates.
Serotyping and determination of resistance genes.
Pneumococcal serotyping was carried out at the central laboratory by the Quellung reaction for the main prevalent serotypes using the Statens Serum Institute (Copenhagen, Denmark) pool typing sera for S. pneumoniae. Serotyping was performed again at the National Reference Laboratory of S. pneumoniae (Instituto de Salud Carlos III, Majadahonda, Madrid, Spain) on those strains that did not typed or that did it only with pooled serum but not to any of the main serotypes. Resistance genes ermB and mefA were searched by PCR in all S. pneumoniae with an MIC to erythromycin ≥0.5 μg/ml (Instituto Valenciano de Microbiología, Valencia, Spain) using the primers already described (13, 25).
Statistical analyses.
Differences in the prevalence of antibiotic resistance between different groups were assessed by the Fisher exact test. Associations were determined by calculation of odd ratios with 95% confidence intervals. P < 0.05 was considered statistically significant. Statistical analyses were performed using EPI-Info version 6.04 and the SPSS 11.5 release. Besides, for calculation of the regional mean prevalence, the point prevalence of each of the regions by age group and by antibiotic was considered a single cluster. For the assessment of geographical coresistance and coupled resistance, one-tailed Pearson correlations were calculated when the provincial prevalences of resistance to different antibiotics in the same or in different species were plotted against each other.
RESULTS
Streptococcus pneumoniae.
The sources of the 2,721 S. pneumoniae isolates were potentially contaminated respiratory samples such as sputa (63.8%), otic samples (9.7%), and potentially sterile samples (blood, pleural fluid, and bronchial telescoped catheter: 26.5%).
Intrinsic activity in terms of MIC50 and MIC90 and the corresponding microbiological and pharmacokinetic/pharmacodynamic interpretative standards are shown for every antibiotic tested in Table 1. Only 5.7% of the isolates had a penicillin MIC ≥4 μg/ml.
TABLE 1.
MIC50, MIC90, and prevalence of resistance for S. pneumoniae (n = 2,721)
| Antibiotic | MIC50 (μg/ml) | MIC90 (μg/ml) | NCCLS susceptibility (% of strains) | Pharmocokinetics/pharmacodynamicsa | |||
|---|---|---|---|---|---|---|---|
| Susceptible | Intermediate | Resistant | Breakpoint (μg/ml) | Susceptibility (% of strains) | |||
| Penicillin | 0.03 | 2 | 56.1 | 23.9 | 20.0 | NA | NA |
| Amoxiclav | 0.03 | 2 | 92.2 | 3.4 | 4.4 | ≤2 (≤4) | 92.2 (95.6) |
| Cefaclor | 1 | ≥16 | 61.7 | 2.4 | 36.0 | ≤0.5 | 40.5 |
| Cefuroxime-axetil | ≤0.12 | 8 | 67.4 | 7.0 | 25.6 | ≤1 | 67.4 |
| Cefotaxime | 0.03 | 1 | 96.7 | 2.9 | 0.4 | ≤1 | 96.7 |
| Erythromycin | 0.06 | ≥64 | 64.8 | 0.7 | 34.5 | ≤0.25 | 64.8 |
| Azithromycin | 0.12 | ≥16 | 64.7 | 0.7 | 34.5 | ≤0.12 | 60.2 |
| Ciprofloxacin | 1 | 2 | NA | NA | 4.6b | ≤1 | 77.8 |
Table 2 shows the different prevalences of resistance to penicillin, erythromycin, and ciprofloxacin among the different geographical regions, overall and by age group. In contrast with former studies, penicillin resistance did not vary significantly by age group, 476/2,348 (20.3%) for adults versus 69/373 (18.5%) for children (P = 0.416). However, it showed significant age variation regarding resistance to erythromycin, 760 out of 2,348 (32.4%) among adults versus 178 out of 373 (47.7%) among children (P < 0.001), and resistance to ciprofloxacin, 120/2,348 (5.1%) in adults versus 5/373 (1.3%) in children (P = 0.001).
TABLE 2.
Prevalence of resistance of S. pneumoniae to penicillin, erythromycin, and ciprofloxacin by geographic region and age groupa
| Region | No. of subjects | % Resistant isolates | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Penicillin | Erythromycin | Ciprofloxacin | ||||||||||
| Adults | Children | All | Adults | Children | All | Adults | Children | All | Adults | Children | All | |
| Basque Country | 531 | 95 | 626 | 15.8 | 7.5 | 14.5 | 24.3 | 47.4 | 27.8 | 5.5 | 0 | 4.6 |
| Cantabria | 114 | 36 | 150 | 21.9 | 16.7 | 20.7 | 36.0 | 16.7 | 31.3 | 4.4 | 0 | 3.3 |
| Galicia | 122 | 20 | 142 | 16.4 | 25.0 | 17.6 | 39.3 | 55.0 | 41.5 | 6.6 | 0 | 5.6 |
| Andalusia | 208 | 40 | 248 | 23.1 | 20.0 | 22.6 | 38.9 | 55.0 | 41.5 | 8.2 | 5.0 | 7.7 |
| Catalonia | 441 | 41 | 482 | 19.0 | 19.5 | 19.1 | 32.2 | 39.0 | 32.8 | 4.1 | 0 | 3.7 |
| Castile-Leon | 105 | 1a | 106 | 26.7 | 100a | 27.4 | 27.6 | 100a | 28.3 | 10.5 | 0a | 10.4 |
| Aragon | 36 | 0a | 36 | 19.4 | —b | 19.4 | 38.9 | — | 38.9 | 2.8 | — | 2.8 |
| Valencia | 179 | 30 | 209 | 18.4 | 16.7 | 18.2 | 35.2 | 53.3 | 37.8 | 2.8 | 0 | 2.4 |
| Madrid | 291 | 72 | 363 | 24.4 | 25.0 | 24.5 | 32.6 | 45.8 | 35.3 | 4.5 | 0.7 | 4.1 |
| Canary Islands | 110 | 0a | 110 | 11.8 | — | 11.8 | 25.5 | — | 25.5 | 3.6 | — | 3.6 |
| Castile-La Mancha | 36 | 2a | 38 | 27.8 | 100a | 31.6 | 41.7 | 50.0a | 42.1 | 5.6 | 0a | 5.3 |
| Balearic Islands | 97 | 8 | 105 | 27.8 | 12.5 | 26.7 | 26.8 | 100 | 32.4 | 3.1 | 0 | 2.9 |
| Murcia | 78 | 28 | 106 | 33.3 | 28.6 | 32.1 | 62.8 | 67.9 | 64.2 | 5.1 | 1.3 | 3.8 |
| Total pooled | 2,348 | 373 | 2,721 | 20.3 | 18.5 | 20.0 | 32.4 | 47.7 | 34.5 | 5.1 | 1.3 | 4.6 |
| Mean % regional resistance (95% CI) | 22.0 (18.4-25.6) | 19.1 (14.0-24.1) | 22.0 (18.3-25.8) | 35.5 (29.5-41.6) | 53.3 (36.1-70.6) | 36.9 (30.9-42.9) | 5.1 (3.8-6.5) | 0.8 (0.0-2.0) | 4.6 (3.3-6.0) |
Erythromycin resistance in children compared to their adult counterparts was very high in respiratory samples (52.6% versus 33.4%; P < 0.001) and sterile fluids (blood, pleural fluid, and bronchial telescoped catheter) (49.5% versus 28.3%; P < 0.001). In otic samples erythromycin resistance was equally high, around 44%, among adults and children.
Besides the pooled percentage of resistance, the calculation of the regional means of resistance allows for a more balanced representation of the whole country, avoiding potential over or underestimation, and gives more realistic intervals of confidence. Mean overall resistance to penicillin was 22.0% (95% confidence interval: 18.3% to 25.8%), without differences between adults (22.0%) and children (19.1%). Mean overall regional erythromycin resistance was 36.9% (95% confidence interval: 30.9% to 42.9%), but in this case resistance in adults was 35.5% (95% confidence interval: 29.5% to 41.6%) compared to 53.3% in children (95% confidence interval: 36.1% to 70.6%), with several regions having a prevalence above 50%. Particularly remarkable was the prevalence of resistance from children obtained in Murcia (68%) and Balearic Islands (100%). Resistance to ciprofloxacin was 4.6% (95% confidence interval: 3.3% to 6.0%) with a clear-cut difference in adults (5.1%) versus children (0.8%), as expected.
The phenotype of resistance among the 957 pneumococcal isolates with decreased susceptibility to erythromycin was mostly macrolide-lincosamide-streptogramin B (MLSB) (89.9%) as opposed to 9.7% with the macrolide (M) phenotype. All isolates with the MLSB resistance phenotype had the gene ermB alone, whereas all those displaying M phenotype had the gene mef. No strain harboring both ermB and mefA genes was identified. Four strains (0.4%) with a repeated MIC of 0.5 μg/ml did not harbor ermB or mefA.
Table 3 shows the frequency of the different pneumococcal serogroups, where SG-19 (15.4%), SG-6 (12.7%), SG-23 (9.5%), SG-14 (8.8%), SG-3 (8.2%), nontypeable (6.4%), and SG-9 (6.3%) comprised more than two-thirds of the total. Relative resistance to penicillin, erythromycin, and ciprofloxacin is also shown for every serogroup.
TABLE 3.
Distribution of pneumococcal (n = 2,721) serogroups and their prevalence of resistance to penicillin, erythromycin, and ciprofloxacin
| Serogroup | No. of strains | % of strains | % Resistant to: | ||
|---|---|---|---|---|---|
| Penicillin | Erythromycin | Ciprofloxacin | |||
| 19 | 418 | 15.4 | 16.0 | 57.9 | 5.5 |
| 6 | 346 | 12.7 | 27.5 | 71.1 | 4.6 |
| 23 | 258 | 9.5 | 35.7 | 44.2 | 3.9 |
| 14 | 240 | 8.8 | 68.8 | 40.8 | 6.3 |
| 3 | 223 | 8.2 | 0.0 | 2.7 | 1.8 |
| NT | 174 | 6.4 | 10.9 | 33.9 | 7.5 |
| 9 | 171 | 6.3 | 48.0 | 9.9 | 5.3 |
| 11 | 90 | 3.3 | 1.1 | 15.6 | 2.2 |
| 10 | 72 | 2.6 | 1.4 | 16.7 | 0.0 |
| 4 | 68 | 2.5 | 2.9 | 16.2 | 7.4 |
| 35 | 61 | 2.2 | 14.8 | 3.3 | 6.6 |
| 15 | 61 | 2.2 | 3.3 | 47.5 | 6.6 |
| 1 | 57 | 2.1 | 0.0 | 10.5 | 1.8 |
| Othersa | 482 | 17.7 | 2.1 | 17.0 | 3.9 |
Streptococcus pyogenes.
Among the 3,174 pharyngeal S. pyogenes isolates, pooled erythromycin resistance was 24.3% (27.6% in adults and 22.9% in children; P = 0.0043) (Table 4). However, two regions that contributed with a large proportion of isolates (56.2%), the Basque Country and Cantabria, both in the north, had the lowest prevalence of resistance (around 20%) and therefore underestimate the overall prevalence in the rest of the country. When mean regional prevalence was calculated, then resistance to erythromycin rose by 37%, reaching 33.2% (95% confidence interval: 24.7% to 41.8%), and with a slightly higher prevalence of 36.0% (95% confidence interval: 24.2% to 47.8%) in children.
TABLE 4.
Prevalence of erythromycin resistance among pharyngeal Streptococcus pyogenes strains (n = 3,174) from adults and childrena
| Region | No. of subjects | Erythromycin resistance (% of strains) | ||||
|---|---|---|---|---|---|---|
| Adults | Children | All | Adults | Children | All | |
| Basque Country | 130 | 775 | 905 | 15.4 | 21.7 | 20.8 |
| Cantabria | 51 | 829 | 880 | 17.6 | 18.9 | 18.9 |
| Galicia | 51 | 12 | 63 | 21.6 | 41.7 | 25.4 |
| Andalusia | 225 | 59 | 284 | 28.4 | 37.3 | 30.3 |
| Catalonia | 4a | 13 | 17 | 50.0a | 30.8 | 35.3 |
| Castile-Leon | 50 | 45 | 95 | 30.0 | 48.9 | 38.9 |
| Aragon | 38 | 2a | 40 | 28.9 | 50.0a | 30.0 |
| Valencia | 241 | 45 | 286 | 31.1 | 33.3 | 31.5 |
| Madrid | 139 | 389 | 528 | 25.9 | 24.7 | 25.0 |
| Canary Islands | 1a | 0a | 1a | 100a | —b | 100a |
| Castile-La Mancha | 11 | 8 | 19 | 54.5 | 75.0 | 63.2 |
| Balearic Islands | 0a | 2a | 2a | NA | 50.0a | 50.0a |
| Murcia | 29 | 25 | 54 | 62.1 | 28.0 | 46.3 |
| Total pooled | 970 | 2,204 | 3,174 | 27.6 | 22.9 | 24.3 |
| Mean % regional resistance (95% CI) | 31.6 (20.7-42.4) | 36.0 (24.2-47.8) | 33.2 (24.7-41.8) |
The phenotype of resistance of the 772 resistant strains was mostly M-efflux (86.0%). The MLSB phenotype represented 14% of the resistance to erythromycin (62% was constitutive and 38% inducible).
Haemophilus influenzae.
As for the 2,645 H. influenzae, the majority were isolated from respiratory samples, mostly sputa (2,166/2,645; 81.9%), and the rest were from otic origin (180/2,645; 6.8%) and from sterile sites such as blood, pleural fluid or other bronchial telescoped catheter (294/2,645; 11.1%). Prevalence of beta-lactamase producing isolates was 20.1%, 25.6%, and 17.7%, respectively, (P = 0.112).
Table 5 depicts the MIC50 and MIC90, along with the prevalence of both microbiological and pharmacokinetic/pharmacodynamic interpretative categories, where the higher impact of pharmacokinetic/pharmacodynamic breakpoints compared with S. pneumoniae is remarkable. Except for ampicillin and probably clarithromycin and cefaclor, all the other antibiotics might offer adequate coverage for this pathogen in view of their microbiological breakpoints. However, by applying pharmacokinetic/pharmacodynamic breakpoints, neither cefaclor, clarithromycin, nor azithromycin can be further considered as suitable in view of their coverage of less than 3%. The pharmacokinetic/pharmacodynamic breakpoints place cefuroxime-axetil at the same level as ampicillin. The only antibiotics that retain full activity are amoxicillin/clavulanate and ciprofloxacin.
TABLE 5.
MIC50, MIC90, and prevalence of resistance for H. influenzae (n = 2,645)
| Antibiotic | MIC50 (μg/ml) | MIC90 (μg/ml) | Susceptibility (%) | PK/PDa breakpoints (μg/ml) | |
|---|---|---|---|---|---|
| NCCLS | PK/PD | ||||
| Ampicillin | ≤0.25 | ≥32 | 74.9 | 76.0 | ≤2 |
| Amoxicillin-clavulanate | 0.5 | 2 | 99.9 | 96.0 (99.9)b | ≤2 (≤4)b |
| Cefaclor | 4 | ≥16 | 82.1 | 1.4 | ≤0.5 |
| Cefuroxime-axetil | 1 | 2 | 100 | 72.8 | ≤1 |
| Azithromycin | 1 | 2 | 100 | 2.2 | ≤0.12 |
| Clarithromycin | 8 | 16 | 72.3 | 1.2 | ≤0.25 |
| Ciprofloxacin | ≤0.5 | ≤0.5 | 100 | 100 | ≤1 |
Although differences can be seen among regions (Table 6) in both the prevalence of beta-lactamase production and the beta-lactamase-negative, ampicillin-resistant phenotype, mean regional overall production of beta-lactamase was 20.3% (95% confidence interval: 17.3% to 23.4%), and that of the beta-lactamase-negative, ampicillin-resistant phenotype was 4.4% (95% confidence interval: 2.2% to 6.6%). Therefore, mean resistance to ampicillin was 24.0% (95% confidence interval: 20.9% to 28.7%). No clear differences in production of beta-lactamase and/or frequency of the beta-lactamase-negative, ampicillin-resistant phenotype were apparent between adults and children.
TABLE 6.
Prevalence of beta-lactamase phenotypes by region among H. influenzae isolates (n = 2,645)a
| Region | No. of subjects | Prevalence of resistance (%)b | Ampicillin | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| BLA | BLNAR | |||||||||
| Adults | Children | All | Adults | Children | All | Adults | Children | All | ||
| Basque Country | 327 | 51 | 378 | 14.4 | 29.4 | 16.4 | 3.7 | 9.8 | 4.5 | 20.9 |
| Cantabria | 151 | 42 | 193 | 25.8 | 19.0 | 24.4 | 3.3 | 0 | 2.6 | 26.9 |
| Galicia | 124 | 33 | 157 | 16.1 | 15.2 | 15.9 | 1.6 | 0 | 1.3 | 17.2 |
| Andalusia | 248 | 41 | 289 | 16.1 | 14.6 | 15.9 | 2.8 | 2.4 | 2.8 | 18.7 |
| Catalonia | 293 | 48 | 341 | 14.3 | 22.9 | 15.5 | 5.5 | 6.3 | 5.6 | 21.1 |
| Castile-Leon | 136 | 10 | 146 | 22.8 | 10.0 | 21.9 | 1.5 | 0 | 1.4 | 23.3 |
| Aragon | 115 | 1a | 116 | 15.7 | 0a | 15.5 | 2.6 | 0a | 2.6 | 18.1 |
| Valencia | 255 | 25 | 280 | 31.8 | 24.0 | 31.1 | 5.9 | 4.0 | 5.7 | 36.8 |
| Madrid | 300 | 98 | 398 | 20.3 | 24.5 | 21.4 | 2.7 | 3.1 | 2.8 | 24.1 |
| Canary Islands | 96 | 0a | 96 | 17.7 | —c | 17.7 | 5.2 | — | 5.2 | 22.9 |
| Castile-La Mancha | 28 | 2a | 30 | 21.4 | 0a | 20.0 | 17.9 | 0a | 16.7 | 36.7 |
| Balearic Islands | 87 | 15 | 102 | 18.4 | 13.3 | 17.6 | 1.1 | 13.3 | 2.9 | 20.6 |
| Murcia | 88 | 31 | 119 | 30.7 | 32.3 | 31.1 | 2.2 | 9.7 | 4.2 | 35.3 |
| Total pooled | 2,248 | 397 | 2,645 | 19.8 | 22.2 | 20.2 | 3.7 | 4.5 | 3.8 | 24.0 |
| Mean % regional resistance (95% CI) | 20.4 (17.1-23.6) | 20.6 (16.0-25.3) | 20.3 (17.3-23.4) | 4.3 (1.8-6.7) | 4.1 (1.8-7.8) | 4.4 (2.2-6.6) | 24.8 (20.9-28.7) |
Coresistance and coupled resistance between antibiotics and bacterial species.
Coresistance in S. pneumoniae involves cross-resistance between penicillin, erythromycin, and ciprofloxacin. We use the term coupled resistance (also named coselection of resistance by some authors) to describe the mutual relation between the prevalence of resistance to these antibiotics in different species, S. pneumoniae, and S. pyogenes, or between each of them and ampicillin resistance in H. influenzae.
For S. pneumoniae, resistance to erythromycin in penicillin resistant (MIC ≥ 2 μg/ml) isolates was 55.8% compared to 29.1% in penicillin nonresistant (MIC ≤ 1 μg/ml) strains (odds ratio = 2.97; 95% confidence interval: 2.44 to 3.62; P ≪ 0.001). Comparing penicillin-nonsusceptible (MIC ≥ 0.12 μg/ml) versus -susceptible (MIC ≤ 0.06 μg/ml) isolates we found a prevalence of resistance to erythromycin of 60.2% versus 15.6% (odds ratio = 8.17; 95% confidence interval: 6.80 to 9.83; p≪0.001). The prevalence of resistance to ciprofloxacin (MIC ≥ 4 μg/ml) among penicillin resistant pneumococci was 8.3% versus 3.7% for penicillin nonresistant (odds ratio = 2.36; 95% confidence interval: 1.59 to 3.50; P < 0.001).
The prevalence of resistance to ciprofloxacin was 6.8% for penicillin-nonsusceptible versus 2.9% for penicillin-susceptible strains (odds ratio = 2.45; 95% confidence interval: 1.66 to 3.63; P < 0.001). Up to 7% of pneumococcal isolates displaying nonsusceptibility to erythromycin (MIC ≥ 0.5 μg/ml) were also resistant to ciprofloxacin, compared to only 3.3% ciprofloxacin resistance among erythromycin-susceptible isolates (odds ratio = 2.21; 95% confidence interval: 1.52 to 3.23; P < 0.001). In the case of complete susceptibility to penicillin, any beta-lactam antibiotic should be considered as susceptible. However, resistance to macrolides and ciprofloxacin reached 15% and 3%, respectively, in these penicillin-susceptible isolates.
For isolates with an intermediate susceptibility to penicillin, only amoxicillin/clavulanate and cefotaxime retained high activity, as opposed to erythromycin or azithromycin, with 62.0% resistance, and cefaclor, with 70.0% resistance. Cefuroxime-axetil and ciprofloxacin resistance rose to 24.9% and 5.5% among these intermediate susceptible pneumococci. Pneumococcal isolates fully resistant to penicillin had 83.5% and 63.1% complete susceptibility to cefotaxime and amoxicillin-clavulanate whereas resistance to both cefaclor and cefuroxime-axetil was above 95%. Resistance to macrolides and ciprofloxacin was 55.8% and 8.3%, respectively (Table 7).
TABLE 7.
Prevalence of S. pneumoniae resistance to several antimicrobials according to susceptibility to penicillin
| Penicillin resistance (no. of strains) | Antimicrobial | % of strains | ||
|---|---|---|---|---|
| Susceptible | Intermediate | Resistant | ||
| Susceptible | Amoxicillin/clavulanate | 100 | 0 | 0 |
| (1,526) | Cefaclor | 98.8 | 1.0 | 0.1 |
| Cefuroxime-axetil | 99.7 | 0.1 | 0.2 | |
| Cefotaxime | 100 | 0 | 0 | |
| Erythromycin | 84.4 | 0.5 | 15.1 | |
| Azithromycin | 84.1 | 0.6 | 15.3 | |
| Ciprofloxacin | 2.9 | |||
| Intermediate | Amoxicillin/clavulanate | 98.1 | 1.9 | 0 |
| (650) | Cefaclor | 21.9 | 8.1 | 70.0 |
| Cefuroxime-axetil | 47.7 | 27.4 | 24.9 | |
| Cefotaxime | 99.8 | 0 | 0.2 | |
| Erythromycin | 36.3 | 1.7 | 62.0 | |
| Azithromycin | 36.6 | 1.4 | 62.0 | |
| Ciprofloxacin | 5.5 | |||
| Resistant | Amoxicillin/clavulanate | 63.1 | 14.9 | 22.0 |
| (545) | Cefaclor | 0.4 | 0 | 99.6 |
| Cefuroxime-axetil | 0.2 | 2.2 | 97.6 | |
| Cefotaxime | 83.5 | 14.5 | 2.0 | |
| Erythromycin | 44.0 | 0.2 | 55.8 | |
| Azithromycin | 43.9 | 0.4 | 55.8 | |
| Ciprofloxacin | 8.3 |
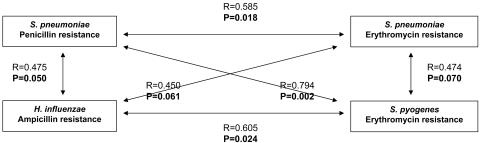
Figure 2 illustrates the concepts of selection of coresistance and of coupled resistance from a geographical perspective. All correlations showed a positive sign and had P values below or very close to 0.05. As far as selection of coresistance was concerned, both resistance to penicillin and erythromycin resistance in S. pneumoniae correlated quite well (R = 0.585; P = 0.018). The remaining correlations concerned different examples of coupled resistance between different species. Penicillin resistance in S. pneumoniae did also present statistical significant correlation with erythromycin resistance in S. pyogenes (R = 0.794; P = 0.002) and with ampicillin resistance in H. influenzae (R = 0.475; P = 0.050). Geographical erythromycin resistance in S. pneumoniae was also very close to statistical significance (R = 0.474; P = 0.07) when plotted against erythromycin resistance in S. pyogenes, and against ampicillin resistance in H. influenzae (R = 0.450; P = 0.061). Finally, erythromycin resistance in S. pyogenes trended to vary in parallel to ampicillin resistance in H. influenzae (R = 0.605; P = 0.024).
FIG. 2.
Geographical selection of coresistance and coselection of resistance.
DISCUSSION
The 25 participating hospitals comprised a total of 24,356 beds, 15% of all the existing hospital beds in Spain. On the other hand, the 8,540 valid isolates represent around 21.5 isolates/100,000 inhabitants. In order to obtain a similar population-adjusted collection in a country like the United States with a population of 290 million, more than 62,000 isolates would have to be recovered in the same time period.
In order to represent more accurately the complex Spanish demography, we tried to have representatives of all the possible regions we could, and then we monitored 13 of the 17 Spanish Autonomous Communities. The four unmonitored autonomous communities represent only 7.4% of the Spanish population.
We found a prevalence of resistance to penicillin in S. pneumoniae of only 20.3% in adults and 18.5% in children, which is good news, keeping in mind the elevated rates reported several years ago. There is a consistent trend to decreasing rates of penicillin resistance in S. pneumoniae in Spain during the last five years (6). It can be suggested that the use of conjugate vaccine and the more rational use of antimicrobial agents along with the introduction of new drugs such as new formulations of beta-lactams or quinolones with improved activity on pneumococci have contributed to this important phenomenon.
Of the different antibiotics commonly used to treat respiratory infections, cefotaxime (or ceftriaxone) appeared as the most potent antipneumococcal agent. Next and very close, amoxicillin (usually given empirically as amoxicillin-clavulanate) was able to display a significant activity both in terms of MIC and in prevalence of resistance, with the advantage of oral administration.
Regarding overall coresistance, the susceptibility to penicillin was an important correlate of the prevalence of resistance to other drugs. Hence, for penicillin susceptible isolates, beta-lactams are adequate therapeutic choices; in contrast, up to 15% were already resistant to erythromycin. For penicillin-intermediate isolates, only cefotaxime and amoxicillin ensured a right coverage. For penicillin-resistant strains, both cefaclor and cefuroxime-axetil lacked activity, whereas cefotaxime only held 2.2% resistance. Up to 62% of these penicillin-resistant isolates were susceptible to conventional amoxicillin-clavulanate. This proportion is likely to increase due to the higher activity of new amoxicillin-clavulanate formulations (1, 5, 10, 14) with a probable susceptibility breakpoint of at least 4 μg/ml, although this breakpoint has not been approved by National Committee for Clinical Laboratory Standards or the Food and Drug Administration yet.
Differences in resistance to penicillin and erythromycin existed according to kind of sample and age, as frequently reported in the literature.
It is noteworthy that serogroups 14 and 9 had a 69% and 50% resistance to penicillin, whereas serogroups 6 and 19 presented 71% and 58% resistance to erythromycin. From an strict epidemiological view, each serogroup seems to behave with a completely different dynamic regarding penicillin and erythromycin resistance (15). Although the spread of conjugate antipneumococcal vaccine should prompt to a decrease in the circulation of these serogroups, a decrease in the prevalence of resistance is not likely to occur unless antibiotic pressure is levered down (18).
Geographical differences of resistance to penicillin, erythromycin and ciprofloxacin were also seen in S. pneumoniae with a trend to being higher in the south relative than in the north.
Regarding S. pyogenes, high resistance rates to erythromycin were consistently seen across regions (Table 4).
There were large discrepancies between microbiological and pharmacokinetic/pharmacodynamic susceptibility breakpoints in H. influenzae for certain antibiotics (Table 5). Ciprofloxacin and amoxicillin-clavulanate covered the almost totality of the strains. Susceptibility to cefaclor shifted from 82% to 1.4% that of azithromycin did from 100% to 2.2%, and that of clarithromycin from 72.3% to 1.2% when pharmacokinetic/pharmacodynamic breakpoints were used, in the case of macrolides probably as a result of the presence of efflux pumps in virtually any H. influenzae strains. This should prompt to reconsider current National Committee for Clinical Laboratory Standards breakpoints for H. influenzae (23).
Geographical differences in production of beta-lactamase and in the beta-lactamase-negative, ampicillin-resistant phenotype in H. influenzae were not so large. To date, high rates of beta-lactamase-negative, ampicillin-resistant strains have only been reported in Japan, and this increased rate in Spain is cause for serious concern. In fact, our group has already noted increasing rates (16). Some authors have recently emphasized an increasing role for H. influenzae in an era of herd immunity to the current drug-resistant S. pneumoniae serotypes by immunization with the seven-valent pneumococcal conjugate vaccine (2, 3), available in Spain since 2001.
A geographical correlation of resistances was consistently found between different antibiotics in the same and in different microorganisms (Fig. 2). As an example of geographical selection of coresistance, the region-by-region prevalence of resistance to penicillin and erythromycin were directly linked (R = 0.585; P = 0.018). As examples of geographical coupled resistance, that is, the coincidental selection of strains of two different species resistant to a given antibiotic, penicillin resistance in S. pneumoniae was also linked with erythromycin resistance in S. pyogenes and to ampicillin resistance in H. influenzae. Resistance to erythromycin in S. pneumoniae missed statistical significance by very little in its linkage to erythromycin resistance in S. pyogenes and to ampicillin resistance in H. influenzae. Resistance to erythromycin in S. pneumoniae was in turn associated with ampicillin resistance in H. influenzae. Although geographical associations of resistance had already been reported between S. pneumoniae and S. pyogenes and erythromycin (8), penicillin and erythromycin in S. pneumoniae (20), and penicillin and ampicillin in S. pneumoniae and H. influenzae (12, 16), to our knowledge this is the first time that so many intertwined associations are described simultaneously in a single country.
The different antibiotic pressure in each region is likely to be the main cause behind these findings. However, it seems hard to put all the blame on antibiotic pressure as being solely responsible for these differences, even though the most recent studies seem to point in this direction in detriment to the hypotheses of independent clonal spread (18). Different clones spreading with different degrees of success would therefore be the answer to the biological stress of a given level of changing antibiotic use. Therefore, it might be worth studying the reasons for different antibiotic use in different regions or time points in order to try to find potential modifiable factors (7).
Acknowledgments
This study was supported in part by a grant from GlaxoSmithKline S.A., Tres Cantos, Spain.
REFERENCES
- 1.Aguilar, L., M. J. Gimenez, C. Garcia-Rey, and J. E. Martin. 2002. New strategies to overcome antimicrobial resistance in Streptococcus pneumoniae with beta-lactam antibiotics. J. Antimicrob. Chemother. Suppl. C**:**93-100. [DOI] [PubMed] [Google Scholar]
- 2.Block, S. L., J. Hedrick, C. J. Harrison, R. Tyler, A. Smith, R. Findlay, and E. Keegan. 2004. Community-wide vaccination with the heptavalent pneumococcal conjugate significantly alters the microbiology of acute otitis media. Pediatr. Infect. Dis. J. 23**:**829-833. [DOI] [PubMed] [Google Scholar]
- 3.Casey, J. R., and M. E. Pichichero. 2004. Changes in frequency and pathogens causing acute otitis media in 1995-2003. Pediatr. Infect. Dis. J. 23**:**824-828. [DOI] [PubMed] [Google Scholar]
- 4.Cornaglia, G., W. Hryniewicz, V. Jarlier, G. Kahlmeter, H. Mittermayer, L. Stratchounski, and F. Baquero. 2004. European recommendations for antimicrobial resistance surveillance. Clin. Microbiol. Infect. 10**:**349-383. [DOI] [PubMed] [Google Scholar]
- 5.File, T. M., M. R. Jacobs, M. D. Poole, and B. Wynne. 2002. Outcome of treatment of respiratory tract infections due to Streptococcus pneumoniae, including drug-resistant strains, with pharmacokinetically enhanced amoxycillin/clavulanate. Int. J. Antimicrob. Agents. 20**:**235. [DOI] [PubMed] [Google Scholar]
- 6.Garcia-Rey, C., E. Bouza, L. Aguilar, J. Garcia-de-Lomas, and F. Baquero. 2003. Evolution of penicillin and erythromycin coresistance in Streptococcus pneumoniae in Spain. Int. J. Antimicrob. Agents 22**:**541-544. [DOI] [PubMed] [Google Scholar]
- 7.Garcia-Rey, C., A. Fenoll, L. Aguilar, and J. Casal. 2004. Effect of social and climatological factors on antimicrobial use and Streptococcus pneumoniae resistance in different provinces in Spain. J. Antimicrob. Chemother. 54**:**465-471. [DOI] [PubMed] [Google Scholar]
- 8.Gomez-Lus, R., J. J. Granizo, L. Aguilar, E. Bouza, A. Gutierrez, and J. Garcia-de-Lomas. 1999. Is there an ecological relationship between rates of antibiotic resistance of species of the genus Streptococcus? The Spanish Surveillance Group for Respiratory Pathogens. J. Clin. Microbiol. 37**:**3384-3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Granizo, J. J., L. Aguilar, J. Casal, C. Garcia-Rey, R. Dal-Re, and F. Baquero. 2000. Streptococcus pneumoniae resistance to erythromycin and penicillin in relation to macrolide and beta-lactam consumption in Spain (1979-1997). J. Antimicrob. Chemother. 46**:**767-773. [DOI] [PubMed] [Google Scholar]
- 10.Jacobs, M. R. 2004. Building in efficacy: developing solutions to combat drug-resistant S. pneumoniae. Clin. Microbiol. Infect. 10(Suppl. 2)**:**18-27. [DOI] [PubMed] [Google Scholar]
- 11.Jacobs, M. R., D. Felmingham, P. C. Appelbaum, and R. N. Gruneberg. 2003. The Alexander Project 1998-2000: susceptibility of pathogens isolated from community-acquired respiratory tract infection to commonly used antimicrobial agents. J. Antimicrob. Chemother. 52**:**229-246. [DOI] [PubMed] [Google Scholar]
- 12.Jones, M. E., J. A. Karlowsky, R. Blosser-Middleton, I. Critchley, C. Thornsberry, and D. F. Sahm. 2002. Relationship between antibiotic resistance in Streptococcus pneumoniae and that in Haemophilus influenzae: evidence for common selective pressure. Antimicrob. Agents Chemother. 46**:**3106-3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kataja, J., P. Huovinen, M. Skurnik, and H. Seppala. 1999. Erythromycin resistance genes in group A streptococci in Finland. The Finnish Study Group for Antimicrobial Resistance. Antimicrob. Agents Chemother. 43**:**48-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaye, C. M., A. Allen, S. Perry, M. McDonagh, M. Davy, K. Storm, N. Bird, and O. Dewit. 2001. The clinical pharmacokinetics of a new pharmacokinetically enhanced formulation of amoxicillin/clavulanate. Clin. Ther. 23**:**578-584. [DOI] [PubMed] [Google Scholar]
- 15.Marco, F., E. Bouza, J. Garcia-de-Lomas, and L. Aguilar. 2000. Streptococcus pneumoniae in community-acquired respiratory tract infections in Spain: the impact of serotype and geographical, seasonal and clinical factors on its susceptibility to the most commonly prescribed antibiotics. The Spanish Surveillance Group for Respiratory Pathogens. J Antimicrob. Chemother. 46**:**557-564. [DOI] [PubMed] [Google Scholar]
- 16.Marco, F., J. Garcia-de-Lomas, C. Garcia-Rey, E. Bouza, L. Aguilar, and C. Fernandez-Mazarrasa. 2001. Antimicrobial susceptibilities of 1,730 Haemophilus influenzae respiratory tract isolates in Spain in 1998-1999. Antimicrob. Agents Chemother. 45**:**3226-3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matic, V., B. Bozdogan, M. R. Jacobs, K. Ubukata, and P. C. Appelbaum. 2003. Contribution of beta-lactamase and PBP amino acid substitutions to amoxicillin/clavulanate resistance in beta-lactamase-positive, amoxicillin/clavulanate-resistant Haemophilus influenzae. J. Antimicrob. Chemother. 52**:**1018-1021. [DOI] [PubMed] [Google Scholar]
- 18.McCormick, A. W., C. G. Whitney, M. M. Farley, R. Lynfield, L. H. Harrison, N. M. Bennett, W. Schaffner, A. Reingold, J. Hadler, P. Cieslak, M. H. Samore, and M. Lipsitch. 2003. Geographic diversity and temporal trends of antimicrobial resistance in Streptococcus pneumoniae in the United States. Nat. Med. 9**:**424-430. [DOI] [PubMed] [Google Scholar]
- 19.National Committee for Clinical Laboratory Standards. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 6th ed. Approved standard M7-A6. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 20.Perez-Trallero, E., C. Fernandez-Mazarrasa, C. Garcia-Rey, E. Bouza, L. Aguilar, J. Garcia-de-Lomas, and F. Baquero. 2001. Antimicrobial susceptibilities of 1,684 Streptococcus pneumoniae and 2,039 Streptococcus pyogenes isolates and their ecological relationships: results of a 1-year (1998-1999) multicenter surveillance study in Spain. Antimicrob. Agents Chemother 45**:**3334-3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perez-Trallero, E., M. Urbieta, M. Montes, I. Ayestaram, and J. M. Marimon. 1998. Emergence of Streptococcus pyogenes strains resistant to erythromycin in Gipuzkoa, Spain. Eur. J. Clin. Microbiol. Infect. Dis. 17**:**25-31. [DOI] [PubMed] [Google Scholar]
- 22.Perez-Trallero, E., J. M. Marimon, A. Gonzalez, C. Garcia-Rey, and L. Aguilar. 2003. Genetic relatedness of recently collected Spanish respiratory tract Streptococcus pneumoniae isolates with reduced susceptibility to amoxicillin. Antimicrob. Agents Chemother. 47**:**3637-3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peric, M., B. Bozdogan, M. R. Jacobs, and P. C. Appelbaum. 2003. Effects of an efflux mechanism and ribosomal mutations on macrolide susceptibility of Haemophilus influenzae clinical isolates. Antimicrob. Agents Chemother. 47**:**1017-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seppala, H., A. Nissinen, Q. Yu, and P. Huovinen. 1993. Three different phenotypes of erythromycin-resistant Streptococcus pyogenes in Finland. J. Antimicrob. Chemother. 32**:**885-891. [DOI] [PubMed] [Google Scholar]
- 25.Shortridge, V. D., G. V. Doern, A. B. Brueggemann, J. M. Beyer, and R. K. Flamm. 1999. Prevalence of macrolide resistance mechanisms in Streptococcus pneumoniae isolates from a multicenter antibiotic resistance surveillance study conducted in the United States in 1994-1995. Clin. Infect. Dis. 29**:**1186-1188. [DOI] [PubMed] [Google Scholar]