Hes1 Directly Controls Cell Proliferation through the Transcriptional Repression of p27Kip1 (original) (raw)
Abstract
A transcriptional regulator, Hes1, plays crucial roles in the control of differentiation and proliferation of neuronal, endocrine, and T-lymphocyte progenitors during development. Mechanisms for the regulation of cell proliferation by Hes1, however, remain to be verified. In embryonic carcinoma cells, endogenous Hes1 expression was repressed by retinoic acid in concord with enhanced p27Kip1 expression and cell cycle arrest. Conversely, conditional expression of a moderate but not maximal level of Hes1 in HeLa cells by a tetracycline-inducible system resulted in reduced p27Kip1 expression, which was attributed to decreased basal transcript rather than enhanced proteasomal degradation, with concomitant increases in the growth rate and saturation density. Hes1 induction repressed the promoter activity of a 5′ flanking basal enhancer region of p27Kip1 gene in a manner dependent on Hes1 expression levels, and this was mediated by its binding to class C sites in the promoter region. Finally, hypoplastic fetal thymi, as well as livers and brains of _Hes1_-deficient mice, showed significantly increased p27Kip1 transcripts compared with those of control littermates. These results have suggested that Hes1 directly contributes to the promotion of progenitor cell proliferation through transcriptional repression of a cyclin-dependent kinase inhibitor, p27Kip1.
Hes1 (Hairy and enhancer of split homolog-1) is a basic helix-loop-helix (bHLH) transcriptional regulator and represses the transcription of target genes by several distinct mechanisms, including interference with B-class bHLH transcription activators via heterodimerization (24), direct binding to the specific DNA sequences, class C site and N box, in the promoter regions of target genes, such as human ASH1 (achaete-scute homolog 1) and Hes1 itself (3, 29), and repression in collaboration with other transcription regulators, such as Myb (1). Hes1 is expressed in developing neuroectodermal and endodermal endocrine tissues, and _Hes1_-deficient (_Hes1_−/−) embryos show severe defects in the neuronal development, as well as pancreatic hypoplasia (10). Since the cells in these tissues of _Hes1_−/− embryos showed premature expression of B-class bHLH factors as well as terminal differentiation markers, the phenotypes were attributed to the accelerated formation of postmitotic neuronal or endocrine cells recruited from the precursor epithelia, resulting in the depletion of the progenitor cell pool (13).
_Hes1_−/− embryos also showed profound thymic hypoplasia, with the numbers of thymocytes being markedly reduced (30). In a normal embryonic thymus, an explosive expansion of the thymic immigrant progenitors occurs preceding the initiation of a series of differentiation processes to ensure sufficient clonal diversity (33). Unlike in the neuronal tissues, however, differentiation of the _Hes1_−/− thymocytes was almost completely arrested at the most immature stages, implying that the impaired proliferation of thymocytes was a primary consequence of Hes1 deficiency rather than a secondary consequence of their accelerated differentiation and maturation (30). Cell transfer studies confirmed that the _Hes1_−/− T-cell progenitors were defective for the proliferation even in the normal thymic environment, and thus, the defect was progenitor-cell autonomous (30). Hes1 is a major target gene activated downstream of Notch family receptors (17). It was reported that conditional Notch1 gene-targeted mice also showed severe thymic hypoplasia essentially identical to that in _Hes1_−/− mice (23), suggesting that promotion of the early T-cell progenitor expansion by Notch1 signaling in the thymus was mediated by Hes1. More recently, it was suggested that Hes1 might mediate the proliferation of granule neuron precursors by granule neuron precursor mitogens such as Jagged 1 and Sonic Hedgehog (25). It was also reported that Hes1 gene transduction could promote the expansion of hematopoietic stem cells without exhausting stem cell activity (17). While all these observations have indicated that Hes1 plays a crucial role in the promotion and/or maintenance of progenitor cell proliferation in various developing tissues, possible mechanisms for that remain largely unknown.
Attempts to directly examine the function of Hes1 using cell line models in vitro were hampered by the failure to obtain stable cell lines expressing Hes1, due to an apparent “toxic” effect of Hes1 overexpression causing cell death by undefined reasons (27). In the present study, we have analyzed the effects of Hes1 on cell proliferation by using a tetracycline-inducible system in HeLa cells and provide evidence that Hes1 at a controlled expression level directly promotes the cell proliferation by repressing the expression of a cyclin-dependent kinase (CDK) inhibitor, p27Kip1, independently of a cell differentiation program. It was suggested that Hes1 repressed the basal transcription of p27Kip1 gene by directly binding to the consensus class C sites in the 5′ flanking enhancer region. It was further confirmed that T-cell progenitors in the hypoplastic thymi of _Hes1_−/− embryos showed significantly elevated levels of p27Kip1 gene transcript compared with those in the control littermates in vivo. Present results may reveal a key molecular mechanism for Hes1 to directly control the developmentally programmed proliferation of progenitor cells.
MATERIALS AND METHODS
Mice.
Hes1 gene-targeted mice were reported before (10) and maintained in the specific-pathogen-free condition in our animal facility. Heterozygous mice (Hes1+/−) were mated, and the embryos were analyzed at embryonic day 14.5 (E14.5). The genotype of each embryo was determined by genomic PCR as described previously (8).
Plasmids, cells, and cultures.
OTF9 (F9) embryonic carcinoma (EC) and HeLa Tet-off cell lines were obtained from the American Type Culture Collection and BD Biosciences (San Jose, CA), respectively, and maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum. C3H10T1/2 and 293T cell lines were also cultured in the same medium. An N-terminally FLAG-tagged Hes1 cDNA in a Tet-off expression vector, pTRE-N-FLAG-Hes1, was constructed by subcloning a BamHI fragment of pLXSG-N-FLAG-Hes1 (11) into a pTRE (pUHD-10.3) vector provided by S. Takeda, Kyoto University, Kyoto, Japan. For the luciferase promoter assay, a pSRα-Hes1 plasmid was constructed by subcloning a BamHI fragment of pLXSG-N-FLAG-Hes1 (11) into a pcDL-SRα-296 vector. A ΔWRPW-Hes1 plasmid was constructed by subcloning a BamHI-XhoI fragment of pTRE-N-FLAG-Hes1 deleted of WRPW domain, obtained by PCR with primers 5′-GGATCCATGGACTACAAGGACGAC-3′ (sense) and 5′-CTCGAGTCACATGGAGTCCGCAGT-3′ (antisense), into a pcDL-SRα-296 vector. HeLa Tet-off cells were cotransfected with pTRE-N-FLAG-Hes1 (5 μg) and pLSV-hygB (0.4 μg; provided by T. Sudo, Toray Inc., Kamakura, Japan) by a calcium phosphate precipitation method and cultured in complete DMEM supplemented with 10 ng/ml doxycycline (Dox), 300 μg/ml hygromycin B, and 100 μg/ml G418. Two weeks later, cells that survived were collected and cloned. Expression of Hes1 in the absence and presence of 10 ng/ml Dox was assessed by immunoblotting with both anti-Hes1 and anti-FLAG antibodies, and clones expressing Hes1 (HeLa/Tet-Hes1) with the least leaky expression of Hes1 in the presence of Dox were selected and maintained in DMEM supplemented with 10 ng/ml Dox and 100 μg/ml G418. To assess the growth of HeLa/Tet-Hes1 cells, the cells were plated in six-well culture plates at 1 × 105 cells/well in the absence or presence of various doses of Dox, and the viable cell numbers were counted every day for 6 days. The cells were fed with fresh DMEM containing the corresponding doses of Dox every other day. Proteasome inhibitor MG132 (Z-Leu-Leu-Leu-H) and calpain inhibitor MG′ (Z-Leu-Leu-H) purchased from Peptide Institute, Osaka, Japan, were added into the culture at final 10 μM in 10 μl dimethyl sulfoxide 2 h before the cell harvest. To induce the differentiation of F9 cells, retinoic acid (RA; Sigma-Aldrich, St. Louis, MO) was added in the monolayer culture at 1 μM.
Cell cycle analysis.
F9 cells cultured for 4 days in the presence or absence of RA were collected by the treatment with trypsin-EDTA. The cells were washed with phosphate-buffered saline (PBS), resuspended in 0.2% Triton X-100 in PBS, filtered through a nylon mesh to remove debris, and treated with 0.5% RNase A, followed by staining with 50 μg/ml propidium iodide (Sigma-Aldrich). After the washing, DNA contents of the cells were analyzed by using FACSCalibur flow cytometry with a cycle test software (BD Biosciences).
Immunoblotting and immunostaining.
F9 cells were harvested, and either total cell or nuclear lysate was prepared. To obtain the nuclear lysate, cells were washed with cold PBS, resuspended in hypotonic buffer (10 mM HEPES, pH 8.0, 50 mM NaCl, 0.5 M sucrose, 1 mM EDTA, 0.5 mM spermidine, 0.15 mM spermine, 0.1% NP-40, protease inhibitor cocktail, 1 mM phenylmethylsulfonyl fluoride [PMSF], 7 mM 2-mercaptoethanol), incubated for 10 min on ice, and spun down at 800 × g for 10 s. The pellets were resuspended in lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.5% deoxycholate, 1% NP-40, 0.1% sodium dodecyl sulfate [SDS], protease inhibitor cocktail, 0.1 mM PMSF), incubated for 15 min on ice, and centrifuged. Immunoblotting of the lysates was performed as described before (31), using rabbit anti-Hes1 (provided by T. Sudo), anti-p27Kip1 (BD Biosciences), anti-p21Cip1 (Santa Cruz Biotechnology, Santa Cruz, CA), or anti-FLAG M2 (Sigma-Aldrich) antibody. Immunofluorescence staining was performed as described previously (6). Briefly, F9 cells cultured on glass coverslips were fixed with 3% formaldehyde in PBS for 15 min at room temperature, treated with 0.2% Triton X-100 in PBS for 15 min, blocked in 1% bovine serum albumin in PBS, and incubated with anti-Hes1, anti-p27Kip1, or anti-claudin-4 antibody (provided by S. Tsukita, Kyoto University, Kyoto, Japan), followed by the corresponding Alexa488- or Cy3-conjugated secondary antibody along with DAPI (4′,6′-diamidino-2-phenylindole). They were embedded in Mowiol mounting medium (Calbiochem, Bad Soden, Germany) and analyzed with a fluorescence microscope (Carl Zeiss, Jena, Germany).
RT-PCR and Southern blotting.
Total RNA was extracted with ISOGEN reagent (Nippon Gene, Toyama, Japan), and the cDNA was synthesized with reverse transcriptase (RT) SuperScript III (Invitrogen BV, Carlsbad, CA) according to the manufacturer's protocol. The primer sequences were as follows: for p27Kip1, 5′-AGCCTGGAGCGGATGGACGCCA-3′ (sense) and 5′-AACCGTCTGAAACATTTTATTATGTT-3′ (antisense); for human p21Cip1, 5′-CTCCAAGAGGAAGCCCTAATCC-3′ (sense) and 5′-TTTGATGATGCCCCCACTCG-3′ (antisense); for mouse p21Cip1, 5′-TCCACAGCGATATCCAGACA-3′ (sense) and 5′-CAGGGCAGAGGAAGTACTGG-3′ (antisense); and for glyceraldehyde-3-phosphate dehydrogenase (GAPDH), 5′-TTCATTGACCTCAACTACATGGT-3′ (sense) and 5′-TTGTCATACCAGGAAATGAGCTT-3′ (antisense). The amount of each template was adjusted to give a PCR signal strength in exponential phase comparable to that of GAPDH after 25 cycles. Amplified PCR products were electrophoresed on 2% agarose gel, blotted onto nylon membranes (Amersham Biosciences, Piscataway, NJ), and hybridized with 32P-labeled full-length p27Kip1 or GAPDH cDNA probe as described previously (7). The intensities of autoradiographic signals were quantified by using NIH Image 1.62 software. Quantitative PCR for p27Kip1 was performed by using TaqMan 6-carboxyfluorescein probes and PCR primer mix for p27Kip1 (TaqMan gene expression assays; Applied Biosystems, Foster City, CA) on an Mx3000P real-time PCR system (Stratagene, La Jolla, CA) according to the manufacturer's instruction. For the endogenous control, the following primers for 36B4 human rRNA and mouse cyclophilin were used in HeLa cells and mouse tissues, respectively: for human 36B4, 5′-TGCATCAGTACCCCATTCTATCA-3′ (sense) and 5′-AAGGTGTAATCCGTCTCCACAGA-3′ (antisense); and for mouse cyclophilin, 5′-TGGAGAGCACCAAGACAGACA-3′ (sense) and 5′-TGCCGGAGTCGACAATGAT-3′ (antisense). The quantity of each transcript was determined by the standard curve using serially diluted cDNA samples, and that of p27Kip1 transcript was normalized to the endogenous control.
Luciferase reporter assay.
A 3.5-kb-long 5′ flanking DNA of human p27Kip1 gene (p27PF; −12 to −3568) and its ApaI fragment (p27PF/ApaI; −12 to −774) subcloned in front of the luciferase reporter gene in a pGVB2 vector were reported before (20), and a Renilla plasmid, pRL-TK, was purchased (Promega, Madison, WI). HeLa/Tet-Hes1 cells were seeded into six-well culture plates (1 × 105 cells/well) and, 24 h later, cotransfected with 2 μg reporter plasmid and 0.1 μg pRL-TK as a transfection efficiency control by using FuGENE6 transfection reagent (Roche Diagnostics, Indianapolis, IN). After incubation for 4 h, the medium and precipitates were removed, and the cells were cultured in fresh complete media containing various concentrations of Dox. After 48 h, the cell monolayers were washed with PBS, harvested by scraping, and resuspended in PLB buffer (Promega). Firefly and Renilla luciferase activities were assayed with the Dual-Luciferase assay kit (Promega), and former luciferase activity was corrected by the transfection efficiency by using the control Renilla luciferase activity in each sample. C3H10T1/2 cells were also seeded into six-well culture plates (1 × 105 cells/well) and, 24 h later, transfected with 0.5 μg pSRα-neo, pSRα-N-FLAG Hes1, or pSRα-N-FLAG ΔWRPW-Hes1 along with 1 μg reporter and 0.1 μg pRL-TK plasmid.
EMSA.
Nuclear protein extracts were prepared from HeLa/Tet-Hes1 cells cultured for 2 days in the absence or presence of Dox (10 ng/ml) as described above. Five micrograms of nuclear extract was incubated with 2 μg of poly(dI-dC) and double-stranded oligonucleotides labeled with [γ-32P]ATP in a final volume of 20 μl incubation buffer (10 mM HEPES, pH 7.9, 50 mM NaCl, 0.1 mM EDTA, 25% glycerol) for 30 min at room temperature. The protein-DNA mixtures were separated on nondenaturing 5% polyacrylamide gels at 4°C in Tris-borate EDTA. Unlabeled wild-type or mutated oligonucleotides (10 or 50 pmol) were included as competitors to confirm the specificities of the complexes formed. The following oligonucleotides in the murine p27Kip1 promoter region were used as probes: ApaI/C-site wild-type 5′-CAGCAGTCACGCGACCA-3′ (sense) and 5′-TGGTCGCGTTGACTGCTG-3′ (antisense), ApaI/C-site-mutated (C-Mu) 5′-CAGCAGTGTCGAGACCA (sense) and 5′-TGGTCTCGACACTGCTG-3′ (antisense), KpnI/C site (proximal) 5′-GTTCCCACACGCAGCCA-3′ (sense) and 5′-TGGCTGCGTGTGGGAAC-3′ (antisense), and KpnI/C site (distal) 5′-CGCAGACCACGAGGTGG-3′ (sense) and 5′-CCACCTCGTGGTCTGCG-3′ (antisense), specific motives being underlined. For the supershift assay, the nuclear extracts from 293T cells transfected with C-FLAG Hes1 cDNA in a pSRα vector were incubated with labeled DNA oligomers for 30 min and then with anti-FLAG antibody for 60 min followed by electrophoretic mobility shift assay (EMSA).
ChIP assay.
The chromatin immunoprecipitation (ChIP) assay was done according to the method of Szak et al. (28) with modifications. Briefly, HeLa cells transfected with Hes1 or empty vector 48 h before were fixed with 1% formaldehyde for 10 min, washed, and extracted with SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-Cl, pH 8.1, protease inhibitors). The lysates were diluted with dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-Cl, pH 8.1, 167 mM NaCl, 1 mM PMSF, 1 μg/ml aprotinin, 1 μg/ml pepstatin A, 1 μg/ml leupeptin), sonicated, diluted again, precleared with protein A Sepharose beads, and immunoprecipitated with anti-Hes1 or normal rabbit immunoglobulin G followed by protein A Sepharose beads. Immunocomplexes were washed with radioimmunoprecipitation assay buffer (150 mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% SDS, 50 mM Tris-HCl, pH 8.0, 5 mM EDTA) followed by incubation in cross-linking reversal buffer (125 mM Tris-HCl, pH 6.8, 10% 2-mercaptoethanol, 4% SDS). After boiling for 30 min, DNAs were extracted from the samples, followed by PCR. The primer set for the p27Kip1 gene promoter including the most proximal class C site was 5′-CGGCCGTTTGGCTAGTTTGTTTGT-3′ (sense; position, −2823) and 5′-AACCCAGCCGCTCTCCAAACCTTG-3′ (antisense; position, −3246), and that for the p27Kip1 gene-coding region intron is described above.
Statistical analysis.
Statistical analysis was done by Student's t test.
RESULTS
Retinoic acid induces down-regulation of Hes1 and increased p27Kip1 expression in concordance with differentiation-linked growth arrest in EC cells.
Treatment of undifferentiated F9 EC cells with RA resulted in significant growth arrest at G1 phase in 4 days (Fig. 1A). RA-treated F9 cells showed a marked accumulation of CDK inhibitor p27Kip1, while the expression level of another CDK inhibitor, p21Cip1, was largely unchanged (Fig. 1B), suggesting that p27Kip1 played a role in the RA-induced growth arrest of F9 cells. On the contrary, expression of Hes1 was markedly repressed in the RA-treated F9 cells, while uninduced F9 cells significantly expressed it (Fig. 1B). The reduced Hes1 and increased p27Kip1 expression following RA treatment was confirmed by using nuclear extracts (Fig. 1B). As shown in Fig. 1C, the level of p27Kip1 transcript was also increased significantly after RA treatment, suggesting that the augmented p27Kip1 expression in part reflected transcriptional activation. These results were confirmed by immunostaining analysis. Thus, Hes1 was detected as a fine granular staining in both the cytosol and nuclei of the vast majority of untreated F9 cells, while it was reduced markedly after RA treatment (Fig. 2). In contrast, the proportion of the cells strongly expressing nuclear p27Kip1 was greatly increased after RA treatment (Fig. 2). A portion of RA-treated but not untreated F9 cells expressed a tight junction-related protein, claudin-4, indicative of the differentiation into typical epithelial cells (Fig. 2).
FIG. 1.
Down-regulation of the endogenous Hes1 in concord with the increased p27Kip1 expression and arrest of cell cycle progression by retinoic acid treatment in F9 cells. (A) F9 EC cells were cultured in the absence or presence of 1 μM RA for 4 days, and DNA contents were analyzed by flow cytometry. (B) Aliquots of the cells described above were lysed, and either total cell or nuclear extracts were immunoblotted with the indicated antibodies. (C) RNAs were extracted from the aliquots described above, cDNAs were synthesized, and RT-PCR was performed for the indicated genes. Amplified products were Southern blotted with corresponding 32P-labeled cDNA probes. Relative densitometric intensities of RA-treated samples compared to the control samples are indicated.
FIG. 2.
Immunostaining analysis for the expression of Hes1 and p27Kip1 in the F9 cells treated with RA. F9 cells were cultured in the absence (above) or presence (below) of 1 μM RA for 4 days, fixed, permeabilized, blocked, and doubly stained with anti-Hes1 (green) and anti-p27Kip1 (red) antibodies, followed by DAPI staining. Aliquots of the cultured cells were also stained with anti-claudin-4 antibody.
Conditional expression of a moderate level of Hes1 in HeLa cells represses p27Kip1 expression and enhances proliferation with higher saturation density.
In order to directly investigate a possible link between Hes1 expression level and cell cycle control via p27Kip1 independently of cell differentiation, we then developed a HeLa cell line stably transfected with Hes1 in a tetracycline-inducible system (HeLa/Tet-Hes1). Parental HeLa cells as well as HeLa/Tet-Hes1 cells maintained in the presence of 10 ng/ml Dox (D10) exhibited undetectable Hes1. However, HeLa/Tet-Hes1 cells were induced to express Hes1 strongly within 24 h after the shift to Dox-free medium lasting for over 2 days, although the evaluation became hard later on due to the progressive cell death in this condition (see below) (Fig. 3A). As also shown in Fig. 3A, the expression level of Hes1 was dependent on the dose of Dox in the medium. Although HeLa/Tet-Hes1 D10 cells showed a growth rate comparable to that of the parental HeLa cells, the growth of HeLa/Tet-Hes1 cells in Dox-free medium (D0) was inhibited strongly (Fig. 3B). It was noted that HeLa/Tet-Hes1 D0 cells revealed progressive cell death after day 3, and the apparent growth inhibition was actually due to the reduced viability. In contrast, HeLa/Tet-Hes1 cells at 0.1 ng/ml Dox (D0.1) showed a significantly enhanced growth rate compared with that of the D10 or parental cells. After 6 days, both HeLa/Tet-Hes1 D0.1 and D10 cells were detached from the plates due to the physical overcrowdedness, but the former attained a higher saturation density than the latter. HeLa/Tet-Hes1 D10 cells expressed no detectable Hes1 throughout the culture period, and the expression level of p27Kip1 was increased progressively as the cell density increased (Fig. 3C), like in the wild-type HeLa cells (data not shown). HeLa/Tet-Hes1 D0.1 cells, on the other hand, did show moderate levels of Hes1 expression, peaking at day 4 after the induction, the reason for reduced Hes1 on day 6 being unknown. In these cells, the expression levels of p27Kip1 remained very low throughout the culture period in spite of the accelerated increase in cell density (Fig. 3C). Expression of p21Cip1 also tended to be increased slightly but much less extensively than p27Kip1 in HeLa/Tet-Hes1 D10 cells, and its level also remained low in HeLa/Tet-Hes1 D0.1 cells (Fig. 3C). These results indicated that expression of a controlled level of Hes1 suppressed the expression of endogenous p27Kip1 protein, leading to an accelerated growth rate, as well as increased saturation density, independent of cell differentiation process.
FIG. 3.
Effects of the conditional expression of Hes1 in HeLa cells on cell proliferation and CDK inhibitors. (A) HeLa/Tet-Hes1 cells maintained in the medium containing Dox (10 ng/ml) were shifted to fresh medium without Dox, and Hes1 expression was examined after indicated periods by immunoblotting (upper panel). HeLa/Tet-Hes1 cells were shifted to fresh media containing indicated doses of Dox, and 2 days later, Hes1 expression was examined by immunoblotting (lower panel). (B) HeLa/Tet-Hes1 cells were cultured at 1 × 105 cells/well of six-well plates, in medium containing 10 ng/ml (○), 0.1 ng/ml (□), or 0 ng/ml (▪) Dox, and the viable cells were counted at the indicated days. The culture medium was replaced with the fresh media containing the corresponding doses of Dox every other day. As a control, parental HeLa cells were cultured similarly in Dox-free medium (•). The means and standard errors (SE) of triplicate cultures are indicated. Asterisks indicate that the majority of cells were dying and detached from the dish. (C) Aliquots of HeLa/Tet-Hes1 cells in the cultures described above were lysed at the indicated days and immunoblotted with the indicated antibodies. The arrowhead indicates the Hes1 band.
Reduced p27Kip1 by Hes1 expression is attributed to transcriptional repression rather than to the enhanced proteasomal degradation.
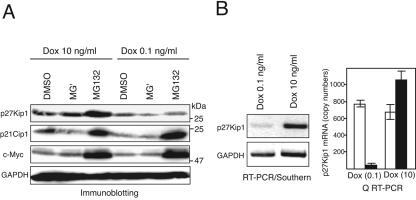
Levels of p27Kip1 protein in cycling cells are controlled by posttranscriptional regulation coupled with the cell cycle stages, with the basal level of the transcript being largely unchanged. We therefore investigated the possible contribution of protein degradation to the Hes1-induced p27Kip1 down-regulation in HeLa/Tet-Hes1 cells. In subconfluent HeLa/Tet-Hes1 D10 cells, the amount of endogenous p27Kip1 protein was increased significantly by treatment with a specific proteasome inhibitor (MG132), but it was not increased with a calpain inhibitor (MG′) (Fig. 4A) like in the parental HeLa cells (data not shown). In quite a contrast, similarly subconfluent HeLa/Tet-Hes1 D0.1 cells expressing a moderate level of Hes1 showed a reduced amount of p27Kip1 protein, and this was barely restored by the treatment with either MG132 or MG′ (Fig. 4A). Thus, the difference in the levels of p27Kip1 protein between HeLa/Tet-Hes1 cells at D0.1 and D10 in the presence of a proteasome inhibitor was much more striking than in the absence of one. The amounts of both p21Cip1 and c-Myc in HeLa/Tet-Hes1 D0.1 cells, on the other hand, were increased by MG132 treatment comparably to those in HeLa/Tet-Hes1 D10 cells (Fig. 4A). Thus, the Hes1-induced reduction of p27Kip1 was unlikely to be due to enhanced proteasomal degradation, although possible degradation via other mechanisms remained to be ruled out. We next examined the transcript of p27Kip1. As shown in Fig. 4B, basal p27Kip1 transcript was reduced markedly in HeLa/Tet-Hes1 D0.1 cells compared with HeLa/Tet-Hes1 D10 cells at the comparable cell confluence as judged by both RT-PCR followed by Southern blotting and quantitative real-time RT-PCR analysis. Thus, the results strongly suggested that the reduction of p27Kip1 protein by a moderate level of Hes1 expression was primarily ascribed to the repression of basal p27Kip1 transcription.
FIG. 4.
Reduced p27Kip1 protein by Hes1 expression is attributed to the repressed basal transcript rather than the enhanced proteasomal degradation. (A) HeLa/Tet-Hes1 cells were cultured in the presence of 0.1 ng/ml or 10 ng/ml Dox for 4 days, and a proteasome inhibitor MG132 (10 μM), a calpain inhibitor MG′ (10 μM), or a vehicle (dimethyl sulfoxide, 10 μl) was added for 2 h before the harvest. Cells were lysed and immunoblotted with the indicated antibodies. (B) (Left panel) HeLa/Tet-Hes1 cells were cultured in the presence of 0.1 ng/ml or 10 ng/ml Dox for 4 days, RNAs were extracted, and RT-PCR was performed for p27Kip1 and GAPDH. Amplified products were Southern blotted using corresponding 32P-labeled cDNA probes followed by autoradiography. (Right panel) Aliquots of the RNAs described above were amplified in real-time PCR using specific primer sets for p27Kip1 (solid bars) and 36B4 rRNA as an endogenous control (open bars). The mean copy numbers and SE are indicated.
Hes1 represses p27Kip1 gene transcription by binding to class C sites in the 5′ flanking regulatory region.
To investigate the direct effect of Hes1 on p27Kip1 transcription, a promoter assay was performed using a 3.5-kb-long 5′ flanking DNA of the p27Kip1 gene containing the basal regulatory region, p27PF, ligated with a luciferase reporter (Fig. 1A). It was reported previously that a CCAAT box at −522 to −518, to which ubiquitous transcription factor NF-Y bound, was essential for basal promoter activity (14). A sequence survey revealed that p27PF contained multiple potential Hes1 binding sites upstream of the CCAAT box, including four sequences compatible with class C sites and three with N box (Fig. 5A). HeLa/Tet-Hes1 cells were transfected with p27PF or p27PF/ApaI that contained a single class C site most proximal to the CCAAT box and cultured in the presence of various doses of Dox for 2 days. By use of aliquots of the cells, it was confirmed that Hes1 expression was progressively increased as the Dox concentrations were reduced, whereas expression of the endogenous p27Kip1 was decreased in an exactly converse manner in these particular experiments (Fig. 5B). As shown in Fig. 5B, it was indicated that the promoter activities of both p27PF and p27PF/ApaI were reduced comparably as the expression level of Hes1 was increased, resulting in nearly 80% suppression in the absence of Dox. To examine the involvement of the class C site in repression, mutations were introduced in the class C site of p27PF/ApaI (p27PF/ApaI-Mu). In contrast to p27PF/ApaI, the promoter activity of p27PF/ApaI-Mu was not affected, even by the maximal expression of Hes1 in the absence of Dox, indicating the involvement of the class C site in repression (Fig. 5C). Similar results were obtained using an independent cell line, C3H10T1/2, in that the cotransfection of Hes1 markedly repressed the promoter activity of p27PF/ApaI but not of p27PF/ApaI-Mu (data not shown). Repressor activity of Hes1 via N box is shown to be dependent on the C-terminal WRPW domain, through which repressor cofactor Groucho (TLE1) is recruited (19). Hes1 deleted of a WRPW domain (Hes1ΔWRPW), however, repressed the promoter activity of p27PF comparably to the wild-type Hes1, suggesting that the repression of p27Kip1 by Hes1 was independent of Groucho (Fig. 5D).
FIG. 5.
Hes1 expression represses the promoter activity of the 5′ flanking regulatory region of p27Kip1 gene in a dose-dependent manner. (A) A schematic representation of a 3.5-kb 5′ flanking DNA upstream of the translation start site of the p27Kip1 gene (p27PF). There are multiple potential Hes1 binding sites, three sites compatible with N box and four sites compatible with class C site. Two positive elements, Sp1 and CTF sites, to which transcription factors Sp1 and NF-Y have been shown to bind and enhance the promoter activity, respectively, are also indicated. An ApaI fragment of p27PF (p27PF/ApaI) contains a single class C site (CACGCG), which has been mutated to GTCGAC by site-directed mutagenesis (p27PF/ApaI-Mu). (B) HeLa/Tet-Hes1 cells were transfected with p27PF (solid columns) or p27PF/ApaI (grey columns) plasmid and cultured in the absence or presence of various concentrations of Dox for 2 days, and the luciferase activity was assayed and corrected for the transfection efficiency. The activities were normalized to that of the cells at 10 ng/ml Dox, and the means and SE of three independent experiments are indicated. The mean relative light units (RLUs) of p27PF and p27PF/ApaI at 10 ng/ml Dox were 5.7 × 105 and 45.2 × 105, respectively. Asterisks indicate statistically significant suppression (P < 0.01). Aliquots of the cells were lysed and immunoblotted with anti-Hes1 and anti-p27Kip1 antibodies to ensure their expression profiles in the particular experiments. (C) HeLa/Tet-Hes1 cells were transfected with p27PF/ApaI (open columns) or p27PF/ApaI-Mu (filled columns) plasmid, cultured in the absence of Dox and in the presence of various concentrations of Dox for 2 days, and the luciferase activity was assayed as described above. The activities were normalized to that of the cells at 10 ng/ml Dox, and the means and SE of three independent experiments are indicated. The mean RLUs of p27PF/ApaI and p27PF/ApaI-Mu at 10 ng/ml Dox were 3.6 × 105 and 1.7 × 105, respectively. Asterisks indicate statistically significant suppression (P < 0.05). (D) C3H10T1/2 cells were transfected with a plasmid containing neo, Hes1, or _Hes1_ΔWRPW cDNA, along with p27PF and pRL-TK plasmid, and 2 days later, the luciferase activity was determined. Means of RLUs in triplicate culture corrected for the transfection efficiency are indicated.
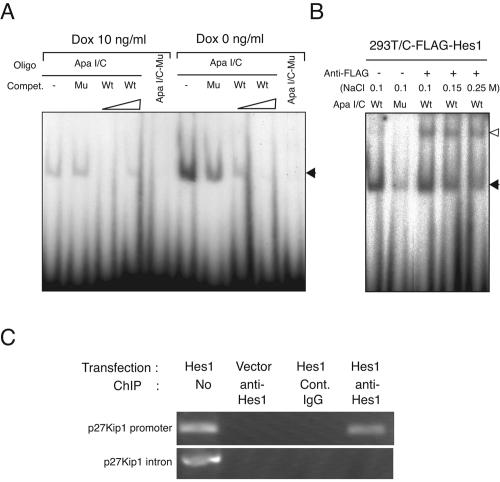
We then examined the direct binding of Hes1 to the class C sites of p27PF by EMSA. As shown in Fig. 6A, a specific protein binding to the DNA oligomer containing a class C site of p27PF/ApaI (ApaI/C) was detected in the nuclear extract of HeLa/Tet-Hes1 D0 cells, while significant protein binding was barely found in the extract of HeLa/Tet-Hes1 D10 cells. Although the specific protein binding was competitively inhibited by cold, excessive ApaI/C oligomer, an oligomer with site-directed mutations within the class C site (ApaI/C-Mu) affected the binding only marginally if at all (Fig. 6A). Consistent with this, no protein binding to the ApaI/C-Mu oligomer was detected by using the nuclear extract of HeLa/Tet-Hes1 D0 cells (Fig. 6A). Although not shown, similar results were obtained by using two oligomers including more-upstream class C sites of p27PF (Fig. 5A). Because available anti-Hes1 and anti-FLAG antibodies reacted poorly to the native Hes1 and N-terminally FLAG-tagged Hes1, respectively (Murata et al., unpublished observation), we attempted the supershift assay by using C-FLAG Hes1 and anti-FLAG antibody. Nuclear extracts of 293T cells transfected with C-FLAG Hes1 cDNA were incubated with labeled ApaI/C oligomer, followed by anti-FLAG antibody. As shown in Fig. 6B, a significant proportion of the oligomer-bound band was supershifted by anti-FLAG antibody. To further confirm the binding of Hes1 to p27Kip1 promoter in vivo, a ChIP assay was performed. In HeLa cells transfected with Hes1 cDNA, DNA of the p27Kip1 promoter region including a proximal C site but not that of the p27Kip1 gene intron could be specifically detected in the Hes1-immunoprecipitated DNA complex from formaldehyde-treated cells, indicating Hes1 occupancy at the p27Kip1 promoter in vivo (Fig. 6C). These results collectively indicated that Hes1 repressed the transcription of p27Kip1 gene via direct binding to class C sites in the promoter region.
FIG. 6.
Hes1 binds to the 5′ flanking regulatory region of p27Kip1 gene via class C sites. (A) HeLa/Tet-Hes1 cells were cultured in the absence or presence of Dox (10 ng/ml) for 2 days, and the nuclear extracts were prepared. Although not shown, it was confirmed that the extract in the absence of Dox contained Hes1, and the extract in the presence of Dox barely did. Each extract was incubated with a 32P-labeled class C site-containing DNA oligomer of the p27PF/ApaI or the oligomer with mutation within the C site, and EMSA was performed. As competitors, excess doses (100- and 200-fold) of cold ApaI/C or ApaI/C-Mu oligomer were included. The solid arrowhead indicates the mobility-shifted oligomer band. (B) 293T cells were transfected with C-terminally FLAG-tagged Hes1 cDNA, and the nuclear extracts were incubated with labeled ApaI/C or ApaI/C-Mu oligomer, followed by anti-FLAG antibody in various salt concentrations. The open arrowhead indicates the supershifted band; the solid arrowhead indicates the original mobility-shifted oligomer band. (C) HeLa cells were transfected with Hes1 cDNA or vector alone, cross-linked with formaldehyde, and immunoprecipitated with anti-Hes1 or control rabbit immunoglobulin G (Cont. IgG). DNAs were extracted from the immunocomplexes and PCR amplified by the primer set of p27Kip1 promoter region or a p27Kip1 gene intron. DNA without ChIP served as a control (the first lane).
Progenitors in _Hes1_-deficient embryos express elevated levels of basal p27Kip1 transcript compared with those in control littermates.
We finally examined the expression of p27Kip1 in the thymocytes of _Hes1_−/− embryos. As shown in Fig. 7A, on E14.5, _Hes1_−/− mice, which typically lacked eyes, showed much smaller thymic lobes, and the thymocyte numbers were less than 10% of the controls on average. RT-PCR analysis followed by Southern blotting using a 32P-labeled p27Kip1 cDNA probe indicated that p27Kip1 transcript was significantly increased in the thymocytes of _Hes1_−/− embryos compared with those of Hes1+/− littermate embryos as standardized by GAPDH transcript (Fig. 7B). The experiments were repeated using four _Hes1_−/− and four Hes1+/− embryos in two littermates, and the results indicated a significant difference between them (Fig. 7B). Similar results were also obtained in hematopoietic fetal liver cells and brains by quantitative real-time RT-PCR, in that the progenitor cells of _Hes1_−/− embryos exhibited significantly more p27Kip1 transcript than those of Hes1+/− embryos (Fig. 7C). The results thus confirmed that Hes1 deficiency in vivo indeed resulted in an increased basal level of p27Kip1 transcript in the tissue progenitors and strongly suggested that defective p27Kip1 repression was responsible at least in part for the impaired expansion of _Hes1_−/− T-cell progenitors in the thymus.
FIG. 7.
Increased p27Kip1 transcript in the thymocytes of _Hes1_−/− embryos compared with Hes1+/− littermates. (A) Hes1+/− mice were mated, and embryos at E14.5 were genotyped. Thymic lobes of the _Hes1_−/− embryo, which lacked eyes (arrows), were severely hypoplastic compared with those of Hes1+/− littermates (inserts). The thymocyte numbers in the former were less than 10% of the latter. (B) (Left panel) RNAs were extracted from the thymocytes of Hes1+/− and _Hes1_−/− embryos, and RT-PCR was performed for Hes1, p27Kip1, and GAPDH, followed by Southern blotting for p27Kip1 and GAPDH, using corresponding 32P-labeled cDNA probes. (Right panel) Means and SE of the radiointensities of p27Kip1 RT-PCR products following Southern blotting normalized by those of GAPDH in the thymocytes of four Hes1+/− and four _Hes1_−/− embryos are indicated. (C) RNAs were extracted from the liver cells and brain tissues of Hes1+/− and _Hes1_−/− embryos, and quantitative real-time RT-PCR was performed using a p27Kip1 primer set. The mean copy numbers and SE of p27Kip1 transcript normalized to those of control cyclophilin transcripts are indicated.
DISCUSSION
Hes1 is one of the major target genes in Notch signaling, and it plays an essential role in the embryonic development of neuronal and endocrine tissues (13, 18). While Hes1 controls cell differentiation by transcriptionally repressing the genes related to tissue specification, it is also suggested that Hes1 may be involved in maintaining the proliferation potential of progenitors (25). The probable mechanisms for control of cell proliferation by Hes1, however, remain largely unknown. In the present study, we first examined the expression profile of endogenous Hes1 in undifferentiated EC cells following RA treatment. Hes1 expression was down-regulated by RA treatment in concord with the arrest of cell cycle progression, which was associated with a marked increase in CDK inhibitor p27Kip1. The increase in p27Kip1 was at least in part due to the increase in its basal transcript, prompting us to investigate the possibility that Hes1 might control cell proliferation through the regulation of p27Kip1 gene expression.
The growth arrest of EC cells by RA was tightly linked to cell differentiation, and the effect described above could be a secondary consequence of differentiation program. We then employed HeLa cells that expressed no endogenous Hes1 to examine the effect of Hes1 expression on cell proliferation independently of differentiation. Since stable Hes1-transfectant lines were not obtained by use of regular high-expression vectors as reported in other cell lines (27), we produced a HeLa cell line stably transfected with Hes1 in a tetracycline-inducible system. The HeLa/Tet-Hes1 cells were conditionally induced to express various amounts of Hes1, depending on the doses of Dox. Maximal expression of Hes1 in the absence of Dox resulted in progressive cell death as expected, exact mechanisms for which remained to be seen. On the other hand, expression of a moderate level of Hes1 in the presence of a low dose of Dox (0.1 ng/ml) significantly enhanced cell proliferation and increased the saturation density, reminiscent of compromised contact inhibition, compared to those with undetectable Hes1 expression in the presence of 10-ng/ml Dox or parental HeLa cells. The enhanced proliferation by Hes1 expression was associated with the reduced expression of endogenous p27Kip1, which paralleled the decline of basal p27Kip1 transcript. Such repression was specific for p27Kip1, since the expression of another CDK inhibitor, p21Cip1, was largely unaffected. The p27Kip1 level in cycling cells is controlled posttranscriptionally by regulated protein degradation via ubiquitin/proteasome pathway (22), and consistently, the amount of p27Kip1 protein in HeLa/Tet-Hes1 D10 cells was increased markedly by treatment with a specific proteasome inhibitor. In HeLa/Tet-Hes1 D0.1 cells, however, the reduced p27Kip1 expression was barely restored by the proteasome inhibitor. The results strongly suggested that the reduction of p27Kip1 by Hes1 expression was primarily attributed to the repression of its basal transcript, although additional involvement of other posttranscriptional regulation remains possible.
It was confirmed by an in vitro reporter assay that the basal promoter activity of a 5′ flanking regulatory region of p27Kip1 gene (p27PF) in HeLa/Tet-Hes1 cells was repressed in a manner dependent on the expression levels of Hes1. Hes1 has been shown to act as a transcriptional repressor by directly binding to N box sequences (CACNAG) in the rodent Hes1 gene (24, 29) or a similar sequence called class C site (CACGCG) in the human ASH1 gene (3). It was found that p27PF contained multiple motifs compatible with the consensus N box or class C site, one of the class C sites (ApaI/C) being located in the close vicinity of the CCAAT box responsible for the basal promoter activity of p27Kip1 gene. EMSA analysis indicated that the nuclear extract of HeLa/Tet-Hes1 D0.1 cells, but not of HeLa/Tet-Hes1 D10 cells, contained a protein that was specifically bound to the class C site-containing DNA oligomers of p27PF, including an ApaI/C oligomer. The protein binding was not detected for ApaI/C-Mu, and ApaI/C-Mu did not inhibit the protein binding to the wild-type oligomer, indicating that binding was indeed mediated by the class C site. By supershift assay using C-terminally FLAG-tagged Hes1 and anti-FLAG antibody, it was strongly suggested that Hes1 could bind to the ApaI/C of p27PF. Finally, it was confirmed by ChIP assay that Hes1 was indeed bound selectively to the p27Kip1 gene promoter region including a proximal class C site in the HeLa cells expressing Hes1. Collectively, it was indicated that Hes1 repressed the p27Kip1 transcription by directly binding to the class C sites in the 5′ flanking regulatory region. This repressive activity was suggested to be independent of repressive cofactor Groucho, because a Hes1 deleted of C-terminal WPRW domain required for the Groucho recruitment (5) was still capable of repressing the p27Kip1 promoter activity comparably to the wild-type Hes1, and this exact mechanism remains to be investigated.
Transcriptional regulation of the p27Kip1 gene is shown to play roles in cell proliferation, differentiation, and survival in several cellular systems. For instance, vitamin D3 induces the terminal differentiation of myeloid leukemia cell lines into monocytes/macrophages via activation of p27Kip1 gene transcription, which is mediated by ubiquitous transcription factors Sp1 and NF-Y (9). Cytokine-mediated proliferation and survival of hematopoietic cells, on the other hand, are regulated by the down-regulation of p27Kip1 transcription, which results from the inhibited transcriptional activity of folkhead transcription factor FKHR-L1 through PI3K-dependent phosphorylation (4). It is also reported that tissue toxicity caused by dioxin is mediated through the direct transcriptional activation of p27Kip1 gene by Ah receptor, a member of the bHLH/PAS (period-AhR/ARNT-Sim) transcription factor family, resulting in impaired cell proliferation (16). These results indicate that distinct transcription factors positively control the p27Kip1 gene transcription, depending on the contexts of cells and stimuli. Present results, on the other hand, strongly suggest that Hes1 represents one of the first transcriptional repressors for the p27Kip1 gene, thereby directly promoting cell proliferation.
Hes1 deficiency in vivo results in the impaired expansion of progenitors in various developing tissues (30), and this may make a good contrast to p27 Kip1 deficiency in vivo, which leads to the hyperplasia of many tissues (21). Thus, it is implied that a physiological role of Hes1 may be in part related to the direct promotion and maintenance of cell proliferation in the progenitor cells. Hes1 is a major downstream target of Notch signaling (13), and there is significant evidence that Notch family receptors contribute to the regulation of cell proliferation directly rather than as a consequence of differentiation. Typically, constitutive active forms of Notch have been shown to be oncogenic in acute lymphoblastic leukemia (T-ALL) and other tumors (32). It was indicated that Notch inhibitors induced the G1 arrest of the cell cycle and suppressed the proliferation of Notch-associated T-ALL cells (34). Since the Notch inhibitors were shown to suppress Hes1 expression rapidly and completely in the T-ALL cells (34), it might be possible that Hes1 was in part responsible for their continuous proliferation. While expression of active Notch rather induced cell cycle arrest in small cell lung cancer cells, this activity was shown to be independent of Hes1 (26). With PC12 cells, however, it was also reported that conditional Hes1 expression rather inhibited the nerve growth factor-induced cell proliferation as well as the differentiation, along with the paradoxically reduced p21Cip1 (2). On the other hand, it was reported that Hes1 repression in neuronal stem cells in vitro caused reduced proliferation and accelerated differentiation with concomitant increase in p21Cip1 (12). Thus, Hes1 may also affect the expression of p21Cip1 either directly or indirectly by influencing the differentiation in certain cells, and its involvement in the control of proliferation by Hes1 remains to be verified.
During embryonic development, primitive T-cell progenitors in the liver migrate into the thymus and extensively expand preceding the rearrangement of antigen receptor genes (33). In Notch1 or Hes1 deficiency, the thymocyte expansion was arrested almost completely at the earliest stages (30). The arrest of proliferation of _Hes1_−/− T-cell progenitors was not accompanied by detectable differentiation or maturation, suggesting that the effect of Hes1 was primary rather than secondary to the differentiation (29). Present results confirmed that the basal p27Kip1 transcript was increased significantly in the _Hes1_−/− thymocytes in vivo compared with those in the Hes1+/− littermates. _Hes1_−/− fetal hematopoietic liver cells as well as brains also exhibited increased basal p27Kip1 transcript, although fetal liver cell numbers were largely unchanged. It is of note that one of the most prominent tissue toxicities by dioxin in mice is thymic hypoplasia, and p27Kip1-deficient thymic glands are much less sensitive to the toxic effect than those of wild-type mice (16). Thus, it may be suggested that transcriptional repression of the p27Kip1 gene is particularly important in sustaining the extensive progenitor expansion in certain tissues such as thymus. Interestingly, while _Hes1_−/− thymocytes were almost totally arrested for proliferation in vivo when transferred into _Rag2_−/− mice (30), they could considerably grow and expand in the serum-containing organ culture system (15), indicating that their arrested proliferation in vivo was at least in part reversible in vitro. We presume that potent serum factors in the culture can induce effective protein degradation of the increased p27Kip1 in _Hes1_−/− thymocytes to resume their proliferation in the culture, and this reversibility may explain significant variety in the extents of thymic hypoplasia of individual _Hes1_−/− embryos (30). In any case, present results strongly suggest that Hes1 plays a crucial role in promoting and maintaining the developmentally programmed proliferation of thymic T-cell progenitors and possibly of the progenitors in other systems, via constitutive repression of the basal p27Kip1 gene transcription.
Acknowledgments
We thank T. Sudo, S. Takeda, and S. Tsukita for providing vectors and antibody and Y. Hamazaki for helping immunostaining.
This work was supported by grant-in-aid from the Ministry of Education, Science, Sports, and Culture of Japan.
REFERENCES
- 1.Allen, R. D., III, H. K. Kim, S. D. Sarafova, and G. Siu. 2001. Negative regulation of CD4 gene expression by a HES-1-c-Myb complex. Mol. Cell. Biol. 21**:**3071-3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Castella, P., S. Sawai, K. Nakao, J. A. Wagner, and M. Caudy. 2000. HES-1 repression of differentiation and proliferation in PC12 cells: role for the helix 3-helix 4 domain in transcription repression. Mol. Cell. Biol. 20**:**6170-6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, H., A. Thiagalingam, H. Chopra, M. W. Borges, J. N. Feder, B. D. Nelkin, S. B. Baylin, and D. W. Ball. 1997. Conservation of the Drosophila lateral inhibition pathway in human lung cancer: a hairy-related protein (HES-1) directly represses achaete-scute homolog-1 expression. Proc. Natl. Acad. Sci. USA 94**:**5355-5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dijkers, P. F., R. H. Medema, C. Pals, L. Banerji, N. S. Thomas, E. W. Lam, B. M. Burgering, J. A. Raaijmakers, J. W. Lammers, L. Koenderman, and P. J. Coffer. 2000. Forkhead transcription factor FKHR-L1 modulates cytokine-dependent transcriptional regulation of p27(KIP1). Mol. Cell. Biol. 20**:**9138-9148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grbavec, D., and S. Stifani. 1996. Molecular interaction between TLE1 and the carboxyl-terminal domain of HES-1 containing the WRPW motif. Biochem. Biophys. Res. Commun. 223**:**701-705. [DOI] [PubMed] [Google Scholar]
- 6.Hamazaki, Y., M. Itoh, H. Sasaki, M. Furuse, and S. Tsukita. 2002. Multi-PDZ domain protein 1 (MUPP1) is concentrated at tight junctions through its possible interaction with claudin-1 and junctional adhesion molecule. J. Biol. Chem. 277**:**455-461. [DOI] [PubMed] [Google Scholar]
- 7.Hattori, M., N. Tsukamoto, M. S. Nur-e-Kamal, B. Rubinfeld, K. Iwai, H. Kubota, H. Maruta, and N. Minato. 1995. Molecular cloning of a novel mitogen-inducible nuclear protein with a Ran GTPase-activating domain that affects cell cycle progression. Mol. Cell. Biol. 15**:**552-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hirata, H., K. Tomita, Y. Bessho, and R. Kageyama. 2001. Hes1 and Hes3 regulate maintenance of the isthmic organizer and development of the mid/hindbrain. EMBO J. 20**:**4454-4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inoue, T., J. Kamiyama, and T. Sakai. 1999. Sp1 and NF-Y synergistically mediate the effect of vitamin D(3) in the p27(Kip1) gene promoter that lacks vitamin D response elements. J. Biol. Chem. 274**:**32309-32317. [DOI] [PubMed] [Google Scholar]
- 10.Ishibashi, M., S. L. Ang, K. Shiota, S. Nakanishi, R. Kageyama, and F. Guillemot. 1995. Targeted disruption of mammalian hairy and Enhancer of split homolog-1 (HES-1) leads to up-regulation of neural helix-loop-helix factors, premature neurogenesis, and severe neural tube defects. Genes Dev. 9**:**3136-3148. [DOI] [PubMed] [Google Scholar]
- 11.Ishibashi, M., K. Moriyoshi, Y. Sasai, K. Shiota, S. Nakanishi, and R. Kageyama. 1994. Persistent expression of helix-loop-helix factor HES-1 prevents mammalian neural differentiation in the central nervous system. EMBO J. 13**:**1799-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kabos, P., A. Kabosova, and T. Neuman. 2002. Blocking HES1 expression initiates GABAergic differentiation and induces the expression of p21CIP1/WAF1 in human neural stem cells. J. Biol. Chem. 277**:**8763-8766. [DOI] [PubMed] [Google Scholar]
- 13.Kageyama, R., and S. Nakanishi. 1997. Helix-loop-helix factors in growth and differentiation of the vertebrate nervous system. Curr. Opin. Genet. Dev. 7**:**659-665. [DOI] [PubMed] [Google Scholar]
- 14.Kamiyama, J., T. Inoue, N. Ohtani-Fujita, S. Minami, H. Yamagishi, and T. Sakai. 1999. The ubiquitous transcription factor NF-Y positively regulates the transcription of human p27Kip1 through a CCAAT box located in the 5-upstream region of the p27Kip1 gene. FEBS Lett. 455**:**281-285. [DOI] [PubMed] [Google Scholar]
- 15.Kaneta, M., M. Osawa, K. Sudo, H. Nakauchi, A. G. Farr, and Y. Takahama. 2000. A role for pref-1 and HES-1 in thymocyte development. J. Immunol. 164**:**256-264. [DOI] [PubMed] [Google Scholar]
- 16.Kolluri, S. K., C. Weiss, A. Koff, and M. Gottlicher. 1999. p27(Kip1) induction and inhibition of proliferation by the intracellular Ah receptor in developing thymus and hepatoma cells. Genes Dev. 13**:**1742-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kunisato, A., S. Chiba, E. Nakagami-Yamaguchi, K. Kumano, T. Saito, S. Masuda, T. Yamaguchi, M. Osawa, R. Kageyama, H. Nakauchi, M. Nishikawa, and H. Hirai. 2003. HES-1 preserves purified hematopoietic stem cells ex vivo and accumulates side population cells in vivo. Blood 101**:**1777-1783. [DOI] [PubMed] [Google Scholar]
- 18.Lewis, J. 1996. Neurogenic genes and vertebrate neurogenesis. Curr. Opin. Neurobiol. 6**:**3-10. [DOI] [PubMed] [Google Scholar]
- 19.McLarren, K. W., F. M. Theriault, and S. Stifani. 2001. Association with the nuclear matrix and interaction with Groucho and RUNX proteins regulate the transcription repression activity of the basic helix loop helix factor Hes1. J. Biol. Chem. 276**:**1578-1584. [DOI] [PubMed] [Google Scholar]
- 20.Minami, S., N. Ohtani-Fujita, E. Igata, T. Tamaki, and T. Sakai. 1997. Molecular cloning and characterization of the human p27Kip1 gene promoter. FEBS Lett. 411**:**1-6. [DOI] [PubMed] [Google Scholar]
- 21.Nakayama, K., N. Ishida, M. Shirane, A. Inomata, T. Inoue, N. Shishido, I. Horii, and D. Y. Loh. 1996. Mice lacking p27(Kip1) display increased body size, multiple organ hyperplasia, retinal dysplasia, and pituitary tumors. Cell 85**:**707-720. [DOI] [PubMed] [Google Scholar]
- 22.Pagano, M., S. W. Tam, A. M. Theodoras, P. Beer-Romero, G. Del Sal, V. Chau, P. R. Yew, G. F. Draetta, and M. Rolfe. 1995. Role of the ubiquitin-proteasome pathway in regulating abundance of the cyclin-dependent kinase inhibitor p27. Science 269**:**682-685. [DOI] [PubMed] [Google Scholar]
- 23.Radtke, F., A. Wilson, G. Stark, M. Bauer, J. van Meerwijk, H. R. MacDonald, and M. Aguet. 1999. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity 10**:**547-558. [DOI] [PubMed] [Google Scholar]
- 24.Sasai, Y., R. Kageyama, Y. Tagawa, R. Shigemoto, and S. Nakanishi. 1992. Two mammalian helix-loop-helix factors structurally related to Drosophila hairy and enhancer of split. Genes Dev. 6**:**2620-2634. [DOI] [PubMed] [Google Scholar]
- 25.Solecki, D. J., X. L. Liu, T. Tomoda, Y. Fang, and M. E. Hatten. 2001. Activated Notch2 signaling inhibits differentiation of cerebellar granule neuron precursors by maintaining proliferation. Neuron 31**:**557-568. [DOI] [PubMed] [Google Scholar]
- 26.Sriuranpong, V., M. W. Borges, R. K. Ravi, D. R. Arnold, B. D. Nelkin, S. B. Baylin, and D. W. Ball. 2001. Notch signaling induces cell cycle arrest in small cell lung cancer cells. Cancer Res. 61**:**3200-3205. [PubMed] [Google Scholar]
- 27.Strom, A., P. Castella, J. Rockwood, J. Wagner, and M. Caudy. 1997. Mediation of NGF signaling by post-translational inhibition of HES-1, a basic helix-loop-helix repressor of neuronal differentiation. Genes Dev. 11**:**3168-3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szak, S. T., D. Mays, and J. A. Pietenpol. 2001. Kinetics of p53 binding to promoter sites in vivo. Mol. Cell. Biol. 21**:**3375-3386.11313463 [Google Scholar]
- 29.Takebayashi, K., Y. Sasai, Y. Sakai, T. Watanabe, S. Nakanishi, and R. Kageyama. 1994. Structure, chromosomal locus, and promoter analysis of the gene encoding the mouse helix-loop-helix factor HES-1. Negative autoregulation through the multiple N box elements. J. Biol. Chem. 269**:**5150-5156. [PubMed] [Google Scholar]
- 30.Tomita, K., M. Hattori, E. Nakamura, S. Nakanishi, N. Minato, and R. Kageyama. 1999. The bHLH gene Hes1 is essential for expansion of early T cell precursors. Genes Dev. 13**:**1203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsukamoto, N., M. Hattori, H. Yang, J. L. Bos, and N. Minato. 1999. Rap1 GTPase-activating protein SPA-1 negatively regulates cell adhesion. J. Biol. Chem. 274**:**18463-18469. [DOI] [PubMed] [Google Scholar]
- 32.Uyttendaele, H., G. Marazzi, G. Wu, Q. Yan, D. Sassoon, and J. Kitajewski. 1996. Notch4/int-3, a mammary proto-oncogene, is an endothelial cell-specific mammalian Notch gene. Development 122**:**2251-2259. [DOI] [PubMed] [Google Scholar]
- 33.von Boehmer, H. 1994. Positive selection of lymphocytes. Cell 76**:**219-228. [DOI] [PubMed] [Google Scholar]
- 34.Weng, A. P., Y. Nam, M. S. Wolfe, W. S. Pear, J. D. Griffin, S. C. Blacklow, and J. C. Aster. 2003. Growth suppression of pre-T acute lymphoblastic leukemia cells by inhibition of notch signaling. Mol. Cell. Biol. 23**:**655-664. [DOI] [PMC free article] [PubMed] [Google Scholar]