Production of infectious hepatitis C virus in tissue culture from a cloned viral genome (original) (raw)
. Author manuscript; available in PMC: 2010 Aug 9.
Published in final edited form as: Nat Med. 2005 Jun 12;11(7):791–796. doi: 10.1038/nm1268
Abstract
Hepatitis C virus (HCV) infection causes chronic liver diseases and is a global public health problem. Detailed analyses of HCV have been hampered by the lack of viral culture systems. Subgenomic replicons of the JFH1 genotype 2a strain cloned from an individual with fulminant hepatitis replicate efficiently in cell culture. Here we show that the JFH1 genome replicates efficiently and supports secretion of viral particles after transfection into a human hepatoma cell line (Huh7). Particles have a density of about 1.15–1.17 g/ml and a spherical morphology with an average diameter of about 55 nm. Secreted virus is infectious for Huh7 cells and infectivity can be neutralized by CD81-specific antibodies and by immunoglobulins from chronically infected individuals. The cell culture–generated HCV is infectious for chimpanzee. This system provides a powerful tool for studying the viral life cycle and developing antiviral strategies.
HCV is a major cause of chronic liver diseases1,2. Development of selectable drugs and efficient vaccines has been hampered by poor virus growth in cell culture3. Although subgenomic replicons replicate efficiently in cultured cells4, for unknown reasons infectious viral particles are not produced5,6. Subgenomic replicons of the HCV genotype 2a JFH1 strain cloned from an individual with fulminant hepatitis7 replicate efficiently in Huh7 cells and do not require cell culture–adaptive mutations8–10. In this study, we show that transfection of _in vitro_–transcribed full-length JFH1 RNA into Huh7 cells results in secretion of viral particles that are infectious for cultured cells and a chimpanzee.
Results
Virus production from cells transfected with full-length JFH1 RNA
We transfected _in vitro_–transcribed RNAs corresponding to the full-length JFH1 genome (Fig. 1a) and a replication-incompetent mutant (JFH1/GND) into Huh7 cells. Total RNAs from cells harvested at different time points were analyzed by northern hybridization (Fig. 1b). Up to 12 h after transfection, we detected only degraded input RNA. But genome-length RNA was found in JFH1-transfected cells after 24 h, and remained detectable up to 72 h. The same was true for expression of viral proteins (Fig. 1c and Supplementary Fig. 1 online). Immunofluorescence analysis of JFH1-transfected cells showed that 70–80% of the cells were positive for core and NS3 proteins at 72 h after transfection (Fig. 1d), indicating that this genome replicates to high levels in transfected Huh7 cells.
Figure 1.
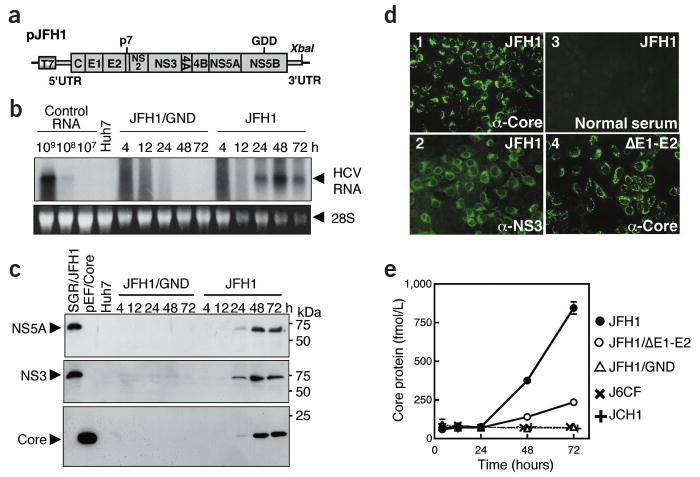
Transient replication of JFH1 RNA in transfected Huh7 cells. (a) Organization of the full-length HCV construct pJFH1. Open reading frames (thick boxes) are flanked by the 5′- and 3′-UTRs (thin boxes). T7, T7 RNA polymerase promoter; GDD, active-site motif of NS5B polymerase; XbaI, restriction site. (b) Northern blot analysis of total RNA prepared from cells transfected with full-length JFH1 and JFH1/GND RNA. Control RNA, given numbers of synthetic HCV RNA; Huh7, RNA isolated from naive cells. Arrowheads indicate full-length HCV RNA and 28S ribosomal RNA (28S). Upper panel, northern blot; lower panel, ethidium bromide staining. (c) Western blot analysis of transfected cells for HCV proteins NS5A, NS3 and core. Lysates of SGR/JFH1-RNA8 or pEF/Core29 DNA– transfected Huh7 cells and naive Huh7 cells served as positive and negative controls, respectively. (d) Immunofluorescence assay of cells fixed 72 h after transfection with JFH1 or JFH1/ΔE1-E2 RNA. Magnification, ×200. (e) HCV core protein secretion into culture medium. HCV RNAs were transfected into Huh7 cells, culture media were harvested at given time points, and HCV core protein concentrations were determined.
As a surrogate for virus production, we quantified secretion of core protein into the culture medium of cells transfected with JFH1 mutants JFH/GND and JFH1/ΔE1-E2 or full-length genomes of different origin (J6CF11 or JCH1 (ref. 7)). Core protein was secreted efficiently from JFH1 RNA but much less efficiently from JFH1/ΔE1-E2 RNA–transfected cells (Fig. 1e), despite comparable core protein levels. No core protein was secreted from the other RNA-transfected cells, consistent with their lack of replication (data not shown). These results suggest that core secretion depends on HCV RNA replication and that envelope glycoproteins are required, although the RNA segment deleted in JFH1/ΔE1-E2 may contain signal(s) required for core secretion.
To determine whether JFH1 RNA–transfected cells can sustain continuous HCV replication, we serially passaged cells. Cells transfected with JFH1/ΔE1-E2 RNA or a subgenomic replicon (SGR-JFH1)8 lacking the core to NS2 region served as controls. We determined HCV RNA titer using real-time detection reverse transcription (RTD)-PCR12. In JFH1-transfected cells, viral RNA and core protein titers in the medium increased rapidly at 5 d after transfection, and remained high for the next 7 d, followed by a slow decrease (Fig. 2a). In contrast, RNA levels in supernatant of control cells gradually decreased with increasing passage. At day 30, background levels were reached, which were about 100-fold lower as compared to JFH1 (Fig. 2a). During the first two passages, transfected cells had similar levels of intracellular HCV RNA, but they declined much more rapidly in the controls (Fig. 2a). We also determined levels of core protein and obtained similar results (Supplementary Fig. 2 online). Immunofluorescence assay showed that 3 d after transfection, JFH1 RNA and JFH1/ΔE1-E2 RNA transfection gave similar numbers of HCV-positive cells (Fig. 1d). After 13 d, 50–60% of JFH1 RNA–transfected cells were still positive for core protein, but only a few with the mutant (Fig. 2b). These results indicate a limited spread of infection. Alternatively, JFH1 RNA–replicating cells may have a longer half-life than cells containing JFH1/ΔE1-E2 or SGR-JFH1 RNAs.
Figure 2.
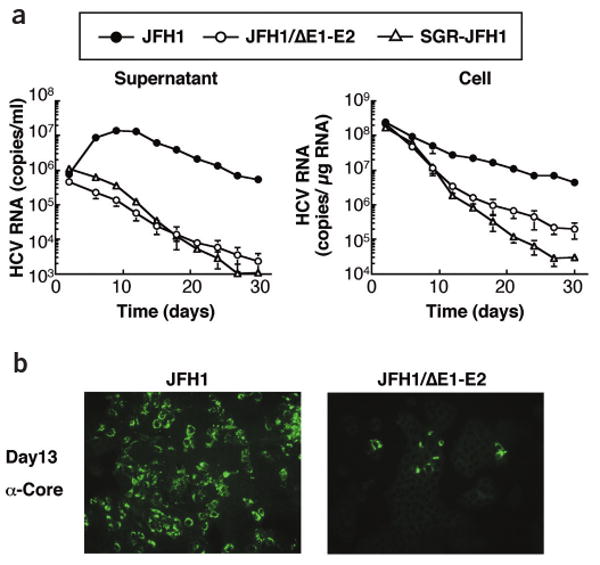
Time course of HCV RNA replication and core protein expression in transfected cells. (a) Levels of HCV RNA in transfected cells (right) and corresponding culture supernatants (left). Huh7 cells were transfected with given HCV RNAs and passaged up to 30 d. Viral RNA was determined 2, 5, 8, 12, 15, 18, 21, 24, 27 and 30 d after transfection. (b) Immunofluorescence assay of HCV core protein in passaged cells transfected with JFH1 and JFH1/ΔE1-E2 RNA. Transfected cells were harvested 1 d after the fourth passage (day 13 after transfection). Magnification, ×100.
Biophysical properties of HCV particles
To characterize secreted viral particles, we analyzed supernatant of JFH1-transfected cells using a sucrose density gradient. Core protein and nuclease-resistant HCV RNA sedimented to the same density of 1.17 g/ml (Supplementary Fig. 3 online). To show co-sedimentation of all structural proteins, we generated a JFH1-E2HA construct containing a hemagglutinin tag replacing part of the E2 hypervariable region (HVR). HCV lacking the HVR is infectious in chimpanzee13, and JFH1-E2HA virus remained infectious for Huh7 cells. Similar to the wild type, viral particles of this mutant had a density of ∼1.15 g/ml (Fig. 3a). Moreover, we detected core, E1 and E2 proteins in the same peak fraction of the sucrose density gradient (Fig. 3b), indicating the production of complete viral particles.
Figure 3.
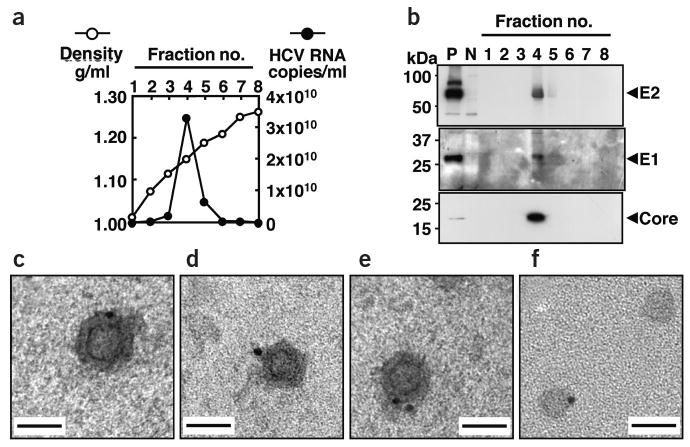
Density gradient and electron microscope analysis of recombinant HCV particles. (a,b) Co-sedimentation of viral RNA and structural proteins. (a) Concentrated culture medium collected from JFH1/E2HA RNA–transfected cells was fractionated using a 10–60% sucrose density gradient. HCV RNA titer in each fraction was determined. (b) Density gradient fractions were further concentrated and analyzed by western blotting for core, E1 or E2-hemagglutinin. P, cell lysate prepared from JFH1/E2HA RNA–transfected Huh7 cells; N, cell lysate from untransfected Huh7 cells. Arrowheads indicate positions of HCV proteins. (c–f) Electron micrograph of spherical structures shown by immunogold labeling. Grids were incubated with a concentrated JFH1 virus stock and then with the E2 monoclonal antibody CBH5 (ref. 14). Bound antibodies were detected with Protein A coupled to gold particles 10 nm in diameter. (c–e) Three representative examples showing the same structure. (f) Control grid coated with concentrated cell-free supernatant derived from mock-transfected cells. In rare cases, we observed gold particles attached to unstructured protein aggregates. Scale bar, 50 nm.
Detection of HCV particles by electron microscopy
Cell culture–derived JFH1 particles were visualized by immunoelectron microscopy, using an E2-specific antibody (CBH5)14. We detected gold-labeled spherical structures with an electron-dense inner core using a concentrated virus preparation (Fig. 3c–e), whereas we found only unstructured aggregates with the mock-transfected control (Fig. 3f). The inner ring has a slightly angular morphology and a diameter of 30–35 nm, consistent with nucleocapsids15. The overall diameter of the structures (50–65 nm) is compatible with the predicted size of HCV15–18.
Infectivity of HCV particles
We inoculated naive Huh7 cells with supernatant harvested from JFH1 RNA– or JFH1/ΔE1-E2 RNA–transfected cells, and 48 h later we double-stained cells for core and NS5A (Fig. 4a). Only cells inoculated with JFH1 medium were positive for these proteins (10– 20 cells/cover slip), with the number increasing about 20-fold when using 30-fold concentrated medium (∼0.5% of inoculated cells; Fig. 4a). To exclude the possibility that residual in vitro transcripts were captured by inoculated cells, we prepared supernatant from cells treated with the same amount of JFH1 RNA but without the electroporation. Upon inoculation of naive Huh7 cells, we observed no core protein–positive cells (Fig. 4a), as was the case with supernatant from JFH1/ΔE1-E2 RNA–transfected cells. Furthermore, ultraviolet irradiation of the inoculum substantially reduced the number of positive cells. Finally, we found no productive infection with HepG2, IMY-N9 and HeLa cells (Fig. 4a), consistent with their lower permissiveness for HCV RNA replication9,10.
Figure 4.

Infectivity of viral particles and neutralization of infection. (a, insert) Immunofluorescence analysis of cells infected with viral particles. Huh7 cells inoculated with virus-containing medium were stained simultaneously for core (left) and NS5A (right). (a) Cell lines specified on the bottom were also inoculated with a 30-fold concentrated supernatant from full-length JFH1 RNA– or JFH1/ΔE1-E2 RNA– transfected cells (Sup). In some experiments, culture supernatant of nontransfected cells was used (EP−), or culture supernatant was irradiated with ultraviolet light before inoculation of cells (UV+). We stained cells 2 d after infection for core protein and counted positive cells. (b) Comparison of infectivity of culture supernatant from JFH1 RNA– and JFH1/ΔE1-E2 RNA– transfected cells. Supernatants containing about 108 RNA copies/ml were used for inoculation of Huh7 cells and amounts of cell-associated RNA were determined 0, 12, 24 and 48 h after inoculation. (c) Inhibition of infection by CD81-specific antibody. We used 20-fold concentrated supernatants from cells transfected with given genomes for infection of Huh7 cells in the presence of a CD81-specific (α-CD81, black bars) or a control antibody (Ctrl Ab, gray bars), or in the absence of antibody (Ab(−), white bars). Inoculated cells were analyzed by RTD-PCR. (d) Production of infectious HCV particles carrying the firefly luciferase reporter gene and neutralization of infection by sera from infected individuals. Upper panel, schematic representation of Luc-JFH1 construct with luciferase (Luc) reporter gene (Supplementary Fig. 6 online). E-I, EMCV-IRES. Bottom left panel, neutralization of Luc-JFH1 virus by sera from infected individuals. Culture supernatants containing Luc-JFH1 viral particles were mixed with given dilutions of sera from healthy donor (Control), or sera from individuals infected with given genotypes (lanes 1–9). Results of CD81-specific antibody neutralization are shown in the right; black bar, 2 μg/ml; gray bar, 0.4 μg/ml; white bar, 0.08 μg/ml. Luciferase activity was determined 72 h later and is expressed relative to the values obtained with control serum A. Bottom right panel, neutralization by immunoglobulins purified from infected individuals' sera. Luc-JFH1 virus–containing supernatant was mixed with 2 mg total protein contained in control serum B or patient serum 3 (serum, black bars). Addition of this amount of serum protein is equivalent to a final serum dilution of approximately 1:20. Alternatively, virus preparation was mixed with 2 mg protein of the same sera depleted of immunoglobulins (Serum Ig(−), open bars), or 130 μg of corresponding purified immunoglobulins (Ig, gray bars). Infectivity is expressed relative to control serum B.
To compare infectivity of culture supernatant of JFH1 RNA– and JFH1/ΔE1-E2 RNA–transfected cells, we inoculated Huh7 cells with concentrates containing equivalent RNA copy numbers (1.17 × 108 copies/ml). We detected similar amounts of RNA in cells after an adsorption period of 3 h and found similar decreases up to 12 h (Fig. 4b). But 12 h later, RNA titers increased only in JFH1-inoculated cells, suggesting that productive infection depends on HCV envelope glycoproteins (Fig. 4b). This conclusion was supported by results obtained with single E-gene deletions (Supplementary Fig. 4 online). JFH1-E2HA particles were also infectious for Huh7 cells, but the RNA titer in infected cells was approximately 10 times lower compared to JFH1 virus infection (data not shown). Finally, HCV can be passaged by infection but virus titers decrease upon serial passages (data not shown).
Neutralization of HCV infection by CD81-specific antibody
CD81 was shown to be involved in HCV entry using HCV pseudoparticles19,20. To determine whether authentic particles follow the same entry route and to confirm specificity of uptake, we incubated Huh7 cells with JFH1 or JFH1/ΔE1-E2 inocula in the presence of CD81-specific or nonspecific antibodies. We scored infection 48 h later, using NS3-specific immunofluorescence (Supplementary Fig. 5 online) or RTD-PCR (Fig. 4c). CD81-specific antibodies reduced infected cells about tenfold and HCV RNA about 100-fold as compared to control antibody, confirming specificity of the infection and the important role of CD81 in HCV entry.
Neutralization of infection by patient sera
To facilitate quantification of infection, we generated a bicistronic JFH1 luciferase reporter construct (Fig. 4d and Supplementary Fig. 6 online). Infectious titers attainable by JFH1 and Luc-JFH1 genomes are similar, indicating that added sequences do not markedly affect production of infectious particles (data not shown). Taking advantage of this system, we performed neutralization experiments using sera of individuals infected with HCV genotype 2 or 1. Culture supernatants containing Luc-JFH1 virus particles were mixed with serum dilutions and infection was determined by luciferase assay (we considered a reduction to at least 50% significant). At a dilution of 1:20, all HCV sera showed neutralizing activity comparable to 0.08 μg/ml CD81-specific antibody (Fig. 4d). Neutralization was dose dependent and highest with sera 3 and 4. None of the serum samples inhibited entry of HIV-based pseudoparticles bearing MLV-derived envelope proteins into Huh7 (data not shown). Finally, antibodies purified from patient 3 but not from control serum B inhibited infection with similar efficiency as the original serum, whereas immunoglobulin depletion prevented neutralization (Fig. 4d). These data show that antibodies in sera from infected individuals are capable of neutralizing JFH1 viruses.
Infectivity of cell culture–derived HCV particles in vivo
To show the infectivity of cell culture–grown JFH1 virus in vivo, we intravenously inoculated medium from JFH1 RNA transfected cells into a chimpanzee. The inoculum contained 7.65 × 106 RNA copies/ml and had a core protein concentration of 4,630 fmol/L. To ensure that the HCV RNA remaining in the medium after transfection did not cause infection, we collected medium from a mock-transfected culture, in which HCV RNA was added to the cells without transfection. This sample contained 3.47 × 104 RNA copies/ml and had undetectable levels of core protein. Chimpanzee X0215 was inoculated with 1 ml of undiluted control medium and showed no signs of infection for 6 weeks (Fig. 5). Thereafter, we inoculated the chimpanzee with 1 ml of a 104 dilution of supernatant from JFH1 RNA–transfected cells. After 6 weeks of monitoring with no signs of infection, we re-inoculated the chimpanzee with 1 ml of a 103 dilution. HCV RNA became detectable in the serum at week 2, persisted until week 5, and thereafter became undetectable. Viremia was low with highest HCV RNA titer of 2.04 × 103 copies/ml at week 4. Infection was cleared without HCV-specific seroconversion, elevation of alanine aminotransferase or histological evidence of liver injury (Fig. 5). We sequenced parts of the 5′-untranslated region (UTR; nucleotides 128–331), the E2 HVR (nucleotides 1,438–1,828) and NS5B (nucleotides 9,049–9,382) of the circulating viral RNA at week 4 after the last inoculation. The sequences were identical to the JFH1 strain. These results show the in vivo infectivity of JFH1 virus produced in culture.
Figure 5. In vivo.
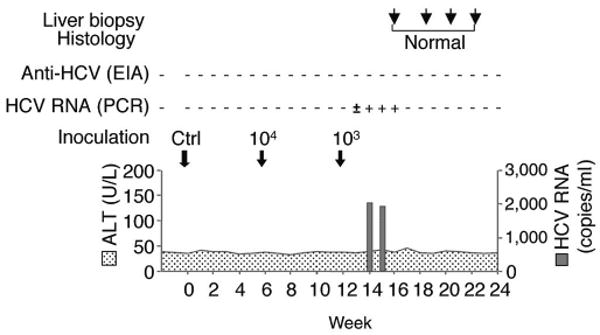
infectivity of JFH1 virus produced In tissue culture. Chimpanzee X0215 was first inoculated with 1 ml of the undiluted culture medium from mock-transfected Huh7 cells (Ctrl). We re-inoculated the chimpanzee 6 weeks later with 1 ml of a 104 dilution of culture medium from full-length JFH1 RNA-transfected cells (104). After 6 more weeks, we repeated inoculatlon with 1 ml of a 103 dilution (103). The course of infection is shown with arrows indicating the three inoculations. HCV RNA (copies/ml) and ALT (IU/L) levels are plotted; HCV-specific, HCV RNA and liver biopsy results are shown above the graph.
Discussion
This study shows that recombinant HCV particles are produced and secreted from JFH1 RNA–transfected cells, and secreted viruses are infectious for both Huh7 cells and a chimpanzee. Biochemical analyses show that cell culture–grown virus particles have a density of 1.15–1.17 g/ml and are spherical in size, with an average outer diameter of about 55 nm. Infectivity can be neutralized by CD81-specific antibodies, supporting the importance of CD81 in HCV entry19,20. Neutralization and cross-neutralization were achieved with immunoglobulins from infected individuals' sera. Cross-neutralization was also found with chimeric viruses composed of the core to NS2 region of the Con1 isolate (genotype 1) and the JFH1 replicase (T.P., G. Koutsoudakis, S. Kallis, T.K., T.W & R.B., unpublished data), indicating conserved epitopes involved in HCV entry. The possibility to create chimeric infectious virus broadens the scope of this system and allows (cross-)neutralization to be addressed in more detail.
It is unclear why the spread of infection in Huh7 cells is limited. Activation of innate immunity in transfected or infected cells is one possibility. Alternatively, JFH1 RNA–containing cells may have a growth disadvantage and therefore be displaced during passage. Moreover, formation of assembly-competent envelope protein complexes and virus release may be a rate-limiting step, reducing virus production to a level insufficient to sustain infection.
Earlier attempts to infect Huh7 cells with sera from infected individuals were not successful. But recombinant viral particles have a homogeneous density, whereas HCV in human sera shows much lower and heterogeneous densities, suggesting an association with cellular components, especially lipoproteins21, that may interfere with infection. Further studies are required to analyze the mechanism underlying the different infectivities of recombinant and serum-derived virus particles, especially using the serum from JFH1 virus–infected chimpanzee.
Culture-grown JFH1 viral particles are infectious in chimpanzee. The ratio of RNA titer versus infectious titer of the culture medium is about 1,000, which is lower than reported for infectious human and chimpanzee serum (10–100)22,23. Possible explanations are more defective viruses produced by the JFH1 strain or overestimation of viral RNA because of the input RNA used for transfection. Notably, human sera with lower infectivity have a density of about 1.17 g/ml, whereas those with higher infectivity sediment primarily at a lower density24.
The transient course of infection of the JFH1 strain differs from most infectious clones22,25,26, usually causing higher viremia and occasionally HCV-specific seroconversion or hepatocellular injury. This may result from the inoculum (culture grown virus versus inoculation of genomic in vitro transcripts) or the rather old age of the chimpanzee (20–30 years). Alternatively, the JFH1 strain, although replicating efficiently in vitro8–10, may be less infectious in vivo27. Nevertheless, this is the first report describing the production of HCV in cell culture, shich can infect both cells and primates.
Methods
For details of Methods, please see Supplementary Methods online.
Plasmid construction
Based on the consensus sequence of JFH1 (ref. 7), we assembled plasmid pJFH1 containing the full-length JFH1 cDNA downstream of the T7 RNA promoter. We generated the following mutants of pJFH1: pJFH1/GND carrying a mutation in the NS5B GDD motif, which abolishes RNA polymerase activity4,5; pJFH1/ΔE1-E2 with a deletion of 351 amino acids (amino acids 217–567); pJFH1/E2HA with the hemagglutinin tag (YPYDVPDYA) replacing part of the E2 HVR (amino acids 394–02). pJ6CF is the prototype genotype 2a clone11. We constructed plasmid pJCH1 analogous to pJFH1 (ref. 7). In plasmid pFK-Luc-JFH1, the 5′-UTR and part of the core region were fused to the firefly luciferase gene. The second cistron is expressed through the encephalomyocarditis virus IRES and encodes the complete JFH1 polyprotein.
RNA transfection and analysis of transfected cells
In vitro synthesis of HCV RNA, electroporation and northern blot analysis were performed as described previously4,8. For detection of HCV proteins by western blot NS5A (ref. 9), we used NS3 (ref. 8) and core-specific (clone 2H9) antibodies and peroxidase-labeled rabbit-specific goat immunoglobulin (Biosource) or mouse-specific sheep immunoglobulin (Amersham Pharmacia).E1 and E2-specific polyclonal antibodies were raised by immunization of rabbits with synthetic peptides. We used rat monoclonal hemagglutinin-specific antibody (Roche) and peroxidase-labeled rat-specific goat IgG to detect hemagglutinin-tagged E2 protein. Immunofluorescence was performed using the same primary antibodies.
Quantification of HCV core protein and RNA
We quantified HCV core protein in culture supernatant or cell lysate using a new immunoassay described previously28. Total RNA was isolated from cell lysates or culture media by Isogen (Nippon Gene). We determined RNA copy numbers of HCV by RTD-PCR as described12.
Sucrose density gradient analysis
We cleared culture medium collected 6 d after transfection using low-speed centrifugation, and passed it through a 0.45-μm filter. We layered filtrate on a sucrose gradient (60% to 10%, wt/vol) and centrifuged it for 16 h. We harvested and analyzed fractions for HCV RNA titers using RTD-PCR
Infection of cells with secreted HCV
We collected culture medium 72 h after transfection, cleared it using low-speed centrifugation and passed it through a 0.45-μm filter. Part of the filtrate was concentrated 30-fold using an Amicon Ultra-15 (cut off: 1 × 105 Da; Millipore). We seeded cells 24 h before infection at a density of 5 × 104 cells/well in a 12-well plate, or at 1 × 105 cells/well in a 6-well plate. We infected cells with 100 μl of inoculum for 3 h, washed them, added complete medium and cultured cells for 12, 24, 48, 72 and 96 h. We performed immunofluorescence 2 d after infection.
Electron microscopy
We concentrated supernatant harvested from JFH1-transfected Huh7 cells and mock-transfected cells using ultracentrifugation and adsorbed it onto carbon-coated grids. Grids were fixed with 3% paraformaldehyde, and blocked in a solution of 0.8% BSA, 1% coldwater fish skin gelatin (Sigma) and 20 mM glycine. We performed immunogold labeling with an E2-specific antibody (CBH5) and Protein A coupled to 10-nm gold particles. After extensive washing, we stained grids with 1.8% methylcellulose and 0.3% uranyl acetate.
Virus neutralization assays
Target cells were infected with culture supernatants supplemented with JS-81 (BD Biosciences) or Mab46D2 (Dengue type 2–specific antibody) at a final concentration of 10 μg/ml (unless otherwise stated). After inoculation, we supplemented cells with fresh medium. We lysed cells 72 h after infection for RTD-PCR or luciferase assays.
For neutralization with sera from infected individuals, we mixed virus-containing supernatants with dilutions of heat-inactivated serum. After incubation for 1 h, we added mixtures to target cells and measured infection. Immunoglobulins contained in human sera were purified by using a HiTrap Affinity Protein G column (Amersham Pharmacia).
In vivo infection
The chimpanzee experiment was conducted in an American Association of Laboratory Animal Care–accredited animal facility under approved animal protocol. We collected culture medium from Huh7 cells transfected with the JFH1 genome and cleared them by centrifugation before inoculation into chimpanzee X0215. Serum samples were tested for alanine aminotransferase levels, HCV-specific antibodies (EIA2.0, Abbott Laboratories) and HCV RNA that was quantified by the Roche Amplicor Cobas Monitor II (Roche). We collected liver biopsies for histological analysis.
Supplementary Material
Supplemental Fig.1
Supplemental Fig.2
Supplemental Fig.3
Supplemental Fig.4
Supplemental Fig.5
Supplemental Fig.6
Supplemental Methods
Acknowledgments
This study was supported by a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science, by a Grant from Toray Industries, Inc., by the Program for Promotion of Fundamental Studies in Health Sciences of the Pharmaceuticals and Medical Devices Agency (PMDA), by the Research on Health Sciences focusing on Drug Innovation from the Japan Health Sciences Foundation, by the NHLBI contract N01-HB-27091 for the use of chimpanzees, by a grant from the European Community (QLK2-CT-2002-01329), and by a grant from the Deutsche Forschungsgemeinschaft (SFB 638; Teilprojekt A5). T.K. was partially supported by Hepatitis Virus Research Foundation of Japan. The authors are grateful to J. Bukh for providing the pJ6CF plasmid, to S. Foung for provision of the antibody CBH5, to S. Koike for his comments, to K. Yasui, S. Sone and J.-I. Tanabe for their support, to G. Koutsoudakis and E. Steinmann, and to S. Kallis and U. Herian for technical assistance. We are also grateful to K. Takagi, K. Hiramatsu and members of Central Clinical Laboratory in Nagoya City University Hospital, R. Sapp of the Liver Diseases Branch, H. McClure of the Southwest Foundation for Biomedical Research for their technical assistance, H. Barth, B. Bartosch, T. Baumert, J. Encke and C. Sarrazin for providing human sera.
Footnotes
Accession numbers. The Genbank accession numbers for the consensus sequence of JFH1 is AB047639 and for pJFH1 it is AB047640.
Competing Interests Statement: The authors declare that they have no competing financial interests.
References
- 1.Choo QL, et al. Isolation of a cDNA clone derived from a blood-borne non-A non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 2.Kiyosawa K, et al. Interrelationship of blood transfusion, non-A, non-B hepatitis and hepatocellular carcinoma: analysis by detection of antibody to hepatitis C virus. Hepatology. 1990;12:671–675. doi: 10.1002/hep.1840120409. [DOI] [PubMed] [Google Scholar]
- 3.Bartenschlager R, Lohmann V. Replication of hepatitis C virus. J Gen Virol. 2000;81:1631–1648. doi: 10.1099/0022-1317-81-7-1631. [DOI] [PubMed] [Google Scholar]
- 4.Lohmann V, et al. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 5.Ikeda M, Yi M, Li K, Lemon SM. Selectable subgenomic and genome-length dicistronic RNAs derived from an infectious molecular clone of the HCV-N strain of hepatitis C virus replicate efficiently in cultured Huh7 cells. J Virol. 2002;76:2997–3006. doi: 10.1128/JVI.76.6.2997-3006.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pietschmann T, et al. Persistent and transient replication of full-length hepatitis C virus genomes in cell culture. J Virol. 2002;76:4008–4021. doi: 10.1128/JVI.76.8.4008-4021.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kato T, et al. Sequence analysis of hepatitis C virus isolated from a fulminant hepatitis patient. J Med Virol. 2001;64:334–339. doi: 10.1002/jmv.1055. [DOI] [PubMed] [Google Scholar]
- 8.Kato T, et al. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology. 2003;125:1808–1817. doi: 10.1053/j.gastro.2003.09.023. [DOI] [PubMed] [Google Scholar]
- 9.Date T, et al. Genotype 2a hepatitis C virus subgenomic replicon can replicate in HepG2 and IMY-N9 cells. J Biol Chem. 2004;279:22371–22376. doi: 10.1074/jbc.M311120200. [DOI] [PubMed] [Google Scholar]
- 10.Kato T, et al. Non-hepatic cell lines HeLa and 293 cells support efficient replication of hepatitis C virus genotype 2a subgenomic replicon. J Virol. 2005;79:592–596. doi: 10.1128/JVI.79.1.592-596.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yanagi M, Purcell RH, Emerson SU, Bukh J. Hepatitis C virus: an infectious molecular clone of a second major genotype (2a) and lack of viability of intertypic 1a and 2a chimeras. Virology. 1999;262:250–263. doi: 10.1006/viro.1999.9889. [DOI] [PubMed] [Google Scholar]
- 12.Takeuchi T, et al. Real-time detection system for quantification of hepatitis C virus genome. Gastroenterology. 1999;116:636–642. doi: 10.1016/s0016-5085(99)70185-x. [DOI] [PubMed] [Google Scholar]
- 13.Forns X, et al. Hepatitis C virus lacking the hypervariable region 1 of the second envelope protein is infectious and causes acute resolving or persistent infection in chimpanzees. Proc Natl Acad Sci USA. 2000;97:13318–13323. doi: 10.1073/pnas.230453597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hadlock KG, et al. Human monoclonal antibodies that inhibit binding of hepatitis C virus E2 protein to CD81 and recognize conserved conformational epitopes. J Virol. 2000;74:10407–10416. doi: 10.1128/jvi.74.22.10407-10416.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takahashi K, et al. p26 protein and 33-nm particle associated with nucleocapsid of hepatitis C virus recovered from the circulation of infected hosts. Virology. 1992;191:431–434. doi: 10.1016/0042-6822(92)90204-3. [DOI] [PubMed] [Google Scholar]
- 16.He LF, et al. Determining the size of non-A, non-B hepatitis virus by filtration. J Infect Dis. 1987;156:636–640. doi: 10.1093/infdis/156.4.636. [DOI] [PubMed] [Google Scholar]
- 17.Kaito M, et al. Hepatitis C virus particle detected by immunoelectron microscopic study. J Gen Virol. 1994;75:1755–1760. doi: 10.1099/0022-1317-75-7-1755. [DOI] [PubMed] [Google Scholar]
- 18.Shimizu YK, Feinstone SM, Kohara M, Purcell RH, Yoshikura H. Hepatitis C virus: Detection of intracellular virus particles by electron microscopy. Hepatology. 1996;23:205–209. doi: 10.1002/hep.510230202. [DOI] [PubMed] [Google Scholar]
- 19.Bartosch B, Dubuisson J, Cosset FL. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J Exp Med. 2003;197:633–642. [Google Scholar]
- 20.Hsu M, et al. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc Natl Acad Sci USA. 2003;100:7271–7276. doi: 10.1073/pnas.0832180100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomssen R, et al. Association of hepatitis C virus in human sera with beta-lipoprotein. Med Microbiol Immunol (Berl) 1992;181:293–300. doi: 10.1007/BF00198849. [DOI] [PubMed] [Google Scholar]
- 22.Thomson M, et al. The clearance of hepatitis C virus infection in chimpanzees may not necessarily correlate with the appearance of acquired immunity. J Virol. 2003;77:862–870. doi: 10.1128/JVI.77.2.862-870.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Major ME, et al. Previously infected and recovered chimpanzees exhibit rapid responses that control hepatitis C virus replication upon rechallenge. J Virol. 2002;76:6586–6595. doi: 10.1128/JVI.76.13.6586-6595.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hijikata M, et al. Equilibrium centrifugation studies of hepatitis C virus: evidence for circulating immune complexes. J Virol. 1993;67:1953–1958. doi: 10.1128/jvi.67.4.1953-1958.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kolykhalov AA, et al. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science. 1997;277:570–574. doi: 10.1126/science.277.5325.570. [DOI] [PubMed] [Google Scholar]
- 26.Yanagi M, Purcell RH, Emerson SU, Bukh J. Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc Natl Acad Sci USA. 1997;94:8738–8743. doi: 10.1073/pnas.94.16.8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bukh J, et al. Mutations that permit efficient replication of hepatitis C virus RNA in Huh-7 cells prevent productive replication in chimpanzees. Proc Natl Acad Sci USA. 2002;99:14416–14421. doi: 10.1073/pnas.212532699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aoyagi K, et al. Development of a simple and highly sensitive enzyme immunoassay for hepatitis C virus core antigen. J Clin Microbiol. 1999;37:1802–1808. doi: 10.1128/jcm.37.6.1802-1808.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kato T, et al. Processing of hepatitis C virus core protein is regulated by its C-terminal sequence. J Med Virol. 2003;69:357–366. doi: 10.1002/jmv.10297. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Fig.1
Supplemental Fig.2
Supplemental Fig.3
Supplemental Fig.4
Supplemental Fig.5
Supplemental Fig.6
Supplemental Methods