Systems Biology for the Virtual Plant (original) (raw)
WHAT IS SYSTEMS BIOLOGY?
Systems thinking is used in a variety of scientific and technological fields. Indeed, this paradigm has proved indispensable in disciplines as disparate as commerce, production, and the aviation industry. Aleksander Bogdanov (1873–1928) was probably the first exponent of systems thinking. In his “Tektology: Universal Organization Science” (1913–1922), Bogdanov ambitiously proposed that all physical, biological, and human sciences could be unified by treating them as sets of relationships and by seeking the organizational principles that underlie all systems (Poustilnik, 1998). The foundation of general systems theory was later developed extensively by the biologist Ludwig von Bertalanffy (for review, see von Bertalanffy, 1968). Bertalanffy's statements on the topic appeared as early as the mid-1920s. Why then is it only now in the genomic era that it is so “hip” to talk about systems biology? Analogous to past developments in other scientific disciplines, biologists in the post-genomic era are challenged with huge volumes of data (e.g. genome sequences, expression data), originating from heterogeneous technologies (e.g. microarray, yeast two-hybrid, ChIP-chip) and representing innumerable states of the system (experimental conditions). This massive influx of information and the desire to make it biologically coherent has forced us to think not in terms of single molecules but in terms of “systems.”
Although ecologists and physiologists have been using a systems approach to study plants for many years, a systems biology approach that reaches to and includes molecular details is only feasible now with the advent of genomic technologies. Thus, the exciting prospect of the post-genomic era is for the first time to be able to integrate knowledge across different levels of biological organization and to anchor this at the molecular level. Systems biology is sometimes loosely associated with the use of genomic technologies to understand specific biological processes. We believe systems biology has a larger and more ambitious scope, and we advocate a definition anchored in the general systems theory put forth by Bogdanov and Bertalanffy: The exercise of integrating the existing knowledge about biological components, building a model of the system as a whole and extracting the unifying organizational principles that explain the form and function of living organisms. The practical implementation of this definition is addressed in the next section.
It should be emphasized that the intention of this article is not to be a literature review or update on plant systems biology, which is indeed in a nascent state, but rather to provide our “vision” for systems biology in plant research—and the path that will take us there.
SYSTEMS BIOLOGY AT WORK
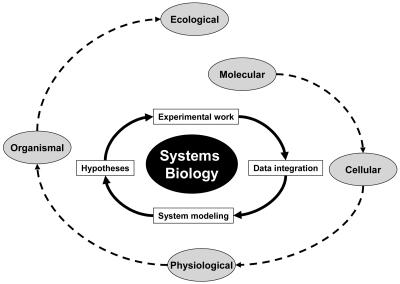
Practically speaking, a systems approach to understanding biology can be described as an iterative process that includes (1) data collection and integration of all available information (ideally all components and their relationships in the organism), (2) system modeling, (3) experimentation at a global level, and (4) generation of new hypotheses (modified from Ideker et al., 2001a; Kitano, 2001; Palsson, 2004; Fig. 1). The promise of systems biology is that by using this approach we will greatly increase our understanding as well as obtain a holistic view of the form and function of biological systems.
Figure 1.
Systems biology for the Virtual Plant. The ultimate goal of systems biology applied to plant research is to generate a model of the plant as a whole that describes processes across all layers of biological organization (molecular, cellular, physiological, organismal, and ecological; peripheral spiral, dotted arrows). Systems biology embodies an iterative process of experimentation at a global level, data integration, system modeling, and generation of hypotheses (internal cycle, solid arrows). These hypotheses lead to the design of new experiments that start a new round of the cycle. Each iteration refines the model and deepens our biological understanding of the system. Ultimately, such models may be used in a predictive mode and applied to improving traits associated with agriculture.
One of the best recent examples of the application of a systems approach to biology using genomic techniques is the work of Ideker, Thorsson, and coworkers (Ideker et al., 2001b). As a proof of principle, Ideker, Thorsson, and coworkers applied the systems approach to the yeast (Saccharomyces cerevisiae) galactose utilization (GAL) pathway, a process that has been studied extensively during the past 30 years using a one gene/protein at a time or reductionist approach. Ideker, Thorsson, and coworkers created knockout strains for the nine genes involved in the GAL pathway. These genes code for four enzymes, one transporter, and five transcription factors. Global mRNA levels were then examined in the mutant strains and compared to wild type both in the presence and absence of the “signal” galactose. By combining expression data in mutant and wild-type strains, protein level measurements and protein:protein and protein:DNA interaction data, they were able to identify a number of processes regulated in the treatments, and to identify their interconnections to each other and to potential regulatory proteins. These results provided insight into the regulation of metabolic pathways and the way distinct pathways are connected to each other, and to other cellular processes in the yeast cell. Furthermore, by integrating the protein and RNA measurements into a single model and identifying agreements and discrepancies between RNA and protein levels, the authors were able to hypothesize transcriptional versus posttranscriptional regulation. Finally, disagreements between the observed expression patterns and those predicted by the initial model led to the generation of new hypotheses about the regulation of the GAL pathway. In one striking example, the model generated with the knowledge prior to this work predicted that mutations in the genes encoding enzymes would not disturb the expression of other GAL genes. However, mutation of the GAL10 and GAL7 genes, coding for galactose metabolism enzymes, did affect the expression of other GAL genes. This prompted a new hypothesis that a metabolite could act as a signal to control the regulation of some of the genes in the pathway. Further experiments verified this hypothesis, hence refining the model for the regulation of the GAL pathway.
In this single landmark systems study in yeast, which involved integrating the existing knowledge about biological components, building a model of the system, and developing biological hypotheses for further experimentation, led to novel insights about a pathway that had been studied for decades. For example, it revealed how the GAL pathway is regulated and how its regulation is interconnected with other processes in the yeast cell. Other examples of systems biology approaches to research have been published and are reviewed elsewhere (e.g. Ideker et al., 2001a).
Similar to the example above, systems biology applied to plant research would help us identify the gene networks that are important, for example, for plant development, metabolism, or that are implicated in the plant response to biotic or abiotic stress. Furthermore, the systems approach would help us find the connections between these different gene networks. This is important because it would help us to cross the artificial boundaries that have been created in plant biology, in which discrete processes like development are mostly studied in isolation and segregated from other processes such as metabolism or responses to biotic and abiotic stress. In addition to increasing our understanding of plants, we believe the integrated models produced by the systems approach should help us make predictions, for example, about the effect of a perturbation (e.g. gene mutation) in traits of interest, such as seed yield or plant growth, or to determine the conditions that would optimize the growth of the plant. Such predictions and in general the new discoveries obtained with the systems approach will certainly find important practical applications.
AN INFRASTRUCTURE TO SUPPORT SYSTEMS BIOLOGY RESEARCH IN PLANTS
What do we need to profit from the powerful systems biology approach in plants? To successfully iterate through the cycle described in Figure 1, we need high-quality quantitative genomic data (described below) and a flexible software platform that integrates arbitrary data types and that is coupled to data visualization and analysis tools. Integrating the current and future genomic data in biologically meaningful ways and with the means to explore it is essential to carry out efficient and productive analysis of the large data structures typical of post-genomic biological research. Below we address the four activities (Fig. 1) that are encountered during the exercise of systems biology research.
New Integrated Database Systems for Heterogenous Data Types
The first logical step to address the molecular mechanisms underlying the biological associations of genes in gene lists generated using genomic techniques is to analyze a gene's function, gene product associations, and activities in the context of known biological pathways. Unfortunately, this task is becoming increasingly difficult. Aside from the sheer volume of knowledge accumulated in the literature, genomic data come from a large number of experimental approaches and an even larger number of laboratories. It is challenging to integrate this scale and diversity of genomic information in biologically meaningful ways. One layer of difficulty stems from the fact that information is stored in numerous databases and is encoded in various formats and database schemas. Indeed, integrating heterogeneous molecular biological databases has been acknowledged as one of the most important ongoing tasks in bioinformatics (Stevens et al., 2001). Different approaches to integrating biological databases have been employed in the past (for example, see Karp, 1996; Wilkinson et al., 2003). In our view, an integrated software platform to support systems biology research should satisfy two minimum requirements. (1) The researcher must have fast access to computer representations of biological molecules, their known properties (e.g. function, activity, domain structure), interrelationships (e.g. physical interactions, regulatory interactions), and states (e.g. mRNA levels in different organs) in a simple yet flexible way. (2) The software must accommodate exploratory queries. The inherent need to access heterogeneous data provided by different labs underscores the importance of standardizing the terms and schemas used to represent data in public databases and computational models of biological process.
A relatively new and attractive alternative to database integration is based on the idea that knowing where to find data and how to act on information is almost always better than trying to move everything into one place. We believe that the ideal software platform should NOT store all the data, but rather would selectively store a subset of genome-wide information that would support modeling efforts (see below). To maximize molecular biology database interoperability, the software should also “know” where to find and how to access a wide range of biological data and services. Efforts in this direction are ongoing (e.g. BioMOBY; Wilkinson et al., 2003).
System Modeling for Predictive Value
Computer-generated models can enable one to visualize and analyze biological data from a systems perspective (Ideker et al., 2001a). Several environments have been developed in the past years that permit data integration and modeling. Such software allows a detailed mathematical representation of cellular processes as well as qualitative representations of cellular components and their interactions. Generally, quantitative models are directed toward a specific cellular process of interest and are built by representing existing literature as a set of mathematical equations. Quantitative models are powerful because they describe a biological process in detail. Unfortunately, they require a very detailed understanding of the process under scrutiny: knowledge of kinetic parameters for enzymatic reactions, binding affinities of interacting molecules, etc. This information is not readily obtained for every component in a biological process, and even less so in every model organism. In fact, there are still many gaps in our qualitative understanding of plants. For example, only about 20% of all genes in Arabidopsis (Arabidopsis thaliana) had been experimentally characterized. Indeed, the crucial first stage in building quantitative models is constructing a map of the network to be analyzed. As a first step to fill this gap, our lab is building a high-level representation of plant cellular components and interactions to create a qualitative multinetwork model of the molecular data in the context of a plant cell. Integrating what is known about the plant cell molecular interactions from various data sources (enzyme:metabolite, protein:protein, protein:DNA, RNA:RNA) into a single coherent multinetwork model will provide a framework onto which one can analyze and integrate experimental measurements. Such a qualitative model will be of great help to identify target pathways for intervention and to enable the results of such interventions (e.g. genetic modifications) to be evaluated in a holistic way. Such a multinetwork model will also constitute the initial scaffold required to support the development of a “Virtual Plant” software platform. Naturally, as additional molecular details become available, these multinetwork models should incorporate quantitative as well as spatial and temporal aspects of the biological processes.
Because models without predictive power are not very useful, we believe that a significant amount of energy should be devoted to the development of new statistical and machine learning methods that allow the use of genomic data in a predictive mode. Thus, an important aspect of developing a Virtual Plant software platform should be the ability to use the data to model and predict molecular network states under untested conditions. For example, our lab has used the multinetwork model (above) to identify the subnetworks of genes whose expression is controlled by nitrate and carbon treatments. To predict how these gene networks respond under untested conditions, we are using statistical methods that describe the behavior of the genes in the multinetwork in response to various nutrient regimes. This description will enable the generation of predictive models that may also be used for designed genetic engineering as mentioned earlier.
Creating New Visualization and Data Analysis Tools to Enable the Formulation of Novel Hypotheses
Whereas statistical techniques are vital for discovering quantitative distinctions in datasets (e.g. which genes have the highest probability of being key regulators), scientists' pattern recognition skills often lead to the most enduring qualitative biological insights. To support those skills in a data-rich environment, we believe the development of new visualization tools is vital. Therefore, efforts should be directed toward the development of tools that allow the visual analysis of genomic datasets in a dynamic fashion. The novel visualization techniques should render the multivariate information in visual formats that facilitate extraction of biological concepts. Such new visualization directions should go beyond current approaches that use network graphs and include dynamic visualization aspects.
In addition, for effective research from a systems perspective, these new visualization tools should be supported by robust statistical measures and in a common software environment. The combination of visualization and math/statistic analysis tools should greatly ease the generation of biological hypotheses. Only then will biologists be able to effortlessly navigate the available genomic data and understand, for example, how internal and external perturbations affect processes, pathways, and networks controlling plant growth and development. Such a software platform is essential for a systems analysis of the genomic data available and would provide a framework for the analysis of future high-throughput data.
One of the best available software platforms for network data visualization and analysis is Cytoscape (Shannon et al., 2003). Cytoscape provides various layouts for network data visualization and plug-ins for data analysis. In addition, its flexible architecture allows the addition of new functionalities and thus constitutes an excellent platform onto which to develop new computational methods.
Genomic Experimentation and Experimental Design
As indicated in the report to the Department of Energy Workshop on Plant Systems Biology (Minorsky, 2003), to support the modeling efforts in systems biology research it is necessary to attain high-quality quantitative measurements of the levels of constituents (e.g. metabolites, RNAs, proteins) of the plant cell with dynamic (treatment and developmental time courses) and spatial resolution (single-cell level). Efforts toward these goals are under way (Birnbaum et al., 2003; Weigel et al., 2005). However, in our view, this constitutes only part of the story. To create models of the cell, it is at least equally and perhaps more essential to acquire information about molecular interactions of the cellular constituents. This includes, but is not limited to, protein:protein, protein:DNA, and RNA:RNA interactions, e.g. microRNA:target interactions. And it is in this area where plant research lags considerably behind research in other model organisms. Defining the Arabidopsis orfeome is an important first step to be able to carry out high-throughput yeast two-hybrid analysis of Arabidopsis proteins. A high-throughput approach for ChIP-chip data would also benefit from having the Arabidopsis orfeome, and it will help provide the transcriptional wiring for the entire genome. Defining the RNA:RNA interactions is important to understanding the posttranscriptional regulatory networks. To do this, a detailed characterization of the Arabidopsis small RNAs and predictions of their targets are necessary. The initial analysis of this interaction data will provide binary readouts in the form these two molecules do (not) interact. Data of this type would be extremely useful to develop multinetwork models of the plant cell (above). We believe it is essential that major efforts be directed toward obtaining high-throughput “interaction” data of this sort.
In addition, it does not suffice to simply obtain measurements of this kind under only one reference condition. To obtain an accurate picture of the organism, it is important to have snapshots under a variety of growth conditions and developmental states. Integrating the effects of these “external” signals with different states of development is a huge endeavor. Because the number of variables that biologists would like to study is quite large (different nutrients, water, light, temperature, biotic and abiotic stress, etc.), and most of them are continuous in nature, it is impossible in practice to obtain snapshots for all combinations of variables at all possible values because the experimental space is infinite. To efficiently explore the large experimental space generated by these conditions that are relevant to plant biology, it is necessary to devise methods to systematically and efficiently explore large experimental spaces. Biological intuition will certainly play an important role in constraining the experimental spaces, and hypothesis-driven perturbations will indicate the dimensions to explore. Mathematical or statistical methods, such as adaptive combinatorial design (Lejay et al., 2004), may then provide the basis for systematic and efficient exploration of these experimental spaces. Alternatively, and especially if the experimental space is large, random sampling may be considered. Another option is to use the conditions that nature provides in the field. This “ecological genomics” approach has the advantage of analyzing the combinations of all variables that are relevant to plant growth and development and to which plants have adapted. Adding this ecological dimension will ultimately be necessary to help us understand how developmental, metabolic, and other gene networks are modulated and interact in response to the “real-life” conditions.
SYSTEMS BIOLOGY AS A COMMUNITY EFFORT
To succeed, systems biology must be a collaborative and cross-disciplinary/organismal endeavor. Systems biology efforts in plant research must be led by the plant community, but must also be performed in close collaboration with scientists in disciplines such as computer science, statistics, and information visualization. The synergy between the different areas of expertise should power a new integrative and interdisciplinary approach to biological research concreted through the Virtual Plant software platform. Moreover, collaborations across groups working in various model organisms should enable the development of approaches that are agnostic to the source of the data.
FINAL REMARKS
At the core of living organisms there is a finite code: the genome. We now have the full genome of several plant species, including Arabidopsis, rice (Oryza sativa), and poplar (Populus trichocarpa). The challenge ahead is to decode this information into the components (genes) and their relationships (e.g. regulatory, physical interactions) and build an in silico model of the plant that encompasses all levels of organization (Fig. 1). To achieve this goal, high-throughput experimental data are required, and a new software platform must be developed that emphasizes conceptual integration of genomic data and that focuses on physical and regulatory relationships among plant gene products rather than on the individual components—and to do this in a dynamic fashion. Succeeding in this endeavor would be a major step toward understanding the genetic blueprint for form and function of plants and would provide a working example for research in other organisms.
Acknowledgments
We would like to acknowledge Dr. Ken Birnbaum, Dr. Fabio Piano, and Dr. Miriam Gifford for critical reading of this manuscript and helpful discussions.
1
This work was funded by grants from the National Science Foundation (IIS–9988345 and 0115586) to D.E.S. and grants from the National Science Foundation N2010 (IBN0115586), National Institutes of Health (GM3277), and Department of Energy (DEFG02–89ER14034) to G.M.C.
References
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN (2003) A gene expression map of the Arabidopsis root. Science 302**:** 1956–1960 [DOI] [PubMed] [Google Scholar]
- Ideker T, Galitski T, Hood L (2001. a) A new approach to decoding life: systems biology. Annu Rev Genomics Hum Genet 2**:** 343–372 [DOI] [PubMed] [Google Scholar]
- Ideker T, Thorsson V, Ranish JA, Christmas R, Buhler J, Eng JK, Bumgarner R, Goodlett DR, Aebersold R, Hood L (2001. b) Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science 292**:** 929–934 [DOI] [PubMed] [Google Scholar]
- Karp PD (1996) A strategy for database interoperation. J Comput Biol 2**:** 573–586 [DOI] [PubMed] [Google Scholar]
- Kitano H (2001) Foundations of Systems Biology. MIT Press, Cambridge, MA
- Lejay LV, Shasha DE, Palenchar PM, Kouranov AY, Cruikshank AA, Chou MF, Coruzzi GM (2004) Adaptive combinatorial design to explore large experimental spaces: approach and validation. Syst Biol 2**:** 1–7 [DOI] [PubMed] [Google Scholar]
- Minorsky PV (2003) Achieving the in silico plant. Systems biology and the future of plant biological research. Plant Physiol 132**:** 404–409 [PMC free article] [PubMed] [Google Scholar]
- Palsson BO (2004) In silico biotechnology: era of reconstruction and interrogation. Curr Opin Biotechnol 15**:** 50–51 [DOI] [PubMed] [Google Scholar]
- Poustilnik S (1998) Biological ideas in Tektology. In J Biggart, P Dudley, F King, eds, Alexander Bogdanov and The Origins of Systems Thinking in Russia. Ashgate Publishing, Brookfield, VT, pp 63–73
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13**:** 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens R, Goble C, Baker P, Brass A (2001) A classification of tasks in bioinformatics. Bioinformatics 17**:** 180–188 [DOI] [PubMed] [Google Scholar]
- von Bertalanffy L (1968) General System Theory: Foundations, Development, Applications. George Braziller, New York
- Weigel D, Altmann T, Nover L (2005) AtGenExpress. http://web.uni-frankfurt.de/fb15/botanik/mcb/AFGN/atgenex.htm (March 14, 2005)
- Wilkinson MD, Gessler D, Farmer A, Stein L (2003) The BioMOBY project explores open-source, simple, extensible protocols for enabling biological database interoperability. Proc Virt Conf Genom and Bioinf 3**:** 17–27 [Google Scholar]