The net of life: Reconstructing the microbial phylogenetic network (original) (raw)
Abstract
It has previously been suggested that the phylogeny of microbial species might be better described as a network containing vertical and horizontal gene transfer (HGT) events. Yet, all phylogenetic reconstructions so far have presented microbial trees rather than networks. Here, we present a first attempt to reconstruct such an evolutionary network, which we term the “net of life.” We use available tree reconstruction methods to infer vertical inheritance, and use an ancestral state inference algorithm to map HGT events on the tree. We also describe a weighting scheme used to estimate the number of genes exchanged between pairs of organisms. We demonstrate that vertical inheritance constitutes the bulk of gene transfer on the tree of life. We term the bulk of horizontal gene flow between tree nodes as “vines,” and demonstrate that multiple but mostly tiny vines interconnect the tree. Our results strongly suggest that the HGT network is a scale-free graph, a finding with important implications for genome evolution. We propose that genes might propagate extremely rapidly across microbial species through the HGT network, using certain organisms as hubs.
Following the legacy of Darwin's Origin of Species (Darwin 1859), most current methods for phylogenetic reconstruction depict evolutionary history of organisms as a tree. Phylogenetic trees have been derived from compositional signatures (Fox et al. 1980), sequence alignments (Doolittle 1981), or alignments of artificially concatenated conserved orthologs (Brown et al. 2001; Rokas et al. 2003). With genome sequencing technology, methods based on complete genome sequences appeared, including trees based on gene content (Fitz-Gibbon and House 1999; Snel et al. 1999; Tekaia et al. 1999; Lin and Gerstein 2000; Korbel et al. 2002;), gene order (Korbel et al. 2002), average ortholog similarity (Clarke et al. 2002), and genome conservation—a novel genome-based method combining gene content and sequence similarity (Kunin et al. 2005).
All these tree-like representations of evolution have an inherent drawback, dealing solely with vertical inheritance (Bapteste et al. 2004). Yet, a well-established consensus between evolutionary biologists is that the genomic history of most microbial species is mosaic, with a significant amount of horizontal gene transfer (HGT) present (Boucher et al. 2003). Although the quantification of the evolutionary effect of the HGT is still a subject of an ongoing debate (Snel et al. 2002; Kunin and Ouzounis 2003a), its existence is not questioned. The strong influence of HGT led to a proposal that presentation of microbial phylogeny as a tree is inaccurate as instances of HGT are not recorded in this presentation (Doolittle 1999; Martin 1999), and a correct representation should reflect HGT events.
Attempts to deal with this issue include algorithmic solutions for network-like tree reconstruction, mostly addressing recombination (but not HGT) as a form of nonvertical inheritance (Wang et al. 2001; Gusfield et al. 2004), and topological analyses of tree structure (Piel et al. 2003; Makarenkov and Legendre 2004). Thus, the widely accepted view that the phylogenetic history of genomes should be represented as a network rather than a tree has not been realized yet.
Here we present a first attempt to reconstruct the history of the microbial world, recording both horizontal and vertical gene transfer. For a scaffold depicting vertical gene transfer we use established tree reconstruction methods, on which we document the instances of horizontal transfer that intertwine the tree. We discuss the major properties of this complex phylogenetic network based on a multitude of genome comparisons; demonstrate its scale-free, small-world nature; and discuss the patterns of gene propagation through the network.
Results
Data
To ensure that our results are not affected solely by the orthology data (see Methods), we used two data sets: OFAM (see Methods) and groups of orthologs defined by STRING (von Mering et al. 2003). Similarly, to avoid possible bias from a single tree reconstruction method, we derived genomic trees with three independent methods: gene content, average ortholog similarity, and genome conservation (see Methods) for OFAM data and gene content for STRING data.
The summary of the evolutionary events reconstructed with each method is presented in Table 1. It is evident that although HGT is readily detectable, the bulk of the genes are still transferred by vertical gene transfer, which is the most prevailing mode of inheritance (Kunin and Ouzounis 2003a). In analogy, the net of life is not a grid, where all edges are of a similar strength, but more like a tree, with robust branching stems connected by thin climbing vines.
Table 1.
Summary of settings and results from various experimental designs
| Orthology data | Tree reconstruction method | Organisms | HGT events | Gene loss | Vertical transfers |
|---|---|---|---|---|---|
| OFAM | Average ortholog similarity | 165 | 39,005 | 88,834 | 640,328 |
| OFAM | Gene content | 165 | 36,385 | 89,951 | 646,791 |
| OFAM | Genome conservation | 165 | 39,589 | 84,630 | 635,056 |
| STRING | Gene content | 98 | 9968 | 32,943 | 288,225 |
HGT vine width distribution
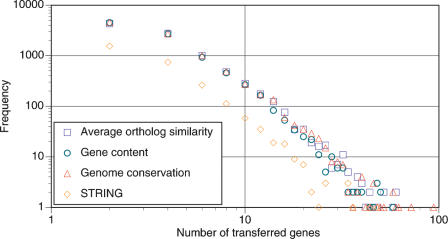
We define the HGT vine width as a summary of all horizontal transfer events between two nodes on the tree, subsequently fixated within the genome. The distribution of HGT vine widths, or number of genes transferred between any two nodes on the tree, is shown in Figure 1. All data sets and trees produce virtually identical frequency distribution (Fig. 1), following a power law (Table 2A), with the STRING data shifted by an order of magnitude, due to lower coverage of genomes (Table 1).
Figure 1.
Distribution of HGT vine widths.
Table 2.
Parameters for the power-law distribution (b, k) for (A) HGT vine widths (Fig. 1) and (B) the connectivity of the network (Fig. 2), according to the four methods used
| y = a * xk; where a = exp(b) = eb | |||
|---|---|---|---|
| Method | b | K | R2 |
| A | |||
| Average ortholog similarity | 11.8 | —2.88 | 0.95 |
| Gene content | 11.8 | —2.93 | 0.96 |
| Genome conservation | 11.7 | —2.84 | 0.94 |
| STRING | 9.9 | —2.68 | 0.95 |
| B | |||
| Average ortholog similarity | 6.7 | —1.93 | 0.68 |
| Gene content | 7.4 | —2.25 | 0.77 |
| Genome conservation | 5.8 | —1.54 | 0.72 |
| STRING | 6.0 | —2.55 | 0.83 |
Connectivity of the network
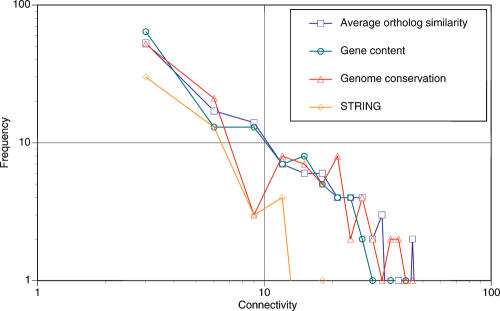
To investigate the properties of the HGT network, we removed the underlying (vertical inheritance) tree from the net of life. Since our inference of HGT vine widths is probabilistic (see Methods), we had to select a meaningful threshold to depict the inferred events. Thus, to investigate the connectivity of the HGT network, we experimented with several thresholds, namely, one (a single HGT), five, and 10. Irrespectively of the tree used and data set, the HGT network displays small-world behavior, with the diameter of the network fluctuating between five and six.
When higher thresholds are chosen for the analysis, the network also demonstrates power law distribution of connectivity of nodes (Table 2B), once again irrespectively of the data set or the tree used (Fig. 2). This power-law signal is obscured at the lowest thresholds, where many nodes appear to have high connectivity. We suggest that this deviation from the power law is a result of noise inevitable when a probability model is examined at low thresholds, namely, possibly containing more false-positive instances. Our usage of thresholds higher than one for evidence of HGT is indeed reinforced by biological observations that genes often travel between organisms as groups rather than singletons (Boucher et al. 2003). We thus conclude that the HGT network is likely to have a power-law distribution of connectivity, and thus be scale-free.
Figure 2.
Connectivity of the HGT network.
HGT champions
We aimed at investigating the HGT network in search of hubs and the widest HGT vines. Unlike the global properties of the network, which are virtually identical and independent on the data set, the exact number of predicted gene transfers between two nodes is highly dependent on the tree structure. Incorrect tree architecture can cause the mistaken inference of high amounts of HGT, particularly when two related organisms are positioned distantly on a tree. We thus aimed to exclude tree architecture bias from our analysis and examined results consistent between different tree architectures. Also, since the tree architectures are different, inner nodes (i.e. ancestral states) are often incomparable, and thus we limited the analysis to the leaves (terminal nodes) of the tree, i.e., the sequenced genomes from contemporary species.
When examining 165 microbial genomes for the network hubs, certain species came out on the top of the connectivity list with a remarkable consistency between the results obtained from different trees and data sets (Table 3). We found Pirellula sp., Bradyrhizobium japonicum, and Erwinia carotovora always at the top of the list of (terminal) nodes with the largest number of HGT partners. Interestingly, the original genome report for Pirellula sp. provides certain hints for HGT events in this species (Glockner et al. 2003). Furthermore, there is evidence for HGT between B. japonicum and E. carotovora in the literature (Streit et al. 2004). In conclusion, these hubs can serve as bacterial “gene banks,” providing a medium to acquire and redistribute genes in the microbial communities, caused either by specific genetic mechanisms or by virtue of their close proximity to and interaction with other species in their environmental niches.
Table 3.
The list of species representing the major hubs in the HGT network and their connectivity ranking in the three trees considered
| Organism | Average ortholog similarity | Gene content | Genome conservation | STRING |
|---|---|---|---|---|
| Pirellula sp. | 2 | 1 | 1 | Absent |
| Bradyrhizobium japonicum | 3 | 3 | 2 | 4 |
| Erwinia carotovora | 5 | 2 | 4 | Absent |
| Clostridium acetobutylicum | 4 | 4 | 10 | 5 |
| Chromobacterium violaceum | 6 | 10 | 9 | Absent |
We have also examined HGT vines that are reported to be wide and consistent across data sets and trees. One of the widest HGT vines is observed between the Bradyrhizobium genus (or sometimes the broader Rhizobiales group) of Alpha Proteobacteria and the Beta Proteobacterium Ralstonia solanacearum. Phylogenetically distant, both these species are soil bacteria, penetrating plant roots and forming—symbiotic in case of Bradyrhizobium (Kiers et al. 2003) and parasitic in case of Ralstonia (Alfano and Collmer 2004; Genin and Boucher 2004)—relationships with plants. Both cause tumor-like structures, and possess complex molecular mechanisms to interact with the host plants (Sawada et al. 2003). Both bacteria are reported to have acquired large number of genes horizontally (Kaneko et al. 2002; Salanoubat et al. 2002). Careful analysis of the genes that are transferred between the two bacteria can help to understand the mechanisms of pathogen–host interactions in these species, as well in other cases of HGT detected between species with similar life styles.
Discussion
The strongest limitation of the types of the network reconstruction presented here is the inability of the ancestral state inference methods to precisely establish the donor organism for a HGT event. Often a HGT event is inferred across nodes of the tree that existed at different time periods. In this case, GeneTrace determines the donor group of organisms rather than a particular donor species, and the prediction should be read as “the donor is a progeny of the node.” Although this effect might influence the character of the inferred network, the consistency between the results of medium- and high-confidence HGT vine width thresholds, as well any input data used in this study, indicates that the properties of the phylogenetic network reported here are genuine and realistic. A method to correctly infer HGT donors should greatly improve reconstruction of the network.
The GeneTrace method applied here uses phylogenetic distribution as a marker of HGT events. However, HGT more often occurs between related organisms, followed by homologous recombination (Vulic et al. 1997). Rather than introducing new protein families into a genome, this type of HGT causes orthologous gene replacement. In this study, we did not address this mechanism, we focus instead on events that introduce novel protein families into genomes. We are currently working on incorporation of detecting homologous HGT events in the phylogenetic network.
Another limitation is our inability to determine the correct path across organisms when multiple HGT events happened. Although the probabilistic schema described in the Methods section was designed to reduce the impact of this phenomenon, identification of the exact order and direction of HGT events would drastically improve reconstruction of the network.
The hubs of the HGT network presented here might partially result from the phylogenetic coverage of the sequenced species. When the coverage is low, multiple HGT events accumulate on long branches, and an artificial “hub” might appear. Thus, the reconstruction and understanding of the net of life will improve with better phylogenetic representation of sequenced organisms.
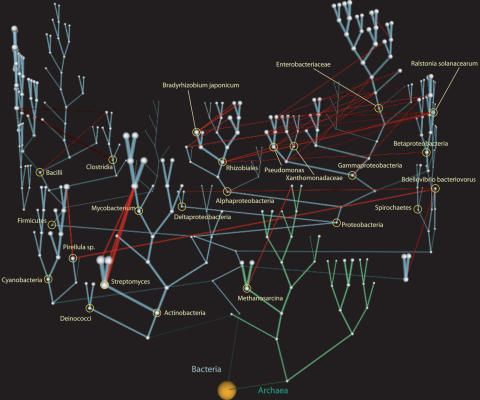
The currently acceptable representation of phylogenetic data is in the form of a tree-like structure in a two-dimensional space, often referred to as a “dendrogram” (meaning tree-graph in Greek). This presentation has the limitation of an inherent inability to depict HGT events. We propose to represent the phylogenetic data in the form of a three-dimensional tree, where beyond a tree drawn in the conventional two-dimensional space, HGT vines require a third dimension. When convergence of gene content is particularly high, participating nodes can be drawn closer in the third dimension. An example of such drawing is shown in Figure 3, with real data from this study. The full tree is available in VRML format, including all species identifiers (Janssen et al. 2003), as Supplemental material.
Figure 3.
Three-dimensional representation of the net of life. The tree backbone was generated by using the average gene similarity approach (see Methods). The root is represented as a yellow sphere. Bacteria are shown as nodes on cyan branches; Archaea, as nodes on green branches. Red lines correspond to the vines representing HGT. The radius of the nodes is proportional to the estimated gene content size (in terms of number of gene families). Also, the widths of both the vertical inheritance branches and the horizontal inheritance vines correspond to the numbers of gene families transferred by either mechanism. For visualization purposes, only values for HGT vine width >30 are shown. Certain key species and taxa are labeled; for full names, please refer to Supplemental material.
Our results suggest that the connectivity of microbial HGT network has a power-law behavior; i.e., the connectivity distribution appears as a decreasing straight line on a log-log scale (Fig. 2). A network in which connectivity of nodes distributes as a power-law has also scale-free and small-world properties. Scale-free networks display identical properties when any random subset of the complete network is sampled, suggesting that our conclusions should not be strongly affected by an ever-increasing number of genomes.
In a small-world network, the average shortest path between any two of its nodes (termed “network diameter”) involves traversing only relatively few nodes. This has a profound ecological meaning and strong implications for genome evolution. In the context of the HGT network, a small-world structure means that a substantially beneficial gene appearing in any organism can swing across species barriers and reach any other organism via a very small number of HGT events. In fact, this prediction of our hypothesis has an independent verification from the “experiment” of antibiotics-resistance genes that are known to spread extremely rapidly across species (Jacoby 1996), or the preferential involvement of specific functional classes (Nakamura et al. 2004). Although most of the reported instances of drug resistance involve pathogenic bacteria, based on the scale-free model, we predict that the initial donor and final acceptor organisms might have nothing in common in terms of phylogenetic origin, ecological niche, or geographical distribution, and communicate indirectly through the “hubs” in the network of life.
Methods
In order to reconstruct the phylogenetic network of microbial species, we required a data set of orthologs across all currently sequenced species. We used BLASTP (Altschul et al. 1997) to find best bidirectional hits across 165 microbial genomes in COGENT database release 184 (Janssen et al. 2003). To eliminate paralogy, we used only bidirectional best hits across genomes. We then clustered these hits by using Markov clustering algorithm (MCL) (Enright et al. 2002). The exhaustive nature of this schema ensures that all genes that had at least one bidirectional best hit in another organism are represented (L. Goldovsky, P. Jenssen, D. Ahrén, B. Audit, I. Cases, N. Darzentas, A.J. Enright, N. López-Bigas, J.M. Peregrin-Alvarez, M. Smith, et al., in prep.). We call the resulting protein families used for the analysis described herein as the “OFAM” data set. This data set is accessible at http://cgg.ebi.ac.uk/services/ortho-fam/.
To ensure that the results are not an artifact of the orthology definition, we used orthology information for 110 species from the STRING database (von Mering et al. 2003), from which we cross-linked 106 species to COGENT, resulting in 98 prokaryotic species, after excluding Eukaryotes. STRING adopts the definition of orthologs as groups of homologs built from at least one triplet of best-matching pairs of sequences, also known as clusters of orthologous genes (COGs) (Tatusov et al. 1997).
To reconstruct the microbial phylogenetic network, we required a phylogenetic tree. There are many methods for the reconstruction of phylogenetic trees (see Introduction); however, none guarantees 100% accuracy. To avoid biases generated by any single tree, we used three methods of genome-based phylogenetic reconstruction, i.e., gene content (Korbel et al. 2002), average gene similarity (Clarke et al. 2002), and genome conservation (Kunin et al. 2005). The first method derives phylogenetic distances from conservation of gene content, the second uses only sequence similarity between genomes, and the third combines the two measures to achieve maximum precision and contrast (Kunin et al. 2005). While being based on complete genomes, all these methods produce phylogenies that are remarkably similar to the classical 16S rRNA trees. All methods are implemented as it appears on the Genome Phylogeny Server (http://cgg.ebi.ac.uk/cgi-bin/gps/GPS.pl) and described elsewhere (Kunin et al. 2005). Only results consistent across different trees and with consistently high jacknife scores (Kunin and Ouzounis 2003b) are considered robust. For STRING data, we used a gene content tree constructed according to (Korbel et al. 2002).
Just as there are many methods to reconstruct phylogenetic trees, there are several available methods to identify HGT events. We could not use methods that are based on identification of biased GC content or codon usage, as these can only identify recently acquired genes and are not designed to reconstruct early events. We thus used GeneTrace—a method that identifies HGT from the phylogenetic distribution of protein families on the tree of life (Kunin and Ouzounis 2003b). GeneTrace assumes that presence of a gene family in multiple members of a clade reveals its ancestral nature, absence of a gene in some members of a clade indicates gene loss, and patchy presence of the gene family in distantly related clades implies HGT. This method was shown to have at least 90% accuracy on simulated data (Kunin and Ouzounis 2003b) and at least 81% accuracy on biological data (Kunin and Ouzounis 2003a), being capable of reconstructing HGT events on most levels on the tree of life.
A limitation of the GeneTrace approach to reconstructing HGT events is its inability to distinguish between the donor and the acceptor genomes (Kunin and Ouzounis 2003b). Thus, a gene that was extensively transferred horizontally creates links between all lineages that possess the gene, regardless whether they were involved in the particular transfer or not. We thus adopted a schema for normalization of the number of transferred genes, to avoid multiple counts of a single HGT event, as below.
Consider a situation when a protein family appears twice in distant sections of a tree. In this case, at least one HGT event may be necessary to explain the phylogenetic distribution of the family. Consider now a protein family that has three dispersed roots within a tree. Then, at least two horizontal transfer events are necessary to explain the distribution. However, simple linking of all nodes creates three possible edges for horizontal transfer. Assuming equal probability for all possible scenarios, we then assign the value of 2/3 as a probability for each possible event to be depicted correctly (and 1/3 for an incorrect detection). Thus, while the minimal number of edges required to connect all nodes (n) by HGT is _n_–1, the number of all possible connections is n(_n_–1)/2. This gives us the probability that each of the edges describes a valid HGT event as (_n_–1)/(n(_n_–1)/2), or 2/n. Thus, to each node that connects independent origins of a protein family, previously labeled by GeneTrace as arising from HGT, we assign a probability of 2/n.
To describe the inferred sum of all HGT events between two nodes within an evolutionary net, we sum up all probabilities of transfer for each gene family transferred between the two nodes and term the resulting edge as “vine” and the weight of the edge as “vine width.”
Acknowledgments
We thank members of the Computational Genomics Group for useful discussions. CAO acknowledges support from the UK Medical Research Council and IBM Research.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3666505\. Article published online before print in June 2005.
Footnotes
[Supplemental material is available online at www.genome.org.\]
References
- Alfano, J.R. and Collmer, A. 2004. Type III secretion system effector proteins: Double agents in bacterial disease and plant defense. Annu. Rev. Phytopathol. 42**:** 385-414. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25**:** 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bapteste, E., Boucher, Y., Leigh, J., and Doolittle, W.F. 2004. Phylogenetic reconstruction and lateral gene transfer. Trends Microbiol. 12**:** 406-411. [DOI] [PubMed] [Google Scholar]
- Boucher, Y., Douady, C.J., Papke, R.T., Walsh, D.A., Boudreau, M.E., Nesbo, C.L., Case, R.J., and Doolittle, W.F. 2003. Lateral gene transfer and the origins of prokaryotic groups. Annu. Rev. Genet. 37**:** 283-328. [DOI] [PubMed] [Google Scholar]
- Brown, J.R., Douady, C.J., Italia, M.J., Marshall, W.E., and Stanhope, M.J. 2001. Universal trees based on large combined protein sequence data sets. Nat. Genet. 28**:** 281-285. [DOI] [PubMed] [Google Scholar]
- Clarke, G.D., Beiko, R.G., Ragan, M.A., and Charlebois, R.L. 2002. Inferring genome trees by using a filter to eliminate phylogenetically discordant sequences and a distance matrix based on mean normalized BLASTP scores. J. Bacteriol. 184**:** 2072-2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin, C. 1859. On the origin of species by means of natural selection. J. Murray, London.
- Doolittle, R.F. 1981. Similar amino acid sequences: Chance or common ancestry? Science 214**:** 149-159. [DOI] [PubMed] [Google Scholar]
- ———. 1999. Phylogenetic classification and the universal tree. Science 284**:** 2124-2129. [DOI] [PubMed] [Google Scholar]
- Enright, A.J., Van Dongen, S., and Ouzounis, C.A. 2002. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 30**:** 1575-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitz-Gibbon, S.T. and House, C.H. 1999. Whole genome-based phylogenetic analysis of free-living microorganisms. Nucleic Acids Res. 27**:** 4218-4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox, G.E., Stackebrandt, E., Hespell, R.B., Gibson, J., Maniloff, J., Dyer, T.A., Wolfe, R.S., Balch, W.E., Tanner, R.S., Magrum, L.J., et al. 1980. The phylogeny of prokaryotes. Science 209**:** 457-463. [DOI] [PubMed] [Google Scholar]
- Genin, S. and Boucher, C. 2004. Lessons learned from the genome analysis of Ralstonia solanacearum. Annu. Rev. Phytopathol. 42**:** 107-134. [DOI] [PubMed] [Google Scholar]
- Glockner, F.O., Kube, M., Bauer, M., Teeling, H., Lombardot, T., Ludwig, W., Gade, D., Beck, A., Borzym, K., Heitmann, K., et al. 2003. Complete genome sequence of the marine planctomycete Pirellula sp. strain 1. Proc. Natl. Acad. Sci. 100**:** 8298-8303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusfield, D., Eddhu, S., and Langley, C. 2004. Optimal, efficient reconstruction of phylogenetic networks with constrained recombination. J. Bioinform. Comput. Biol. 2**:** 173-213. [DOI] [PubMed] [Google Scholar]
- Jacoby, G.A. 1996. Antimicrobial-resistant pathogens in the 1990s. Annu. Rev. Med. 47**:** 169-179. [DOI] [PubMed] [Google Scholar]
- Janssen, P., Enright, A.J., Audit, B., Cases, I., Goldovsky, L., Harte, N., Kunin, V., and Ouzounis, C.A. 2003. COmplete GENome Tracking (COGENT): A flexible data environment for computational genomics. Bioinformatics 19**:** 1451-1452. [DOI] [PubMed] [Google Scholar]
- Kaneko, T., Nakamura, Y., Sato, S., Minamisawa, K., Uchiumi, T., Sasamoto, S., Watanabe, A., Idesawa, K., Iriguchi, M., Kawashima, K., et al. 2002. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res. 9**:** 189-197. [DOI] [PubMed] [Google Scholar]
- Kiers, E.T., Rousseau, R.A., West, S.A., and Denison, R.F. 2003. Host sanctions and the legume-rhizobium mutualism. Nature 425**:** 78-81. [DOI] [PubMed] [Google Scholar]
- Korbel, J.O., Snel, B., Huynen, M.A., and Bork, P. 2002. SHOT: A web server for the construction of genome phylogenies. Trends Genet. 18**:** 158-162. [DOI] [PubMed] [Google Scholar]
- Kunin, V. and Ouzounis, C.A. 2003a. The balance of driving forces during genome evolution in prokaryotes. Genome Res. 13**:** 1589-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2003b. GeneTRACE: Reconstruction of gene content of ancestral species. Bioinformatics 19**:** 1412-1416. [DOI] [PubMed] [Google Scholar]
- Kunin, V., Ahren, D., Goldovsky, L., Janssen, P., and Ouzounis, C.A. 2005. Measuring genome conservation across taxa: Divided strains and united kingdoms. Nucleic Acids Res. 33**:** 616-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, J. and Gerstein, M. 2000. Whole-genome trees based on the occurrence of folds and orthologs: Implications for comparing genomes on different levels. Genome Res. 10**:** 808-818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarenkov, V. and Legendre, P. 2004. From a phylogenetic tree to a reticulated network. J. Comput. Biol. 11**:** 195-212. [DOI] [PubMed] [Google Scholar]
- Martin, W. 1999. Mosaic bacterial chromosomes: A challenge en route to a tree of genomes. Bioessays 21**:** 99-104. [DOI] [PubMed] [Google Scholar]
- Nakamura, Y., Itoh, T., Matsuda, H., and Gojobori, T. 2004. Biased biological functions of horizontally transferred genes in prokaryotic genomes. Nat. Genet. 36**:** 760-766. [DOI] [PubMed] [Google Scholar]
- Piel, W.H., Sanderson, M.J., and Donoghue, M.J. 2003. The small-world dynamics of tree networks and data mining in phyloinformatics. Bioinformatics 19**:** 1162-1168. [DOI] [PubMed] [Google Scholar]
- Rokas, A., Williams, B.L., King, N., and Carroll, S.B. 2003. Genome-scale approaches to resolving incongruence in molecular phylogenies. Nature 425**:** 798-804. [DOI] [PubMed] [Google Scholar]
- Salanoubat, M., Genin, S., Artiguenave, F., Gouzy, J., Mangenot, S., Arlat, M., Billault, A., Brottier, P., Camus, J.C., Cattolico, L., et al. 2002. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415**:** 497-502. [DOI] [PubMed] [Google Scholar]
- Sawada, H., Kuykendall, L.D., and Young, J.M. 2003. Changing concepts in the systematics of bacterial nitrogen-fixing legume symbionts. J. Gen. Appl. Microbiol. 49**:** 155-179. [DOI] [PubMed] [Google Scholar]
- Snel, B., Bork, P., and Huynen, M.A. 1999. Genome phylogeny based on gene content. Nat. Genet. 21**:** 108-110. [DOI] [PubMed] [Google Scholar]
- ———. 2002. Genomes in flux: The evolution of archaeal and proteobacterial gene content. Genome Res. 12**:** 17-25. [DOI] [PubMed] [Google Scholar]
- Streit, W.R., Schmitz, R.A., Perret, X., Staehelin, C., Deakin, W.J., Raasch, C., Liesegang, H., and Broughton, W.J. 2004. An evolutionary hot spot: The pNGR234b replicon of Rhizobium sp. strain NGR234. J. Bacteriol. 186**:** 535-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov, R.L., Koonin, E.V., and Lipman, D.J. 1997. A genomic perspective on protein families. Science 278**:** 631-637. [DOI] [PubMed] [Google Scholar]
- Tekaia, F., Lazcano, A., and Dujon, B. 1999. The genomic tree as revealed from whole proteome comparisons. Genome Res. 9**:** 550-557. [PMC free article] [PubMed] [Google Scholar]
- von Mering, C., Huynen, M., Jaeggi, D., Schmidt, S., Bork, P., and Snel, B. 2003. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res. 31**:** 258-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vulic, M., Dionisio, F., Taddei, F., and Radman, M. 1997. Molecular keys to speciation: DNA polymorphism and the control of genetic exchange in enterobacteria. Proc. Natl. Acad. Sci. 94**:** 9763-9767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L., Zhang, K., and Zhang, L. 2001. Perfect phylogenetic networks with recombination. J. Comput. Biol. 8**:** 69-78. [DOI] [PubMed] [Google Scholar]
Web site references
- http://cgg.ebi.ac.uk/services/ortho-fam/; OFAM data set.
- http://cgg.ebi.ac.uk/cgi-bin/gps/GPS.pl; Genome Phylogeny Server.