Human POT1 disrupts telomeric G-quadruplexes allowing telomerase extension in vitro (original) (raw)
Abstract
The POT1 (protection of telomeres 1) protein binds the ssDNA overhangs at the ends of chromosomes in diverse eukaryotes. POT1 is essential for chromosome end-protection, as best demonstrated in fission yeast. In human cells, hPOT1 is also involved in telomere-length regulation. We now show that telomeric oligonucleotides, such as d[GGG(TTAGGG)3], which form intramolecular G-quadruplexes through Hoogsteen base-pairing, serve as only marginal primers for extension by recombinant human telomerase; telomerase stalls after every nucleotide addition. Addition of hPOT1 to the reaction restores the normal processive elongation pattern seen with primers that cannot form G-quadruplexes. hPOT1 does not act catalytically but, instead, forms a stoichiometric complex with the DNA, freeing its 3′ tail. An antisense oligonucleotide, which base-pairs near the 5′ end of the telomeric sequence, leaving a telomerase-extendible 3′ tail, duplicates the effect of hPOT1 on activation of G-quadruplex primers. Thus, hPOT1 may function simply by trapping the unfolded forms of these telomeric primers in an equilibrium population. We propose an additional role for hPOT1 in telomere maintenance: disrupting G-quadruplex structures in telomeric DNA, thereby allowing proper elongation by telomerase.
Keywords: DNA-protein complex, G-quartet, OB fold, replication, telomere
In most eukaryotes, the ends of the linear chromosomes are replicated through the action of telomerase, a ribonucleoprotein enzyme. The enzymology of telomerase has been the subject of numerous studies evaluating the sequence specificity, processivity, DNA primer affinity, and nucleotide-binding properties of the enzymes from ciliated protozoa, yeast, and humans (e.g., see refs. 1-6). Almost all of these in vitro studies have used synthetic DNA oligonucleotides as surrogates for the natural DNA primer, which is a DNA-protein complex (Fig. 1_A_).
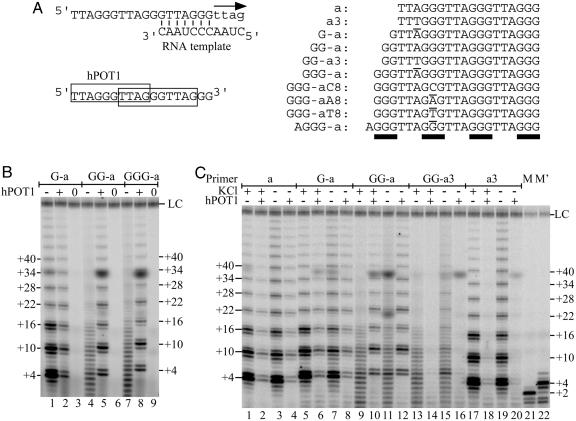
Fig. 1.
Poor primers for telomerase are rescued by hPOT1. (A) Primer a extended by telomerase, which incorporates nucleotides shown as lowercase letters; hPOT1 binds to either of two overlapping binding sites on primer a (Left). Sequence of primers used in this study (Right). Nontelomeric nucleotides are underlined, blocks of Gs are highlighted with bars. (B) Direct telomerase-activity assay performed with 100 nM primer G-a (lanes 1-3), GG-a (lanes 4-6), or GGG-a (lanes 7-9), in the presence of protein buffer (-) or 333 nM hPOT1 (+). Lanes 0, the telomerase assay was performed with immunopurified hTERT without the RNA (hTER) subunit. (C) Extension of primers in the presence and absence of hPOT1 and in the presence and absence of 50 mM KCl. Concentrations of primer and protein are as in A. M and M′, markers synthesized by using the direct telomerase activity assay with primer d(GGTTAG)3 in the presence of dGTP only (+2) or in the presence of dGTP and dTTP (+4). LC, loading control.
Only a few studies have investigated the action of telomerase on a telomeric DNA primer that is bound by an authentic chromosome end-binding protein. Both TEBP, the telomere end-binding protein from hypotrichous ciliates, and hPOT1, the corresponding protein from humans (7), can sequester the telomeric DNA 3′ end and render it unavailable for telomerase extension (8-10). This inhibition is expected from the x-ray crystal structures of these DNA-protein complexes (11, 12). In contrast, when hPOT1 binds to a more internal site on telomeric DNA, leaving a free 3′ tail (Fig. 1 A), the resulting complex is extended by telomerase with modestly increased activity and processivity (10). In the current work, we find that, with longer telomeric oligonucleotides, binding of hPOT1 has a much more dramatic effect, converting these DNAs from marginal to robust substrates for recombinant telomerase in vitro. We provide evidence that activation of these DNAs by hPOT1 occurs because the protein is able to disrupt the highly folded intramolecular G-quadruplex structures.
PolyG and other guanine-rich ssDNAs have long been known to form four-stranded helices (13, 14). In addition, G-rich oligonucleotides containing as few as four telomeric repeats can fold into compact intramolecular structures containing square-planar arrays (“G-quartets”) of four hydrogen-bonded guanines (15-17). These structures are stabilized by certain monovalent cations (Na+ or K+), which bind in the central cavity formed by the square-planar arrays of guanine bases (18). It is challenging to test for the presence of an unusual DNA structure in vivo, and the best evidence, thus far, is that antibodies specific for G-quadruplex DNA react with macronuclei from a hypotrichous ciliate (19). In addition to this limited evidence, the thermodynamic and kinetic stability of these structures suggests that they would necessarily form in vivo in the absence of an active mechanism to prevent their formation or to unfold them, once formed (20-22).
Not surprisingly, stable G-quadruplex structures are poor substrates for interaction with components that must recognize telomeric sequences within ssDNA. Oxytricha telomerase is unable to initiate extension of the K+-stabilized folded form of d(TTTTGGGG)4, the Oxytricha telomeric sequence (23). When telomerase initiates on a shorter oligonucleotide, it tends to dissociate if the portion not base-paired to the telomerase RNA template can form a quadruplex structure. This observation has been made for telomerases from both Oxytricha (23) and human (24). In terms of telomere-binding proteins, Oxytricha TEBP cannot bind the G-quadruplex form of d(TTTTGGGG)4, and binding is limited by the very slow rate of unfolding of the DNA (_t_1/2 = 18 h in 50 mM KCl at 37°C) (25). In a HeLa cell extract, human POT1 and TRF2 proteins bind to human telomeric DNA less efficiently when the single-stranded overhang is in the form of an intramolecular quadruplex (26).
Now, however, we find that the human system acts rather differently from the Oxytricha system in vitro. First, repeats of the human telomeric DNA sequence (TTAGGG) containing four GGGs are not totally inhibited from telomerase extension, although the reaction stalls after each added nucleotide. Second, the hPOT1 protein readily binds to human telomeric DNA that has folded into a quadruplex structure, probably because the folded DNA is in equilibrium with unfolded or partially folded forms (27-29). As a result, hPOT1 dramatically enhances the ability of quadruplex-forming human telomeric DNAs to serve as telomerase substrates.
Methods
hPOT1 Protein. hPOT1 splice version 2, containing the entire DNA-binding domain, was expressed by using baculovirus-infected insect cells. The N-terminal GST tag was removed and hPOT1V2 was purified to homogeneity, as described in ref. 12.
DNA Primer Nomenclature. Primers that include the sequence of primer a (ref. 10; and see Fig. 1 A) are named in a 5′-to-3′ direction; for example, G-a indicates a primer with a single 5′ terminal G preceding the sequence of primer “a.” Nucleotide changes within the primer-a sequence are denoted by the identity and position of the altered nucleotide, counting from the 5′ end. Primer a3 denotes a single-base mutant of primer a that forces hPOT1 to bind at the 3′ site only (10).
In Vitro Reconstitution of Human Telomerase. C-terminal HA-tagged human telomerase reverse transcriptase (TERT) was expressed from phTERT-HA2 and the hTER RNA subunit from phTR by using the TnT quick-coupled transcription/translation system (Promega). Each 500-μl reaction contained 400 μl of TnT-quick mix, 20 μl of PCR enhancer (Promega), 10 μl of 1 mM methionine, 4 μl of [35S]methionine (PerkinElmer), 46 μl of water, and 10 μg each of supercoiled phTERT-HA2 and Fok I-cut phTR plasmid DNA. After incubation at 30°C for 2 h, the reconstituted telomerase complex was affinity-purified on anti-HA F7 Agarose beads (Santa Cruz Biotechnology). Anti-HA F7 Agarose beads (500 μl), washed with 1× telomerase buffer without KCl (see below), were added for immunopurification at 4°C overnight. The beads were washed with 1× telomerase assay buffer with 30% glycerol four times and then resuspended in 1× telomerase assay buffer with 30% glycerol. The quantity of 35S-hTERT was determined. The unused beads were quick-frozen in liquid N2 and stored at -80°C.
Telomerase-Activity Assay. Activity of the immunopurified human telomerase complex reconstituted in vitro was determined by a direct assay modified from a published protocol (30). Briefly, the reaction mixture (20 μl) contained 1× telomerase assay buffer (50 mM Tris·HCl, pH 8.0/50 mM KCl/1 mM MgCl2/5 mM 2-mercaptoethanol/1 mM spermidine), 100 nM telomeric DNA primer, 0.5 mM dATP, 0.5 mM dTTP, 2 μM dGTP, and 1.25 μM [α-32P]dGTP [800 Ci/mmol (1 Ci = 37 GBq)] with 6 μl of immunopurified telomerase complex (20-40 nM telomerase). The reaction was incubated at 30°C for 1 h and stopped with the addition of 100 μl of 3.6 M NH4OAc, containing 20 μg of glycogen and 1,000 cpm of a 32P end-labeled DNA loading control. Ethanol (500 μl) was added for precipitation. After incubation at -80°C for 1 h, samples were centrifuged at 4°C for 20 min, and the pellets were washed with 70% ethanol and resuspended in 10 μl of H2O followed by 10 μl of 2× gel-loading buffer (94% formamide/0.1× TBE/0.1% bromophenol blue/0.1% xylene cyanol). The heat-denatured samples were loaded onto a 10% polyacrylamide/1× TBE/7 M urea denaturing gel for electrophoresis. After electrophoresis, the gel was dried and quantified by using a PhosphorImager (Molecular Dynamics).
Native Gels. Samples for analysis by native gel electrophoresis were prepared as follows: varying concentrations (0.5-600 nM) of 5′ 32P-labeled DNA were annealed by heating to 95°C for 5 min, snap-cooled on ice for 5 min, and added to 1× telomerase buffer (see above), 5% glycerol, 0.1× protein buffer (2.5 mM Tris·HCl, pH 8.0/15 mM NaCl/1 mM DTT). After 5 min at room temperature, a 3-fold excess of hPOT1, a 5-fold excess of antisense DNA (CCTAACCCTAACC), or nothing was added, and the complexes were allowed to form at 30°C for 60 min. At the end of the incubation, the 10-μl reaction was loaded onto an 8% polyacrylamide/0.5× TBE gel for electrophoresis at 5°C. After electrophoresis, the gel was dried and quantified by using a PhosphorImager.
Snake-Venom Phosphodiesterase I (SVPI) Digestion. SVPI digestion was performed as described in ref. 10. hPOT1-primer complex or antisense DNA-primer complex was preformed for 60 min at 30°C in a 9-μl solution containing 1.1× telomerase assay buffer (see above), 0.11× protein buffer (see above), 112 nM 5′-labeled 32P-DNA (annealed as described above), and 370 nM hPOT1 or 556 nM antisense DNA. The digest was started by the addition of 0.3 μg of SVPI in 1 μl of stock solution (100 mM Tris·HCl, pH 8.9/100 mM NaCl/15 mM MgCl2). The reaction was incubated at 30°C for 5 min and then stopped by the addition of 1 μl of 100 mM EDTA. After heat-inactivation at 95°C for 2 min, 10 μl of 2× gel-loading buffer (see above) was added to the sample. 10 μl of the final mixture was loaded onto a 20% polyacrylamide/1× TBE/7 M urea denaturing gel for electrophoresis.
Results
Certain Poor Primers for Telomerase Are Rescued by hPOT1. Biochemical studies of human telomerase have revealed that it can extend a wide variety of ssDNAs with telomeric or related sequence. Primers can be as short as a 6-mer or 7-mer (10, 31) or can be partially double-stranded, as long as they have at least a 5- to 6-nt 3′ tail (10, 32). We were, therefore, initially surprised to find that, whereas the telomeric 19-mer (primer G-a) gave the now-classical hexanucleotide ladder of telomerase extension products, the addition of a single G to its 5′ end (primer GG-a) resulted in a broad distribution of poorly extended products (Fig. 1 A and B). Addition of purified hPOT1 protein at a concentration sufficient to drive essentially all of the DNA molecules into DNA-protein complexes reduced the amount of primer G-a that was extended. This result was expected, as hPOT1, which binds approximately two telomeric repeats (Fig. 1 A), preferentially binds to 3′-terminal repeats, inhibiting telomerase extension, but, in a minority of cases, hPOT1 binds to the 5′ repeats, allowing telomerase extension (10). In marked contrast, addition of hPOT1 to primer GG-a resulted in a dramatic rescue of the normal telomerase extension pattern (compare lanes 4 and 5 of Fig. 1_B_). The experiments shown here utilize only the DNA-binding domain of hPOT1, but the full-length protein had indistinguishable activity (data not shown).
We considered that the aberrant pattern of telomerase extension of primer GG-a in the absence of hPOT1 might result from telomerase “stuttering” and adding some nontelomeric sequence, such as strings of Gs. However, two lines of evidence indicated that the normal telomeric sequence was being added. First, extension required all three nucleotide substrates, dGTP, dTTP, and dATP (data not shown). Second, electrophoretic mobility on these polyacrylamide sequencing gels is extremely sensitive to base composition, so the precise comigration of the products of reactions with and without hPOT1 provided a strong argument that the same sequence was being added. Thus, we conclude that telomerase stalls after every nucleotide addition when it extends primer GG-a. Remarkably, telomerase appears to be stalling without dissociating, because increasing the DNA primer concentration from 0.1 μM to 1.0 or 10 μM had no effect on the extension pattern (data not shown). If telomerase were dissociating, it would have been able to add only a single nt to a given primer at the highest concentration.
Stalled extension by telomerase was not restricted to primer GG-a. Primers GGG-a and GG-a3 gave similar results (Fig. 1_B_, lane 7 and 1_C_, lanes 13, 15). Addition of hPOT1 rescued the extension of primer GGG-a (Fig. 1_B_, lane 8). As expected, the addition of hPOT1 completely inhibited the extension of primer GG-a3 (Fig. 1_C_, lanes 14, 16); the single-nucleotide mutation present in the a3 series of primers forces hPOT1 to bind to the 3′ end of the primer, occluding interaction with telomerase (10). One possible reason for incomplete extension might be too low a nucleotide concentration; however, the stalled extension of primers GG-a, GGG-a, and GG-a3 could not be rescued by increasing the limiting nucleotide concentration in the in vitro telomerase assay (from 2 to 16 μM dGTP; data not shown). Minimizing the salt concentration during telomerase extension (+ and - KCl in Fig. 1_C_) partially restored the normal six-base ladder of products (compare lanes 13 and 15), a result that will be discussed below.
The Poor Primers Form Intramolecular G-Quadruplexes. Particularly puzzling was the fact that a small change in the number of guanines at the 5′ end of the oligonucleotide, far away from the site of extension at the 3′ end, switched a primer from being normal to being aberrant. A feature shared by all of the stalling primers, but none of the good primers, was the presence of four blocks of 2-3 guanines. Given that four repeats of the Oxytricha telomeric sequence TTTTGGGG fold into an intramolecular G-quadruplex that cannot be extended by Oxytricha telomerase (23), we hypothesized that those human telomeric primers containing four blocks of Gs were, similarly, forming G-quadruplexes that were poor telomerase substrates.
Native gel electrophoresis, under the ionic conditions of the telomerase enzyme assay, showed that primer G-a migrated slightly more slowly than primer a, as expected from their relative chain lengths; but the even-longer primers GG-a and GGG-a had abnormally fast electrophoretic mobilities, indicating a compact conformation (Fig. 2_A_, lanes 1-4). The compact structures were formed quickly, because boiling the oligonucleotides before electrophoresis had no effect, and their formation was concentration-independent over a wide range (0.5-600 nM; data not shown). These properties were consistent with intramolecular G-quadruplex formation.
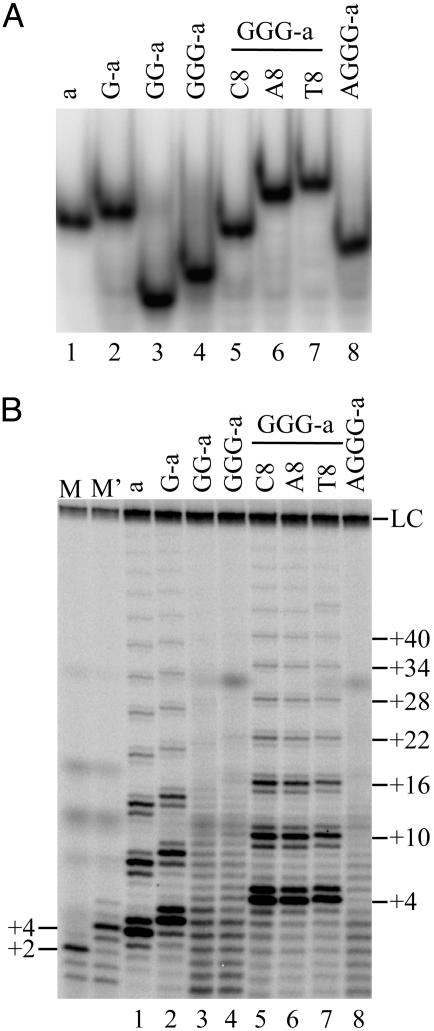
Fig. 2.
Poor primers form intramolecular G-quadruplexes. (A) Native gel analysis of primers. (B) Direct telomerase activity assay using the same primers (100 nM). Lanes M and M′ are as described in Fig. 1.
As a further test of this hypothesis, we designed a set of single-base mutants of oligonucleotide GGG-a to inhibit G-quadruplex formation. The second GGG block was mutated to GNG, where N = C, T, or A. These mutations had a dramatic effect on native gel mobility; none of the mutants showed the fast mobility seen with GGG-a, but, instead, GGG-aA8 and GGG-aT8 migrated as expected for unfolded 21-mers (Fig. 2 A, lanes 6-7). Primer GGG-aC8 had an intermediate mobility, consistent with either a partially folded form or rapid equilibrium between folded and unfolded states (Fig. 2 A, lane 5). There was, once again, a perfect correlation between gel mobility and telomerase extension; all of the mutants were good telomerase primers, even in the absence of hPOT1 (Fig. 2_B_, lanes 5-7). In contrast, a single-base change outside the G-blocks, as in primer GG-a3, did not disrupt the compact folded form and did not prevent telomerase stalling (Fig. 1_C_).
Intramolecular G-quadruplexes are stabilized by specific cations, such as Na+ and K+ (15, 16). It therefore seemed possible that reducing the cation concentration to the minimum required for telomerase activity might activate the folded primers. As shown in Fig. 1_C_, lane 15, lowering the KCl concentration gave partial restoration of the activity of primer GG-a3; a clear 6-nt ladder of extension products was superimposed on the heterogeneous distribution of stalled products.
hPOT1 Forms Stable Complexes with the G-Quadruplex-Forming DNAs. Electrophoretic mobility-shift analysis showed that all of the G-quadruplex oligonucleotides formed complexes with hPOT1 indistinguishable from those formed by unfolded oligonucleotides (Fig. 3_A_). In previous work, these complexes have been characterized as 1:1 complexes of hPOT1:ssDNA (12). This result supported the argument against the possibility that hPOT1 might be acting catalytically: opening the G-quadruplex structures and then departing. Furthermore, addition of hPOT1 to the GG-a3 oligonucleotide (whose mutation prevents its 5′ end from binding in the DNA-binding cleft of hPOT1) did not activate it for extension (Fig. 1_C_), again supporting the idea that a stable DNA-protein complex was necessary for telomerase action.
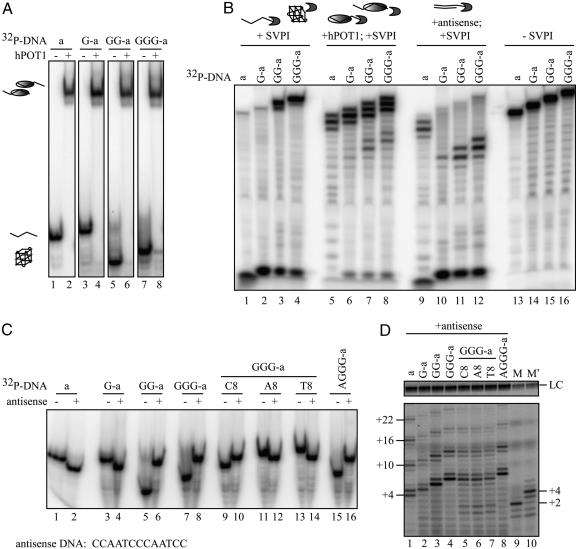
Fig. 3.
hPOT1 forms stable complexes with G-quadruplex-forming primers. (A) Native gel analysis of 100 nM primers in the presence (+) or absence (-)of333 nM hPOT1. (B) Denaturing gel electrophoresis of SVPI digests of primers alone (lanes 1-4), in complex with hPOT1 (lanes 5-8), or in complex with antisense DNA (lanes 9-12); no SVPI, lanes 13-16. (C) Native gel analysis of the primers (100 nM) in the presence (+) or absence (-) of 500 nM antisense DNA. The antisense DNA sequence is written 3′-to-5′. (D) Direct telomerase activity assays. Before adding telomerase, the DNA oligonucleotides (100 nM) were incubated with 500 nM antisense DNA for 10 min at room temperature. Lanes M and M′ are as described in Fig. 1.
We mapped the position of the leading edge of the hPOT1 protein on the DNA by using SVPI, which degrades ssDNA exonucleolytically from the 3′ end. On oligonucleotides with two overlapping copies of the preferred binding site, TTAGGGTTAG, SVPI degraded either 1-2 nt or 6-7 nt, consistent with hPOT1 binding in the 3′ half or the 5′ half of the DNA, respectively (Fig. 3_B_, lanes 5-8). Complexes of hPOT1 with G-quadruplex-forming oligonucleotides (GG-a and GGG-a) had a larger fraction of hPOT1 loaded on their 5′ end (6-7 nt digested) than those formed with unfolded oligonucleotides (a and G-a). In the absence of hPOT1, the G-quadruplex structures were rather resistant to SVPI digestion (Fig. 3_B_, compare lanes 3 and 4 with lanes 1 and 2).
Complementary Antisense Oligonucleotide also Allows Telomerase Extension of G-Quadruplex Primers. The intramolecular G-quadruplex form of an oligonucleotide is in equilibrium with unfolded or partially unfolded forms (33-35). Therefore, hPOT1 may function by simply “waiting” for the G-quadruplex to unfold and then binding and trapping the open form. If this were the mechanism, then an antisense oligonucleotide, whose binding would leave a telomerase-extendable 3′ tail, might substitute for hPOT1 in activating G-quadruplex primers.
To test this idea, we first used native gel electrophoresis to show that binding of the antisense oligonucleotide to G-quadruplexes was quantitative under telomerase buffer conditions (Fig. 3_C_). This experiment provided an additional confirmation of our assignment of folded and open DNA structures: binding of the antisense DNA to unfolded DNA (primer a, G-a, GGG-aA8, or GGG-aT8) increased its electrophoretic mobility, as expected, because more negative charge is added, but the end-to-end length is not greatly changed. On the other hand, binding of the antisense DNA to a G-quadruplex (primer GG-a, GGG-a, or AGGG-a) trapped it in an unfolded form, no longer having the unusually high mobility of the compact form. The partially folded primer GGG-aC8 showed an intermediate effect, being shifted to lower mobility but not as large a shift as with GG-a or GGG-a (Fig. 3_C_). SVPI digestion confirmed that the antisense oligonucleotides bound to their complementary sequence on the telomeric primer (Fig. 3_B_, lanes 9-12). Interestingly, the preferred binding site was near the 5′ end of telomeric primers GG-a and GGG-a, perhaps providing an indication that the 5′ end of the G-quadruplex unfolded first.
Finally, we added the antisense oligonucleotide to the G-quadruplexes and then tested for telomerase extension. The normal extension pattern was restored (Fig. 3_D_). From these studies, we conclude that hPOT1 and antisense DNA share the ability to disrupt human DNA G-quadruplexes and restore normal telomerase extension. They may very well act passively, binding to unfolded or partially folded intermediates that are in dynamic equilibrium with the G-quadruplex structures (Fig. 4).
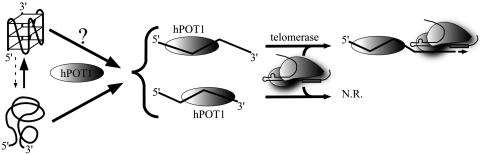
Fig. 4.
Model for hPOT1 disruption of intramolecular G-quadruplex DNAs, allowing their extension by telomerase. Quadruplex form is shown in equilibrium with partially or completely unstructured forms, which bind hPOT1. Our data do not rule out a more active opening of the quadruplex structure by hPOT1, indicated by?. When hPOT1 binds near the 5′ end of the primer, leaving an 8-nt tail, it can be extended by telomerase (10). When hPOT1 binds near the 3′ end of the primer, leaving a 2-nt tail, there is no reaction (N.R.).
Discussion
The ssDNA tail at the ends of human chromosomes is 130-210 nt long, which amounts to 21-35 repeats of d(TTAGGG) (36, 37). However, studies of human telomerase activity in vitro have typically used synthetic DNA primers, containing no more than three repeats. We now find that, as soon as the primer includes a fourth block of Gs, as in d[GGG(TTAGGG)3], telomerase extension stalls after every nucleotide added, because of intramolecular G-quadruplex formation. Addition of the human chromosome end-binding protein hPOT1 disrupts the G-quadruplexes, forming a stable ssDNA-hPOT1 complex and rescuing extension by recombinant telomerase. The DNA-binding domain of hPOT1 is sufficient for this activity, because the protein used in most of our studies contained only the N-terminal half of hPOT1 with the two oligonucleotide/oligosaccharide binding folds. Full-length hPOT1 had the same activity.
Several heterogeneous nuclear ribonucleoproteins (hnRNP A1, its shortened derivative UP1, and hnRNP D), which bind both single-stranded RNA and DNA, also disrupt human telomeric DNA quadruplexes (38-40). However, in marked contrast to the hPOT1 complex, the hnRNP-DNA complex is not a telomerase substrate (41, 42). Thus, the telomere-specific hPOT1 protein appears to have an activity quite distinct from the activity of these more general nucleic-acid-binding proteins.
The mechanism and kinetics by which hPOT1 disrupts intramolecular G-quadruplexes remain to be investigated. The protein could act passively, capturing the small proportion of unfolded DNA in the equilibrium population (27-29), thereby driving the binding to completion (Fig. 4). However, the observation that both hPOT1 and the antisense DNA bind more toward the 5′ ends of G-quadruplex-forming DNAs than with nonquadruplex DNAs is interesting. This finding may be providing an initial hint that the binding target is, in fact, a partially unfolded telomeric DNA species, open at its 5′ end. In addition, the current study is limited to recombinant telomerase core enzyme (hTERT protein and hTER RNA), and it will be important to test whether the natural telomerase holoenzyme acts any differently.
In addition to hPOT1, other factors disrupt G-quadruplexes. The yeast Sgs1 helicase and the human helicases that are products of the Bloom's- and Werner's-syndrome genes all unwind G-quadruplexes in an ATP-dependent reaction (20-22). This activity has been proposed to be important for proper telomere function in vivo (43).
Our findings may have implications for the use of G-quadruplex-interacting ligands as telomerase inhibitors, a possibility that is being intensely pursued as a potential anticancer chemotherapeutic approach (e.g., see refs. 44-47 and references therein). First, our observation that intramolecular G-quadruplexes of the human telomeric sequence are just on the edge of being reasonable telomerase substrates supports the idea that compounds that stabilize the folded form would make the DNA recalcitrant to telomerase extension. Second, our finding that hPOT1 disrupts the intramolecular quadruplex, forming a primer DNA-protein complex that is readily extended by telomerase, suggests that a combination therapy, targeting hPOT1 and stabilizing G-quadruplexes, might have synergistic efficacy.
Acknowledgments
We thank M. Lei (present address, University of Michigan, Ann Arbor) for establishing the hPOT1 expression and purification system, J. L. Chen (Arizona State University, Tempe) and C. W. Greider (The Johns Hopkins University School of Medicine, Baltimore) for generously providing the hTERT and hTER plasmids, and C. W. Greider and S. Neidle for helpful critique of the manuscript. This work was supported by the Howard Hughes Medical Institute.
Author contributions: A.J.Z., E.R.P., and T.R.C. designed research; A.J.Z. and E.R.P. performed research; and T.R.C. wrote the paper.
Abbreviations: TERT, telomerase reverse transcriptase; SVPI, snake venom phospodiesterase I.
References
- 1.Greider, C. W. (1991) Mol. Cell. Biol. 11**,** 4572-4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collins, K. & Greider, C. W. (1995) EMBO J. 14**,** 5422-5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gilley, D. & Blackburn, E. H. (1996) Mol. Cell. Biol. 16**,** 66-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hammond, P. W. & Cech, T. R. (1997) Nucleic Acids Res. 25**,** 3698-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hammond, P. W. & Cech, T. R. (1998) Biochemistry 37**,** 5162-5172. [DOI] [PubMed] [Google Scholar]
- 6.Bryan, T. M., Goodrich, K. J. & Cech, T. R. (2000) Mol. Cell 6**,** 493-499. [DOI] [PubMed] [Google Scholar]
- 7.Baumann, P. & Cech, T. R. (2001) Science 292**,** 1171-1175. [DOI] [PubMed] [Google Scholar]
- 8.Froelich-Ammon, S. J., Dickinson, B. A., Bevilacqua, J. M., Schultz, S. C. & Cech, T. R. (1998) Genes Dev. 12**,** 1504-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kelleher, C., Kurth, I. & Lingner, J. (2005) Mol. Cell. Biol. 25**,** 808-818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lei, M., Zaug, A. J., Podell, E. J. & Cech, T. R. (2005) J. Biol. Chem. 280**,** 20449-20456. [DOI] [PubMed] [Google Scholar]
- 11.Horvath, M. P., Schweiker, V. L., Bevilacqua, J. M., Ruggles, J. A. & Schultz, S. C. (1998) Cell 95**,** 963-974. [DOI] [PubMed] [Google Scholar]
- 12.Lei, M., Podell, T. R. & Cech, T. R. (2004) Nat. Struct. Mol. Biol. 11**,** 1223-1229. [DOI] [PubMed] [Google Scholar]
- 13.Zimmerman, S. B., Cohen, G. H. & Davies, D. R. (1975) J. Mol. Biol. 92**,** 181-192. [DOI] [PubMed] [Google Scholar]
- 14.Sen, D. & Gilbert, W. (1988) Nature 334**,** 364-366. [DOI] [PubMed] [Google Scholar]
- 15.Sundquist, W. I. & Klug, A. (1989) Nature 342**,** 825-829. [DOI] [PubMed] [Google Scholar]
- 16.Williamson, J. R., Raghuraman, M. K. & Cech, T. R. (1989) Cell 59**,** 871-880. [DOI] [PubMed] [Google Scholar]
- 17.Wang, Y. & Patel, D. J. (1993) Structure (London) 1**,** 263-282. [DOI] [PubMed] [Google Scholar]
- 18.Parkinson, G. N., Lee, M. P. H. & Neidle, S. (2002) Nature 417**,** 876-880. [DOI] [PubMed] [Google Scholar]
- 19.Schaffitzel, C., Berger, I., Postberg, J., Hanes, J., Lipps, H. J. & Pluckthun, A. (2001) Proc. Natl. Acad. Sci. USA 98**,** 8572-8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun, H., Bennett, R. J. & Maizels, N. (1999) Nucleic Acids Res. 27**,** 1978-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mohaghegh, P., Karow, J. K., Brosh, R. M., Jr., Bohr, V. A. & Hickson, I. D. (2001) Nucleic Acids Res. 29**,** 2843-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huber, M. D., Lee, D. C. & Maizels, N. (2002) Nucleic Acids Res. 30**,** 3954-3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zahler, A. M., Williamson, J. R., Cech, T. R. & Prescott, D. M. (1991) Nature 350**,** 718-720. [DOI] [PubMed] [Google Scholar]
- 24.Fletcher, T. M., Sun, D., Salazar, M. & Hurley, L. H. (1998) Biochemistry 37**,** 5536-5541. [DOI] [PubMed] [Google Scholar]
- 25.Raghuraman, M. K. & Cech, T. R. (1990) Nucleic Acids Res. 18**,** 4543-4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yanez, G. H., Khan, S. J., Locovei, A. M., Pedroso, I. M. & Fletcher, T. M. (2004) Biochem. Biophys. Res. Commun. 328**,** 49-56. [DOI] [PubMed] [Google Scholar]
- 27.Mergny, J. L., Phan, A. T. & Lacroix, L. (1998) FEBS Lett. 435**,** 74-78. [DOI] [PubMed] [Google Scholar]
- 28.Balagurumoorthy, P. & Brahmachari, S. K. (1994) J. Biol. Chem. 269**,** 21858-21869. [PubMed] [Google Scholar]
- 29.Zhao, Y., Kan, Z.-X., Hao, Y.-H., Chen, H. & Tan, Z. (2004) J. Am. Chem. Soc. 126**,** 13255-13264. [DOI] [PubMed] [Google Scholar]
- 30.Chen, J. L. & Greider, C. W. (2003) EMBO J. 22**,** 304-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morin, G. B. (1989) Cell 59**,** 521-529. [DOI] [PubMed] [Google Scholar]
- 32.Rivera, M. A. & Blackburn, E. H. (2004) J. Biol. Chem. 279**,** 53770-53781. [DOI] [PubMed] [Google Scholar]
- 33.Li, W., Wu, P., Ohmichi, T. & Sugimoto, N. (2002) FEBS Lett. 526**,** 77-81. [DOI] [PubMed] [Google Scholar]
- 34.Phan, A. T. & Mergny, J.-L. (2002) Nucleic Acids Res. 30**,** 4618-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Risitano, A. & Fox, K. R. (2003) Biochemistry 42**,** 6507-6513. [DOI] [PubMed] [Google Scholar]
- 36.Makarov, V. L., Hirose, Y. & Langmore, J. P. (1997) Cell 88**,** 657-666. [DOI] [PubMed] [Google Scholar]
- 37.Wright, W. E., Tesmer, V. M., Huffman, K. E., Levene, S. D. & Shay, J. W. (1997) Genes Dev. 11**,** 2801-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fukuda, H., Katahira, M., Tsuchiya, N., Enokizono, Y., Sugimura, T., Nagao, M. & Nakagama, H. (2002) Proc. Natl. Acad. Sci. USA 99**,** 12685-12690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Enokizono, Y., Matsugami, A., Uesugi, S., Fukuda, H., Tsuchiya, N., Sugimura, T., Nagao, M., Nakagama, H. & Katahira, M. (2003) Nucleic Acids Res. Suppl. 2003, 231-232. [DOI] [PubMed]
- 40.Myers, J. C., Moore, S. A. & Shamoo, Y. (2003) J. Biol. Chem. 278**,** 42300-42306. [DOI] [PubMed] [Google Scholar]
- 41.Dallaire, F., Dupuis, S., Fiset, S. & Chabot, B. (2000) J. Biol. Chem. 275**,** 14509-14516. [DOI] [PubMed] [Google Scholar]
- 42.Enokizono, Y., Konishi, Y., Nagata, K., Ouhashi, K., Uesugi, S., Ishikawa, F. & Katahira, M. (2005) J. Biol. Chem. 280**,** 18862-18870. [DOI] [PubMed] [Google Scholar]
- 43.Crabbe, L., Verdun, R. E., Haggblom, C. I. & Karlseder, J. (2004) Science 306**,** 1951-1953. [DOI] [PubMed] [Google Scholar]
- 44.Kim, M. Y., Gleason-Guzman, M., Izbicka, E., Nishioka, D. & Hurley, L. H. (2003) Cancer Res. 63**,** 3247-3256. [PubMed] [Google Scholar]
- 45.Gomez, D., Paterski, R., Lemarteleur, T., Shin-ya, K., Mergny, J.-L. & Riou, J.-F. (2004) J. Biol. Chem. 279**,** 41487-41494. [DOI] [PubMed] [Google Scholar]
- 46.Burger, A. M., Dai, F., Schultes, C. M., Reszka, A. P., Moore, M. J., Double, J. A. & Neidle, S. (2005) Cancer Res. 65**,** 1489-1496. [DOI] [PubMed] [Google Scholar]
- 47.Pennarun, G., Granotier, C., Gauthier, L. R., Gomez, D., Hoffschir, F., Mandine, E., Riou, J.-F., Mergny, J.-L., Mailliet, P. & Boussin, F. D. (2005) Oncogene 24**,** 2917-2928. [DOI] [PubMed] [Google Scholar]