Interactions between Hairy/Enhancer of Split-related proteins and the pancreatic transcription factor Ptf1-p48 modulate function of the PTF1 transcriptional complex (original) (raw)
Abstract
In the developing pancreas, the onset of exocrine differentiation is driven by the activity of the PTF1 (pancreas transciption factor 1) transcriptional complex, which is comprised of the class II bHLH (basic helix–loop–helix) protein, Ptf1-p48 [also known as Ptf1a (pancreas specific transcription factor 1a)], and a class I E-box binding partner. Activity of the PTF1 complex is normally inhibited by the Notch signalling pathway, a process mediated by Notch effector proteins in the HES (Hairy/Enhancer of Split) family of bHLH transcriptional repressors. In the present study, we show that this inhibitory effect occurs through direct interaction between HES family members and Ptf1-p48. The HES family members Hey1 (hairy/enhancer-of-split related with YRPW motif 1) and Hey2 co-immunoprecipitate with Ptf1-p48, and Ptf1-p48 binding by Hes1 is also evident in yeast two-hybrid and GST (glutathione S-transferase) pull-down assays. The ability of Hes1 to interact with Ptf1-p48 resides within a fragment comprised of the bHLH, Orange and C-terminal domains, and does not require the N-terminal or WRPW elements. The ability of truncated versions of Hes1 to bind Ptf1-p48 correlates with their ability to down-regulate the activity of the PTF1 transcriptional complex, defining Ptf1-p48 binding as the most likely mechanism by which Notch effector proteins delay exocrine pancreatic differentiation.
Keywords: development, differentiation, Hairy/Enhancer of Split (HES), Notch, pancreas, pancreas transciption factor 1 (PTF1)
Abbreviations: bHLH, basic helix–loop–helix; E13.5, embryonic day 13.5 (etc.); GST, glutathione S-transferase; HES, Hairy/Enhancer of Split; Hey1, hairy/enhancer-of-split related with YRPW motif 1; Hey2, hairy/enhancer-of-split related with YRPW motif 2; MyoD, myogenic differentiation 1; PTF1, pancreas transciption factor 1; Ptf1-p48, pancreas specific transcription factor 1a (Ptfla); Sc, Scute; X-Gal, 5-bromo-4-chloroindol-3-yl β-D-galactopyranoside
INTRODUCTION
During foregut development in the mouse, expression of the bHLH (basic helix–loop–helix) transcription factor Ptf1-p48 [also known as Ptf1a (pancreas specific transcription factor 1a)] distinguishes nascent pancreas from adjacent gut and liver. Mice with targeted inactivation of Ptf1-p48 exhibit severely aborted pancreatic morphogenesis, as well as substantial reductions in both exocrine and endocrine cell types [1,2]. Detailed lineage tracing studies have confirmed that Ptf1-p48-positive progenitor cells ultimately give rise to all differentiated pancreatic cell types. This confirms a central role for Ptf1-p48 in the specification and expansion of a pancreatic progenitor pool [1]. Ptf1-p48 is also known to be required later in development, where it acts to guide terminal differentiation and cell-type-specific gene expression in pancreatic acinar cells [3,4]. In addition to its central role in pancreatic development, Ptf1-p48 is also expressed in developing retina, hindbrain and spinal cord [5,6], and has recently been implicated as an important regulator of cerebellar development [7].
Although the interacting proteins and relevant target genes mediating the early effects of Ptf1 have not yet been identified, the late influence of Ptf1-p48 on acinar cell differentiation is known to occur through the multiprotein PTF1 (pancreas transciption factor 1) transcriptional complex. This complex includes the class II HLH Ptf1-p48 protein, as well as a class I E-box binding partner [3]. The PTF1 complex binds directly to common promoter elements in acinar cell-specific genes, resulting in acinar-cell differentiation and associated cell cycle exit [3,8]. Recent evidence suggests that the Notch signalling pathway represents a critical switch in the regulation of the early compared with the late effects of Ptf1-p48 [9–11]. Although the early pancreas-promoting effects of Ptf1-p48 occur in the setting of an active Notch pathway, Notch signalling appears to be silenced with the onset of acinar cell differentiation. Notch has been shown further to actively inhibit the late effects of Ptf1-p48 by preventing the PTF1 transcriptional complex from binding to target DNA [11]. In so doing, Notch effectively delays the onset of exocrine differentiation, thereby preserving an appropriate pancreatic progenitor pool that is required for normal growth and morphogenesis. The observation of frequent Notch pathway activation in human pancreatic cancer suggests that this effect may also play a role in pancreatic tumorigenesis [12].
Although these recent studies have provided important information regarding Notch regulation of the pancreatic progenitor pool, the mechanism by which Notch exerts its influence on Ptf1-p48 function has not yet been elucidated. Based on previous studies suggesting that Notch and its effector proteins are capable of influencing the function of the PTF1 transcriptional complex without altering Ptf1-p48 expression [11], we have investigated the possibility that Notch effector proteins in the HES (Hairy/Enhancer of Split) family, including Hes1 (hairy and enhancer of split 1), Hey1 (hairy/enhancer-of-split related with YRPW motif 1) and Hey2, are capable of directly interacting with Ptf1-p48 in a manner that inhibits function of the PTF1 transcriptional complex.
MATERIALS AND METHODS
Plasmid constructs
FLAG epitope-tagged full-length and truncated rat Hes1 expression vectors were generated by PCR amplification of the appropriate fragments, followed by in-frame assembly in pCMV-Tag1 (Stratagene). The sequences of all constructs were verified and protein expression was confirmed by Western blotting using anti-FLAG antisera (Stratagene). Expression vectors encoding human E47 and rat Ptf1-p48 were obtained from Dr Michael Chin (Vascular Medicine Research, Brigham and Women's Hospital, Harvard Medical School, Harvard University, Cambridge, MA, U.S.A.) and Dr Francisco Real (Institut Munincipal d'Investigacio Medica, Universitat Pompeu Fabra, Barcelona, Spain). For production of GST (glutathione S-transferase)-fusion proteins, full-length and truncated rat Ptf1-p48 coding sequences were PCR amplified and cloned in-frame into pGEX-4t-1 (Amersham Biosciences). Ptf1-p48 elements used for construction of GST-fusion proteins included full-length (amino acids 1–326), N-terminal (amino acids 1–170), N-terminal+bHLH (amino acids 1–196), bHLH+C-terminal (amino acids 171–326) and C-terminal alone (amino acids 189–326). To create V5 epitope-tagged Ptf1, Ptf1-p48 coding sequence was PCR amplified and cloned in-frame in pcDNA3.1/V5-His (Invitrogen) to obtain pcDNA-Ptf1-V5. A previously characterized PTF1-responsive luciferase reporter containing four tandem repeats of the PTF1 binding site from the rat chymotrypsinogen promoter [5] was obtained from Dr Masashi Kawaichi (Nara Institute of Science and Technology, Nara, Japan). The sequences of all the clones were verified prior to use.
Immunoprecipitation and Western blotting
COS7 cells seeded at (2–2.5)×106 per 10-cm-diameter culture dish were transfected with 15 μg of appropriate expression plasmids using Lipofectamine 2000 (Invitrogen), according to the manufacturer's protocol. At 2 days after transfection, the cells were washed and lysed in 1 ml of ice-cold lysis buffer [1× PBS (pH 7.4), 5 mM EDTA, 0.5% (v/v) Triton X-100 and 0.5 mM PMSF, supplemented with 10 μg/ml of the following protease inhibitors: leupeptin, aprotinin and pepstatin]. The cell suspensions were sonicated, for 10 s, six times and the extract was clarified by centrifugation at 16000 g for 30 min at 4 °C. Total protein (1 mg) was pre-cleared with anti-(mouse IgG)–agarose beads (catalogue number A6531; Sigma–Aldrich) for 10 min at 4 °C. After centrifugation at 735 g for 2 min, the supernatant was incubated overnight at 4 °C with either a monoclonal anti-FLAG antibody (1:500 dilution; Sigma) or monoclonal anti-V5 (1:500 dilution; Invitrogen). Anti-(mouse IgG)–agarose beads were added and incubated for 1 h at 25 °C and then washed four times in lysis buffer, before resuspending in 40 μl of 1× SDS sample loading buffer [50 mM Tris/HCl (pH 6.8), 100 mM dithiothreitol, 2% (v/v) SDS, 0.1% (w/v) Bromophenol Blue and 10% (v/v) glycerol]. Samples were then subjected to electrophoresis on 10% (w/v) denaturing polyacrylamide gels. Western blot analysis was performed using either a monoclonal anti-FLAG antibody (1:1000 dilution; Stratagene) or a rabbit anti-Ptf1-p48 (1:1000 dilution; provided by Dr Francisco Real). Immune complexes were detected using either peroxidase-conjugated anti-mouse (1:5000 dilution) or anti-rabbit (1:5000 dilution) IgG (Santa Cruz), and protein bands were visualized using the Enhanced Chemiluminescence System (Pierce). To ensure loading of equivalent amounts of protein, protein concentrations of cell lysates were quantified using the Bradford method (Bio-Rad Laboratories), and the relative amounts were further verified by SDS/10% PAGE and staining with Coomassie Blue.
Yeast two-hybrid assay
Yeast two-hybrid assays were performed using the Yeast Two-Hybrid System 3 (Clontech). Full-length and truncated rat Ptf1-p48 elements were PCR amplified from a full-length Ptf1-p48 cDNA (provided by Dr Francisco Real), using primers with appropriate restriction enzyme sites, and inserted in-frame into the pGBKT7 vector to construct pGBK-Ptf1 bait plasmids. Ptf1-p48 elements utilized included full-length (amino acids 1–326), bHLH+C-terminal (amino acids 171–326) and C-terminal alone (amino acids 189–326). The coding region of full-length rat Hes1 was inserted in-frame into pGADT7 to construct the pGAD-Hes1 prey plasmid. In this system, the pGADT7 vector provides a Leu2 nutritional marker, the pGBKT7 vector provides a TRP1 marker and reconstitution of Gal4 activity by bait–prey interaction results in activation of UAS (upstream activating sequence)-responsive HIS3 and lacZ markers. Bait and prey plasmids were confirmed by DNA sequencing and were then used to transform the yeast strain AH109, using the lithium acetate method [12a]. Following initial expansion on media lacking leucine and tryptophan, yeast clones were plated on to selective media containing X-Gal (5-bromo-4-chloroindol-3-yl β-D-galactopyranoside) and lacking leucine, tryptophan and histidine, or combinations thereof.
GST pull-down assays
GST pull-down assays were performed as follows. Briefly, GST fusion proteins were expressed in Escherichia coli strain BL21, which was grown in super broth medium (32 g/l tryptone, 20 g/l yeast extract, 5 g/l NaCl, 5 mM NaOH), at 37 °C, to mid-log phase and then expression was induced by 0.1 mM IPTG (isopropyl β-D-thiogalactopyranoside) for another 4 h at 37 °C. Bacteria were harvested by centrifugation at 4500 g for 15 min at 4 °C and lysed by sonication in STE buffer [10 mM Tris (pH 8.0), 1 mM EDTA and 150 mM NaCl]. Lysates were clarified by centrifugation at 10000 g for 30 min at 4 °C, and then mixed with glutathione–Sepharose 4B beads for 30 min at 25 °C in PBS. Glutathione beads containing the appropriate fusion proteins were incubated in a binding buffer [50 mM Tris (pH 7.5), 100 mM NaCl, 0.1% (v/v) Triton X-100, 1 mM dithiothreitol and 10% (v/v) glycerol] supplemented with complete protease inhibitors (Sigma). Binding assays were carried out in 1 ml of binding buffer containing pre-loaded beads, along with 20 μl of [35S]methionine-labelled Hes1 protein [transcribed and translated in vitro using the TNT coupled transcription/translation system (Promega)]. Hes1 elements used included full-length (amino acids 1–282), N-terminal+bHLH (amino acids 1–93), Orange+C-terminal+WRPW (amino acids 109–282), bHLH+Orange+C-terminal (amino acids 36–276) and bHLH+Orange+C-terminal+WRPW (amino acids 36–282). In each case, the quantity and quality of _in vitro_-synthesized protein was monitored using SDS/15% PAGE followed by PhosphorImager (Molecular Dynamics) analysis of the dried gel. After overnight incubation, the beads were collected by centrifugation at 500 g for 5 min at 25 °C, washed four times in binding buffer and boiled in 2× SDS loading buffer. Bound proteins were resolved by electrophoresis on either 10 or 15% polyacrylamide gels. After electrophoresis, the gels were dried and imaged using a PhosphorImager.
Luciferase assays
COS7 cells were transfected in 24-well plates with 0.5 μg/well of the PTF1-luc firefly luciferase reporter plasmid, in combination with the indicated expression vectors. In addition, 0.025 μg/well of the pRL-TK Renilla luciferase reporter plasmid (Promega) was added to control for transfection efficiency. Total DNA per transfection was kept constant at 1 μg/well by adding vector DNA as needed. After 24 h, cells were harvested using the dual-luciferase reporter assay system (Promega). For each condition, sequential measurement of firefly and Renilla luciferase was performed, and normalized luciferase activity was reported as the ratio of firefly to Renilla luciferase activity. The data reported represent results from three independent experiments, each performed in triplicate. Appropriate expression from all of the constructs was confirmed by Western blotting using the following antibodies and dilutions: rabbit anti-V5 antibody (Invitrogen), 1:5000; rabbit anti-E47 anitbody (Santa Cruz), 1:1000; mouse monoclonal anti-FLAG antibody (Stratagene) 1:1000.
RESULTS
HES family members co-immunoprecipitate with Ptf1-p48 in transient transfection assays
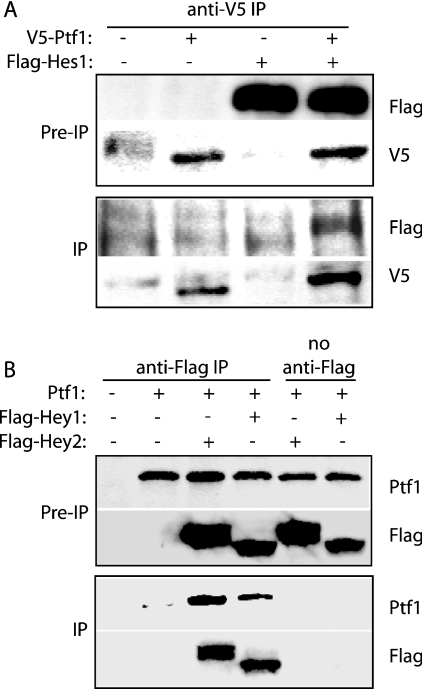
We have determined previously [11] that Notch signalling inhibits function of the PTF1 transcriptional complex, independent of changes in expression levels of PTF1 component proteins. We therefore hypothesized that this inhibition might be mediated by physical interactions between Ptf1-p48 and members of the HES family of bHLH transcriptional repressors. To evaluate this possibility, we co-transfected COS7 cells with FLAG-tagged full length Hes1 and V5-tagged Ptf1-p48, either alone or in combination. At 48 h following transfection, cell extracts were immunoprecipitated with anti-V5 antisera, and then subjected to immunoblotting with anti-FLAG antisera (Figure 1A). Immunoreactive FLAG–Hes1 was detected only in samples derived from V5–Ptf1-p48 transfected cells, suggesting a physical interaction between Hes1 and Ptf1-p48.
Figure 1. HES-related proteins and Ptf1-p48 co-immunoprecipitate in COS7 cell lysates.
(A) Association between Ptf1-p48 and Hes1. COS7 cells were transfected with expression vectors encoding V5 epitope-tagged Ptf1-p48 and FLAG-tagged Hes1, either alone or in combination. Immunoblot analysis of cell lysates prior to IP (immunoprecipitation) confirms appropriate expression following transient transfection. Following IP with the anti-V5 antibody, FLAG–Hes1 was detectable in the immunoprecipitate only in the presence of V5–Ptf1-p48. (B) Association of Ptf1-p48 with Hey1 and Hey2. COS7 cells were transfected with expression vectors encoding Ptf1-p48, FLAG–Hey1 and FLAG–Hey2, either alone or in combination. Pre-IP samples confirmed appropriate expression. Following IP with the anti-FLAG antibody, immunoreactive Ptf1 was present in the immunoprecipitate only in the presence of either Hey1 or Hey2. Pre-IP samples represented one-tenth of the total cell lysates used in the IP reaction.
In order to determine whether the ability to interact with Ptf1-p48 represented a common feature of HES-related proteins, similar experiments were performed using FLAG-tagged full-length Hey1 and Hey2 [13]. For this section of the present study, immunoprecipitation was performed using anti-FLAG antisera, and immunoblots were performed with anti-Ptf1-p48 antisera. As in the case of Hes1, transfection with either FLAG–Hey1 or FLAG–Hey2 resulted in co-immunopreciptation of immunoreactive Ptf1-p48 (Figure 1B).
Hes1 and the C-terminus of Ptf1-p48 physically interact in a yeast two-hybrid assay
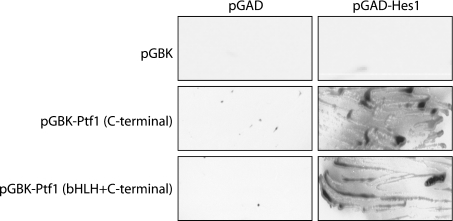
To confirm further the interaction between Hes1 and Ptf1-p48, we employed a yeast two-hybrid approach. Initial studies suggested that the N-terminal domain of Ptf1-p48 possessed an intrinsic transactivation domain. This was demonstrated by the ability of N-terminal elements placed in-frame with the Gal4 DNA-binding domain to generate colonies on restrictive media, either alone or in combination with an empty vector containing the Gal4 transactivation domain (results not shown). In contrast, the C-terminal domain of Ptf1-p48, either alone or in combination with the bHLH domain, had no intrinsic activity. These elements, cloned in-frame with the Gal4 DNA-binding domain, were therefore used as bait and full-length Hes1, cloned in-frame with the Gal4 transactivation domain, was used as prey. As shown in Figure 2, both the Ptf1-p48 C-terminal fragment, as well as a longer fragment including the bHLH domain, were able to interact with full-length Hes1, as demonstrated by the formation of β-galactosidase-positive colonies on synthetic drop-out media lacking leucine, tryptophan and histidine. No significant growth was observed when the bait plasmids were co-transformed along with an empty vector containing the Gal4 activation domain (pGAD-T7), or when pGAD–Hes1 was co-transformed with an empty vector containing the Gal4 DNA-binding domain (Figure 2).
Figure 2. Hes1 and the C-terminal domain of Ptf1-p48 interact in a yeast two-hybrid assay.
Colonies from SD (synthetic drop-out) media lacking leucine and tryptophan were streaked on X-Gal-containing SD plates lacking leucine, tryptophan and histidine. Vectors containing the Gal4 DNA-binding domain in-frame with either the C-terminal domain of Ptf1-p48 (pGBK-Ptf1 C-terminal) or with a combined bHLH/C-terminal domain (pGBK-Ptf1 bHLH+C-terminal) did not produce viable colonies when combined with an empty Gal4 activation vector (pGAD), but did produce blue colonies when combined with the Gal4 activation domain in-frame with Hes1. No viable colonies were observed when pGAD-Hes1 was transformed with an empty pGBK vector.
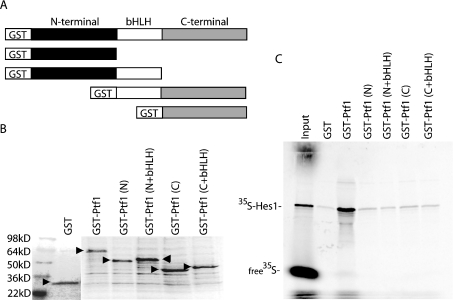
Full-length Ptf1-p48 protein binds to Hes1 in vitro
Results from the co-immunoprecipation and yeast two-hybrid studies suggested that Hes1 was capable of physically interacting with Ptf1-p48, either directly or in combination with an adaptor protein. To determine if Hes1 and Ptf1 were capable of direct interaction in the absence of other associated proteins, we constructed GST-fusion proteins, encoding either full-length or truncated versions of Ptf1-p48 (Figure 3A), for use in GST pull-down assays with _in vitro_-synthesized 35S-labelled Hes1. As shown in Figure 3(B), these constructs allowed for in vitro synthesis of GST-fusion proteins of the appropriate molecular masses. When equal amounts of either GST alone or the various GST–Ptf1-p48 fusion proteins were loaded on to glutathione beads, only full-length GST–Ptf1-p48 was able to bind 35S-labelled Hes1 at a level exceeding that of GST alone (Figure 3C). These results suggest a direct interaction between Hes1 and Ptf1-p48, without the need for associated adaptor proteins. In contrast with the ability of an isolated C-terminal fragment of Ptf1-p48 to interact with Hes1 in the context of a yeast two-hybrid assay, the requirement for full-length Ptf1-p48 in the GST pull-down assay may reflect differences between yeast and in vitro systems with respect to post-translational protein modification and folding.
Figure 3. Hes1 and Ptf1-p48 interact directly in GST pull-down assays.
(A) Schematic representation of GST–Ptf1-p48 fusion proteins utilized in the GST pull-down assays. (B) Coomassie-Blue-stained SDS/15%PAGE gel showing purified GST–Ptf1-p48 fusion proteins (arrowheads), confirming equal loading of proteins on glutathione beads. (C) Retention of 35S-labelled in vitro transcribed and translated Hes1 on glutathione beads pre-loaded with full-length GST–Ptf1-p48, as detected by PhosphorImager detection following resolution on SDS/10% PAGE gel. Truncated versions of Ptf1-p48 involving GST fused to either N-terminal alone [GST–Ptf1(N)], N-terminal + bHLH [GST–Ptf1(N+bHLH)], C-terminal alone [GST–Pft1(C)], or C-terminal + bHLH [GST–Pft1(C+bHLH)] do not retain 35S-labelled Hes1 at a level exceeding that by GST alone. The input lane was loaded with one tenth of the total 35S-labelled Hes1 used in each pull-down assay.
C-terminal and bHLH domains of Hes1 are necessary for interaction with Ptf1-p48
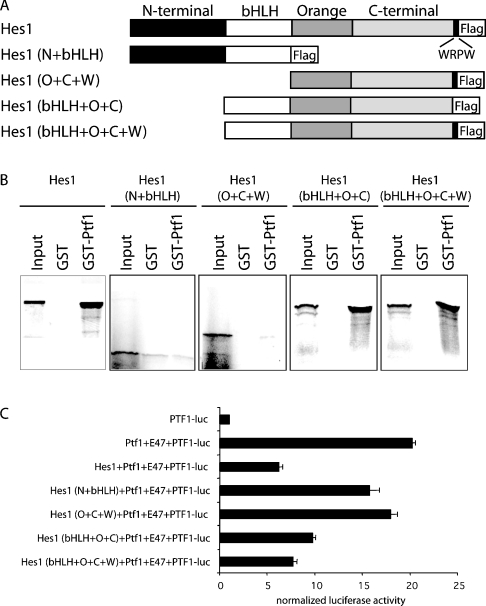
We next determined the specific Hes1 protein domains required for interaction with Ptf1-p48 and correlated these findings with the ability of truncated Hes1 fragements to down-regulate the activity of the PTF1 transcriptional complex. The domain structure of HES family members has been elucidated by extensive structure–function analyses. HES-related proteins contain a distinctive proline-rich bHLH domain, an Orange domain and a WRPW motif at the C-terminus. The bHLH domain confers DNA binding specificity [14,15], whereas the Orange domain mediates interaction with other transcriptional activators and is essential for full transcriptional repression [16]. The WRPW motif is critical for recruiting members of the Groucho family of co-repressors [17].
In an attempt to map the site of interaction of Ptf1-p48 on Hes1, a deletion series of FLAG-tagged constructs was generated (Figure 4A). This series included full-length Hes1, the N-terminal and bHLH domains (N+bHLH), the Orange, C-terminal and WRPW domains (O+C+W), and the bHLH, Orange and C-terminal domains, with and without the WRPW motif (bHLH+O+C and bHLH+O+C+W respectively). Each of these fragments was synthesized in vitro an 35S-labelled protein and then added to glutathione beads loaded previously with either GST alone or a full-length GST–Ptf1-p48 fusion protein. In this section of the present study, only fragments containing the bHLH, Orange and C-terminal domains of Hes1 were able bind Ptf1-p48, defining these domains as requisite elements mediating the Hes1/Ptf1-p48 interaction (Figure 4B).
Figure 4. Hes1 binding to Ptf1-p48 correlates with functional inhibition of the PTF1 transcriptional complex.
(A) Schematic representation of in vitro transcribed and translated 35S-labelled Hes1 proteins used in the GST pull-down assays with full-length GST–Ptf1-p48. (B) Retention of full-length and truncated versions of 35S-labelled Hes1 by GST–Ptf1-p48 and GST alone, as determined by Phosphorimager analysis following resolution on SDS/15% PAGE gel. The input lanes were loaded with one tenth of the total 35S-labelled Hes1 used in each pull-down assay. Full-length Hes1, Hes1 (bHLH+O+C) and Hes1 (bHLH+O+C+W) were retained by GST–Ptf1-p48, whereas versions of Hes1 lacking either the C-terminal [Hes1 (N+bHLH)] or bHLH [Hes1 (O+C+W)] domains were not retained. (C) Corresponding effects of full-length and truncated versions of Hes1 on the activity of the PTF1 transcriptional complex were assessed by activation of PTF-responsive luciferase reporter (PTF1-luc). Maximal inhibition of PTF1 activity was achieved by full-length Hes1, Hes1 (bHLH+O+C), and Hes1 (bHLH+O+C+W), whereas versions of Hes1 unable to bind Ptf1-p48 had a lesser effect.
Ptf1-p48 binding capacity is required for Hes1 to down-regulate the activity of the PTF1 transcriptional complex
We have demonstrated previously [11] the ability of HES family members, including Hes1, Hey1 and Hey2, to down-regulate the activity of the PTF1 transcriptional complex. Based on our delineation of truncated Hes1 protein elements capable of physically interacting with Ptf1-p48, we next correlated the ability of Hes1 fragments to bind Ptf1-p48 with the ability of these fragments to down-regulate the activity of the PTF1 transcriptional complex. For this section of the present study, activity of the PTF1 transcriptional complex was determined by the ability of Ptf1-p48 and E47 to co-operatively activate a PTF1-responsive luciferase reporter containing a 4× tandem repeat of the PTF1 binding element from the rat chymotrypsinogen promoter (PTF1-luc). COS7 cells were transfected with pcDNA-based expression vectors encoding Ptf1-p48 and E47, in combination with PTF1-luc and either full-length or truncated Hes1 (Figure 4C). As expected, COS7 cells exhibited little endogenous PTF1 activity, reflecting their non-pancreatic identity. Co-transfection of Ptf1-p48 and E47 resulted in a 20-fold activation of the PTF1 reporter, and this activation was dramatically inhibited by the addition of full-length Hes1. When the N+bHLH, O+C+W, bHLH+O+C and bHLH+O+C+W fragments of Hes1 were evaluated in this assay, only those fragments shown in the present study (Figure 4B) to physically interact with Ptf1-p48 (bHLH+O+C and bHLH+O+C+W) were able to recapitulate the inhibitory influence of full-length Hes1 on PTF1 activity. These results demonstrate a strong correlation between the ability of Hes1 to bind Ptf1-p48 and its ability to down-regulate the activity of the PTF1 transcriptional complex. This suggests that the influence of Notch on exocrine pancreatic differentiation may be dependent upon direct interactions between Ptf1-p48 and HES-related proteins.
DISCUSSION
During pancreatic development, Notch signalling appears to modulate a critical switch between early Ptf1-p48 functions, related to specification and expansion of pancreatic progenitor cells, and late Ptf1-p48 functions, related to terminal acinar cell differentiation. In the present study, we define a potential molecular mechanism for this effect, involving physical interaction between Ptf1-p48 and Notch effector proteins in the HES family of bHLH transcriptional repressors. We have documented this interaction by several complementary methods, including co-immunoprecipitation, yeast two-hybrid and GST pull-down assays. Although the necessary reagents for evaluating interactions between endogenous proteins (i.e. antibodies suitable for immunoprecipitation) are not currently available, the specificity of the Ptf1-p48/Hes1 interaction is supported further by a requirement for discrete protein domains and by a strict functional correlation between the ability of truncated versions of Hes1 to bind Ptf1-p48 and their corresponding ability to alter function of the PTF1 transcriptional complex.
Early in pancreatic development, Notch signalling is activated in a dedicated exocrine pancreatic progenitor pool, as demonstrated by widespread co-expression of Hes1 and Ptf1-p48 in E13.5 (embryonic day 13.5) pancreatic epithelium [11]. With the focal onset of acinar cell differentiation on E14.5, Hes1 expression is silenced in nascent Ptf1-p48-positive acinar cells, whereas persistent Hes1 expression is observed in the immediately adjacent undifferentiated epithelium. More direct evidence of the negative influence of Notch on acinar cell differentiation is provided by the observation that ectopic expression of either activated Notch or Notch target genes in explant cultures of E10.5 dorsal pancreatic buds results in cell-autonomous inhibition of acinar cell differentiation, even in the face of ongoing Ptf1-p48 protein expression [11]. Similar results are observed in vivo following expression of an activated Notch transgene [9,10]. In addition, inactivation of Notch signalling in zebrafish embryos, either by mutation at the mindbomb locus or by injection of a dominant-negative Suppressor of Hairless RNA, results in acceleration of exocrine differentiation [11]. This defines inhibition of the PTF1 transcriptional complex as a normal function of Notch signalling in developing pancreas.
In many settings, Notch effector proteins in the HES family act to prevent or delay differentiation by suppressing expression of lineage-specifying bHLH transcription factors, including Ngn3 (neurogenin 3) [18] and MyoD (myogenic differentiation 1) [19,20], reflecting the ability of many HES-related proteins to act as transcriptional repressors. In the case of Ptf1-p48, however, this type of influence would not only prevent PTF1-dependent exocrine differentiation, but also prevent the early pancreas-promoting and morphogenetic influences of Ptf1-p48, which may be mediated without formation of a typical PTF1 transcriptional complex. Instead, the ability of HES-related proteins to alter Ptf1-p48 function in the face of ongoing expression allows for the early morphogenetic effects of Ptf1-p48 to be accomplished in the context of active Notch signalling, even while induction of acinar cell differentiation is prevented.
Several other class II bHLH proteins appear to be functionally modulated by direct interaction with HES-related proteins. Hey1/CHF2 is capable of forming complexes with MyoD, and has been shown to impair myogenesis by inhibiting binding of MyoD/E47 heterodimers to DNA [21]. In Drosophila, Enhancer of Split [E(spl)]-related proteins have similarly been shown to form complexes with Sc (Scute), a proneural bHLH transcription factor, impairing the ability of Sc-containing complexes to activate target genes [22]. Unlike many protein–protein interactions involving bHLH proteins, the interactions described above all occur without a requirement for binding by adjacent HLH domains. For both Ptf1-p48 and Sc, association with HES-related proteins occurs even in the case of isolated C-terminal domains lacking HLH elements (present study and [22]). Similarly, truncated versions of Hey1/CHF2 involving isolated C-terminal elements lacking the bHLH domain are fully able to suppress MyoD function [21]. In addition to interactions with class II bHLH proteins, Hes1 has also been shown to modulate JAK/STAT (signal transducer and activator of transcription) signalling through a direct interaction with STAT3 [23] and to similarly alter the activity of Runt/Cbfa1 (runt related transcription factor 2) [24].
Together with previous work [11], the present study supports a model in which Notch-dependent Hes1 expression acts as a critical switch during exocrine pancreatic development, allowing for orderly transition between the early morphogenetic influence of Ptf1-p48 and later induction of acinar cell differentiation. Prior to E14.5, Ptf1-p48 appears to be expressed in the context of an active Notch pathway, in which the morphogenetic effects of Ptf1-p48 are permitted, whereas the PTF1-dependent induction of acinar cell differentiation is delayed. Based on the results from the present study, it appears that this early inhibition of the PTF1 transcriptional complex is at least partially mediated by physical interaction between Hes1 and Ptf1-p48. Although previous studies have shown that Hes1 prevents binding of the PTF1 complex to its target DNA [11], it remains to be determined whether complexes involving Hes1 and Ptf1-p48 are capable of binding to alternative DNA targets, as well as whether such complexes are involved in the active promotion of pancreatic morphogenesis.
Acknowledgments
We thank Dr Ray MacDonald (The University of Texas Southwestern Medical Center, Dallas, TX, U.S.A.) for many helpful discussions, as well as Dr Francisco Real, Dr Michael Chin, and Dr Masashi Kawaichi for generously sharing valuable reagents. This work was supported by National Institutes of Health grants DK61215 and DK56211 (to S.D.L.), and a Pilot Project Grant from the Johns Hopkins Department of Surgery (to B.G.). S.D.L. is also supported by the Paul K. Neumann Professorship at Johns Hopkins University.
References
- 1.Kawaguchi Y., Cooper B., Gannon M., Ray M., MacDonald R. J., Wright C. V. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat. Genet. 2002;32:128–134. doi: 10.1038/ng959. [DOI] [PubMed] [Google Scholar]
- 2.Krapp A., Knofler M., Ledermann B., Burki K., Berney C., Zoerkler N., Hagenbuchle O., Wellauer P. K. The bHLH protein PTF1-p48 is essential for the formation of the exocrine and the correct spatial organization of the endocrine pancreas. Genes Dev. 1998;12:3752–3763. doi: 10.1101/gad.12.23.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rose S. D., Swift G. H., Peyton M. J., Hammer R. E., MacDonald R. J. The role of PTF1-P48 in pancreatic acinar gene expression. J. Biol. Chem. 2001;276:44018–44026. doi: 10.1074/jbc.M106264200. [DOI] [PubMed] [Google Scholar]
- 4.Adell T., Gomez-Cuadrado A., Skoudy A., Pettengill O. S., Longnecker D. S., Real F. X. Role of the basic helix–loop–helix transcription factor p48 in the differentiation phenotype of exocrine pancreas cancer cells. Cell Growth Differ. 2000;11:137–147. [PubMed] [Google Scholar]
- 5.Obata J., Yano M., Mimura H., Goto T., Nakayama R., Mibu Y., Oka C., Kawaichi M. p48 subunit of mouse PTF1 binds to RBP-Jκ/CBF-1, the intracellular mediator of Notch signalling, and is expressed in the neural tube of early stage embryos. Genes Cells. 2001;6:345–360. doi: 10.1046/j.1365-2443.2001.00422.x. [DOI] [PubMed] [Google Scholar]
- 6.Lin J. W., Biankin A. V., Horb M. E., Ghosh B., Prasad N. B., Yee N. S., Pack M. A., Leach S. D. Differential requirement for ptf1a in endocrine and exocrine lineages of developing zebrafish pancreas. Dev. Biol. 2004;270:474–486. doi: 10.1016/j.ydbio.2004.02.023. [DOI] [PubMed] [Google Scholar]
- 7.Sellick G. S., Barker K. T., Stolte-Dijkstra I., Fleischmann C., Coleman R. J., Garrett C., Gloyn A. L., Edghill E. L., Hattersley A. T., Wellauer P. K., et al. Mutations in PTF1A cause pancreatic and cerebellar agenesis. Nat. Genet. 2004;36:1301–1305. doi: 10.1038/ng1475. [DOI] [PubMed] [Google Scholar]
- 8.Rodolosse A., Chalaux E., Adell T., Hagege H., Skoudy A., Real F. X. PTF1α/p48 transcription factor couples proliferation and differentiation in the exocrine pancreas. Gastroenterology. 2004;127:937–949. doi: 10.1053/j.gastro.2004.06.058. [DOI] [PubMed] [Google Scholar]
- 9.Murtaugh L. C., Stanger B. Z., Kwan K. M., Melton D. A. Notch signalling controls multiple steps of pancreatic differentiation. Proc. Natl. Acad. Sci. U.S.A. 2003;100:14920–14925. doi: 10.1073/pnas.2436557100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hald J., Hjorth J. P., German M. S., Madsen O. D., Serup P., Jensen J. Activated Notch1 prevents differentiation of pancreatic acinar cells and attenuates endocrine development. Dev. Biol. 2003;260:426–437. doi: 10.1016/s0012-1606(03)00326-9. [DOI] [PubMed] [Google Scholar]
- 11.Esni F., Ghosh B., Biankin A. V., Lin J. W., Albert M. A., Yu X., MacDonald R. J., Civin C. I., Real F. X., Pack M. A., et al. Notch inhibits Ptf1 function and acinar cell differentiation in developing mouse and zebrafish pancreas. Development. 2004;131:4213–4224. doi: 10.1242/dev.01280. [DOI] [PubMed] [Google Scholar]
- 12.Miyamoto Y., Maitra A., Ghosh B., Zechner U., Argani P., Iacobuzio-Donahue C. A., Sriuranpong V., Iso T., Meszoely I. M., Wolfe M. S., et al. Notch mediates TGFα-induced changes in epithelial differentiation during pancreatic tumourigenesis. Cancer Cell. 2003;3:565–576. doi: 10.1016/s1535-6108(03)00140-5. [DOI] [PubMed] [Google Scholar]
- 12a.Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iso T., Kedes L., Hamamori Y. HES and HERP families: multiple effectors of the Notch signalling pathway. J. Cell. Physiol. 2003;194:237–255. doi: 10.1002/jcp.10208. [DOI] [PubMed] [Google Scholar]
- 14.Sasai Y., Kageyama R., Tagawa Y., Shigemoto R., Nakanishi S. Two mammalian helix-loop-helix factors structurally related to Drosophila hairy and Enhancer of split. Genes Dev. 1992;6:2620–2634. doi: 10.1101/gad.6.12b.2620. [DOI] [PubMed] [Google Scholar]
- 15.Fisher A., Caudy M. The function of hairy-related bHLH repressor proteins in cell fate decisions. Bioessays. 1998;20:298–306. doi: 10.1002/(SICI)1521-1878(199804)20:4<298::AID-BIES6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 16.Dawson S. R., Turner D. L., Weintraub H., Parkhurst S. M. Specificity for the hairy/enhancer of split basic helix–loop–helix (bHLH) proteins maps outside the bHLH domain and suggests two separable modes of transcriptional repression. Mol. Cell. Biol. 1995;15:6923–6931. doi: 10.1128/mcb.15.12.6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fisher A. L., Caudy M. Groucho proteins: transcriptional corepressors for specific subsets of DNA-binding transcription factors in vertebrates and invertebrates. Genes Dev. 1998;12:1931–1940. doi: 10.1101/gad.12.13.1931. [DOI] [PubMed] [Google Scholar]
- 18.Lee J. C., Smith S. B., Watada H., Lin J., Scheel D., Wang J., Mirmira R. G., German M. S. Regulation of the pancreatic pro-endocrine gene neurogenin3. Diabetes. 2001;50:928–936. doi: 10.2337/diabetes.50.5.928. [DOI] [PubMed] [Google Scholar]
- 19.Kuroda K., Tani S., Tamura K., Minoguchi S., Kurooka H., Honjo T. Delta-induced Notch signalling mediated by RBP-J inhibits MyoD expression and myogenesis. J. Biol. Chem. 1999;274:7238–7244. doi: 10.1074/jbc.274.11.7238. [DOI] [PubMed] [Google Scholar]
- 20.Umbhauer M., Boucaut J. C., Shi D. L. Repression of XMyoD expression and myogenesis by Xhairy-1 in Xenopus early embryo. Mech. Dev. 2001;109:61–68. doi: 10.1016/s0925-4773(01)00517-2. [DOI] [PubMed] [Google Scholar]
- 21.Sun J., Kamei C. N., Layne M. D., Jain M. K., Liao J. K., Lee M. E., Chin M. T. Regulation of myogenic terminal differentiation by the hairy-related transcription factor CHF2. J. Biol. Chem. 2001;276:18591–18596. doi: 10.1074/jbc.M101163200. [DOI] [PubMed] [Google Scholar]
- 22.Giagtzoglou N., Koumbanakis K. A., Fullard J., Zarifi I., Delidakis C. Role of the Sc C terminus in transcriptional activation and E(spl) repressor recruitment. J. Biol. Chem. 2005;280:1299–1305. doi: 10.1074/jbc.M408949200. [DOI] [PubMed] [Google Scholar]
- 23.Kamakura S., Oishi K., Yoshimatsu T., Nakafuku M., Masuyama N., Gotoh Y. Hes binding to STAT3 mediates crosstalk between Notch and JAK-STAT signalling. Nat. Cell Biol. 2004;6:547–554. doi: 10.1038/ncb1138. [DOI] [PubMed] [Google Scholar]
- 24.McLarren K. W., Lo R., Grbavec D., Thirunavukkarasu K., Karsenty G., Stifani S. The mammalian basic helix loop helix protein HES-1 binds to and modulates the transactivating function of the runt-related factor Cbfa1. J. Biol. Chem. 2000;275:530–538. doi: 10.1074/jbc.275.1.530. [DOI] [PubMed] [Google Scholar]