Loss of Bif-1 Suppresses Bax/Bak Conformational Change and Mitochondrial Apoptosis (original) (raw)
Abstract
Bif-1, a member of the endophilin B protein family, interacts with Bax and promotes interleukin-3 withdrawal-induced Bax conformational change and apoptosis when overexpressed in FL5.12 cells. Here, we provide evidence that Bif-1 plays a regulatory role in apoptotic activation of not only Bax but also Bak and appears to be involved in suppression of tumorigenesis. Inhibition of endogenous Bif-1 expression in HeLa cells by RNA interference abrogated the conformational change of Bax and Bak, cytochrome c release, and caspase 3 activation induced by various intrinsic death signals. Similar results were obtained in Bif-1 knockout mouse embryonic fibroblasts. While Bif-1 did not directly interact with Bak, it heterodimerized with Bax on mitochondria in intact cells, and this interaction was enhanced by apoptosis induction and preceded the Bax conformational change. Moreover, suppression of Bif-1 expression was associated with an enhanced ability of HeLa cells to form colonies in soft agar and tumors in nude mice. Taken together, these findings support the notion that Bif-1 is an important component of the mitochondrial pathway for apoptosis as a novel Bax/Bak activator, and loss of this proapoptotic molecule may contribute to tumorigenesis.
Apoptosis or programmed cell death plays a vital role in the normal development and maintenance of tissue homeostasis. Defects in this cell suicide pathway facilitate the accumulation of excess and/or abnormal cells in the body, resulting in cancer development (13, 45). The apoptotic process is executed by a family of cysteine proteases which specifically cleave their substrates at aspartic acid residues. These proteases, known as caspases, are mainly activated through two major pathways: extrinsic and intrinsic. The extrinsic pathway is mediated by ligation of the TNFR1, CD95/Fas, and TRAIL death receptors, while the intrinsic pathway is initiated by formation of the cytosolic apoptosome composed of Apaf-1, procaspase 9, and the cytochrome c released from mitochondria. In addition to cytochrome c, a variety of other proteins, including AIF, Smac/Diablo, Omi/HtrA2, and Endo G, are also released from the mitochondrial intermembrane space by various death signals as a result of outer mitochondrial membrane permeabilization (OMMP), and many of them are actively involved in the process of caspase-dependent and/or -independent cell death (13, 55).
The Bcl-2 family of proteins plays a central role in the intrinsic pathway of apoptosis by controlling OMMP (5, 13). The antiapoptotic members of the Bcl-2 family, such as Bcl-2 and Bcl-XL, prevent the release of mitochondrial apoptogenic factors, whereas the proapoptotic members of this family, which can be further separated into two subgroups, the multidomain BH1-3 proteins (Bax, Bak, and Bok) and the BH3-only proteins (e.g., Bad, Bid, Bim, and Puma), trigger this event. Genetic studies show that the multidomain proapoptotic proteins Bax and Bak, which partially complement one another, are required for OMMP and apoptosis induced by many types of death stimulation (7, 31, 57, 59, 65). In healthy cells, Bax is distributed in the cytoplasm or loosely attached to membranes (58, 61), whereas Bak is mainly located on the outer mitochondrial membrane (OMM) (14). The solution structure of Bax indicates that the C-terminal transmembrane domain is hidden in the hydrophobic pocket of this protein (52). Apoptotic signals induce conformation changes in the Bax and Bak proteins, leading to Bax translocation to OMM and the formation of membrane-integrated homo-oligomers of Bax and Bak, which results in OMMP (5, 13, 50). The antiapoptotic Bcl-2 family members, such as Bcl-2 and Bcl-XL, inhibit Bax and Bak activation, whereas the BH3-only proteins promote it. Most of these BH3-only proteins appear to function as transdominant inhibitors that activate Bax and Bak by binding to and antagonizing antiapoptotic Bcl-2 family proteins, with the exception of tBid and BimS or BimAD, which are capable of inducing OMMP by direct binding to and activation of Bax and Bak (26, 28, 34, 53, 61). Although BH3-only proteins are essential for mitochondrial apoptosis, the precise mechanism underlying both Bax and Bak activation remains far from clear.
One of the best-studied Bax activators is the BH3-only protein Bid, which binds proapoptotic Bax and Bak, as well as antiapoptotic Bcl-2 and Bcl-XL (54, 56). In response to certain apoptotic signals, Bid is cleaved by caspase 8, and the resulting truncated Bid (also called tBid) translocates from the cytosol to mitochondrial membranes, where it binds the mitochondrion-specific lipid cardiolipin (30, 32, 33). Indeed, Bax oligomerization and insertion into OMM can be triggered by tBid (9). Although tBid and Bax fail to permeabilize cardiolipin-free endoplasmic reticulum (ER) membranes, they act on cardiolipin-containing liposomes or outer mitochondrial membrane vesicles (27), suggesting that cardiolipin is a critical component for activating Bax. However, tBid-induced Bax oligomerization in mitochondrial membranes is inhibited by pretreatment of mitochondria with protease K (46), indicating that another unidentified OMM protein(s) is also required for Bax oligomerization. Consistently, tBid and Bax can completely release preloaded dextran from outer mitochondrial membrane vesicles compared to chemically defined protein-free liposomes (27), suggesting that some mitochondrial protein(s) indeed contributes to the permeabilization reaction.
Several non-Bcl-2-related proteins, including Bif-1 (6), Ku70 (47), 14-3-3 theta (40), Humanin (HN) peptide (15), ASC (42), and p53 (4, 8, 29, 35, 60), have recently been shown to be involved in the regulation of Bax and/or Bak activity. Bif-1 and ASC bind to and activate Bax, whereas Ku70, 14-3-3, and HN inhibit Bax activation by direct interaction with this proapoptotic protein in cells. Apoptotic signals disrupt the interaction between Bax and 14-3-3, thus unleashing Bax to translocate to mitochondria by caspase-dependent and -independent mechanisms (40). In contrast, immunoblot analyses suggest that apoptotic stimuli release Bax from the Ku70/Bax protein complex by induction of Ku70 protein degradation (47). In Ku70 knockout cells, however, Bax did not exhibit translocation to mitochondria in the absence of apoptotic stimuli (47). These findings suggest that there could be additional factors that prevent relocation of Bax under conditions in which Bax should be kept inactive or that are necessary to actively induce Bax conformational change after Ku70 degradation in response to apoptotic signals (41). In the case of p53, besides its transcriptional control of proapoptotic genes, this tumor suppressor has been demonstrated to translocate from the nucleus to mitochondria, where it directly binds to Bcl-XL or Bak and induces Bax/Bak activation and cytochrome c release (4, 8, 29, 35, 60).
To gain insights into Bax function, we (6) and others (44) independently have identified a novel Bax-binding protein termed Bif-1 (Bax-interacting factor 1) and SH3GLB1 (SH3 domain GRB2-like endophilin B1), respectively, by yeast two-hybrid screens using Bax as the bait. Interestingly, the interaction of Bif-1 with Bax in mammalian cells appears to be specifically enhanced by apoptotic stimulation, such as interleukin 3 (IL-3) withdrawal or microtubule damage, which is accompanied by a conformational change in the Bax protein (6). Ectopic expression of Bif-1 in FL5.12 cells promotes IL-3 deprivation-induced conformational change in the Bax protein, caspase activation, and apoptotic cell death (6). Bif-1 is also known as endophilin B1 (21), a member of the evolutionarily conserved endophilin B family, which contains an N-BAR (Bin-amphiphysin-Rvs) domain and a C-terminal SH3 domain but shares no significant homology with members of the Bcl-2 family. Unlike endophilin A1, which is essential for synaptic vesicle endocytosis (21), Bif-1/endophilin B1 is associated with intracellular membranes (6, 10, 37) and does not appear to operate in endocytosis at the plasma membrane (37). Interestingly, it has been shown that Bif-1/endophilin B1 is involved in the regulation of morphological dynamics of mitochondria (23), and a significant portion of Bif-1 translocates to mitochondria in response to apoptotic signals (6, 23). These findings suggest that Bif-1 may represent a new type of Bax activator that controls the mitochondrial pathway of apoptosis.
In this study, we investigated the importance of endogenous Bif-1 in mitochondrion-mediated apoptosis and tumorigenesis by gene silencing and ablation. Loss of Bif-1 delayed the activation of Bax and Bak, cytochrome c release, caspase activation, and cell death in HeLa and mouse embryonic fibroblasts (MEFs). Importantly, suppression of Bif-1 expression in HeLa cells promoted colony formation in soft agar and tumor growth in nude mice with no significant effect on cell proliferation. Moreover, using the bimolecular fluorescence complementation (BiFC) technique, we demonstrated that the heterodimerization of Bif-1 with Bax on mitochondria is enhanced by apoptotic stimulation. The results presented in this paper further suggest that Bif-1 is a crucial regulator for not only Bax but also Bak activation and may have a role in the suppression of cancer progression.
MATERIALS AND METHODS
Plasmids.
The pBS/U6 plasmid (51) was digested with ApaI, and the 3′ protruding end was converted to a blunt end with Klenow. The plasmid was then digested with EcoRI. Oligonucleotides 5′-TGGTCTTCGTCG-3′ (sense) and 5′-AATTCGACGAAGACCA-3′ (antisense) (the underlining indicates BbsI recognition sequence) were annealed and inserted between the blunt end and the EcoRI site to make the pBSU6-BbsI vector. To construct the Bif-1 small interfering RNA (siRNA) expression plasmid pBSU6-siBif-1, oligonucleotides 5′-TTTGCACCAAGTCGTATAAACATTCAAGAGATGTTTATACGACTTGGTGCTTTTTTG-3′ (sense) and 5′-AATTCAAAAAAGCACCAAGTCGTATAAACATCTCTTGAATGTTTATACGACTTGGTG-3′ (antisense), containing the target sequences of human Bif-1 (indicated by underlining), a 9-nucleotide spacer, and six Ts, were annealed and inserted into the BbsI and EcoRI sites of the pBSU6-BbsI vector. Bak siRNA expression using the sequence 5′-GGGGACGACATCAACCGACGCTATGACTC-3′ and Bax siRNA expression using the sequence 5′-GTAACATGGAGCTGCAGAGGATGATTGCC-3′ were performed using the pREP4 vector and hygromycin selection procedure described previously (23). For the BiFC assay, two cDNA fragments that encode residues 1 to 154 and 155 to 238 of the EYFP protein were amplified by PCR using pBiFC-YN155 and pBiFC-YC155 (20) as templates. These fragments were subcloned into the KpnI and BamHI sites of pcDNA3 to produce the pcDNA3-YN154 and pcDNA3-YC155 vectors. The cDNA fragments encoding mouse Bax and human Bif-1 were then subcloned into the EcoRI and XhoI sites of pcDNA3-YN154 and pcDNA3-YC155. The linker sequences between the EYFP fragments and Bif-1 or Bax were NPLVTAASVLEF and NPLVTAASVLEFAA, respectively.
Cell culture and transfection.
HeLa cells were cultured in Dulbecco's modified Eagle's medium supplemented with 5% fetal bovine serum, 100 μg/ml streptomycin, and 100 U/ml penicillin. For RNA interference experiments, the 21-nucleotide siRNA duplexes were synthesized and purified by Dharmacon Research (Boulder, CO). The target sequences of human Bif-1 siRNAs were as follows: siRNA-1, 5′-AAGACAGAAUUGGAUGCUCAC-3′; siRNA-2, 5′-AAGCUCCAAGUCGUAUAAACA-3′; and siRNA-3, 5′-AACAAGUGGCCUAGUAAUCAC-3′. The siRNA targeting green fluorescent protein (GFP) was described previously (18). To achieve transient suppression of Bif-1 expression, the duplex siRNAs were transfected into HeLa cells with oligofectamine reagent (Invitrogen) as described previously (18). To stably suppress the expression of Bif-1, HeLa cells were seeded at ∼70% confluence in a 10-cm plate and cultured overnight. The next day, the cells were transfected with 50 μl of Lipofectamine (Invitrogen) mixed with 5 μg of the Bif-1 siRNA expression plasmid pBSU6-siBif-1 plus 0.5 μg of pBabe-Puro vector. At 24 h after transfection, the cells were trypsinized, divided into 10 10-cm plates, and selected in 1 μM of puromycin (Sigma). Primary MEFs were generated from 12.5-day-postcoitus embryos. To obtain immortalized fibroblasts, primary MEFs (P1) were routinely passaged by following the 3T3 protocol. The cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 100 μg/ml streptomycin, and 100 U/ml penicillin.
Generation of Bif-1-deficient mice.
The mouse bif-1 gene was disrupted by homologous recombination in embryonic stem (ES) cells using a targeting vector that eliminated exon 1 and the translation initiation codon of the bif-1 gene. Briefly, a 1.3-kb PstI fragment of the 5′-flanking region and a 5.3-kb KpnI-SalI fragment spanning the first intron of the mouse bif-1 gene obtained by screening of the 129/SvJ mouse lambda FIX II library (Stratagene) were subcloned with the neomycin resistance gene and herpes simplex virus thymidine kinase gene into the pBluescript vector (Stratagene). The targeting vector was then linearized at a unique NotI site and electroporated into R1 ES cells. After positive and negative selection with G418 (200 μg/ml) and ganciclovir (2 μM), 32 independent homologous recombinants were obtained and confirmed by Southern blot hybridization. Eight of the targeted ES clones were microinjected into C57BL/6 mouse blastocysts, and chimeric mice derived from two of the clones transmitted the targeted allele through the germ line when crossed with C57BL/6 mice. To produce homozygous Bif-1-deficient (_bif-1_−/−) mice, heterozygous mice were intercrossed, and their offspring were analyzed by PCR and Southern blotting. Immunoblot analysis confirmed the lack of Bif-1 protein expression in the homozygous mutants.
Detection of Bax and Bak conformational changes.
Total cellular lysates were prepared in CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate} lysis buffer (150 mM NaCl, 10 mM HEPES, pH 7.4, 1% CHAPS) containing protease inhibitors and subjected to immunoprecipitation with either anti-Bax 6A7 monoclonal antibody (Sigma) or anti-Bak (Ab-2) monoclonal antibody (Oncogene) as described previously (6). The resulting immunoprecipitates containing the conformationally changed Bax or Bak protein were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE)-immunoblot analysis with anti-Bax N20 (Santa Cruz) or anti-Bak (Upstate) polyclonal antibody. For immunofluorescence analysis, cells grown on coverslips were fixed in 3.7% formaldehyde, permeabilized in 0.2% CHAPS, stained with anti-Bax N20 rabbit antiserum, and detected by fluorescein isothiocyanate-conjugated secondary antibody (Chemicon International, Inc.). The coverslips were then mounted with DAPI (4′,6′-diamidino-2-phenylindole)-containing mount media (Vector Laboratories, Inc.) and analyzed by fluorescence microscopy.
Release of cytochrome c from isolated mitochondria.
Mitochondria were isolated from mouse livers, kept on ice, and used within 3 h of preparation as described previously (60). The isolated mitochondria (250 μg of protein) were incubated at 30°C with recombinant proteins in 50 μl of mitochondrial assay buffer (60) for 30 min. Reaction mixtures were centrifuged at 13,000 × g for 10 min, and the resulting supernatant and pellet fractions were subjected to immunoblot analysis with anti-cytochrome c monoclonal antibody (BD PharMingen). Caspase 8-cleaved Bid (tBid) protein was purchased from Sigma, and recombinant full-length Bif-1 protein was purified using the IMPACT system (New England Biolabs). In brief, the Bif-1 open reading frame was amplified by PCR and subcloned into the pTYB1 vector (New England Biolabs) with NdeI and SapI sites. The resulting plasmid, which expresses a Bif-1 and intein tag fusion protein, was transformed into Escherichia coli BL21. Recombinant proteins were isolated by chitin affinity chromatography according to the manufacturer's protocol. The Bif-1 protein was cleaved off from the intein tag by dithiothreitol and dialyzed in 10 mM HEPES (pH 7.4), 100 mM NaCl, 0.2 mM EDTA.
BiFC assay.
COS7 cells grown on coverslips were transiently transfected with pcDNA3-YN154-Bif-1 and pcDNA3-YC155-Bax or pcDNA3-YC155-Bif-1 plasmid by Lipofectamine (Invitrogen). The transfectants were incubated at 37°C for 21 h and switched to 30°C for 3 h to promote yellow fluorescent protein (YFP) fluorophore maturation. After incubation with 20 nM MitoTracker Red CMSRos for 30 min, the cells were exposed to 1 μM staurosporine (STS) for 3 h in the presence of 50 μM z-VAD-fmk. Then, the cells were washed three times with phosphate-buffered saline (PBS), fixed in 3.7% formaldehyde, mounted with DAPI-containing mount medium (Vector Laboratories, Inc.), and analyzed with a Zeiss laser scanning confocal microscope.
Soft-agar assay.
HeLa cells were suspended in 0.3% agar in growth medium and seeded at 125 to 1,000 cells/well in triplicate in six-well plates precoated with 0.6% agar in growth medium. The cells were then fed with growth medium (0.1 ml/well) once a week until the colonies grew to a suitable size for observation (2 to 3 weeks). After being stained with 1 mg/ml 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) in growth medium, the colonies were photographed and counted.
Tumor xenograft.
HeLa cells were harvested, suspended in PBS, and subcutaneously injected into the right and left flanks (10 × 106 cells/flank) of 4- to 5-week-old female nude mice (five mice/cell line). The tumor size was determined by measuring the length (l) and the width (w) and calculating the volume (V = _lw_2/2).
BrdU incorporation assay.
HeLa cells were incubated with 30 μg/ml bromodeoxyuridine (BrdU) for 1 h at 37°C. The cells were then harvested, fixed in 70% ethanol, washed once with PBS, and incubated in 0.1% HCl containing 0.04% pepsin for 1 h at 37°C. After neutralization by adding 0.1 M sodium borate, the cells were washed once with PBTP (0.5% Tween 20, 0.5% bovine serum albumin in PBS) and then incubated in 200 μl PBTP along with 15 μl fluorescein isothiocyanate-conjugated anti-BrdU antibody for 1 h in the dark. After being washed with PBTP, the cells were incubated in a 10-μg/ml propidium iodide solution (containing RNase A) for 30 min at 37°C in the dark. BrdU incorporation and DNA content were analyzed on a Becton Dickinson FACScan.
RESULTS
Loss of Bif-1 suppresses Bax conformational change, cytochrome c release, and caspase 3 activation.
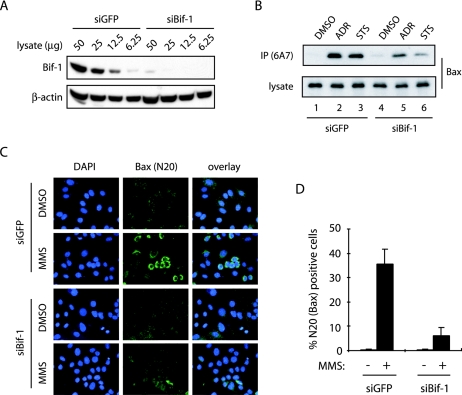
In our previous report (6), we showed that overexpression of Bif-1 promotes Bax conformational change, caspase 3 activation, and apoptosis in FL5.12 cells following IL-3 withdrawal. To further investigate the role of Bif-1 in Bax-mediated apoptosis, we first employed siRNAs to suppress the expression of endogenous Bif-1 and examined its effect on Bax conformational change, cytochrome c release, and caspase activation in response to apoptotic signals. As shown by immunoblotting (Fig. 1A) with anti-Bif-1 monoclonal antibody, transfection of Bif-1 siRNA-1 (siBif-1) resulted in a dramatic suppression of Bif-1 expression in HeLa cells compared with cells transfected with the control GFP-specific siRNA (siGFP). To examine Bax conformational change, we performed both immunoprecipitation (Fig. 1B) and immunofluorescence (Fig. 1C) experiments with antibodies specific for the conformationally changed Bax protein as described previously (6, 63). Immunoprecipitation with anti-Bax 6A7 antibody showed that treatment of HeLa cells with adriamycin (ADR) or STS induced the exposure of the Bax N-terminal epitope 6A7 (Fig. 1B), which is hidden in undamaged cells and becomes exposed upon a conformational change in the Bax protein in response to apoptotic signals (19). Compared with control siGFP-transfected cells, ADR- or STS-induced 6A7 epitope exposure was suppressed in cells transfected with siBif-1 (Fig. 1B). These results were confirmed by immunofluorescence staining with anti-Bax N20 polyclonal antibody, which recognizes a Bax N-terminal epitope similar to 6A7. As shown in Fig. 1C and D, the immunoreactivity of anti-Bax N20 antibody was dramatically reduced in HeLa cells transfected with siBif-1 compared to siGFP following treatment with the DNA-damaging agent methyl methanesulfonate.
FIG. 1.
Knockdown of Bif-1 prevents Bax conformational change. HeLa cells were transfected with siRNAs targeting Bif-1 (siBif-1) or control GFP (siGFP) and subjected to the following analyses. (A) Immunoblot analysis with anti-Bif-1 monoclonal antibody (Imgenex) revealed that Bif-1 protein expression was significantly reduced by transfection with siBif-1 duplexes. (B to D) SiRNA-transfected cells were exposed to either 1 μM ADR for 12 h, 0.5 μM STS for 6 h, or 1 mM methyl methanesulfonate (MMS) for 10 h. Bax conformational change was determined by immunoprecipitation (IP) with anti-Bax 6A7 monoclonal antibody (B), as well as by immunofluorescence staining (green) with anti-Bax N20 polyclonal antibody (C). The N20-positive cells in panel C were counted under fluorescence microscopy (D). The data shown are means plus standard deviations (n = 3). The nuclear morphology was examined by DAPI (blue) staining.
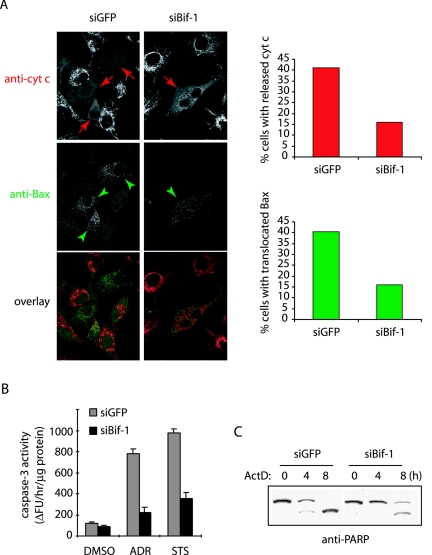
Since activated Bax triggers the OMMP, leading to the release of cytochrome c and subsequent downstream caspase cascade activation, we reasoned that siBif-1-mediated inhibition of Bax conformational change should contribute to suppression of these events induced by intrinsic death signals. To test this hypothesis, we monitored the intracellular redistribution of Bax and cytochrome c by immunostaining in HeLa cells transfected with GFP- and Bif-1-silencing vectors (23) after actinomycin D (ActD) treatment. As expected, the inhibition of Bif-1 expression resulted in the suppression of both mitochondrial translocation of Bax and cytochrome c release following ActD treatment (Fig. 2A). Notably, all cells with mitochondrion-associated Bax displayed diffuse staining of cytochrome c (Fig. 2A), confirming that Bif-1 acts upstream of cytochrome c release in the activation of apoptosis.
FIG. 2.
Knockdown of Bif-1 inhibits Bax translocation to mitochondria, cytochrome c release, caspase 3 activation, and PARP cleavage. (A) HeLa cells transfected with siGFP or siBif-1 were pretreated with 75 μM z-VAD-fmk for 30 min before incubation with 20 μM ActD for 4 h. The cells were stained with anti-cytochrome c (anti-cyt c) monoclonal antibody (red) plus anti-Bax polyclonal antiserum (green), and cells with released cytochrome c (indicated by red arrows) and mitochondrion-translocated Bax (indicated by green arrowheads) were counted (n > 500) under fluorescence microscopy and shown in bar graphs. (B) The siRNA-transfected HeLa cells were exposed to either DMSO as a control, 1 μM ADR for 12 h, or 0.5 μM STS for 6 h, and the caspase 3-like activity was measured by using the caspase 3 fluorometric assay kit (Sigma) according to the manufacturer's protocol. (C) HeLa cells with siGFP or siBif-1 were treated with 20 μM ActD for the indicated periods and subjected to SDS-PAGE/immunoblot analysis with anti-PARP antibody.
We next compared the caspase activity in HeLa cells transfected with siBif-1 versus siGFP after exposure to ADR, STS, or dimethyl sulfoxide (DMSO) as a control. Consistent with the Bax activation and cytochrome c release assays, both ADR and STS induced caspase 3-like activity, which was dramatically inhibited in cells transfected with siBif-1 compared to siGFP (Fig. 2B). Moreover, the cleavage of PARP, a well-known caspase 3 substrate, was significantly delayed by Bif-1 silencing (Fig. 2C).
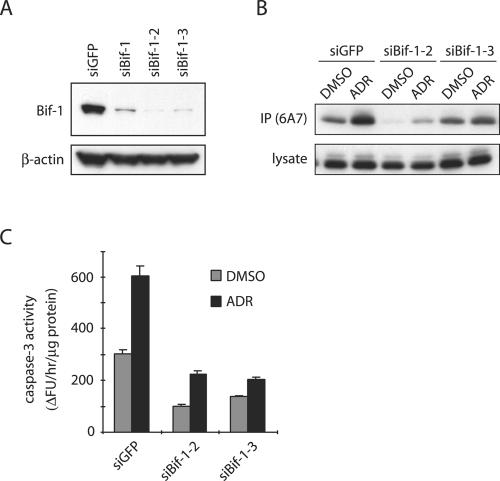
To confirm the specificity of siBif-1, we repeated the experiments with two additional siRNAs, siRNA-2 (siBif-1-2) and siRNA-3 (siBif-1-3), which target different regions of the Bif-1 mRNA in HeLa cells. Consistent with siBif-1, inhibition of Bif-1 expression by either siBif-1-2 or siBif-1-3 (Fig. 3A) also prevented ADR-induced Bax conformational change (Fig. 3B) and caspase 3 activation (Fig. 3C), indicating that the observed phenotype was most likely attributable to the reduced function of Bif-1 and not caused by off-target effects.
FIG. 3.
Knockdown of Bif-1 suppresses Bax conformational change and caspase 3 activity in ADR-treated HeLa cells. (A) HeLa cells were transfected with siRNAs targeting Bif-1 or GFP for 48 h and subjected to SDS-PAGE/immunoblot analysis. (B and C) HeLa cells transfected with siRNAs targeting Bif-1 or GFP were exposed to DMSO or 1 μM ADR for 12 h and subjected to immunoprecipitation (IP) with anti-Bax 6A7 monoclonal antibody and to caspase 3 activity assay. The error bars indicate standard deviations.
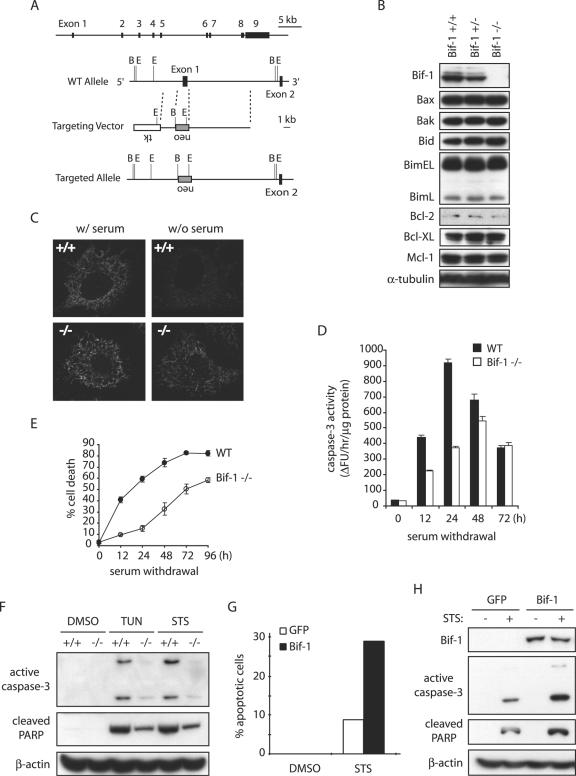
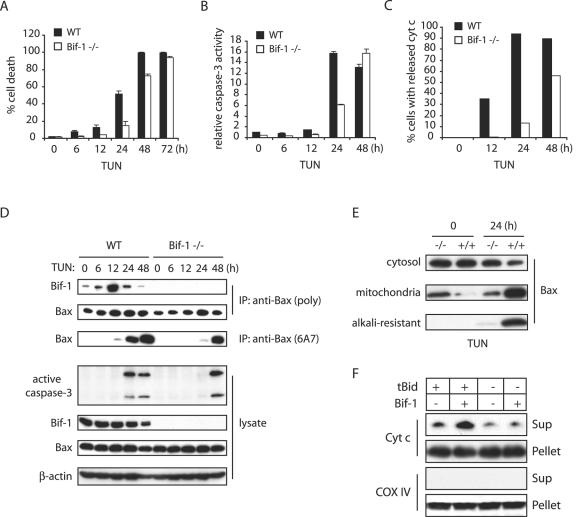
To further study the cellular function of Bif-1, we disrupted the bif-1 gene in mouse ES cells by homologous recombination, eliminating the first exon and the basal promoter region of the bif-1 gene (Fig. 4A) and established MEFs from the resulting Bif-1 knockout, as well as wild-type, embryos (Fig. 4B). Knockout of Bif-1 did not significantly affect the protein levels of Bcl-2 family proteins, including Bax, Bak, Bid, Bim, Bcl-2, Bcl-XL, and Mcl-1, in mouse embryos (Fig. 4B). The _bif-1_−/− mice were born in the expected Mendelian frequency and appeared indistinguishable from their littermates at birth. The physiological importance of Bif-1 for mouse development and tissue homeostasis will be characterized and described elsewhere. To determine whether Bif-1 knockout cells are resistant to intrinsic death signals, we first examined cytochrome c release (Fig. 4C), caspase 3 activation (Fig. 4D), and cell death (Fig. 4E) in Bif-1−/− and Bif-1+/+ MEFs after serum deprivation. As expected, Bif-1-null MEFs displayed a significant delay of these apoptotic events in response to growth factor (serum) withdrawal. Similarly, the ablation of Bif-1 suppressed caspase 3 activation and PARP cleavage in MEF cells induced by tunicamycin (TUN) or STS (Fig. 4F).
FIG. 4.
Knockout of Bif-1 in MEFs delays mitochondrial apoptosis. (A) Genomic organization of the mouse bif-1 gene, targeting construct, and targeted locus. The restriction enzyme sites shown are B (BamHI) and E (EcoRI). WT, wild type. (B) Immunoblot analysis of whole-cell lysates from Bif-1+/+, Bif-1+/−, and Bif-1−/− embryonic day 12.5 mouse embryos. (C) Bif-1+/+ and Bif-1−/− MEFs were cultured in the presence or absence of serum for 6 h and applied to immunostaining with anti-cytochrome c antibody. (D and E) Bif-1−/− and WT MEFs were cultured with or without serum for various times before caspase 3 activity analysis (D) and trypan blue dye exclusion assay (E). The error bars indicate standard deviations. (F) Bif-1−/− and Bif-1+/+ MEFs were treated with 1 μg/ml TUN for 24 h, 1 μM STS for 12 h, or DMSO as a control. Cell lysates were prepared in CHAPS lysis buffer and analyzed by SDS-PAGE/immunoblotting with the indicated antibodies. (G and H) Bif-1−/− MEFs were transfected with 10 μg of pIRES2-EGFP vector or pBif-1-IRES2-EGFP plasmid DNA for 40 h using the Nucleofector technology (Amaxa) and subjected to treatment with DMSO or 1 μM STS for 12 h. The percentage of apoptotic GFP-positive cells was determined by DAPI staining assay (G), and cell lysates were prepared and analyzed by SDS-PAGE/immunoblotting (H).
To confirm that the decreased sensitivity of Bif-1−/− cells to intrinsic death stimuli was due to the absence of Bif-1, we determined whether restoring Bif-1 expression would rescue the apoptosis sensitivity of Bif-1-null cells. Bif-1−/− MEFs were transfected with pIRES2-EGFP (Clontech) or pBif-1-IRES2-EGFP to produce enhanced GFP (EGFP) or Bif-1 plus EGFP, respectively. The pIRES2-EGFP vector permits both the gene of interest and the EGFP gene to be translated from a single bicistronic mRNA for convenient differentiation of successfully transfected cells from those that did not take up and express the plasmid. After 24 h of transfection, cells were trypsinized, seeded into chamber slides, and returned to culture overnight before STS treatment and DAPI staining. As shown in Fig. 4G, STS-induced apoptosis of Bif-1 knockout cells was increased by transfection of the Bif-1 expression plasmid (pBif-1-IRES2-EGFP) compared to the control vector (pIRES2-EGFP). In addition, immunoblot analysis revealed that reexpression of the Bif-1 protein promoted STS-induced caspase 3 activation and PARP cleavage in Bif-1−/− MEFs (Fig. 4H). These results indicate that the reduced sensitivity of Bif-1-null cells to intrinsic death signals is primarily due to the absence of Bif-1.
It has been shown that ER stress agents cause OMMP and cytochrome c release and that activation of Bax appears to be essential for mitochondrial apoptosis caused by ER stress (3, 16, 49, 59, 62). Therefore, we next examined whether Bif-1 contributes to the ER stress-mediated Bax activation and apoptosis. To this end, Bif-1−/− and Bif-1+/+ MEFs were treated with TUN for various periods of time and subjected to immunoprecipitation with anti-Bax 6A7 antibody, caspase 3 processing/activity, and trypan blue dye exclusion analyses. The kinetics of cell death were delayed in Bif-1-deficient MEFs (Fig. 5A), and the processing of pro-caspase 3 to active forms (Fig. 5D) and its enzymatic activity (Fig. 5B) in Bif-1−/− MEFs were suppressed relative to wild-type MEFs in response to ER stress. Loss of Bif-1 also protected MEFs from TUN-induced Bax conformational change (Fig. 5D) and cytochrome c release (Fig. 5C), as demonstrated by anti-Bax 6A7 immunoprecipitation and anti-cytochrome c immunofluorescence staining, respectively.
FIG. 5.
Loss of Bif-1 delays Bax activation, cytochrome c release, caspase 3 activation, and cell death in MEFs induced by TUN. (A to C) Bif-1−/− and wild-type (WT) MEFs were treated with 1 μg/ml TUN for the indicated times and subjected to trypan blue dye exclusion (A) and caspase 3 activity (B) assays and immunofluorescence staining with anti-cytochrome c (cyt c) antibody, and cells with released cytochrome c were counted (n > 300) by fluorescence microscopy (C) analyses. The error bars indicate standard deviations. (D) MEFs were treated with 1 μg/ml TUN for various times prior to preparation of cell lysates in CHAPS lysis buffer and immunoprecipitation (IP) with anti-Bax 6A7 monoclonal antibody or anti-Bax polyclonal rabbit antiserum. The resulting immune complexes and total-cell lysates were analyzed by SDS-PAGE/immunoblotting with the indicated antibodies. (E) Bif-1−/− and Bif-1+/+ MEFs were treated with 1 μg/ml TUN for 0 or 24 h and subjected to subcellular fractionation. The resulting cytosol and mitochondrion fractions were analyzed by SDS-PAGE/immunoblotting with anti-Bax antibody to determine Bax translocation. In addition, the mitochondrion fraction was treated with 0.1 M Na2CO3 (pH 11.5) before SDS-PAGE/immunoblot analysis to examine the membrane-integrated (alkali-resistant) Bax protein. (F) Isolated mouse liver mitochondria were incubated in the absence (−) or presence (+) of 10 nM tBid, 200 nM Bif-1, or a combination of both for 30 min at 30°C. After centrifugation, the resulting supernatant (Sup) and pellet fractions were subjected to SDS-PAGE/immunoblot analysis with antibodies specific for cytochrome c (Cyt c) or COX IV.
The data presented above indicated that the suppression or ablation of Bif-1 inhibits Bax conformational change that is associated with the delay of cytochrome c release, caspase 3 activation, and apoptosis. To examine whether this inhibition affects the association and integration of Bax with mitochondria, Bif-1+/+ and Bif-1−/− cells were treated with TUN for 0 and 24 h, and subcellular fractionation was performed. As shown in Fig. 5E, the majority of Bax was detected in the cytosol in both types of cells before treatment. In contrast, the Bax protein was detected predominantly in the mitochondrial fraction of wild-type cells exposed to TUN. However, the majority of Bax in Bif-1−/− cells remained in the cytosolic fraction after TUN treatment. Moreover, the integration of Bax into the OMM was significantly inhibited in Bif-1-deficient cells, as demonstrated by the resistance of membrane-integrated Bax to alkali extraction (12). These results clearly indicate that, either directly or indirectly, Bif-1 is involved not only in Bax conformational change but also its association and integration with the mitochondria, leading to OMMP during apoptosis.
To determine whether Bif-1 is directly involved in OMMP, we examined the effect of purified recombinant Bif-1 protein on the release of cytochrome c from isolated mitochondria induced by caspase 8-cleaved Bid (tBid) in vitro. It is well characterized that tBid triggers Bax and Bak activation that results in OMMP and cytochrome c release (25). However, several lines of evidence suggest that another, unidentified protein(s) might be required to fully activate this OMMP reaction (27, 46). As shown in Fig. 5F, Bif-1 indeed enhanced tBid-mediated cytochrome c release from isolated mitochondria, suggesting that Bif-1 may be a mitochondrial factor required to permeabilize OMM.
Bif-1 interacts with Bax on mitochondria.
To examine if Bif-1 associates with Bax during apoptosis, we first performed coimmunoprecipitation experiments in MEFs treated with TUN. Intriguingly, an increased association of Bax with Bif-1 was observed at 6 h after exposure to TUN, which reached a maximum at 12 h and decreased after 24 h (Fig. 5D). The induced Bax-Bif-1 complex formation clearly preceded the Bax conformational change (Fig. 5D). These results, together with our previous findings in FL5.12 cells (6), suggest that Bif-1 and Bax form a transitory complex in response to apoptotic induction.
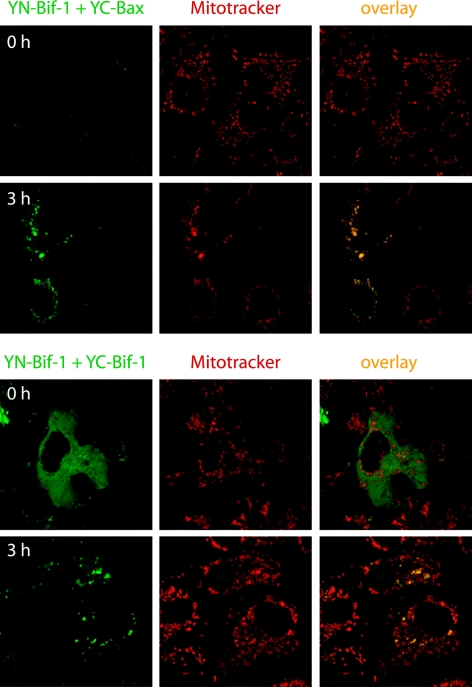
However, this dimerization of Bax with Bif-1 in immunoprecipitation experiments might represent postlysis events induced by detergents. We therefore examined the Bax/Bif-1 dimerization status in intact cells using the BiFC technique, which is based on the formation of a fluorescent complex by fragments of the enhanced YFP brought together by the association of two interaction partners fused to the fragments (20). The most fundamental application of this approach is the direct visualization of the subcellular sites of protein interactions under conditions that closely reflect the normal physiological environment (20). COS-7 cells were cotransfected with pcDNA3-YN154-Bif-1 and pcDNA3-YC155-Bax or pcDNA3-YC155-Bif-1 plasmid at 37°C for 21 h and then switched to 30°C for 3 h to promote YFP fluorophore maturation. As controls, cells were cotransfected with (i) pcDNA3-YN154 and pcDNA3-YC155 empty vectors, (ii) pcDNA3-YN154-Bif-1 and pcDNA3-YC155, and (iii) pcDNA3-YN154 and pcDNA3-YC155-Bax plasmids. After incubation with 20 nM of a mitochondrion-specific dye (MitoTracker Red CMTMRos) for 30 min, the cells were exposed to 1 μM STS in the presence of 50 μM z-VAD-fmk (to prevent cell shrinkage) for 3 h and analyzed by confocal microscopy. As shown in Fig. 6, STS treatment apparently enhanced the YFP signal in cells cotransfected with pcDNA3-YN154-Bif-1 and pcDNA3-YC155-Bax, which was concentrated in punctate foci in the cytosol and colocalized with mitochondria. However, control transfectants displayed nearly undetectable background fluorescence regardless of STS treatment (data not shown). These results strongly indicate that Bif-1 and Bax can interact on mitochondria in intact cells upon apoptotic stimulation. In contrast, the YFP signal in COS-7 cells coexpressing YN154-Bif-1 and YC155-Bif-1 revealed a mostly diffuse pattern throughout the cytosol but became concentrated on mitochondria after STS treatment (Fig. 6), suggesting that Bif-1 forms homodimers in the cytoplasm that translocate to mitochondria during apoptosis.
FIG. 6.
Apoptosis induction enhances Bif-1 interaction with Bax on mitochondria in intact cells as visualized by BiFC analysis. COS-7 cells were cotransfected with pcDNA3-YN154-Bif-1 (YN-Bif-1) and pcDNA3-YC155-Bax (YC-Bax) or pcDNA3-YC155-Bif-1 (YC-Bif-1) plasmid at 37°C for 21 h and then switched to 30°C for 3 h to promote YFP (green) fluorophore maturation. After incubation with 20 nM of MitoTracker (red) for 30 min, the cells were exposed to 1 μM STS in the presence of 50 μM z-VAD-fmk for 3 h and analyzed by confocal microscopy.
Loss of Bif-1 inhibits not only Bax but also Bak activation.
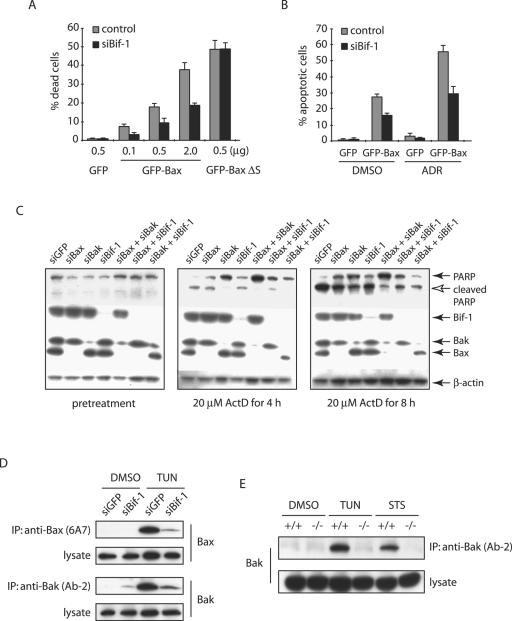
The data presented above clearly demonstrated that loss of Bif-1 delays Bax activation, cytochrome c release, caspase 3 activation, and apoptosis induced by intrinsic death stimuli. To determine whether Bif-1 is important for Bax-mediated apoptosis, we transiently introduced GFP or GFP-Bax into HeLa cells expressing Bif-1 siRNA or control cells. As shown in Fig. 7A, ectopic expression of GFP-Bax caused spontaneous cell death in a concentration-dependent manner, which was significantly suppressed in siBif-1-transfected cells. Similarly, the transfection of GFP-Bax sensitized ADR-induced apoptosis of HeLa cells, and this enhanced sensitivity was attenuated by siRNA to Bif-1 (Fig. 7B). In contrast, the suppression of Bif-1 expression had no effect on the cell death induced by GFP-BaxΔS (Fig. 7A), a conformationally changed and mitochondrion-bound Bax mutant lacking the Ser184 residue (38), supporting our notion that Bif-1 is involved in Bax-mediated apoptosis by controlling its recruitment to mitochondria.
FIG. 7.
Bif-1 contributes to not only Bax, but also Bak activation. (A) siBif-1-expressing HeLa clones or control cells were transfected with the indicated amounts of plasmid DNA encoding GFP, GFP-Bax, or GFP-BaxΔS mutant for 24 h prior to trypan blue dye exclusion assay. The error bars indicate standard deviations. (B) HeLa clones were transfected with 0.5 μg GFP- or GFP-Bax-producing plasmids for 24 h and treated with 1 μM ADR or control DMSO for 8 h. After being stained with DAPI, the apoptotic GFP-positive cells were counted (n > 300). (C) HeLa cells transfected with the indicated siRNAs were treated with 20 μM ActD for the indicated periods of time and subjected to SDS-PAGE/immunoblot analysis with the indicated antibodies. (D) HeLa cells transfected with siGFP or siBif-1 were treated with 4 μg/ml TUN for 24 h prior to the preparation of cell lysates in CHAPS lysis buffer. Immunoprecipitation (IP) was performed with anti-Bak monoclonal antibody (Ab-2) or anti-Bax 6A7 monoclonal antibody, and the resulting immune complexes and total-cell lysates were analyzed by SDS-PAGE/immunoblotting with anti-Bax and anti-Bak polyclonal antibodies. (E) Bif-1−/− and Bif-1+/+ MEFs were treated with 1 μg/ml TUN for 24 h, 1 μM STS for 12 h, or DMSO as a control. Cell lysates were prepared in CHAPS lysis buffer and subjected to immunoprecipitation with anti-Bak monoclonal antibody (Ab-2). The resulting immune complexes, as well as total-cell lysates, were analyzed by SDS-PAGE/immunoblotting with anti-Bak polyclonal antibody.
The genetic studies in mouse models indicated that Bax and Bak are functionally redundant (31, 57). However, Bif-1 selectively interacts with Bax but not Bak (reference 44 and data not shown), and loss of Bif-1 in either HeLa cells or MEFs clearly delays Bax activation, cytochrome c release, caspase 3 activation, and cell death. If the proapoptotic activity of Bif-1 is largely dependent on its ability to activate Bax, we reasoned that cells lacking both Bif-1 and Bak should behave similarly to Bax and Bak double-knockout cells in response to apoptotic signals. To address this issue, we employed an RNA interference approach to alter the expression of Bif-1, Bax, and Bak in HeLa cells and determined their effects on ActD-induced apoptosis as demonstrated by PARP cleavage. As shown in Fig. 7C, the expression of Bax and Bak was significantly suppressed by siBax and siBak, respectively. Inconsistent with the Bax/Bak redundancy in the mouse model (31, 57), human cervical cancer HeLa cells transfected with either siBax, siBak, or siBif-1 significantly reduced PARP cleavage after ActD treatment (Fig. 7C). However, siBak was more potent in suppression of PARP cleavage than siBax, whereas siBif-1 was comparable to siBax, and cells transfected with both siBak and siBif-1 were not as resistant as siBak/siBax double transfectants to ActD-induced PARP cleavage, indicating that Bif-1 is important but not absolutely required for Bax-mediated apoptosis. Interestingly, the PARP cleavage in siBax/siBif-1 double transfectants was suppressed more than that in siBax or siBif-1 singly transfected cells, suggesting that Bax is not the only protein regulated by Bif-1. To determine whether Bif-1 also contributes to the apoptotic activation of Bak, we performed immunoprecipitation with anti-Bak (Ab-2) antibody that recognizes only the conformationally changed Bak protein (14). Similar to Bax, the Bak conformational change induced by TUN in HeLa cells was inhibited by Bif-1 silencing (Fig. 7D). These findings were confirmed in Bif-1 knockout MEFs. As shown in Fig. 7E, Bif-1-deficient MEFs were resistant to Bak conformational change induced by TUN or STS compared to wild-type MEFs. Taken together, these results indicate that Bif-1 promotes not only Bax but also Bak activation during apoptosis.
Suppression of Bif-1 expression promotes anchorage-independent growth and tumorigenesis of HeLa cells.
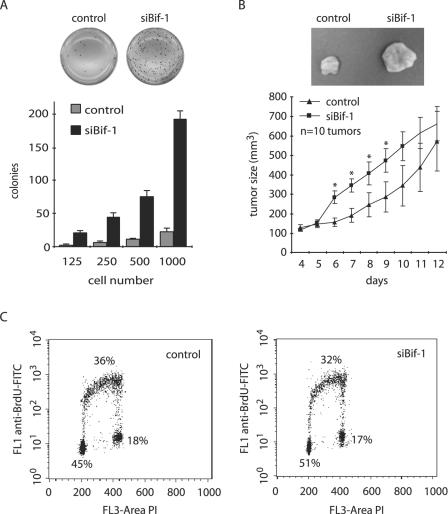
Dysregulation of apoptosis is a major contributor to tumor progression, because defects in this physiological cell death mechanism not only cause tumor cells to accumulate in the body but also contribute to the difficulties of treatment and to metastatic spread (survival without attachment) of the cells. The data presented above, together with our previous report (6), indicate that Bif-1 plays an important role in apoptosis. To determine whether inhibition of Bif-1 expression enhances anchorage-independent growth of cancer cells, stable siBif-1-transfected HeLa or control cells were cultured in soft agar. As shown in Fig. 8A, the number of colonies in soft agar formed by siBif-1-expressing cells increased dramatically compared to control cells. This result clearly indicates that suppression of Bif-1 expression promotes anchorage-independent growth of HeLa cells.
FIG. 8.
Inhibition of Bif-1 expression enhances the tumorigenicity of HeLa cells. (A) siBif-1 stable clones or control HeLa cells were seeded at 125 to 1,000 cells/well in triplicate in six-well plates in 0.3% agar over a 0.6% agar layer and cultured for 2 to 3 weeks. After being stained with 1 mg/ml MTT in growth medium, the colonies were photographed and counted. (B) Stable HeLa transfectants were harvested, suspended in PBS, and injected subcutaneously into the right and left flanks (10 × 106 cells/flank) of female nude mice (five mice/cell line). The tumor size was monitored daily using calipers. Statistical significance was determined at P < 0.05 (*). The error bars indicate standard deviations. (C) Cultures of logarithmically growing HeLa transfectants were subjected to BrdU incorporation and DNA content analysis by flow cytometry. FITC, fluorescein isothiocyanate.
Next, we examined the effect of reduced Bif-1 expression on tumorigenicity of HeLa cells in nude mice. Consistent with the data in soft agar, knockdown of Bif-1 significantly accelerated tumor formation of HeLa cells in nude mice after implantation (Fig. 8B). Taken together, these results demonstrate that Bif-1 may function as a tumor suppressor.
It has become apparent that the successive genetic aberrations acquired by cancer cells result in defects in regulatory signal transduction circuits that govern normal cell proliferation, differentiation, and programmed cell death (17). To determine whether Bif-1 also plays a role in cell proliferation that contributes to the tumorigenicity of HeLa cells, we analyzed BrdU incorporation in HeLa cells stably transfected with siBif-1-expressing plasmid by flow cytometry. As shown in Fig. 8C, knockdown of Bif-1 did not significantly affect the cell cycle progression of this human cervical cell line.
DISCUSSION
The findings reported here suggest a regulatory role for Bif-1 in apoptosis as a new type of Bax/Bak activator. Despite the results showing that Bif-1 does not appear to be absolutely required for apoptosis, loss of Bif-1 significantly delays the activation of Bax and Bak, cytochrome c release, caspase 3 activation, and cell death, suggesting that Bif-1 plays an important regulatory role in apoptosis through the activation of Bax/Bak in concert with other types of proapoptotic molecules, such as BH3-only proteins. Indeed, knockdown of Bif-1 suppresses the death-inducing activity of wild-type, but not a conformationally active, Bax in HeLa cells (Fig. 7A and B), although ectopic overexpression of Bif-1 in HeLa and 293 cells reportedly has a minor effect on cell death (44), probably due to the relatively high levels of endogenous Bif-1 protein in these cells (not shown). Nevertheless, inhibition of Bif-1 expression enhances the anchorage-independent growth and tumorigenicity of HeLa cells without a significant effect on cell proliferation. Collectively, these findings suggest that Bif-1 is an important component of the mitochondrial pathway for apoptosis as a novel Bax/Bak activator that may contribute to suppression of tumorigenesis. Indeed, the downregulation of Bif-1 mRNA was reported in lung adenocarcinomas (2).
The Bif-1 protein contains an N-terminal BAR domain, which is commonly found in proteins implicated in vesicle generation and membrane remodeling (43). Bif-1 forms a homodimer through its coiled-coil domain located within the BAR domain (44), and the N-terminal part (1 to 27 amino acids), but not the C-terminal SH3 domain, is required for Bif-1 binding to Bax (6, 44). Crystallographic studies show that the dimer of the Drosophila amphiphysin BAR domain forms a banana-shaped structure, which binds to negatively charged membranes and induces membrane curvature (43). The N-BAR domains possess an amino-terminal amphipathic α helix that enhances their ability to bind lipids and to induce membranes to form tubes (43). Consistently, the N-BAR-containing protein Bif-1 binds lipid, exhibits lysophosphatidic acid acyl transferase activity, and deforms liposomes into tubules (10, 37). Moreover, it has been shown that Bax-mediated liposome permeabilization occurs through a mechanism sensitive to intrinsic membrane curvature (1). Therefore, it is possible that Bif-1-mediated OMM deformations facilitate and/or stabilize Bax/Bak oligomerization and integration into OMM during apoptosis.
Although the N-BAR domain is important, the C-terminal SH3 domain of Bif-1 may also play a role in membrane dynamics of mitochondria through its interactions with proline-rich molecules involved in membrane remodeling. Changes in mitochondrial morphology, such as mitochondrial fragmentation and crista remodeling, have been described in connection with many modes of apoptosis (11, 24, 48). Interestingly, tBid-treated mitochondria showed an increase in intracrista connectivity and formation of a more highly interconnected crista network (48), whereas overexpression of a dominant-negative mutant (K38A) of Drp1 inhibited apoptotic fragmentation of mitochondria and cytochrome c release (11). This pathway seems to be evolutionarily conserved, because inhibition of Drp1-mediated mitochondrial fragmentation prevents apoptosis in Caenorhabditis elegans as well (22). Drp1 is a mediator of mitochondrial fission that contains the PXXP motif, a potential SH3 domain-binding site, suggesting a possible interaction between the SH3 domain of Bif-1 and the PXXP region of Drp1 for the regulation of mitochondrial fragmentation during apoptosis. In fact, Bif-1 and Drp1 control the same pathway for the regulation of the morphological dynamics of mitochondria, and expression of the N-terminal deletion mutant Bif-1 (60-362ΔN) induces a dominant-negative inhibition of endogenous Bif-1, indicating that the SH3-containing C-terminal region of Bif-1 plays a regulatory role in the maintenance of mitochondrial morphology (23).
Interestingly, we showed that the association of Bif-1 with Bax is dramatically increased in intact cells after induction of apoptosis. This suggests a possibility that the ability of Bif-1 to induce Bax conformational change is regulated by posttranslational modifications, such as phosphorylation. Moreover, a number of retention factors such as Ku70 (47), 14-3-3 (40), and humanin (15), have been reported to bind and keep Bax in an inactive conformation in viable cells. Apoptotic stimuli induce the dissociation of these retention factors, resulting in Bax conformational change, oligomerization, and insertion into OMM (15, 40, 47). Therefore, it will be interesting to see if these suppressors prevent Bif-1 binding and activation of Bax in the cell under conditions in which Bax should be kept inactive.
Numerous studies indicate that evasion of apoptosis is a hallmark of most and perhaps all types of cancer (17). The BH1-3 prodeath proteins Bax and Bak are essential gatekeepers to the mitochondrial apoptosis machinery (57). We showed that silencing of these proapoptotic proteins by siRNAs results in the inhibition of PARP cleavage, and these effects are enhanced by cotransfection with siBif-1. Considering the existence of Bif-1 in Bax/Bak double-knockout cells, which are resistant to intrinsic death signals, Bif-1 must act upstream of Bax/Bak to regulate apoptosis. Indeed, Bif-1 silencing results in the inhibition of Bak conformational change as well as Bax, indicating that Bif-1 is involved in not only Bax but also Bak activation. Since Bif-1 specifically interacts with Bax but not Bak (44; data not shown), the activation of Bak by Bif-1 might occur indirectly through Bax or other mitochondrial proteins. It has been suggested that Bak activation requires Bax (36). The loss of Bax and Bak in transformed BMK cells facilitates tumor formation in nude mice (39). Our results showing that loss of Bif-1 prevents both Bax and Bak activation and apoptosis suggest a potential role for Bif-1 in suppression of tumorigenesis. Indeed, suppression of endogenous Bif-1 expression enhanced the tumorigenicity of HeLa cells, as demonstrated by anchorage-independent growth in soft agar and tumor formation in nude mice. Surprisingly, Bif-1-deficient mouse embryos developed normally and were born at the expected Mendelian frequency with no obvious abnormalities, at least at the time of birth, which is different from the phenotypes reported in Bax/Bak double-knockout mice (31). However, loss of one of the Bax/Bak activators may not be enough to completely inactivate these gatekeepers of the mitochondrial death machinery. Similarly, knockout of Bid, a well-characterized Bax/Bak activator, produced no apparent developmental defects or tissue abnormalities in mice (64). The precise physiological roles of Bif-1 in mammalian development, tissue homeostasis, and tumorigenesis remain to be explored by the analysis of Bif-1-deficient mice.
Acknowledgments
We thank Noreen Luetteke and Hartmut Berns at the Moffitt Cancer Center and Akiyoshi Fukamizu at the University of Tsukuba for excellent assistance in generating Bif-1 targeting vector and knockout mice. We acknowledge the services provided by the Moffitt Mouse Models Core, Molecular Biology Core, Flow Cytometry Core, and Molecular Imaging Core. We are grateful to Yang Shi at the Harvard Medical School for pBS/U6 vector and Tom Kerppola at the University of Michigan Medical School for pBiFC-YN155 and pBiFC-YC155 plasmids.
This work was supported by NIH grant CA82197 and fellowships from the Lymphoma Research Foundation (H.Y.) and the Uehara Memorial Foundation (Y.T.). This work was also supported in part by the Intramural Research Program of the NINDS, NIH. Y.T. is a fellow of the Japan Society for Promotion of Science.
REFERENCES
- 1.Basanez, G., J. C. Sharpe, J. Galanis, T. B. Brandt, J. M. Hardwick, and J. Zimmerberg. 2002. Bax-type apoptotic proteins porate pure lipid bilayers through a mechanism sensitive to intrinsic monolayer curvature. J. Biol. Chem. 277**:**49360-49365. [DOI] [PubMed] [Google Scholar]
- 2.Bonner, A. E., W. J. Lemon, T. R. Devereux, R. A. Lubet, and M. You. 2004. Molecular profiling of mouse lung tumors: association with tumor progression, lung development, and human lung adenocarcinomas. Oncogene 23**:**1166-1176. [DOI] [PubMed] [Google Scholar]
- 3.Boya, P., I. Cohen, N. Zamzami, H. L. Vieira, and G. Kroemer. 2002. Endoplasmic reticulum stress-induced cell death requires mitochondrial membrane permeabilization. Cell Death Differ. 9**:**465-467. [DOI] [PubMed] [Google Scholar]
- 4.Chipuk, J. E., T. Kuwana, L. Bouchier-Hayes, N. M. Droin, D. D. Newmeyer, M. Schuler, and D. R. Green. 2004. Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science 303**:**1010-1014. [DOI] [PubMed] [Google Scholar]
- 5.Cory, S., D. C. Huang, and J. M. Adams. 2003. The Bcl-2 family: roles in cell survival and oncogenesis. Oncogene 22**:**8590-8607. [DOI] [PubMed] [Google Scholar]
- 6.Cuddeback, S. M., H. Yamaguchi, K. Komatsu, T. Miyashita, M. Yamada, C. Wu, S. Singh, and H. G. Wang. 2001. Molecular cloning and characterization of Bif-1: a novel SH3 domain-containing protein that associates with Bax. J. Biol. Chem. 276**:**20559-20565. [DOI] [PubMed] [Google Scholar]
- 7.Degenhardt, K., R. Sundararajan, T. Lindsten, C. Thompson, and E. White. 2002. Bax and Bak independently promote cytochrome C release from mitochondria. J. Biol. Chem. 277**:**14127-14134. [DOI] [PubMed] [Google Scholar]
- 8.Dumont, P., J. I. Leu, A. C. Della Pietra III, D. L. George, and M. Murphy. 2003. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat. Genet. 33**:**357-365. [DOI] [PubMed] [Google Scholar]
- 9.Eskes, R., S. Desagher, B. Antonsson, and J. C. Martinou. 2000. Bid induces the oligomerization and insertion of Bax into the outer mitochondrial membrane. Mol. Cell. Biol. 20**:**929-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farsad, K., N. Ringstad, K. Takei, S. R. Floyd, K. Rose, and P. De Camilli. 2001. Generation of high curvature membranes mediated by direct endophilin bilayer interactions. J. Cell Biol. 155**:**193-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frank, S., B. Gaume, E. S. Bergmann-Leitner, W. W. Leitner, E. G. Robert, F. Catez, C. L. Smith, and R. J. Youle. 2001. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev. Cell 1**:**515-525. [DOI] [PubMed] [Google Scholar]
- 12.Goping, I. S., A. Gross, J. N. Lavoie, M. Nguyen, R. Jemmerson, K. Roth, S. J. Korsmeyer, and G. C. Shore. 1998. Regulated targeting of BAX to mitochondria. J. Cell Biol. 143**:**207-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Green, D. R., and G. I. Evan. 2002. A matter of life and death. Cancer Cell 1**:**19-30. [DOI] [PubMed] [Google Scholar]
- 14.Griffiths, G. J., L. Dubrez, C. P. Morgan, N. A. Jones, J. Whitehouse, B. M. Corfe, C. Dive, and J. A. Hickman. 1999. Cell damage-induced conformational changes of the pro-apoptotic protein Bak in vivo precede the onset of apoptosis. J. Cell Biol. 144**:**903-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo, B., D. Zhai, E. Cabezas, K. Welsh, S. Nouraini, A. C. Satterthwait, and J. C. Reed. 2003. Humanin peptide suppresses apoptosis by interfering with Bax activation. Nature 423**:**456-461. [DOI] [PubMed] [Google Scholar]
- 16.Hacki, J., L. Egger, L. Monney, S. Conus, T. Rosse, I. Fellay, and C. Borner. 2000. Apoptotic crosstalk between the endoplasmic reticulum and mitochondria controlled by Bcl-2. Oncogene 19**:**2286-2295. [DOI] [PubMed] [Google Scholar]
- 17.Hanahan, D., and R. A. Weinberg. 2000. The hallmarks of cancer. Cell 100**:**57-70. [DOI] [PubMed] [Google Scholar]
- 18.Hirai, I., and H. G. Wang. 2002. A role of the C-terminal region of human Rad9 (hRad9) in nuclear transport of the hRad9 checkpoint complex. J. Biol. Chem. 277**:**25722-25727. [DOI] [PubMed] [Google Scholar]
- 19.Hsu, Y. T., and R. J. Youle. 1998. Bax in murine thymus is a soluble monomeric protein that displays differential detergent-induced conformations. J. Biol. Chem. 273**:**10777-10783. [DOI] [PubMed] [Google Scholar]
- 20.Hu, C. D., Y. Chinenov, and T. K. Kerppola. 2002. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 9**:**789-798. [DOI] [PubMed] [Google Scholar]
- 21.Huttner, W. B., and A. Schmidt. 2000. Lipids, lipid modification and lipid-protein interaction in membrane budding and fission—insights from the roles of endophilin A1 and synaptophysin in synaptic vesicle endocytosis. Curr. Opin. Neurobiol. 10**:**543-551. [DOI] [PubMed] [Google Scholar]
- 22.Jagasia, R., P. Grote, B. Westermann, and B. Conradt. 2005. DRP-1-mediated mitochondrial fragmentation during EGL-1-induced cell death in C. elegans. Nature 433**:**754-760. [DOI] [PubMed] [Google Scholar]
- 23.Karbowski, M., S. Y. Jeong, and R. J. Youle. 2004. Endophilin B1 is required for the maintenance of mitochondrial morphology. J. Cell Biol. 166**:**1027-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karbowski, M., Y. J. Lee, B. Gaume, S. Y. Jeong, S. Frank, A. Nechushtan, A. Santel, M. Fuller, C. L. Smith, and R. J. Youle. 2002. Spatial and temporal association of Bax with mitochondrial fission sites, Drp1, and Mfn2 during apoptosis. J. Cell Biol. 159**:**931-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korsmeyer, S. J., M. C. Wei, M. Saito, S. Weiler, K. J. Oh, and P. H. Schlesinger. 2000. Pro-apoptotic cascade activates BID, which oligomerizes BAK or BAX into pores that result in the release of cytochrome c. Cell Death Differ. 7**:**1166-1173. [DOI] [PubMed] [Google Scholar]
- 26.Kuwana, T., L. Bouchier-Hayes, J. E. Chipuk, C. Bonzon, B. A. Sullivan, D. R. Green, and D. D. Newmeyer. 2005. BH3 domains of BH3-only proteins differentially regulate Bax-mediated mitochondrial membrane permeabilization both directly and indirectly. Mol. Cell 17**:**525-535. [DOI] [PubMed] [Google Scholar]
- 27.Kuwana, T., M. R. Mackey, G. Perkins, M. H. Ellisman, M. Latterich, R. Schneiter, D. R. Green, and D. D. Newmeyer. 2002. Bid, Bax, and lipids cooperate to form supramolecular openings in the outer mitochondrial membrane. Cell 111**:**331-342. [DOI] [PubMed] [Google Scholar]
- 28.Letai, A., M. C. Bassik, L. D. Walensky, M. D. Sorcinelli, S. Weiler, and S. J. Korsmeyer. 2002. Distinct BH3 domains either sensitize or activate mitochondrial apoptosis, serving as prototype cancer therapeutics. Cancer Cell 2**:**183-192. [DOI] [PubMed] [Google Scholar]
- 29.Leu, J. I., P. Dumont, M. Hafey, M. E. Murphy, and D. L. George. 2004. Mitochondrial p53 activates Bak and causes disruption of a Bak-Mcl1 complex. Nat. Cell Biol. 6**:**443-450. [DOI] [PubMed] [Google Scholar]
- 30.Li, H., H. Zhu, C. J. Xu, and J. Yuan. 1998. Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell 94**:**491-501. [DOI] [PubMed] [Google Scholar]
- 31.Lindsten, T., A. J. Ross, A. King, W. X. Zong, J. C. Rathmell, H. A. Shiels, E. Ulrich, K. G. Waymire, P. Mahar, K. Frauwirth, Y. Chen, M. Wei, V. M. Eng, D. M. Adelman, M. C. Simon, A. Ma, J. A. Golden, G. Evan, S. J. Korsmeyer, G. R. MacGregor, and C. B. Thompson. 2000. The combined functions of proapoptotic Bcl-2 family members Bak and Bax are essential for normal development of multiple tissues. Mol. Cell 6**:**1389-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo, X., I. Budihardjo, H. Zou, C. Slaughter, and X. Wang. 1998. Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell 94**:**481-490. [DOI] [PubMed] [Google Scholar]
- 33.Lutter, M., M. Fang, X. Luo, M. Nishijima, X. Xie, and X. Wang. 2000. Cardiolipin provides specificity for targeting of tBid to mitochondria. Nat. Cell Biol. 2**:**754-761. [DOI] [PubMed] [Google Scholar]
- 34.Marani, M., T. Tenev, D. Hancock, J. Downward, and N. R. Lemoine. 2002. Identification of novel isoforms of the BH3 domain protein Bim which directly activate Bax to trigger apoptosis. Mol. Cell. Biol. 22**:**3577-3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mihara, M., S. Erster, A. Zaika, O. Petrenko, T. Chittenden, P. Pancoska, and U. M. Moll. 2003. p53 has a direct apoptogenic role at the mitochondria. Mol. Cell 11**:**577-590. [DOI] [PubMed] [Google Scholar]
- 36.Mikhailov, V., M. Mikhailova, K. Degenhardt, M. A. Venkatachalam, E. White, and P. Saikumar. 2003. Association of Bax and Bak homo-oligomers in mitochondria. Bax requirement for Bak reorganization and cytochrome c release. J. Biol. Chem. 278**:**5367-5376. [DOI] [PubMed] [Google Scholar]
- 37.Modregger, J., A. A. Schmidt, B. Ritter, W. B. Huttner, and M. Plomann. 2003. Characterization of Endophilin B1b, a brain-specific membrane-associated lysophosphatidic acid acyl transferase with properties distinct from endophilin A1. J. Biol. Chem. 278**:**4160-4167. [DOI] [PubMed] [Google Scholar]
- 38.Nechushtan, A., C. L. Smith, Y. T. Hsu, and R. J. Youle. 1999. Conformation of the Bax C-terminus regulates subcellular location and cell death. EMBO J. 18**:**2330-2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nelson, D. A., T. T. Tan, A. B. Rabson, D. Anderson, K. Degenhardt, and E. White. 2004. Hypoxia and defective apoptosis drive genomic instability and tumorigenesis. Genes Dev. 18**:**2095-2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nomura, M., S. Shimizu, T. Sugiyama, M. Narita, T. Ito, H. Matsuda, and Y. Tsujimoto. 2003. 14-3-3 Interacts directly with and negatively regulates pro-apoptotic Bax. J. Biol. Chem. 278**:**2058-2065. [DOI] [PubMed] [Google Scholar]
- 41.Nothwehr, S. F., and J. C. Martinou. 2003. A retention factor keeps death at bay. Nat. Cell Biol. 5**:**281-283. [DOI] [PubMed] [Google Scholar]
- 42.Ohtsuka, T., H. Ryu, Y. A. Minamishima, S. Macip, J. Sagara, K. I. Nakayama, S. A. Aaronson, and S. W. Lee. 2004. ASC is a Bax adaptor and regulates the p53-Bax mitochondrial apoptosis pathway. Nat. Cell Biol. 6**:**121-128. [DOI] [PubMed] [Google Scholar]
- 43.Peter, B. J., H. M. Kent, I. G. Mills, Y. Vallis, P. J. Butler, P. R. Evans, and H. T. McMahon. 2004. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science 303**:**495-499. [DOI] [PubMed] [Google Scholar]
- 44.Pierrat, B., M. Simonen, M. Cueto, J. Mestan, P. Ferrigno, and J. Heim. 2001. SH3GLB, a new endophilin-related protein family featuring an SH3 domain. Genomics 71**:**222-234. [DOI] [PubMed] [Google Scholar]
- 45.Reed, J. C. 2003. Apoptosis-targeted therapies for cancer. Cancer Cell 3**:**17-22. [DOI] [PubMed] [Google Scholar]
- 46.Roucou, X., S. Montessuit, B. Antonsson, and J. C. Martinou. 2002. Bax oligomerization in mitochondrial membranes requires tBid (caspase-8-cleaved Bid) and a mitochondrial protein. Biochem. J. 368**:**915-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sawada, M., W. Sun, P. Hayes, K. Leskov, D. A. Boothman, and S. Matsuyama. 2003. Ku70 suppresses the apoptotic translocation of Bax to mitochondria. Nat. Cell Biol. 5**:**320-329. [DOI] [PubMed] [Google Scholar]
- 48.Scorrano, L., M. Ashiya, K. Buttle, S. Weiler, S. A. Oakes, C. A. Mannella, and S. J. Korsmeyer. 2002. A distinct pathway remodels mitochondrial cristae and mobilizes cytochrome c during apoptosis. Dev. Cell 2**:**55-67. [DOI] [PubMed] [Google Scholar]
- 49.Scorrano, L., S. A. Oakes, J. T. Opferman, E. H. Cheng, M. D. Sorcinelli, T. Pozzan, and S. J. Korsmeyer. 2003. BAX and BAK regulation of endoplasmic reticulum Ca2+: a control point for apoptosis. Science 300**:**135-139. [DOI] [PubMed] [Google Scholar]
- 50.Sharpe, J. C., D. Arnoult, and R. J. Youle. 2004. Control of mitochondrial permeability by Bcl-2 family members. Biochim. Biophys. Acta 1644**:**107-113. [DOI] [PubMed] [Google Scholar]
- 51.Sui, G., C. Soohoo, B. el Affar, F. Gay, Y. Shi, W. C. Forrester, and Y. Shi. 2002. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc. Natl. Acad. Sci. USA 99**:**5515-5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suzuki, M., R. J. Youle, and N. Tjandra. 2000. Structure of Bax. Coregulation of dimer formation and intracellular localization. Cell 103**:**645-654. [DOI] [PubMed] [Google Scholar]
- 53.Terrones, O., B. Antonsson, H. Yamaguchi, H. G. Wang, J. Liu, R. M. Lee, A. Herrmann, and G. Basanez. 2004. Lipidic pore formation by the concerted action of proapoptotic BAX and tBID. J. Biol. Chem. 279**:**30081-30091. [DOI] [PubMed] [Google Scholar]
- 54.Wang, K., W.-M. Yin, D. T. Chao, C. L. Milliman, and S. J. Korsmeyer. 1996. BID: a novel BH3 domain-only death agonist. Genes Dev. 10**:**2859-2869. [DOI] [PubMed] [Google Scholar]
- 55.Wang, X. 2001. The expanding role of mitochondria in apoptosis. Genes Dev. 15**:**2922-2933.11711427 [Google Scholar]
- 56.Wei, M. C., T. Lindsten, V. K. Mootha, S. Weiler, A. Gross, M. Ashiya, C. B. Thompson, and S. J. Korsmeyer. 2000. tBID, a membrane-targeted death ligand, oligomerizes BAK to release cytochrome c. Genes Dev. 14**:**2060-2071. [PMC free article] [PubMed] [Google Scholar]
- 57.Wei, M. C., W. X. Zong, E. H. Cheng, T. Lindsten, V. Panoutsakopoulou, A. J. Ross, K. A. Roth, G. R. MacGregor, C. B. Thompson, and S. J. Korsmeyer. 2001. Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science 292**:**727-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wolter, K. G., Y. T. Hsu, C. L. Smith, A. Nechushtan, X. G. Xi, and R. J. Youle. 1997. Movement of Bax from the cytosol to mitochondria during apoptosis. J. Cell Biol. 139**:**1281-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yamaguchi, H., K. Bhalla, and H. G. Wang. 2003. Bax plays a pivotal role in thapsigargin-induced apoptosis of human colon cancer HCT116 cells by controlling Smac/Diablo and Omi/HtrA2 release from mitochondria. Cancer Res. 63**:**1483-1489. [PubMed] [Google Scholar]
- 60.Yamaguchi, H., J. Chen, K. Bhalla, and H. G. Wang. 2004. Regulation of Bax activation and apoptotic response to microtubule-damaging agents by p53 transcription-dependent and -independent pathways. J. Biol. Chem. 279**:**39431-39437. [DOI] [PubMed] [Google Scholar]
- 61.Yamaguchi, H., and H. G. Wang. 2002. Bcl-XL protects BimEL-induced Bax conformational change and cytochrome C release independent of interacting with Bax or BimEL. J. Biol. Chem. 277**:**41604-41612. [DOI] [PubMed] [Google Scholar]
- 62.Yamaguchi, H., and H. G. Wang. 2004. CHOP is involved in endoplasmic reticulum stress-induced apoptosis by enhancing DR5 expression in human carcinoma cells. J. Biol. Chem. 279**:**45495-45502. [DOI] [PubMed] [Google Scholar]
- 63.Yamaguchi, H., and H.-G. Wang. 2001. The protein kinase PKB/Akt regulates cell survival and apoptosis by inhibiting Bax conformational change. Oncogene 20**:**7779-7786. [DOI] [PubMed] [Google Scholar]
- 64.Yin, X. M., K. Wang, A. Gross, Y. Zhao, S. Zinkel, B. Klocke, K. A. Roth, and S. J. Korsmeyer. 1999. Bid-deficient mice are resistant to Fas-induced hepatocellular apoptosis. Nature 400**:**886-891. [DOI] [PubMed] [Google Scholar]
- 65.Zhang, L., J. Yu, B. H. Park, K. W. Kinzler, and B. Vogelstein. 2000. Role of BAX in the apoptotic response to anticancer agents. Science 290**:**989-992. [DOI] [PubMed] [Google Scholar]