Tol-Dependent Macromolecule Import through the Escherichia coli Cell Envelope Requires the Presence of an Exposed TolA Binding Motif (original) (raw)
Abstract
The Tol-Pal proteins of the cell envelope of Escherichia coli are required for maintaining outer membrane integrity. This system forms protein complexes in which TolA plays a central role by providing a bridge between the inner and outer membranes via its interaction with the Pal lipoprotein. The Tol proteins are parasitized by filamentous bacteriophages and group A colicins. The N-terminal domain of the Ff phage g3p protein and the translocation domains of colicins interact directly with TolA during the processes of import through the cell envelope. Recently, a four-amino-acid sequence in Pal has been shown to be involved in Pal's interaction with TolA. A similar motif is also present in the sequence of two TolA partners, g3p and colicin A. Here, a mutational study was conducted to define the function of these motifs in the binding activity and import process of TolA. The various domains were produced and exported to the bacterial periplasm, and their cellular effects were analyzed. Cells producing the g3p domain were tolerant to colicins and filamentous phages and had destabilized outer membranes, while g3p deleted of three residues in the motif was affected in TolA binding and had no effect on cell integrity or colicin or phage import. A conserved Tyr residue in the colicin A translocation domain was involved in TolA binding and colicin A import. Furthermore, in vivo and in vitro coprecipitation analyses demonstrated that colicin A and g3p N-terminal domains compete for binding to TolA.
The Tol-Pal complex of Escherichia coli is composed of five proteins that interact in the cell envelope. The TolA, TolQ, and TolR cytoplasmic membrane proteins interact with each other through their transmembrane segments (23, 35). The outer membrane-anchored Pal lipoprotein interacts with the periplasmic TolB protein (8). A link between inner and outer membranes is mediated by the interaction of the TolA C-terminal domain with Pal and TolB (14, 49). However, while six heterodimeric complexes in the Tol-Pal system have been described so far, no protein complex containing more than two partners has been observed. The Tol-Pal proteins are required for maintaining outer membrane stability, probably through a protein network involving cell wall components such as Lpp, OmpA, and peptidoglycan (16, 18). Deletion or mutation of any of the tol or pal genes results in numerous defects, such as periplasmic leakage, increased susceptibility to many toxic compounds, and formation of outer membrane vesicles (7, 50). The Tol proteins are exploited by filamentous (Ff) bacteriophages and colicins (33, 36, 50) for their import processes. Infection by filamentous bacteriophages is dependent on the minor coat protein, g3p, which is located at one extremity of the phage capsid. Ff phage infection and colicin import mechanisms through the periplasm require, respectively, the N-terminal domain of g3p and the colicin translocation domain (5, 48). These domains interact with the TolA protein during the import process. The TolA protein contains three domains separated by clusters of glycine residues. The N-terminal transmembrane segment which anchors the protein to the cytoplasmic membrane is separated from the C-terminal globular domain by a long α-helical central region (25, 37). The C-terminal domain, called TolAIII, interacts with the translocation domains of colicins and with the N-terminal domain of g3p (g3pN1) (6, 45). The structure of TolAIII had been determined by X-ray crystallography in a cocrystal with g3pN1 (41) and in solution as an isolated domain by heteronuclear magnetic resonance analysis (22). Nuclear magnetic resonance analyses indicated that TolAIII is able to promote folding of the disordered colicin N translocation domain upon interaction (4). TolAIII is a flexible molecule which becomes disordered when bound to the disordered translocation domain of colicin A (21) and undergoes conformational changes upon binding to g3pN1 (22).
We have previously shown that production of TolAIII, colicins, or g3p domains in the periplasm of wild-type (WT) cells confers the whole _tol_-associated phenotype (28), and we have postulated that these domains may alter the outer membrane integrity by modifying the equilibrium of the Tol-Pal interactions (11). Previous analyses have shown that some conserved polypeptide sequences of Pal were involved in maintaining membrane integrity and in the interactions with TolA, TolB, or peptidoglycan and OmpA. The region interacting with TolA is localized in a four-amino-acid sequence located at the C-terminal end of Pal and is called the TolA box. Moreover, analyses of protein patterns indicated that a similar motif is present in the g3p and colicin A N-terminal domains (17) (see Fig. 6). In this study, the protein complexes containing TolA, formed in vitro or in living cells, were analyzed using WT or mutated g3p and colicin A N-terminal domains. Furthermore, the existence of a TolA box in the colicin A and g3p sequences and of trimeric complexes containing TolA associated with imported molecules was determined.
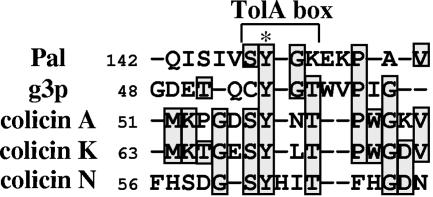
FIG. 6.
Sequence conservation of the TolA box. Sequences containing the TolA boxes of various TolA partners with their five flanking residues were aligned using TCoffee (http://igs-server.cnrs-mrs.fr/Tcoffee/tcoffee_cgi/index.cgi). The positions of the N-terminal residues, the TolA box, and the conserved Tyr residue are indicated.
MATERIALS AND METHODS
Bacterial strains and plasmid constructions.
E. coli strains K-12, W3110, and GM1 (laboratory collection) have been used. pG3 plasmid was constructed by insertion of a synthetic DNA fragment obtained by annealing of STtop (5′-AATTTGGATGGTCTCATCCTCAATTTGAAAAAGGCCTTC) and STbot (5′-AATTGAAGGCCTTTTTCAAATTGAGGATGAGACCATCCA) oligonucleotides encoding Strep-tag II (WSHPQFEK) into the EcoRI site of pINTG3p (28). A plasmid encoding Strep-tagged TolAIII was constructed by using STtop and STbot inserted into the EcoRI site of pINAIII (28). The SacI_-_HindIII DNA fragment of pATh plasmid (21) encoding the N-terminal domain of colicin A devoid of the TolB and TolR binding sequences was inserted into similarly digested pASK-IBA4 plasmid (IBA), giving pTA. pG3 and pTA plasmids derivatives have been constructed by quick-change mutagenesis using primers designed to delete SYNT or PYGR residues of TA and YGT residues of G3. Primers were used for single-codon substitution of Tyr to an Ala residue [corresponding to the modification of two bases: TA(T/C) in GC(T/C)] to construct the pTAy58a, pTAy90a, and pG3y54a plasmids. Briefly, the entire plasmid's DNA was amplified with Pfu turbo DNA polymerase (Stratagene) by using two overlapping mutagenic primers which were divergently oriented. Plasmids encoding colicin A (y58a and y90a) were constructed by quick-change mutagenesis with the corresponding primers by using pColA9 plasmid (38) as template. The various mutations were confirmed by DNA sequencing.
Protein purifications.
E. coli strain W3110 harboring the various plasmids were grown in LB medium supplemented with ampicillin (100 μg · ml−1) to exponential growth phase. Protein production was then induced either with anhydrotetracycline (AHT) (0.1 μg · ml−1) for 3 h at 30°C (TA and TA variants) or with IPTG (isopropyl-β-d-thiogalactopyranoside; 100 μM) overnight at 30°C (G3 and G3 variants). Under these conditions, the recombinant proteins were recovered in large amounts from the cell supernatants owing to outer membrane leakiness. Cells were collected by centrifugation at 6,000 rpm, and proteins present in the supernatants were recovered by precipitation with ammonium sulfate (70%) overnight at 4°C. After centrifugation for 45 min at 6,000 rpm, pellets were resuspended in 20 mM Tris-HCl (pH 8) and 100 mM NaCl and dialyzed against the same buffer before purification. Soluble samples were recovered by ultracentrifugation at 100,000 × g for 1 h. TA and G3 protein variants were purified according to a two-step procedure using immobilized metal affinity chromatography followed by Strep-tactin chromatography. Proteins were dialyzed in 20 mM (pH 6.8) phosphate buffer and 100 mM NaCl. No protein contaminant was detected upon Coomassie blue staining. The protein concentrations were estimated according to the theoretical extinction coefficients (M−1 · cm−1) at 280 nm corresponding to 27.880 for TA; 26.600 for TAΔsynt, TAy58a, TAΔpygr, and TAy90a; 26.840 for G3; and 25.560 for G3Δygt and G3y54a.
Colicins A and its derivatives colicin A (y58a) and colicin A (y90a) were recovered in the cell supernatants 4 h after mitomycin C treatment. The periplasmic region of TolA (TolAII-III [25]), the C-terminal domain of TolA (TolAIII corresponding to previously described TolAIII3 [21]), g3pN1 (G3 protein devoid of the Strep-tag [21]), TolB (1), TR-A (colicin A devoid of the C-terminal catalytic domain [5]), and ATh (the colicin A translocation domain devoid of the Strep-tag [21]) were purified according to the listed references. Strep-tagged TolAIII was purified using Strep-tactin chromatography (IBA). Purified Pal corresponding to the Pal protein in which the first cysteine residue was replaced with an alanine (PalC1A [2]) was a generous gift of Anne Walburger.
Periplasmic leakage and sodium dodecyl sulfate (SDS) sensitivity analyses.
Qualitative analysis of RNase I release was performed using the technique previously described by Lazzaroni and Portalier (34). Briefly, cells were plated on LB agar supplemented with 1% RNA (type VI from Torula yeast; Sigma). After overnight growth, RNA was precipitated with cold 10% trichloroacetic acid, and RNase I leakage was detected by identification of clear zones surrounding colonies.
SDS sensitivities of W3110 cells harboring the pTA or pG3 variants were checked during the exponential growth phase in LB medium. W3110pTA and W3110pG3 cells as well as their derivatives were induced with AHT (20 ng · ml−1) and IPTG (100 μM), respectively, for 30 min, resuspended in LB medium, and further incubated for 90 min in the presence of 1% SDS. The percentage of cell survival was measured from the turbidity ratio of SDS-treated cells to untreated cells and was the average of triplicate assays (±10%).
Colicin sensitivity assays.
Colicin A variant activities were identified by the presence of clear halos on a lawn of sensitive W3110 cells using serial dilutions of the same concentration of various colicins. W3110 cells were grown to exponential growth phase (_A_600, 0.3 to 0.6), and cultures were plated on LB agar. Then, 0.8 μl of serial dilutions of colicins was spotted onto the lawns. After overnight culture at 37°C, bacterial sensitivity was measured by the detection of clear halos. Colicin sensitivity of W3110 cells harboring the pTA or pG3 variants was checked during exponential growth phase in LB medium. W3110pTA and W3110pG3 cells as well as their respective derivatives were induced with AHT (20 ng · ml−1) and IPTG (100 μM), respectively, for 30 min, resuspended in LB medium, and further incubated for 90 min in the presence of various amounts of colicins A, E1, and E9. The percentage of surviving cells was calculated from the turbidity ratio of colicin-treated cells to untreated cells (9), and the lethal doses were deduced from the survival curves.
Filamentous phage infection.
These tests were performed using the tetracycline-resistant Ff phage cloning vector fd-Tc (30). Briefly, GM1 cells expressing the pG3 variants were grown to exponential growth phase and diluted in LB medium in order to count about 100 to 400 colonies per LB plate. Then, fd-Tc phages were added to the cell suspension and incubated for 15 min at 20°C without shaking and then for 30 min at 37°C with shaking. Infection frequencies are reported as the number of CFU of infected bacteria (selected on ampicillin plus tetracycline plates) versus the number of CFU of total bacteria (selected on ampicillin plates). However, we observed that GM1 cells harboring the pIBA plasmid (or the pTA derivatives) were not infected even in the absence of the AHT inducer.
Overlay immunodetection.
One microliter of serial twofold dilutions of purified TolAIII was spotted onto a nitrocellulose membrane. Membranes were blocked for one hour at room temperature in phosphate-buffered saline (PBS) buffer containing 3% bovine serum albumin and 0.5% Tween-20 and washed three times for 5 min with PBS-0.1% Tween. Then, membranes were incubated for 1 h with 200 nM of the protein of interest (TA or G3 variants) in PBS buffer at room temperature (RT) and washed two times in PBS buffer. TA and G3 variant binding activity was revealed by using Strep-tactin alkaline phosphatase (IBA) as recommended by the manufacturer for 1 h in PBS buffer containing 0.1% Tween.
In vivo cross-linking with paraformaldehyde and copurification.
Cells harboring various plasmids were grown to exponential phase in LB medium, and expression of the variants was induced with IPTG (100 μM for pG3 derivatives) at 30°C for 2 h or AHT (20 ng · ml−1 for pTA derivatives) at 37°C for 1 h. Then, cells were washed and resuspended in 10 mM sodium phosphate buffer (pH 6.8) and incubated for 20 min with 1% formaldehyde. Proteins were solubilized in TES (10 mM Tris-HCl, pH 7.2, 5 mM EDTA, 1% SDS) and immunoprecipitated using anti-TolA antibody (14) or adsorbed onto magnetic Strep-tactin beads. For copurification using magnetic Strep-tactin beads (IBA), the soluble TES fraction was diluted 20- to 100-fold in TNET (Tris-HCl 10 mM, pH 7.2, 5 mM EDTA, 150 mM NaCl, 0.1% Triton X-100) and incubated for 30 min at RT with the magnetic beads and then washed three times with Magbuffer W/I (IBA). Strep-tagged proteins and copurified partners were eluted with 20 μl of Magbuffer E (IBA) and analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE). Differently, copurification using magnetic Strep-tactin beads was performed using purified proteins in the absence of cross-linking agent, as recommended for the purification of Strep-tagged proteins (IBA).
SDS-PAGE shift assay.
A gel shift assay was performed as previously described using SDS-PAGE (24) or Tricine-SDS-PAGE (46). Briefly, proteins were incubated in 125 mM Tris-HCl (pH 8.0) and 5% glycerol for 30 min at RT. Samples were directly loaded and separated by electrophoresis (10 V · cm−1). Proteins and complexes of low molecular mobility were detected after Coomassie blue staining.
Attenuated total reflection-FTIR.
Spectra were obtained with a Bruker equinox 55 Fourier transform infrared spectroscopy (FTIR) spectrometer coupled with an attenuated total reflection device. For each protein spectrum (protein concentrations were between 20 and 30 μM), 150 scans were collected at room temperature with a resolution of 2 cm−1. Data were analyzed with OPUS software (Bruker). The buffer solution used for dialysis was measured under identical conditions and subtracted from the individual protein spectra. Spectra were fitted with Lorentzian functions.
RESULTS
Construction and periplasmic production of the colicin A and g3p derivatives.
The translocation domain of colicin A contains three binding sequences involved in interactions with the TolA, TolB, and TolR proteins (31). To get specific information about the TolA binding region, the N-terminal sequence containing the consensus TolB box and the TolR binding sequences was removed (Fig. 1A). The DNA fragment encoding the truncated translocation domain of colicin A (TA) was cloned downstream of the DNA sequence encoding the OmpA signal sequence, giving the pTA plasmid. This protein is produced in the periplasm and still interacts with TolA (see below). Since previous analyses suggested that two TolA binding motifs might be present in the colicin A translocation domain (17), we constructed four derivatives by quick-change mutagenesis. These constructs express the TA domains deleted of the different regions (pTAΔpygr, pTAΔsynt) or carrying point mutations within these motifs (pTAy58a, pTAy90a). Correspondingly, the DNA fragment encoding the g3pN1 domain from M13 phage (G3) was cloned, giving pG3 (Fig. 1B), and two derivatives, pG3Δygt and pG3y54a, encoding G3 with a deletion of the YGT residues and the Y54A point mutation, respectively, were constructed.
FIG. 1.
Amino acid sequences of the G3 and TA peptides. The recombinant colicin A translocation domain (TA) and g3pN1 (G3) are shown in panels A and B, respectively. The N-terminal OmpA signal sequences (lowercase italic letters), the Strep-tagged residues (capital and shaded boldface letters), TolA binding region of TA (31), and the residues of G3 in contact with TolA (41) are indicated (capital boldface letters) together with the first and last residues of the natural sequences of g3pN1 and colicin A translocation domain (capital and shaded letters). The mutations corresponding to residue deletions (capital and underlined boldface letters) and to Tyr-to-Ala mutations are shown (Tyr numbering is relative to the natural sequence of g3p and colicin A). The N-terminal residues of the translocation domain of colicin A, absent from the TA variants, containing the TolR binding sequence (lowercase letters) followed by a TolB box [consensus sequence: DG(T/S)G(S/W)] is shown in the panel A insert.
Initial studies showed that all the variants are detected by using Strep-tactin revelation (Fig. 2A, lower panels). However, the levels of G3 and variants were found to be similar, but the TA level was in excess compared to those of TAy58a, TAy90a, and TAΔpygr, while TAΔsynt was faintly detected. Quantitative estimations using sample dilutions of purified proteins and of bacterial culture extracts revealed that TAy58a and G3Δygt were present at 30,000 to 50,000 copies per cell (not shown). Thus, G3, TA, and their respective variants except TAΔsynt, are found in great excess compared to TolA molecules previously estimated at about 600 copies per cell (37). As expected, fractionation experiments showed that WT G3 and TA domains and their variants are exported in the periplasm (data not shown).
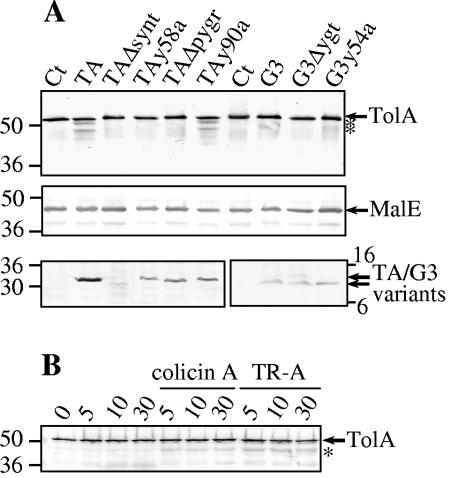
FIG. 2.
In vivo stability of TolA. (A) W3110 cells producing the indicated proteins were loaded on SDS-PAGE gels and transferred onto a nitrocellulose membrane. Immunoblots were revealed with anti-TolAII-III or anti-MalE antibodies or using Strep-tactin colorimetric detection for the TA and G3 variants. (B) W3110 cells were incubated in 10 mM potassium phosphate buffer (pH 7.1) in the absence or presence of colicin A or of the TR-truncated colicin A (TR-A, translocation and reception domain of colicin A) and harvested at the indicated times (min). About 2 × 108 cells were loaded on each lane. Molecular weight markers are indicated together with the degradation products of TolA (*). Ct, control empty vector.
Cellular effects of mutated domains.
We tested the consequences of the periplasmic production of the various g3p peptides (G3, G3Δygt, and G3y54a) by measuring sensitivities to colicins (colicins A, E1, and E9), fd phage infection, release of periplasmic RNase I, and bacterial growth in the presence of SDS. Deoxycholate sensitivity was also found to correlate the SDS results found with the G3 variants (not shown). As shown in Table 1, cells producing the WT G3 domain were tolerant to Tol-dependent colicins except colicin E1 (which used TolC and only the TolA-TolQ proteins), and they presented characteristic tol membrane defects. Interestingly, no effect upon production of the G3Δygt domain was detected, and the G3y54a-producing cells presented phenotypes similar to those of G3-producing cells. Cells remained sensitive to colicin E1 but to a lesser extent in the case of G3- and G3y54a-producing cells. Moreover, we infected the various G3-producing cells with the fd-Tc phage. The infection frequencies, expressed as ratios of infected bacteria (selected on ampicillin-tetracycline plates) to total bacteria harboring the control (pIN), pG3, pG3ygt, and pG3y54a plasmids (selected on ampicillin plates), corresponded to 100, 15, 95, and 75%, respectively. These data suggest that deletion of the YGT motif of G3 abolishes its competitive effect.
TABLE 1.
Cellular effects of G3 and TA variantsd
| Plasmid harbored | Sensitivitya(10−4) to colicin: | RNase I releaseb | Bacterial survival in SDSc(%) | ||
|---|---|---|---|---|---|
| E1 | A | E9 | |||
| pIN | 0.2 | 2.0 | 2.0 | − | 95 |
| pG3 | 0.8 | >1,000 | >1,000 | + | 20 |
| pG3Δygt | 0.2 | 2.0 | 4.0 | − | 90 |
| pG3y54a | 0.8 | >1,000 | >1,000 | + | 20 |
| pIBA | 0.3 | 2.0 | 1.5 | − | 90 |
| pTA | 0.5 | 40 | >1,000 | + | 50 |
| pTAΔsynt | 0.4 | 3.0 | 1.0 | − | 90 |
| pTAy58a | 0.3 | 3.0 | 2.0 | − | 70 |
| pTAΔpygr | 0.5 | 3.0 | 1.5 | − | 75 |
| pTAy90a | 0.5 | 80 | 1,000 | + | 50 |
We also checked the cellular effects of the TA variants (Table 1) and observed that TA and TAy90a have a protective role against colicins E9 and A, while no protection was observed when the two TA deletions and the TAy58a variant were produced. Similarly to G3, no significant protection against colicin E1 was observed from cells producing the TA variants. Hypersensitivity to SDS and RNase I release were also obtained with cells producing the TA and TAy90a variants, intermediate SDS effects were observed in cells producing TAy58a and TAΔpygr, and no effect was observed in TAΔsynt-producing cells. These results demonstrate the importance of both motifs and particularly of the Tyr 58 residue in SYNT. Finally, the two point mutations, Y58A and Y90A, were introduced into the whole colicin A gene. The WT colicin A and the two colicin A derivatives were recovered from cell supernatants, and their killing activities were assessed by spotting serial dilutions on sensitive bacterial lawns. According to the colicin concentrations estimated by SDS-PAGE and Coomassie blue staining, the results demonstrated that colicin A (y90a) was as active as the WT colicin A, while colicin A (y58a) had a 10- to 15-fold lower activity (data not shown). This result agrees with the observed effects upon periplasmic production of the corresponding translocation domain.
In vivo analyses of TolA interactions.
We then tested whether complexes between TolA and the various derivatives were detectable in living cells. For this purpose, we performed both coimmunoprecipitations using anti-TolA antibody and copurification using Strep-tactin affinity beads. As a prerequisite step, whole cells were treated with formaldehyde since we used detergent to solubilize membrane proteins. Then, copurification and coimmunoprecipitation experiments were performed on the total extracted material. First, using the Strep-tactin beads, strep-tagged peptides were precipitated, and cofractionation of TolA was faintly detected upon precipitation of WT TA- or G3-producing cells (not shown). According to the low binding efficiency of TA and G3 cross-linked peptides on Strep-tactin or nickel beads and to nonspecific adsorption of TolA on the beads, the in vivo copurifications of TolA with the TA and G3 variants were not ascertainable. Second, using cells harboring the control plasmid, immunoprecipitation of TolA was found to pull down Pal as previously observed (14). Pal was equally well coimmunoprecipitated with TolA in W3110 cells producing the various peptides. These results indicate that TA, G3, and their variants do not disturb the TolA-Pal interaction (not shown). Using similar analyses, we checked for TA or G3 peptide detection after TolA immunoprecipitation. However, we were not able to detect strep-tagged TA or G3 in the coimmunoprecipitated material (not shown). According to the immunodetections of TolA, it appeared that expression of some peptides induced the apparition of TolA degradation products. Thus, we checked TolA stability in the absence of any cross-linking agent in W3110 cells producing the various TA and G3 variants (Fig. 2A). After 30 min of induction, we observed that TA and TAy90a specifically induced the apparition of TolA degradation products while MalE, as a periplasmic protein control, was not affected. We then checked for the physiological relevance of this observation. TolA stability was followed in living WT W3110 cells incubated with the colicin A or a variant devoid of the C-terminal active domain named TR-A. Figure 2B shows that both molecules induce TolA degradation even after 5 min of incubation.
Peptide purification and FTIR analysis.
The various TA and G3 variants, carrying a hexahistidine tag at the C terminus and an N-terminal Strep-tag, were purified using a two-step (immobilized metal affinity chromatography and Strep-tactin) affinity chromatography procedure. FTIR was performed on the WT and each variant to compare the amide I bands reflecting the protein backbone conformation. Wavelength peaks of the variants obtained by deconvolution analyses were found to be similar to the corresponding WT TA or WT G3 domains except for TAΔsynt (Table 2). These results indicate that the various deletions or point mutations except TAΔsynt do not alter the secondary structures of G3 and TA domains. Because the structure of the g3pN1 domain depends on the formation of two disulfide bridges, we performed mass spectrometry analyses on WT and G3 purified variants. Electrospray analyses gave molecular mass values of 10,423, 10,103, and 10,333 Da, corresponding to the theoretical oxidized masses of G3 (10,425 Da), G3Δygt (10,104 Da), and G3y54a (10,333 Da) monomers, respectively. Thus, we conclude that the different mutations do not modify the global structure of the G3 domains and do not induce multimer formation due to inadequate cysteine oxidization.
TABLE 2.
Results of FTIR analyses of TA and G3 derivativesa
| Peak (cm−1) | Surface area of variant: | |||||||
|---|---|---|---|---|---|---|---|---|
| TA | G3 | |||||||
| WT | Δsynt | y58a | Δpygr | y90a | WT | Δygt | y54a | |
| 1,631-1,637 | 51 | 23 | 46 | 47 | 51 | 39 | 44 | 41 |
| 1,654 | 34 | 63 | 39 | 40 | 33 | 40 | 37 | 38 |
| 1,675 | 15 | 14 | 15 | 14 | 16 | 21 | 19 | 21 |
In vitro interactions of the mutated domains with TolA.
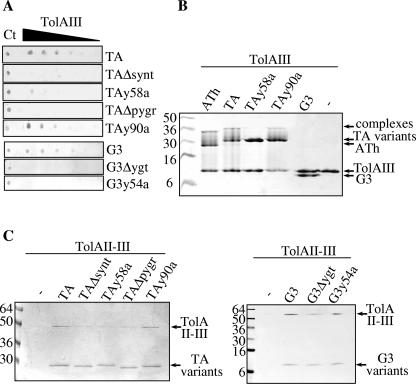
Since no comparison of the interaction of TolA with the N-terminal domains of g3p and colicins has been described, various techniques were used to analyze TolA binding with TA or G3 and their derivatives. First, we used an overlay technique in which one protein is spotted on the nitrocellulose filter and the binding of a second interacting protein is recognized by a specific antibody directed against the latter. Figure 3A shows that TolAIII is detected using TA, TAy90a, and G3 peptide domains but faintly detected using TAy58a. No signal was observed using TAΔpygr, TAΔsynt, and the two G3 variants. Second, we performed gel shift experiments using SDS-PAGE analyses (Fig. 3B). As a control, we observed that ATh (the whole translocation domain containing the TolA, -B, and -R binding regions) forms a complex with TolAIII that slowly dissociates during electrophoresis, as previously described (21). Furthermore, TA and TAy90a form protein complexes with TolAIII, while no complex was detected using TAy58a and G3 domains (Fig. 3B). Similarly, a complex involving TA but no complex with G3 was found using TolAII-III (the whole periplasmic domain of TolA) instead of TolAIII (not shown). Finally, we conducted in vitro pull-down experiments using purified TolAII-III and Strep-tagged domains. TolAII-III was copurified with TA, TAy90a, or G3 peptides, while a weak TolAII-III signal was detected using one of the deleted variants, TAy58a or G3y54a (Fig. 3C). Similar results were obtained when crude extracts of TolAII-III-producing cells were used instead of purified TolAII-III (not shown). In summary, we demonstrated that the Tyr 58 residue of TA is an important determinant for TolA binding since its replacement with an Ala residue abolished the in vitro TolA-TA interaction. Correspondingly, deletion of the YGT motif within G3 affected the interaction with TolA. All these results confirm and extend the findings of the in vivo analyses presented above.
FIG. 3.
In vitro interaction of TA and G3 variants with TolA. (A) Overlay immunodetections of TA and G3 variants bound to immobilized TolAIII. Serial twofold dilutions of TolAIII (3.0, 1.5, 0.8, 0.4, 0.2, and 0.1 μg for TA and 6.0, 3.0, 1.5, 0.8, 0.4, and 0.2 μg for G3) were spotted on a nitrocellulose membrane, incubated with the indicated TA or G3 variants, and further detected using Strep-tactin-coupled alkaline phosphatase. Ct indicates the TA or G3 proteins (1.0 μg for TA and 3.0 μg for G3) used for the control of colorimetric detection levels. (B) Gel shift analyses of TolAIII complexes. Purified TA, TA variants, ATh (the whole translocation domain of colicin A), and G3 were incubated with TolAIII and analyzed by Tricine-SDS-PAGE and Coomassie blue staining. (C) Copurifications of TolAII-III with TA or G3 variants. Copurifications of TolAII-III by Strep-tactin pull-down experiments of TA, G3, and variants were analyzed by Coomassie blue staining of SDS-PAGE (TA) or Tricine-SDS-PAGE (G3). Molecular weight markers are indicated.
In vitro analyses of trimeric complexes containing TolA.
Competitive interactions or trimeric complex formations were assayed by in vitro pull-down experiments in the absence of cross-linker. Copurification experiments were performed using the purified Strep-tag TA or G3 domains with either TolAII-III, TolAIII, g3pN1 (G3 protein devoid of the Strep-tag), and TolB or Pal. Proteins were incubated together at concentrations between 2 and 6 μM (according to their theoretical extinction coefficients), and pull-down experiments using Strep-tactin-coated magnetic beads were performed with the various protein mixtures. Figure 4A shows that Pal has no influence on the TA-TolAII-III complex since a fivefold molar excess of Pal did not prevent the TA-TolAII-III interaction. Similarly, pull-down experiments indicate that the TolAII-III-G3 complex is not influenced by the presence of a fivefold molar excess of Pal (not shown). Using the same purification procedure, we observed that Strep-tagged TolAIII was able to coprecipitate Pal in the absence of TA or G3 peptides (not shown).
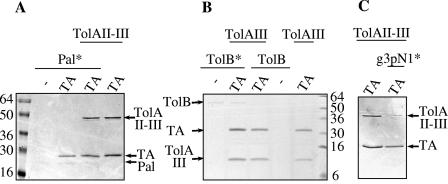
FIG. 4.
TolA-TA complex formation in the presence of excess of Pal, g3pN1, or TolB. TA was incubated with the indicated proteins (concentrations, between 2 and 6 μM). Copurification of TolAII-III (A and C) or TolAIII (B) was assessed after TA pull-down assays using Strep-tag. When indicated, Pal (A), TolB (B), or g3pN1 (G3 devoid of Strep-tag) (C) was added to the mixture. An asterisk indicates that a 5-molar excess of the indicated protein was used. After SDS-PAGE (A and C) or Tricine-SDS-PAGE (B), proteins were detected by Coomassie blue staining. Molecular weight markers are indicated on the left.
TolA also interacts with TolB (49), and both proteins have been shown to interact with the colicin A translocation domain (10). The effect of TolB on TA-TolA complex formation was analyzed. Since TolB migrates exactly at the position of TolAII-III, we used the TolAIII protein for the experiments. Even with a fivefold molar excess of TolB, the TA-TolAIII complex was not displaced (Fig. 4B). We thus conclude that TolB has no competitive effect on the TolAIII-TA complex. Finally, since TA and G3 pull down TolAII-III, we monitored competition experiments. For this purpose, we used the TA, TolAII-III, and g3pN1 domains and observed that the TA-TolAII-III complex is disrupted by an excess of g3pN1. This result showed that TA and G3 do compete for TolA (Fig. 4C).
DISCUSSION
A TolA box is present in colicin A and g3p of Ff phages.
The Tol-Pal system forms a transenvelope bridge linking the inner and outer membranes and the peptidoglycan layer through multiple interactions maintaining cell envelope integrity. As for TolA-Q-R, the TonB-ExbB-D complex links the inner and outer membranes through TonB-receptor interactions (44) to energize the active transport of Fe3+ siderophores and cyanocobalamin. Both systems are able to transduce energy from the inner membrane through the periplasm to outer membrane-associated proteins (15, 27, 39). A conserved pentapeptide sequence has been found in various TonB-dependent outer membrane receptors and in the N-terminal translocation domain of group B colicins (12). This motif, called the TonB box, is involved in the direct interaction of BtuB, FhuA, and colicin D with the TonB protein (13, 42, 43). Similarly, we have previously found a TolA binding motif of four conserved residues, SYGK/E, located in the C-terminal region of Pal (17). A similar motif is present in two other TolA-interacting proteins, g3p and colicin A. In relation to the well-described TonB box found in the TonB-dependent receptors and in the group B colicins (12), we suspected that the TolA motif could be a determinant for Tol-dependent import processes (17). The 3D structure of the g3pN1-TolAIII cocrystal reveals that the YGT residues of the g3p N-terminal domain are in close contact with TolA (Fig. 5C) (41). It is noteworthy that the Tyr residues of the SYGK motif of Pal and of the YGT motif of g3pN1 are located in a beta-strand structure (Fig. 5). Moreover, the side chains of the Tyr residues of Pal and g3p are exposed outside of the molecules and may be available to form hydrogen bonds. In this study, we analyzed the function of the YGT residues of g3pN1 and of the two potential TolA binding motifs of colicin A, SYNT and PYGR, located between residues 52 and 97 of the TolA binding sequence (31). We used G3 or TA variants carrying a deletion of the peptide motifs or a Tyr-to-Ala substitution. Colicin tolerance and outer membrane defects were previously shown to be induced in WT cells by the periplasmic production of the N-terminal domains of colicin A and E3 (9, 10) as well as g3pN1 (28, 48). Here, we observed that in the absence of the TolB box and TolR binding sequence, TA peptide induced tol phenotypes. As previously shown (10), the TolA binding sequence of TA is necessary and sufficient to destabilize the Tol-Pal complex. One important aspect concerns the ability of periplasmic TA to protect cells against colicins, and this peptide was found to be at least 25 times more active to protect cells against colicin E9 than colicin A. This may reflect the affinity of the interaction of the colicin translocation domains to TolA, indicating a higher binding affinity of colicin A than colicin E9 to TolA. Since the effects of TAy90a production were similar to those induced by TA production, our results suggest that TAy90a still interacts with TolA. Consequently, the absence of a cellular effect associated with the periplasmic production of TAy58a reflects the absence of interaction with TolA. This result is in agreement with the observation that TAy58a does not have any effect on TolA stability, while degradation products are observed when TA or TAy90a is produced. Mutations introduced into the whole colicin A molecule further confirmed the role of the Tyr 58 residue in the import process. Similarly, G3 production induced membrane destabilization, and we observed periplasmic release, detergent hypersensitivity together with colicin tolerance, and a decreased level of fd phage infection. Moreover, as observed with TA, the periplasmic production of G3 protects cells from exogenous colicin A and E9. According to these results, G3 induces stronger effects than TA. Production of G3Δygt did not show any effect. This result correlates with previous findings because the YGT motif is located in a region bearing the determinants suspected to confer colicin tolerance and inhibition of phage infection (residues 53 to 99 [48]). However, the Tyr-to-Ala mutation of G3 did not suppress G3 activity, suggesting that G3y54a was still able to bind to TolA. Overall, these data suggest that deletion of specific motifs within TolA partner sequences abolished their interaction with TolA. We thus conducted assays to detect the G3-TolA and TA-TolA interactions by various in vivo or in vitro approaches. In vivo cross-linking experiments and purifications with the Strep-tactin beads were not efficient to confirm the binding to TolA. By a similar approach, anti-TolA antibodies coimmunoprecipitated Pal but were not useful to detect the TA or G3 domains in the eluted material. These latter results may depend on the TolA antibody which could not recognize the TolA-TA or TolA-G3 complexes or may be due to the low amounts of Strep-tagged peptides in interaction with the minor TolA protein. Further in vitro experiments using purified domains were performed and confirmed the interactions between TolA and TA and between G3 and TAy90a and the low binding affinities of the other derivatives. However, by using the gel shift technique, the TA-TolA complex was detected whereas the G3-TolA complex was not, suggesting a stronger affinity for the former complex in the presence of SDS. Kd values between 0.2 and 0.6 μM or between 1.0 and 1.9 μM had been determined by surface plasmon resonance analysis of the whole translocation domains of colicin A (24) or g3pN1 (32) and TolAIII, respectively. However, different binding modes could also explain these results, since both TolA and TA are basic molecules while G3 is acidic. Another example might be the structural disorder induced by the colicin A translocation domain to TolAIII but not by g3p upon complex formation (21). These hypotheses are also consistent with our in vivo observations of TolA stability in G3-producing cells while some degradation of TolA occurs exclusively in cells producing the TA- or TAy90a-interacting variants. According to the various analyses, we conclude that both TA and G3 contain a TolA box in which the Tyr 58 residue of SYNT in TA and the YGT residues in G3 are important determinants. However, our results show that the deletion of the PYGR motif of colicin A produced effects similar to those of the Y58A mutation, while mutation of the Tyr 90 residue has no effect on the Tol system. New analyses, such as alanine scanning mutagenesis and affinity constant determinations, remain to be performed to determine whether residues within both motifs are equally important. According to the recent observation demonstrating a concerted folding of colicin N upon binding to TolA (3, 4), new structural information on the TolA binding region of colicin N will be of interest to understand whether a precise motif is required. Indeed, an SYHIT sequence in which the Tyr residue plays an important role (3) was mapped within the TolA binding region of colicin N (residues 40 to 66). Besides colicin N, the Tol-dependent colicin K contains an SYLT sequence (positions 68 to 71) which could be involved in TolA binding. Figure 6 summarizes TolA box residue conservation between Pal, g3p, and colicins A, N, and K. Further experiments with WT cells producing colicin K and variants will confirm the presence of a TolA box. As previously found with colicins (11), the periplasmic production of G3 made bacteria tolerant to colicins and conferred tol membrane phenotypes, including the formation of elevated amounts of outer membrane vesicles (28). As for TA, the production of G3 did not induce bacterial tolerance to colicin E1. Although previous results demonstrated that the C-terminal domain of TolA is required for colicin E1 translocation (6), it has been shown that TolA interacts differently with colicin E1 than with other group A colicins (47). Accordingly, the continued activity of colicin E1 on cells producing the various peptides may reflect the use of another TolA C-terminal binding sequence.
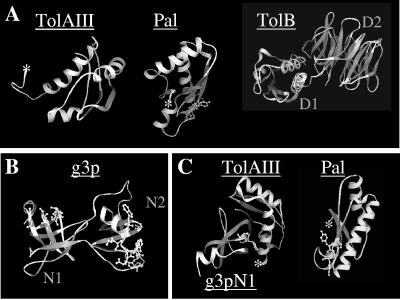
FIG. 5.
Structures of TolAIII and its interacting partners. (A) Crystal structures of TolAIII (Protein Data Bank [PDB] accession number 1S62 [22]), Pal (PDB accession number 1OAP [C. Abergel, A. Walburger, E. Bouveret, and J.-M. Claverie, unpublished results]), and TolB (PDB accession number 1CRZ [1]). The SYGK motif of Pal, involved in its interaction with TolA, is modeled with balls and sticks. (B) Three-dimensional structure of the two N-terminal domains of g3p (PDB accession number 1G3P [40]). Residues of G3pN1 involved in the interaction with TolAIII but not participating in the interaction with G3pN2 (residues E14, NATG42, and GT56 of the YGT motif) and residues of G3pN2 involved in the interaction with the F-pilus are indicated by balls and sticks. (C) The YGT residues of g3pN1 involved in the contact with TolA are shown in the cocrystal structure of the g3pN1-TolAIII complex (PDB accession number 1TOL) together with Pal and its SYGK motif. The N-terminal membrane-anchored sequences of Pal and TolA are indicated by an asterisk. Figures were generated with 3D Mol Viewer (Vector NTI Suite version 8.0).
g3p is a three domain protein in which the receptor binding domain (N2) interacts with the N-terminal domain (N1). Because most of the residues of g3pN1 interact with g3pN2 and also with TolAIII, it was suspected that the g3pN1-N2 interaction dissociates upon binding of g3pN2 to the pilus allowing g3pN1 to freely interact with TolAIII. However, among 21 interacting residues, seven residues interact with TolA exclusively (41). In the 3D structure of g3pN1-N2 (29, 40), these seven residues are exposed and in close contact in a beta-sheet structure. According to our results, we suggest that after g3p-pilus reception, the g3p-TolA interaction, mediated by the exposed Tyr-Gly-Thr residues, may help to dissociate the g3pN1-N2 interdomain interaction favoring formation of the TolA-g3pN1 complex in the periplasm. This model agrees with the mapping of the exposed TolA binding residues of g3pN1 separated from those required for pilus interaction in g3pN2 (20) (Fig. 5B) and with the observation that preincubation of the TolA C-terminal domain with phage particles does not affect the level of phage infection (19). Overall, our data contribute to the understanding of the two-step docking mechanism of Ff phages. However, how g3p crosses the outer membrane to reach TolA remains unknown.
Our results show that the conserved TolA binding motifs of Pal, g3p, and some group A colicins are reminiscent of the well-studied TonB box. Recently, two discriminating mutations within TonB, conferring colicin D tolerance but maintaining the FepA dependent siderophore uptake, were described (42). This result suggests that even with the same box involved in the binding of TonB to colicins and receptors, some specific determinants in TonB affect binding to the different ligands. In this study, we demonstrate that both TA and G3 compete for docking on TolA. Furthermore, as for TonB, different TolA determinants should be involved in TA, G3, or Pal binding. We are currently analyzing the regions of TolA involved in the various interactions to determine the residues of TolAIII required in the multiple binding activities.
Tol complexes and outer membrane integrity.
How does the outer membrane become unstable upon periplasmic production of TA and G3 variants? We can rule out the possibility that membrane instability is the result of TolA degradation. If we observed that TolA is degraded when interacting with TA, TolA remains stable when G3 is produced in the periplasm, which also leads to outer membrane defects. It remains to be understood whether the weak degradation of TolA observed in WT cells incubated with colicin A, or the colicin A devoid of the pore-forming domain, is the result of a direct interaction of imported molecules with TolA. In this study, we demonstrated that Pal does not compete with TA or G3 domains for binding to TolA. Thus, we suppose the existence of a trimeric complex TA(G3)-TolA-Pal in which TA (or G3) may prevent other protein interactions or modify the energy transduction process mediated by TolA. Similarly, we demonstrated that TolB did not inhibit TolAIII-TA complex formation. The presence of a trimeric complex formed with a cross-linked TolB-colicin A translocation domain interacting with TolA has been previously described (10). However, the colicin A translocation domain used in the cited reference possessed the TolB box and the trimeric complex that resulted from the colicin A translocation domain interacting with both TolA and TolB. Since we used TA deleted of the TolB box, the absence of any effect of TolB on the TA-TolA complex suggests that TolB has a lower binding affinity to TolA than TA. Thus, either the TolB-TolA-TA complex is formed but not observed in our analyses or TolB does not compete with TA due to a lower binding affinity. Then, the TolA-TolB interaction previously shown to be involved in cell integrity (26, 49) may be modified in cells producing TA or G3. Pull-down experiments using Strep-tagged TolAIII as well as surface plasmon resonance techniques will help us to confirm the presence of trimeric complexes and to get new information on multimeric complexes containing TolA.
Acknowledgments
We thank Jean-Pierre Duneau for help with the FTIR technique and analyses, Stéphane Bonetto for the gift of the fd-Tc phages, Colin Kleanthous (York, United Kingdom) for the gift of purified colicin E9, Anne Walburger for the gift of purified Pal, Régine Lebrun for matrix-assisted laser desorption ionization-time of flight mass spectrometry analyses, Valérie Prima for DNA sequencing, Denis Duché for careful reading of the manuscript, and anonymous referees for critiques of the manuscript.
This study was supported by the Centre National de la Recherche Scientifique, a grant from Vaincre la Mucoviscidose to R.L., and an MRT fellowship to S.P.
REFERENCES
- 1.Abergel, C., E. Bouveret, J.-M. Claverie, K. Brown, A. Rigal, C. Lazdunski, and H. Bénédetti. 1999. Structure of the Escherichia coli TolB protein determined by MAD methods at 1.95 A resolution. Struct. Fold Des. 7**:**1291-1300. [DOI] [PubMed] [Google Scholar]
- 2.Abergel, C., A. Walburger, S. Chenivesse, and C. Lazdunski. 2001. Crystallization and preliminary crystallographic study of the peptidoglycan-associated lipoprotein from Escherichia coli. Acta Crystallogr. 57**:**317-319. [DOI] [PubMed] [Google Scholar]
- 3.Anderluh, G., Q. Hong, R. Boetzel, C. MacDonald, G. R. Moore, R. Virden, and J. H. Lakey. 2003. Concerted folding and binding of a flexible colicin domain to its periplasmic receptor TolA. J. Biol. Chem. 278**:**21860-21868. [DOI] [PubMed] [Google Scholar]
- 4.Anderluh, G., I. Gokce, and J. H. Lakey. 2004. A natively unfolded toxin domain uses its receptor as a folding template. J. Biol. Chem. 279**:**22002-22009. [DOI] [PubMed] [Google Scholar]
- 5.Baty, D., M. Frenette, R. Lloubès, V. Geli, S. P. Howard, F. Pattus, and C. Lazdunski. 1988. Functional domains of colicin A. Mol. Microbiol. 2**:**807-811. [DOI] [PubMed] [Google Scholar]
- 6.Bénédetti, H., C. Lazdunski, and R. Lloubès. 1991. Protein import into E. coli: colicins A and E1 interact with a component of their translocation system. EMBO J. 10**:**1989-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernadac, A., M. Gavioli, J. C. Lazzaroni, S. Raina, and R. Lloubès. 1998. _Escherichia coli tol_-pal mutants form outer membrane vesicles. J. Bacteriol. 180**:**4872-4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bouveret, E., R. Derouiche, A. Rigal, R. Lloubès, C. Lazdunski, and H. Bénédetti. 1995. Peptidoglycan-associated lipoprotein-TolB interaction. J. Biol. Chem. 270**:**11071-11077. [DOI] [PubMed] [Google Scholar]
- 9.Bouveret, E., A. Rigal, C. Lazdunski, and H. Bénédetti. 1997. The N-terminal domain of colicin E3 interacts with TolB which is involved in the colicin translocation step. Mol. Microbiol. 23**:**909-920. [DOI] [PubMed] [Google Scholar]
- 10.Bouveret, E., A. Rigal, C. Lazdunski, and H. Bénédetti. 1998. Distinct regions of the colicin A translocation domain are involved in the interaction with TolA and TolB proteins upon import into E. coli. Mol. Microbiol. 27**:**143-157. [DOI] [PubMed] [Google Scholar]
- 11.Bouveret, E., L. Journet, A. Walburger, E. Cascales, H. Bénédetti, and R. Lloubès. 2002. Analysis of the E. coli Tol-Pal and TonB systems by periplasmic production of Tol, TonB, colicin, or phage capsid soluble domains. Biochimie 84**:**413-421. [DOI] [PubMed] [Google Scholar]
- 12.Braun, V., S. I. Patzer, and K. Hantke. 2002. Ton-dependent colicins and microcins: modular design and evolution. Biochimie 84**:**365-380. [DOI] [PubMed] [Google Scholar]
- 13.Cadieux, N., and R. J. Kadner. 1999. Site-directed disulfide bonding reveals an interaction site between energy-coupling protein TonB and BtuB, the outer membrane cobalamin transporter. Proc. Natl. Acad. Sci. USA 96**:**10673-10678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cascales, E., M. Gavioli, J. N. Sturgis, and R. Lloubès. 2000. Proton motive force drives the interaction of the inner membrane TolA and outer membrane Pal proteins in Escherichia coli. Mol. Microbiol. 38**:**904-915. [DOI] [PubMed] [Google Scholar]
- 15.Cascales, E., R. Lloubès, and J. N. Sturgis. 2001. The TolQ-TolR proteins energise TolA and share homologies with the flagellar motor proteins MotA-MotB. Mol. Microbiol. 42**:**795-807. [DOI] [PubMed] [Google Scholar]
- 16.Cascales, E., A. Bernadac, M. Gavioli, J. C. Lazzaroni, and R. Lloubès. 2002. Pal lipoprotein plays a major role for outer membrane integrity. J. Bacteriol. 184**:**754-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cascales, E., and R. Lloubès. 2004. Deletion analyses of the peptidoglycan-associated lipoprotein Pal reveals three independent binding sequences including a TolA box. Mol. Microbiol. 51**:**873-885. [DOI] [PubMed] [Google Scholar]
- 18.Clavel, T., P. Germon, A. Vianney, R. Portalier, and J. C. Lazzaroni. 1998. TolB protein of Escherichia coli K-12 interacts with the outer membrane peptidoglycan-associated proteins Pal, Lpp and OmpA. Mol. Microbiol. 29**:**359-367. [DOI] [PubMed] [Google Scholar]
- 19.Click, E. M., and R. E. Webster. 1997. Filamentous phage infection: required interactions with the TolA protein. J. Bacteriol. 179**:**6464-6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deng, L.-W., and R. N. Perham. 2002. Delineating the site of interaction on the pIII protein of filamentous bacteriophage fd with the F-pilus of Escherichia coli. J. Mol. Biol. 319**:**603-614. [DOI] [PubMed] [Google Scholar]
- 21.Deprez, C., L. Blanchard, F. Guerlesquin, M. Gavioli, J. P. Simorre, C. Lazdunski, D. Marion, and R. Lloubès. 2002. Macromolecular import into Escherichia coli: the TolA C-terminal domain changes conformation when interacting with the colicin A toxin. Biochemistry 41**:**2589-2598. [DOI] [PubMed] [Google Scholar]
- 22.Deprez, C., R. Lloubès, M. Gavioli, D. Marion, F. Guerlesquin, and L. Blanchard. 2005. Solution structure of the E. coli TolA C-terminal domain reveals conformational changes upon binding to the phage g3p N-terminal domain. J. Mol. Biol. 346**:**1047-1057. [DOI] [PubMed] [Google Scholar]
- 23.Derouiche, R., H. Bénédetti, J.-C. Lazzaroni, C. Lazdunski, and R. Lloubès. 1995. Protein complex within E. coli inner membrane. TolA N-terminal domain interacts with TolQ and TolR proteins. J. Biol. Chem. 270**:**11078-11084. [DOI] [PubMed] [Google Scholar]
- 24.Derouiche, R., G. Zeder-Lutz, H. Benedetti, M. Gavioli, A. Rigal, C. Lazdunski, and R. Lloubès. 1997. Binding of colicins A and E1 to purified TolA domains. Microbiology 143**:**3185-3192. [DOI] [PubMed] [Google Scholar]
- 25.Derouiche, R., R. Lloubès, S. Sasso, H. Bouteille, R. Oughideni, C. Lazdunski, and E. Loret. 1999. Circular dichroism and molecular modeling of the E. coli TolA periplasmic domains. Biospectroscopy 5**:**189-198. [DOI] [PubMed] [Google Scholar]
- 26.Dubuisson, J. F., A. Vianney, and J.-C. Lazzaroni. 2002. Mutational analysis of the TolA C-terminal domain of Escherichia coli and genetic evidence for an interaction between TolA and TolB. J. Bacteriol. 184**:**4620-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Germon, P., M. C. Ray, A. Vianney, and J.-C. Lazzaroni. 2001. Energy-dependent conformational change in the TolA protein of Escherichia coli involves its N-terminal domain, TolQ, and TolR. J. Bacteriol. 183**:**4110-4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henry, T., S. Pommier, L. Journet, A. Bernadac, J.-P. Gorvel, and R. Lloubès. 2004. Improved methods for producing outer membrane vesicles in gram-negative bacteria. Res. Microbiol. 155**:**437-446. [DOI] [PubMed] [Google Scholar]
- 29.Holliger, P., L. Riechmann, and R. L. Williams. 1999. Crystal structure of the two N-terminal domains of g3p from filamentous phage fd at 1.9 A: evidence for conformational lability. J. Mol. Biol. 288**:**649-657. [DOI] [PubMed] [Google Scholar]
- 30.Hoogenboom, H. R., A. D. Griffiths, K. S. Johnson, D. J. Chiswell, P. Hudson, and G. Winter. 1991. Multi-subunit proteins on the surface of filamentous phage: methodologies for displaying antibody (Fab) heavy and light chains. Nucleic Acids Res. 19**:**4133-4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Journet, L., E. Bouveret, A. Rigal, R. Lloubès, C. Lazdunski, and H. Bénédetti. 2001. Import of colicins across the outer membrane of E. coli involves multiple protein interactions in the periplasm. Mol. Microbiol. 42**:**331-344. [DOI] [PubMed] [Google Scholar]
- 32.Karlsson, F., C. A. Borrebaeck, N. Nilsson, and A. C. Malmborg-Hager. 2003. The mechanism of bacterial infection by filamentous phages involves molecular interactions between TolA and phage protein 3 domains. J. Bacteriol. 185**:**2628-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lazdunski, C., E. Bouveret, A. Rigal, L. Journet, R. Lloubès, and H. Bénédetti. 1998. Colicin import into Escherichia coli cells. J. Bacteriol. 180**:**4993-5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lazzaroni, J.-C., and R. Portalier. 1981. Genetic and biochemical characterization of periplasmic-leaky mutants of Escherichia coli K-12. J. Bacteriol. 145**:**1351-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lazzaroni, J.-C., A. Vianney, J.-L. Popot, H. Bénédetti, F. Samatey, C. Lazdunski, R. Portalier, and V. Geli. 1995. Transmembrane α-helix interactions are required for the functional assembly of the E. coli Tol complex. J. Mol. Biol. 246**:**1-7. [DOI] [PubMed] [Google Scholar]
- 36.Lazzaroni, J.-C., J.-F. Dubuisson, and A. Vianney. 2002. The Tol proteins of Escherichia coli and their involvement in the translocation of group A colicins. Biochimie 84**:**391-397. [DOI] [PubMed] [Google Scholar]
- 37.Levengood, S., W. Beyer, and R. E. Webster. 1991. TolA, a membrane protein involved in colicin uptake contains an extended helical region. Proc. Natl. Acad. Sci. USA 88**:**5939-5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lloubès, R., D. Baty, and C. Lazdunski. 1986. The promoters of the genes for colicin production, release and immunity in the ColA plasmid: effects of convergent transcription and LexA protein. Nucleic Acids Res. 14**:**2621-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lloubès, R., E. Cascales, A. Walburger, A. Bouveret, C. Lazdunski, A. Bernadac, and L. Journet. 2001. The Tol-Pal proteins of the Escherichia coli cell envelope: an energized system required for outer membrane integrity? Res. Microbiol. 152**:**523-529. [DOI] [PubMed] [Google Scholar]
- 40.Lubkowski, J., F. Hennecke, A. Pluckthun, and A. Wlodawer. 1998. The structural basis of phage display elucidated by the crystal structure of the N-terminal domains of g3p. Nat. Struct. Biol. 5**:**140-147. [DOI] [PubMed] [Google Scholar]
- 41.Lubkowski, J., F. Hennecke, A. Plückthun, and A. Wlodawer. 1999. Filamentous phage infection: crystal structure of g3p in complex with its coreceptor, the C-terminal domain of TolA. Structure 7**:**711-722. [DOI] [PubMed] [Google Scholar]
- 42.Mora, L., N. Diaz, R. H. Buckingham, and M. de Zamaroczy. 2005. Import of the transfer RNase colicin D requires site-specific interaction with the energy-transducing protein TonB. J. Bacteriol. 187**:**2693-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peacock, S. R., A. M. Weljie, S. P. Howard, F. D. Price, and H. J. Vogel. 2005. The solution structure of the C-terminal domain of TonB and interaction studies with TonB box peptides. J. Mol. Biol. 345**:**1185-1197. [DOI] [PubMed] [Google Scholar]
- 44.Postle, K., and R. J. Kadner. 2003. Touch and go: tying TonB to transport. Mol. Microbiol. 49**:**869-882. [DOI] [PubMed] [Google Scholar]
- 45.Riechmann, L., and P. Holliger. 1997. The C-terminal domain of TolA is the coreceptor for filamentous phage infection of E. coli. Cell 90**:**351-360. [DOI] [PubMed] [Google Scholar]
- 46.Schagger, H., and G. Von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166**:**369-379. [DOI] [PubMed] [Google Scholar]
- 47.Schendel, S. L., E. M. Click, R. E. Webster, and W. A. Cramer. 1997. The TolA protein interacts with colicin E1 differently than with other group A colicins. J. Bacteriol. 179**:**3683-3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stengele, I., P. Bross, X. Garces, J. Giray, and I. Rasched. 1990. Dissection of functional domains in phage fd adsorption protein. Discrimination between attachment and penetration sites. J. Mol. Biol. 212**:**143-149. [DOI] [PubMed] [Google Scholar]
- 49.Walburger, A., C. Lazdunski, and Y. Corda. 2002. The Tol-Pal system function requires an interaction between the C-terminal domain of TolA and the N-terminal domain of TolB. Mol. Microbiol. 44**:**695-708. [DOI] [PubMed] [Google Scholar]
- 50.Webster, R. 1991. The tol gene products and the import of macromolecules into E. coli. Mol. Microbiol. 5**:**1005-1011. [DOI] [PubMed] [Google Scholar]