Support for the Homeobox Transcription Factor Gene ENGRAILED 2 as an Autism Spectrum Disorder Susceptibility Locus (original) (raw)
Abstract
Our previous research involving 167 nuclear families from the Autism Genetic Resource Exchange (AGRE) demonstrated that two intronic SNPs, rs1861972 and rs1861973, in the homeodomain transcription factor gene ENGRAILED 2 (EN2) are significantly associated with autism spectrum disorder (ASD). In this study, significant replication of association for rs1861972 and rs1861973 is reported for two additional data sets: an independent set of 222 AGRE families (rs1861972_–_rs1861973 haplotype, _P_=.0016) and a separate sample of 129 National Institutes of Mental Health families (rs1861972_–_rs1861973 haplotype, _P_=.0431). Association analysis of the haplotype in the combined sample of both AGRE data sets (389 families) produced a P value of .0000033, whereas combining all three data sets (518 families) produced a P value of .00000035. Population-attributable risk calculations for the associated haplotype, performed using the entire sample of 518 families, determined that the risk allele contributes to as many as 40% of ASD cases in the general population. Linkage disequilibrium (LD) mapping with the use of polymorphisms distributed throughout the gene has shown that only intronic SNPs are in strong LD with rs1861972 and rs1861973. Resequencing and association analysis of all intronic SNPs have identified alleles associated with ASD, which makes them candidates for future functional analysis. Finally, to begin defining the function of EN2 during development, mouse En2 was ectopically expressed in cortical precursors. Fewer _En2_-transfected cells than controls displayed a differentiated phenotype. Together, these data provide further genetic evidence that EN2 might act as an ASD susceptibility locus, and they suggest that a risk allele that perturbs the spatial/temporal expression of EN2 could significantly alter normal brain development.
Introduction
Individuals diagnosed with autism spectrum disorder (ASD [MIM 209850]) exhibit deficiencies in communication and reciprocal social interactions that are often accompanied by restricted or repetitive interests and behaviors. Autopsy and neuroimaging studies suggest that ASD is caused in part by abnormal brain development (Bauman and Kemper 1985, 1986, 1994; Ritvo et al. 1986; Gaffney et al. 1987; Courchesne et al. 1988, 2001, 2003; Murakami et al. 1989; Kleiman et al. 1992; Kemper and Bauman 1993; Hashimoto et al. 1995; Courchesne 1997; Bailey et al. 1998). Twin, family, and disease-modeling studies support a polygenic multifactorial basis in the etiology of ASD (Folstein and Rutter 1977; Ritvo et al. 1985; Risch et al. 1999; Folstein and Rosen 2001; Liu et al. 2001; Alarcon et al. 2002; Auranen et al. 2002).
The CNS structure most consistently affected in individuals with autism is the cerebellum. In 24 of 29 autopsy samples from different individuals, cerebellar defects have been reported, with a decrease in the number of Purkinje cells being present in 23 of these 24 samples. Neurodegenerative signs are mostly absent from these samples, which suggests that these phenotypes are developmental (Bauman and Kemper 1985, 1986, 1994; Ritvo et al. 1986; Kemper and Bauman 1993; Courchesne 1997; Bailey et al. 1998; Palmen et al. 2004). In addition, neuroimaging studies have consistently demonstrated posterior cerebellar hypoplasia (Gaffney et al. 1987; Courchesne et al. 1988, 2001, 2003; Murakami et al. 1989; Kleiman et al. 1992; Hashimoto et al. 1995; Courchesne 1997). Although the cerebellum has been classically considered a motor control center, functional imaging studies indicate that the cerebellum is also active during cognitive tasks that are defective in ASD, including language and attention (Kim et al. 1994; Raichle et al. 1994; Gao et al. 1996; Akshoomoff et al. 1997; Allen et al. 1997; Courchesne 1997; Courchesne and Allen 1997; Allen and Courchesne 2003; Corina et al. 2003; McDermott et al. 2003). Thus, the identified cerebellar defects may contribute directly to some of the behavioral abnormalities associated with ASD. In turn, genetic alterations that perturb cerebellar development may contribute to ASD susceptibility.
ENGRAILED 2 (EN2 [MIM 131310]) was selected as a candidate gene because En2 mouse mutants display anatomical phenotypes in the cerebellum that are similar to those reported for individuals with autism. Two mouse mutants exist for En2: a knockout and a transgenic that causes the developmental misexpression of the gene. In both mutants, adult mice are nonataxic but the cerebellum is hypoplastic, with a decrease in the number of Purkinje cells, indicating that En2 misregulation negatively impacts cerebellar development (Millen et al. 1994, 1995; Kuemerle et al. 1997; Baader et al. 1998, 1999).
EN2 was also selected as a candidate gene because of previous linkage and association studies. EN2 maps to 7q36, a chromosomal region that has provided suggestive evidence for linkage to ASD in Autism Genetic Resource Exchange (AGRE) families that largely overlapped with the pedigrees used for our initial family-based association study (Liu et al. 2001; Alarcon et al. 2002). 7q36 has also yielded suggestive linkage for ASD and dysphasia, another language disorder, in one other study (Auranen et al. 2002). However, most reports either have not demonstrated linkage with 7q36 or have not tested markers spanning telomeric chromosome 7 (Risch et al. 1999; Folstein and Rosen 2001). In addition, significant association between EN2 and autism has been reported previously for a _Pvu_II RFLP mapped by Southern analysis to the 5′ region of the gene (Logan and Joyner 1989; Petit et al. 1995). Thus, both the cerebellar phenotypes of mouse En2 mutants and previous human genetic analysis indicated that EN2 was a suitable candidate gene to test for association with ASD.
Human EN2 spans 8.1 kb of genomic DNA and consists of two exons separated by a single 3.3-kb intron. We have previously tested four SNPs (rs3735653, rs1861972, rs1861973, and rs2361689) that span the majority of the EN2 gene for association with ASD in 167 AGRE families (AGRE I data set) (Gharani et al. 2004). The rs3735653 and rs2361689 SNPs are located in exon 1 and exon 2, respectively, whereas the rs1861972 and rs1861973 SNPs are both located in the single intron. Significant association was observed for the common alleles of rs1861972 and rs1861973, both individually and as a haplotype (Gharani et al. 2004). In contrast, the two exonic SNPs were not associated with ASD. Four-SNP haplotype analysis further revealed that the common alleles of rs1861972 and rs1861973 were not consistently associated with ASD, suggesting that rs1861972 and rs1861973 were nonfunctional polymorphisms in LD with the functional variant(s) located elsewhere in the gene (Gharani et al. 2004).
The current study now extends these observations in three ways. First, replication of the association of rs1861972 and rs1861973 with ASD was tested in two additional samples: 222 additional AGRE families (AGRE II data set) and 129 independent families from the National Institute of Mental Health (NIMH) collection. Second, to define the genomic region associated with ASD and to identify alleles for future functional studies, LD mapping in the AGRE I data set was performed by analyzing 14 additional polymorphisms, which included the previously associated _Pvu_II RFLP. Third, to further investigate the role of EN2 in neuronal development, mouse En2 was ectopically misexpressed in primary rat cortical precursors. These experiments have demonstrated both strong genetic association for EN2 with ASD, further supporting the possibility that EN2 acts as an ASD susceptibility locus, and have provided functional evidence that a risk allele that causes EN2 misexpression could disrupt normal brain development.
Material and Methods
Subjects
Families recruited to AGRE and to the NIMH Center for Collaborative Genetic Studies were used for this study (Risch et al. 1999; Geschwind et al. 2001). All research was approved by the UMDNJ–Robert Wood Johnson Medical School Institutional Review Board.
The AGRE I data set includes 167 families used in our original study (Gharani et al. 2004), and the AGRE II data set includes 222 additional families. AGRE is a central repository of family DNA samples created by the Cure Autism Now Foundation and the Human Biological Data Interchange (see AGRE Web site). Family selection criteria have been described elsewhere (Geschwind et al. 2001; Liu et al. 2001). Families recruited by AGRE comprise at least two affected siblings (diagnosed with autism, Asperger syndrome, or pervasive development disorder–not otherwise specified [PDD-NOS]), one or both parents, and additional affected and unaffected siblings when available. Although unaffected siblings have not undergone an Autism Diagnostic Interview–Revised (ADI-R) evaluation, for the combined AGRE sample, extensive neurological, psychological, and medical evaluations are available for 69 of the 277 unaffected siblings. None of the unaffected siblings display characteristics of a broad autism phenotype. Fragile X information was available for 381 of the 399 AGRE families considered for use in this study. Ten families were removed because at least one individual per family displayed a pre-, intermediate, or full fragile X mutation state. Karyotypic data were available for 109 AGRE families used in this study. Five karyotypically abnormal families with a duplication of SNRPN on chromosome 15q12 (a marker for cytogenic abnormality at the chromosome 15 autism critical region) were also removed.
The NIMH data set is managed and distributed by the NIMH Center for Collaborative Genetic Studies. The center is operated under an NIMH contract to Rutgers University and a subcontract to Washington University (see NIMH Center for Collaborative Genetic Studies Web site). The data set includes 143 families. Of these families, 14 are also included in the AGRE data sets and so were removed from our analysis (see NIMH Web site). These families were originally recruited as part of an NIMH-funded linkage study (grant RO1-MH S2708) at Stanford University School of Medicine. Selection criteria have been described elsewhere (Risch et al. 1999), and anonymous data on family structure, age, and sex, as well as diagnostic interview data and status, are available online (see NIMH Web site). Families in the NIMH data set have at least two affected siblings or more-distantly related individuals (e.g., cousins) with a diagnosis of autism or another pervasive developmental disorder, without any associated primary disorder such as fragile X syndrome. The majority of the families included in the collection are affected-sib multiplex families. Of the 53 unaffected children, 46 have undergone ADI-R evaluation, and these individuals did not meet criteria for ASD (Risch et al. 1999). The diagnosis for six individuals was uncertain, and one individual did not meet ASD criteria by the ADI-R but exhibited behavioral and developmental abnormalities. These seven individuals were excluded from our analysis. No karyotype data were available for the NIMH data set. For our analysis, individuals were considered affected under a narrow diagnostic definition if they were diagnosed with autism, whereas they were considered affected under a broad diagnostic definition if they were diagnosed with autism, Asperger syndrome, or PDD-NOS.
Thirty-two of the families in the new AGRE sample and 20 in the NIMH sample include multiple births. In the AGRE sample, there are 19 families with MZ multiple births (16 MZ twin pairs that are autism:autism concordant, 2 MZ twin pairs that are autism:PDD discordant, and 1 set of MZ quadruplets that are autism:autism concordant), 11 families with DZ multiple births (9 twin pairs and 2 sets of triplets), and 2 additional families with twins of unknown zygosity. In the NIMH sample, 10 families have MZ twins (9 MZ twin pairs that are autism:autism concordant and 1 that is autism:PDD discordant), 9 families have DZ twins, and 1 family has a triplet consisting of an MZ pair and a third DZ sibling (all are autism:autism concordant). DNA is available for all twins in both data sets, and all siblings were genotyped. All DZ siblings are included in the data analysis, but, for MZ siblings, only the first MZ cotwin was selected for analysis.
DNA Analysis
Samples in the AGRE II and NIMH data sets were genotyped for the SNPs rs1861972 and rs1861973 by use of simplex Pyrosequencing assays and the automated PSQ HS 96A platform as described elsewhere (Ronaghi et al. 1998; Ahmadian et al. 2000).
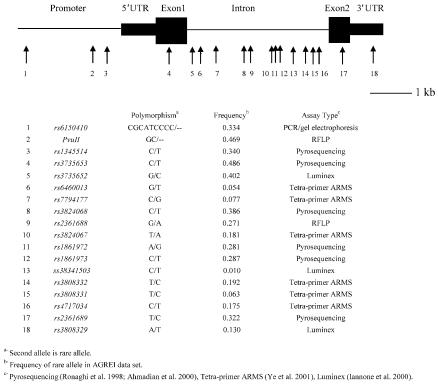
An additional 14 polymorphisms (fig. 1) were genotyped in the AGRE I data set; 12 were identified by dbSNP (see dbSNP Web site). The _Pvu_II RFLP, previously reported to be associated with autism (Petit et al. 1995), was identified as a −/CG insertion/deletion polymorphism by sequence analysis and by comparison with published RFLP reports (Logan and Joyner 1989). An additional intronic SNP (ss38341503) was identified by sequence analysis. Each dbSNP polymorphism was sequence verified using 24 unrelated individuals (23 whites and 1 individual of Hispanic/Latino descent) prior to the design of genotyping assays. Pyrosequencing, the ligase-detection reaction and Luminex cytometry (Iannone et al. 2000), RFLP, or tetra-primer amplification refractory mutation system (ARMS)–PCR assays (see Tetra-primer ARMS-PCR Web site) were used to genotype 13 of the polymorphisms (Ye et al. 2001), whereas a standard PCR and gel electrophoresis assay was used to genotype the rs6150410 insertion/deletion polymorphism (fig. 1). Primers were designed using publicly available software (see Pyrosequencing Technical Support and Primer3 Web sites). The primer sequence and PCR conditions used for each polymorphism are described in appendix B (online only).
Figure 1.
Schematic representation of the human EN2 gene, with the location, name, and MAF of, as well as the genotyping method used for, the polymorphisms tested for association with ASD.
Statistical Analysis
Prior to data analysis, each polymorphism was assessed for deviations from Hardy-Weinberg equilibrium by use of genotype data from all parents and standard formulae. The DNA from the MZ twins was used as a genotyping internal control, with complete genotypic concordance observed for all MZ cotwins. Genotypes were checked for Mendelian inconsistencies by use of the PEDCHECK program, version 1.1 (O’Connell and Weeks 1998), and all identified Mendelian errors were corrected by regenotyping individual samples. In the AGRE I data set, for 9 individuals, genotyping data were missing for four markers. Since recombination events are not expected at a high frequency, given the small (<8 kb) intermarker distances, haplotype inconsistencies were identified by the SIMWALK program, version 2.86 (Weeks and Lathrop 1995). For each of the 14 polymorphisms genotyped in the AGRE I data set, three-marker haplotype analysis with rs1861972 and rs1861973 was performed. Regenotyping of flagged polymorphisms identified only 155 (1.5%) of the 10,360 genotypes that could not be resolved. These genotypes were distributed between the 14 polymorphisms, with a range from 45 genotypes for rs1345514 to 7 genotypes for rs3824068. For rs1861972 and rs1861973, genotyped in the AGRE II and NIMH data sets, two SNP haplotype analyses identified only 12 (0.7%) of the 1,741 genotypes that could not be resolved by regenotyping. The linkage disequilibrium (LD) coefficient (_D_′) for the 18 different polymorphisms was calculated in the AGRE I data set by use of the parental genotypes and the GOLD program, version 1.0 (Abecasis and Cookson 2000).
All single and multilocus association analyses were performed using the program PDTPHASE (version 2.404), which also calculates the corresponding P values. Both haplotype-specific P values and global P values (with adjustments for all possible common haplotypes with a frequency >5%) are calculated by the PDTPHASE program. PDTPHASE is a component of the UNPHASED package of association-analysis programs (Dudbridge 2003). PDTPHASE is a modification of the pedigree-based transmission/disequilibrium test (PDT) (Martin et al. 2000). PDTPHASE, like the PDT, was designed to allow the use of data from related triads and disease-discordant sibships from extended pedigrees when testing for transmission disequilibrium. It determines the presence of association by testing for unequal transmission of either allele from parents to affected offspring and/or unequal sharing of either allele between discordant sibships. Informative extended pedigrees contain at least one informative triad (i.e., an affected child with at least one parent heterozygous at the marker) or discordant sibship (i.e., at least one affected and one unaffected sibling with different marker genotypes). PDTPHASE has a number of advantages over the PDT that increase the statistical power of our analysis. It can handle missing parental data, is able to perform multilocus analysis, and includes an expectation-maximization algorithm that calculates maximum-likelihood gametic frequencies under the null hypothesis, allowing the inclusion of phase-uncertain haplotypes.
The total numbers of families, triads, and discordant sib pairs (DSPs) used for the rs1861972 and rs1861973 replication analyses in the AGRE II and NIMH data sets as well as the extension of the LD map in the original AGRE I data set are listed in table 1.
Table 1.
Data Set Information
| Data Set andDiagnostic Criteria | Familiesa | Triadsa | DSPsa | Subjects |
|---|---|---|---|---|
| AGRE Ib: | ||||
| Narrow | 166 | 262 | 134 | 689 |
| Broad | 167 | 322 | 168 | 750 |
| AGRE IIc: | ||||
| Narrow | 211 | 342 | 225 | 1,033 |
| Broad | 222 | 434 | 269 | 1,071 |
| NIMH: | ||||
| Narrow | 127 | 228 | 71 | 501 |
| Broad | 129 | 237 | 73 | 515 |
In all the haplotype analyses, haplotypes with a frequency <5% were pooled for analysis. PDTPHASE, like the PDT, can calculate two global scores: the PDTsum (which sums the level of significance from all families) and the PDTave (which gives equal weight to all families in a data set). Since most families in our study have similar size and structure and since we observed that the χ2 distribution and P values were similar for both PDT scores, only PDTsum values are reported in the “Results” section.
Using a multiplicative model, haplotype relative risk for the _rs1861972_-rs1861973 A-C haplotype was estimated as the transmission ratio (transmitted/untransmitted [T/U]) (Altshuler et al. 2000) from heterozygous parents to a single affected offspring who was selected randomly from each of the 518 families. TRANSMIT (version 2.5.4) was used for this analysis because it is capable of selecting at random a single affected offspring per family for association analysis. From the output, the number of informative transmissions (i.e., from heterozygous parents) may be derived (refer to Gharani et al. [2004] for details), and the transmission ratio can be estimated. This analysis was repeated 20 times each for the narrow and broad diagnostic definitions, and the mean relative risk was estimated under each diagnosis. The relative risk and haplotype frequency for the A-C haplotype were then used to estimate the population-attributable risk (PAR) by use of the standard formula _PAR_=(_X_-1)/X, where X_=(1-f)2+2_f(1-f)γ+_f_2γ2, f is the haplotype frequency, and γ is the haplotype relative risk.
Sequence Analysis
Four overlapping PCR products of the EN2 intron were PCR amplified from 20 ASD-affected individuals who inherited the _rs1861972_-rs1861973 A-C haplotype from heterozygous parents. Each PCR product was then purified (using QIAGEN-QIAquick PCR Purification Kit) and was sequenced by GENEWIZ on an ABI 3730 DNA analyzer. The sequence was then analyzed for DNA alterations by use of CodonCode Aligner. The primer sequences and PCR conditions used for these experiments are described in appendix B (online only).
En2 Expression Constructs
To misexpress the En2 protein, total RNA was isolated from adult C57BL6/J mouse cerebellum, and a 1,012-bp PCR product that includes the En2 protein-coding sequence was amplified by RT-PCR (Superscript RT) and was then subcloned 3′ of a CMV protein/enhancer. PCR was conducted in a 25-μl reaction containing 0.4 μM of each primer, 0.25 mM dNTP, 1.5 mM MgCl2, 25 mM KCl, and 10 mM Tris-HCl (pH 8.3). Cycling conditions were an initial cycle at 94°C for 4 min; 35 cycles at 94°C for 45 s, 61°C for 1 min, and 74°C for 2 min; and a final step at 74°C for 10 min. The primer sequences were 5′-GTGAAGTATGGAGGAGAAGG-3′ for the forward primer and 5′-CTAAACAGTCCCCTTTGCAG-3′ for the reverse primer. The PCR product was isolated by 0.8% agarose gel electrophoresis, was purified, and was cloned into the pCR2.1 vector (Invitrogen). _Eco_RI restriction-enzyme digest was used to clone the En2 cDNA into the pCMS-EGFP expression vector (Clontech).
Cell Culture and cDNA Transfections
Time-mated pregnant Sprague Dawley rats were obtained from Hilltop Labs. At embryonic day 14.5 (E14.5), embryonic skull and meninges were removed, and dorsolateral cerebral cortex was dissected, was mechanically dissociated, and was plated at 8×104 cells/cm2 on poly-d-lysine (0.1 mg/ml) and laminin (20 μg/ml)–coated 25-mm glass coverslips (VWR) in defined medium, as described elsewhere (Nicot and DiCicco-Bloom 2001). Culture medium consisted of Neurobasal (Gibco) supplemented with 2% B27 and contained glutamine (2 mM), penicillin (50 U/ml), streptomycin (50 μg/ml), BSA (1 mg/ml), and basic fibroblast growth factor (10 ng/ml). Unless stated otherwise, components were obtained from Sigma. Cultures were maintained in a humidified 5% CO2/air incubator at 37°C.
After 24 h in culture, cells were transfected as described elsewhere (Nicot and DiCicco-Bloom 2001) using, for 5 h, Lipofectamine Plus Reagent (BRL) containing one of the following: (1) the mouse En2 enhanced green fluorescent protein (EGFP)–expression plasmid that codes for a full-length En2 protein with 93% nucleotide identity with rat En2; (2) pCMS-EGFP with En2 cloned in the non–protein-coding reverse orientation (REn2); or (3) pCMS-EGFP alone. After an additional day of incubation, cells were fixed with 4% paraformaldehyde and were assessed using phase and fluorescence microscopy.
En2 RT-PCR Expression Analysis
Total RNA was isolated from freshly dissected E14.5 rat cortices and hindbrains, and the expression of En2 was determined by PCR amplification of a 220-bp 3′-UTR PCR product in a 25-μl reaction containing 0.4 μM of each primer, 0.25 mM dNTP, 1.5 mM MgCl2, 25 mM KCl, and 10 mM Tris-HCl at pH 8.3. Cycling conditions were an initial cycle at 94°C for 4 min; 35 cycles at 94°C for 45 s, 56°C for 1 min, and 74°C for 90 s; and a final step at 74°C for 10 min. The primer sequences were 5′-AACCGTGAACAAAAGGCCAGTG-3′ for the forward primer and 5′-CTAAACAGTCCCCTTTGCAG-3′ for the reverse primer.
Immunocytochemistry
Immunocytochemistry was performed as described elsewhere (Nicot and DiCicco-Bloom 2001). The dilutions of primary and secondary antibodies used were polyclonal chicken anti-GFP (1:8,000 [Chemicon]), polyclonal goat anti-En2 (1:500 [Santa Cruz]), anti-mouse βIII-tubulin (1:1,500 [Covance]), Alexa Fluor 488-conjugated rabbit anti-chicken IgG (1:800 [Chemicon]), and Alexa Fluor 594-conjugated rabbit anti-goat or goat anti-mouse IgG (1:1,000 [Vector]).
Results
Replication Studies
Our previous analysis demonstrated that the A allele of rs1861972 and the C allele of rs1861973 were significantly associated with ASD individually and as a haplotype, under both narrow and broad diagnostic criteria (Gharani et al. 2004). Replication of these association results in an additional 222 AGRE families (AGRE II) and in 129 NIMH families was observed in the present study.
For the AGRE II data set, significant (P<.05) evidence of association for the A allele of rs1861972 was observed under the broad diagnostic criteria, whereas a trend toward association was observed under the narrow diagnostic criteria (table 2). For rs1861973, significant association of the C allele was observed under both diagnostic criteria (table 2). Analysis of _rs1861972_-rs1861973 haplotypes demonstrated that the A-C haplotype was significantly overtransmitted to affected offspring under both diagnostic criteria (table 3), whereas the A-T, G-C, and G-T haplotypes were all undertransmitted. Global χ2 tests for all haplotypes yielded significant P values (narrow _P_=.0048; broad _P_=.0016) (table 3) that were similar to those reported previously for the AGRE I data set (Gharani et al. 2004). Thus, replication of our previous results of association of these same alleles of rs1861972 and rs1861973 with ASD is observed in this second AGRE data set.
Table 2.
Association Results for rs1861972 and rs1861973 in Additional Data Sets
| ParentalTransmissionsb | Allele inDSP Siblingsc | |||||
|---|---|---|---|---|---|---|
| Data Set, SNP,aand DiagnosticCriteria | Transmitted | Untransmitted | Affected | Unaffected | χ2Valued | P Valuee |
| AGRE If: | ||||||
| rs1861972: | ||||||
| Narrow | 383 | 354 | 202 | 185 | 4.991 | .0255 |
| Broad | 467 | 436 | 253 | 226 | 5.861 | .0155 |
| rs1861973: | ||||||
| Narrow | 373 | 338 | 199 | 183 | 6.936 | .0084 |
| Broad | 449 | 415 | 242 | 220 | 6.297 | .0121 |
| AGRE IIg: | ||||||
| rs1861972: | ||||||
| Narrow | 438 | 408 | 314 | 297 | 2.997 | .0834 |
| Broad | 555 | 513 | 381 | 359 | 4.730 | .0296 |
| rs1861973: | ||||||
| Narrow | 433 | 395 | 308 | 285 | 4.903 | .0268 |
| Broad | 546 | 500 | 375 | 346 | 6.299 | .0121 |
| NIMH: | ||||||
| rs1861972: | ||||||
| Narrow | 222 | 200 | 113 | 107 | 4.000 | .0455 |
| Broad | 228 | 206 | 115 | 109 | 3.843 | .0500 |
| rs1861973: | ||||||
| Narrow | 227 | 203 | 111 | 104 | 5.139 | .0234 |
| Broad | 234 | 209 | 113 | 105 | 5.585 | .0181 |
Table 3.
Haplotype Association Results for the rs1861972-rs1861973 Haplotype in Additional Data Sets
| ParentalTransmissionsb | Haplotype inDSP Siblingsc | ||||||
|---|---|---|---|---|---|---|---|
| Data Set, Diagnostic Criteria,and Haplotypea | Transmitted | Untransmitted | Affected | Unaffected | Frequency | χ2Valued | _P_Valuee |
| AGRE If: | |||||||
| Narrow: | |||||||
| A-C | 369 | 323 | 195 | 179 | .732 | … | .0024 |
| A-T | 8 | 25 | 5 | 4 | .014 | … | … |
| G-C | 4 | 15 | 4 | 4 | .006 | … | … |
| G-T | 111 | 129 | 52 | 69 | .247 | … | .0714 |
| Global | … | … | … | … | … | 14 | .0009 |
| Broad: | |||||||
| A-C | 444 | 399 | 238 | 216 | .734 | … | .0039 |
| A-T | 11 | 28 | 5 | 4 | .016 | … | … |
| G-C | 5 | 16 | 4 | 4 | .007 | … | … |
| G-T | 140 | 157 | 69 | 92 | .243 | … | .0765 |
| Global | … | … | … | … | … | 13 | .0017 |
| AGRE IIg: | |||||||
| Narrow: | |||||||
| A-C | 429 | 386 | 308 | 285 | .713 | … | .0168 |
| A-T | 4 | 16 | 2 | 5 | .017 | … | … |
| G-C | 2 | 8 | 0 | 0 | .007 | … | … |
| G-T | 161 | 186 | 128 | 148 | .263 | … | .0928 |
| Global | … | … | … | … | … | 11 | .0048 |
| Broad: | |||||||
| A-C | 540 | 487 | 375 | 346 | .721 | … | .0061 |
| A-T | 6 | 18 | 2 | 6 | .014 | … | … |
| G-C | 2 | 11 | 0 | 0 | .006 | … | … |
| G-T | 210 | 242 | 149 | 174 | .259 | … | .0493 |
| Global | … | … | … | … | … | 13 | .0016 |
| NIMH: | |||||||
| Narrow: | |||||||
| A-C | 221 | 198 | 111 | 104 | .676 | … | .0321 |
| A-T | 1 | 2 | 0 | 1 | .007 | … | … |
| G-C | 6 | 5 | 0 | 0 | .025 | … | … |
| G-T | 78 | 101 | 27 | 34 | .292 | … | .0329 |
| Global | … | … | … | … | … | 6.1 | .0463 |
| Broad: | |||||||
| A-C | 227 | 204 | 113 | 105 | .672 | … | .0312 |
| A-T | 1 | 2 | 0 | 2 | .008 | … | … |
| G-C | 7 | 5 | 0 | 0 | .023 | … | … |
| G-T | 81 | 105 | 29 | 35 | .296 | … | .0295 |
| Global | … | … | … | … | … | 6.3 | .0431 |
Since identical criteria have been used to obtain all the AGRE pedigrees, the AGRE I and AGRE II data sets were combined, and association of rs1816972 and rs1861973 with ASD was reanalyzed. Smaller P values were observed in the combined set than in either data set alone (for the haplotype, narrow _P_=.0000067 and broad _P_=.0000033), indicating that the same alleles are associated with ASD in both data sets.
Association of rs1861972 and rs1861973 was then tested in the NIMH data set. Statistically significant association was obtained for the SNPs individually (table 2) and as a haplotype (table 3). As expected, when the NIMH data set was combined with both AGRE data sets and reanalyzed for association, further reduction of the P values was observed (for rs1861972-rs1861973 haplotype, narrow _P_=.00000065 and broad _P_=.00000035). These data represent one of the most significant associations of any gene with ASD. Given the large sample size (518 families), these results implicate an inherited variation in EN2 in susceptibility to ASD.
Since a P value is not a measure of effect size, we used this large combined sample of 518 families to estimate the relative risk and the associated PAR under a multiplicative model for the A-C haplotype. The haplotype relative risk was estimated as ∼1.42 and 1.40 under the narrow and broad diagnoses, respectively. Although this represents a relatively modest increase in individual risk, given the high frequency of this common haplotype (∼67% in the combined sample), relative risks of 1.42 and 1.40 correspond to a large PAR of ∼39.5% and 38% for the narrow and broad diagnoses of ASD, respectively. These data imply that as many as 40% of ASD cases in the population may be influenced by variation in the EN2 gene.
We have previously demonstrated that rs1861972 and rs1861973 are in strong LD with each other (Gharani et al. 2004). Similar results were obtained in both new data sets (AGRE II, _D_′=0.967; NIMH, _D_′=0.977). In addition, the allele frequencies for rs1861972 and rs1861973 in both new data sets are almost identical to each other and to what we reported previously for the AGRE I data set (Gharani et al. 2004) (fig. 1). This, together with the association results obtained in the new samples, suggests that they are likely to be derived from the same population and are therefore likely to share a similar LD relationship with the putative etiological variant.
Extension of LD Map in the AGRE I Data Set
Our previous analysis of the original AGRE I data set suggested that rs1861972 and rs1861973 are nonfunctional polymorphisms in LD with a risk allele(s) located elsewhere in the gene (Gharani et al. 2004). In an attempt to identify this risk allele, the LD map was extended in the original AGRE I data set. This data set was selected for LD mapping because it displays the most significant association for rs1861972 and rs1861973, both individually and as a haplotype, and therefore represents a minimal cost-effective sample set with sufficient power to detect the putative risk allele. Fourteen additional polymorphisms (3 in the promoter, 10 in the intron, and 1 in the 3′ UTR) were tested for association with ASD, making a total of 18 markers typed across the entire gene. Thirteen of these newly typed polymorphisms were identified through dbSNP, whereas one (ss38341503) was identified by resequencing the entire intron in 20 individuals with ASD who inherited the rs1861972- rs1861973 A-C haplotype from heterozygous parents. The location, DNA change, and minor-allele frequency (MAF) of each polymorphism are illustrated in figure 1.
In the absence of knowledge about the exact mode of inheritance at this locus, we may expect the risk allele(s) responsible for rs1861972 and rs1861973 association to exhibit the following inheritance pattern. First, the polymorphism(s) should display strong LD (_D_′>0.70) with both rs1861972 and rs1861973 and, therefore, would be expected to have a similar frequency as the A-C haplotype. Second, the polymorphism(s) should also be associated with ASD individually. If a single polymorphism is responsible for the association of rs1861972 and rs1861973, then this polymorphism should demonstrate at least as significant an association as the A-C haplotype under both the narrow and broad diagnostic criteria. However, if multiple alleles are working in concert and we assume a simple additive model, then each of these polymorphisms should display association with ASD individually because they will be in strong LD with rs1861972 and rs1861973, but their statistical significance may not be as great as when they are analyzed as a haplotype. Third, in multi-SNP haplotype analysis for a single-locus model, the associated allele in conjunction with the A-C haplotype should display at least as significant an association as the A-C haplotype alone. In a multilocus model, only the haplotypes with all or most of the risk alleles will display the greatest statistical significance.
When the 14 additional polymorphisms were tested for association, 12 displayed no evidence of association under both the narrow and broad diagnoses (table 4). Two intronic SNPs, rs3824068 and rs2361688, displayed minimally significant association, but only under one diagnosis (table 4). To investigate whether rs3824068 and rs2361688 could be functioning in a multilocus manner, three- and four-marker haplotype analyses with the _rs1861972_-rs1861973 A-C haplotype were performed. For the _rs3824068_-rs2361688-rs1861972-rs1861973 and the _rs3824068_-_rs1861972_-rs1816973 haplotype analyses, all common core A-C haplotypes (frequency >5%) displayed no association, except for the _rs3824068_-_rs1861972_-rs1816973 T-A-C haplotype, which displayed minimal association under one diagnosis (tables 5 and 6). For the _rs2361688_-_rs1861972_-rs1861973 analysis, the G-A-C haplotype displayed similar statistical significance as the A-C haplotype under the broad diagnosis, but, under the narrow diagnosis, the effect was diluted (table 5). Four other common three-marker A-C haplotypes (_rs6460013_-_rs1861972_-rs1861973 G-A-C, _rs7794177_-_rs1861972_-rs1861973 C-A-C, _rs1861972_-_rs1861973_-ss38341503 A-C-C, and _rs1861972_-rs1861973-rs3808329 A-C-A) displayed similar statistically significant association as the A-C haplotype under at least one diagnostic definition (table 5). However, rs6460013, rs7794177, ss38341503, and rs3808329 were not associated individually with ASD, which indicates that these polymorphisms are not functioning as risk alleles according to our criteria.
Table 4.
Association Analysis for 14 Additional Polymorphisms in the AGRE I Data Seta
| ParentalTransmissionsb | Allele inDSP Siblingsc | |||||
|---|---|---|---|---|---|---|
| Polymorphismand DiagnosticCriteria | Transmitted | Untransmitted | Affected | Unaffected | χ2Valued | P Valuee |
| rs6150410: | ||||||
| Narrow | 317 | 317 | 183 | 189 | .081 | .776 |
| Broad | 394 | 384 | 234 | 239 | .043 | .835 |
| _Pvu_II: | ||||||
| Narrow | 253 | 249 | 127 | 131 | .000 | 1.000 |
| Broad | 306 | 303 | 163 | 162 | .025 | .875 |
| rs1345514: | ||||||
| Narrow | 271 | 278 | 165 | 168 | .220 | .639 |
| Broad | 332 | 331 | 211 | 215 | .015 | .902 |
| rs3735652: | ||||||
| Narrow | 274 | 291 | 156 | 168 | 1.532 | .216 |
| Broad | 350 | 357 | 203 | 220 | .867 | .352 |
| rs6460013: | ||||||
| Narrow | 467 | 466 | 249 | 246 | .364 | .546 |
| Broad | 568 | 569 | 314 | 309 | .381 | .537 |
| rs7794177: | ||||||
| Narrow | 425 | 414 | 239 | 237 | 1.374 | .241 |
| Broad | 519 | 509 | 295 | 299 | .237 | .626 |
| rs3824068: | ||||||
| Narrow | 280 | 316 | 156 | 166 | 4.372 | .036 |
| Broad | 355 | 384 | 205 | 215 | 2.664 | .103 |
| rs2361688: | ||||||
| Narrow | 347 | 328 | 193 | 183 | 2.317 | .128 |
| Broad | 423 | 394 | 243 | 224 | 4.208 | .040 |
| rs3824067: | ||||||
| Narrow | 370 | 357 | 213 | 214 | .581 | .446 |
| Broad | 443 | 438 | 263 | 267 | .003 | .959 |
| ss38341503: | ||||||
| Narrow | 464 | 458 | 266 | 265 | 1.581 | .209 |
| Broad | 565 | 559 | 334 | 332 | 1.684 | .194 |
| rs3808332: | ||||||
| Narrow | 361 | 361 | 208 | 208 | .000 | 1.000 |
| Broad | 433 | 440 | 260 | 261 | .173 | .677 |
| rs3808331: | ||||||
| Narrow | 421 | 417 | 243 | 245 | .040 | .841 |
| Broad | 519 | 510 | 309 | 310 | .525 | .469 |
| rs4717034: | ||||||
| Narrow | 365 | 362 | 205 | 212 | .056 | .812 |
| Broad | 436 | 444 | 257 | 267 | .802 | .370 |
| rs3808329: | ||||||
| Narrow | 424 | 405 | 229 | 243 | .108 | .742 |
| Broad | 516 | 491 | 289 | 303 | .434 | .510 |
Table 5.
Three-Marker Haplotype Analysis of the rs1861972-rs1861973 Haplotype in the AGRE I Data Seta
| Polymorphism, Diagnostic Criteria, and Haplotype | Frequency | P Valueb |
|---|---|---|
| rs1861972-rs1861973: | ||
| Narrow: | ||
| A-C | .732 | .002 |
| Broad: | ||
| A-C | .734 | .004 |
| rs6150410: | ||
| Narrow: | ||
| Ins-A-C | .464 | .108 |
| Del-A-C | .267 | .235 |
| Broad: | ||
| Ins-A-C | .467 | .072 |
| Del-A-C | .265 | .452 |
| _Pvu_II: | ||
| Narrow: | ||
| Ins-A-C | .414 | .335 |
| Del-A-C | .323 | .084 |
| Broad: | ||
| Ins-A-C | .413 | .404 |
| Del-A-C | .325 | .097 |
| rs1345514: | ||
| Narrow: | ||
| C-A-C | .461 | .148 |
| T-A-C | .266 | .339 |
| Broad: | ||
| C-A-C | .465 | .097 |
| T-A-C | .265 | .574 |
| rs3735652: | ||
| Narrow: | ||
| G-A-C | .347 | .292 |
| C-A-C | .382 | .107 |
| Broad: | ||
| G-A-C | .359 | .158 |
| C-A-C | .371 | .289 |
| rs6460013: | ||
| Narrow: | ||
| G-A-C | .690 | .004 |
| T-A-C | .048 | … |
| Broad: | ||
| G-A-C | .690 | .011 |
| T-A-C | .050 | .896 |
| rs7794177: | ||
| Narrow: | ||
| C-A-C | .658 | .004 |
| G-A-C | .070 | .751 |
| Broad: | ||
| C-A-C | .656 | .012 |
| G-A-C | .072 | .901 |
| rs3824068: | ||
| Narrow: | ||
| C-A-C | .359 | .884 |
| T-A-C | .372 | .019 |
| Broad: | ||
| C-A-C | .364 | .504 |
| T-A-C | .372 | .076 |
| rs2361688: | ||
| Narrow: | ||
| G-A-C | .717 | .009 |
| A-A-C | .008 | … |
| Broad: | ||
| G-A-C | .716 | .004 |
| A-A-C | .009 | … |
| rs3824067: | ||
| Narrow: | ||
| T-A-C | .561 | .013 |
| A-A-C | .170 | .654 |
| Broad: | ||
| T-A-C | .560 | .056 |
| A-A-C | .188 | .655 |
| ss38341503: | ||
| Narrow: | ||
| A-C-C | .710 | .001 |
| A-C-T | .005 | … |
| Broad: | ||
| A-C-C | .716 | .001 |
| A-C-T | .009 | … |
| rs3808332: | ||
| Narrow: | ||
| A-C-T | .551 | .022 |
| A-C-C | .185 | .629 |
| Broad: | ||
| A-C-T | .553 | .045 |
| A-C-C | .186 | .505 |
| rs3808331: | ||
| Narrow: | ||
| A-C-T | .672 | .017 |
| A-C-C | .057 | .961 |
| Broad: | ||
| A-C-T | .675 | .012 |
| A-C-C | .057 | .660 |
| rs4717034: | ||
| Narrow: | ||
| A-C-C | .563 | .035 |
| A-C-T | .172 | .506 |
| Broad: | ||
| A-C-C | .557 | .114 |
| A-C-T | .181 | .260 |
| rs3808329: | ||
| Narrow: | ||
| A-C-A | .634 | .012 |
| A-C-G | .105 | .269 |
| Broad: | ||
| A-C-A | .642 | .008 |
| A-C-G | .097 | .652 |
Table 6.
Haplotype Analysis of the rs3824068-rs2361688-rs1861972-rs1861973 Haplotype in AGRE I Data Seta
| Polymorphism, Diagnostic Criteria, and Haplotype | Frequency | P Valueb |
|---|---|---|
| rs3824068-rs2361688-rs1861972-rs1861973: | ||
| Narrow: | ||
| C-G-A-C | .357 | .996 |
| C-A-A-C | .008 | … |
| T-G-A-C | .357 | .054 |
| Broad: | ||
| C-G-A-C | .362 | .361 |
| C-A-A-C | .008 | … |
| T-G-A-C | .356 | .145 |
The intermarker LD relationships for these 14 polymorphisms plus the 4 previously tested SNPs were then examined. All promoter, exonic, and 3′-UTR polymorphisms displayed weak or intermediate LD (_D_′ range 0.024–0.632) with both rs1861972 and rs1861973, providing an explanation as to why they are not associated with ASD (fig. 2A). However, all new intronic SNPs displayed strong LD (_D_′ range 0.720–1.00) with both rs1861972 and rs1861973 (fig. 2A). The lack of association with ASD observed for eight of the intronic SNPs suggests that these intronic variants are in weaker LD with the risk allele than are SNPs rs1861872 or rs1861973. This reduced power to detect LD may be the result of differences in allele frequencies and may reflect the genetic history of when these intronic alleles arose in relation to the risk allele. Evidence for some association of rs2361688 and rs3824068 with ASD suggests that these variants are in stronger LD with the risk allele(s) than the other newly genotyped intronic SNPs.
Figure 2.
Intermarker LD values. A, Tabular representation of the pairwise LD (_D_′) values between the 18 polymorphisms tested for association in the AGRE I data set. The LD relationships of each polymorphism with rs1861972 and rs1861973 are highlighted, with the degree of shading corresponding to the strength of LD: black indicates strong LD (_D_′ > 0.72); gray indicates intermediate/weak LD (_D_′ range 0.024–0.632). B, The EN2 gene is illustrated with the position of all 18 tested polymorphisms demarcated by arrows. The decay of LD of each polymorphism with respect to rs1861972 and rs1861973 is represented by a gradient bar drawn below the gene. The degree of shading corresponds to the strength of LD, as described above. Only intronic polymorphisms are in strong LD with rs1861972 and rs1861973.
One plausible interpretation of the strong LD observed between the intronic SNPs and rs1861972 and rs1861973, as well as the decay of LD for flanking polymorphisms, is that the risk allele is situated at ∼3.0 kb in the intron. Sequence analysis of the intron in 20 individuals affected with ASD who inherited the rs1861972-rs1861973 A-C haplotype from heterozygous parents has identified only one novel SNP (ss383341503) with an MAF of 1%, indicating that additional common intronic polymorphisms are unlikely. We have now tested all intronic SNPs, and only rs1861972 and rs1861973 are consistently associated both individually and as a haplotype under both diagnostic criteria. Together, these analyses suggest that the A allele of rs1861972 and the C allele of rs1861973 may function as risk alleles in cis and suggest that the lack of association of some of the core A-C haplotypes identified in the multi-SNP haplotype analysis is because of other, unidentified epistatic genetic or environmental interactions. Comparative genomic studies of human, chimp, mouse, rat, and dog sequences do not place either SNP within conserved regions (data not shown). However, computer prediction programs have determined that the associated alleles of rs1861972 and rs1861973 are situated within consensus binding sites for the CBP and LvC transcription factors, respectively (see TRANSFAC Web site). For the associated alleles, the match ratio for each transcription factor was 100%. The nonassociated alleles alter a conserved nucleotide in the consensus binding site, and so, when the sequence was reanalyzed, the nonassociated alleles were predicted to abolish the binding of both transcription factors.
Effects of En2 Misexpression on Neurogenesis
Risk alleles of human EN2 associated with ASD may potentially alter the spatial and/or temporal expression of the gene during brain development. To begin examining the consequences of gene misexpression, we transfected the En2 EGFP vectors into cultures of primary neuronal precursors obtained from rat E14.5 cerebral cortex. The expression plasmid alone (pCMS-EGFP) or with En2 cloned in the reverse orientation (REn2) was used as a control. We have previously used this well-characterized model system to define the effects of extracellular signals and transfected genes on cortical neurogenesis (Lu and DiCicco-Bloom 1997; Nicot and DiCicco-Bloom 2001; Carey et al. 2002). En2 is not expressed in E14.5 rat cortical cells, as assessed by RT-PCR (fig. 3A), so our experiments define the effects of En2 misexpression in a naive cell population. At 24 h after transfection, all three vectors generated similar numbers of EGFP-expressing cells (pCMS-EGFP, 174.5 ± 45.1; REn2, 201 ± 20.4; En2, 163.3 ± 11.9 [GFP+ cells ± SEM]; _P_>.05), as detected by EGFP autofluorescence and immunocytochemistry, which suggests that vector expression per se is not deleterious. As expected, En2 protein immunoreactivity was detected only in EGFP-positive, _En2_-transfected cells, but not in _REn2_- or EGFP-transfected cells (data not shown).
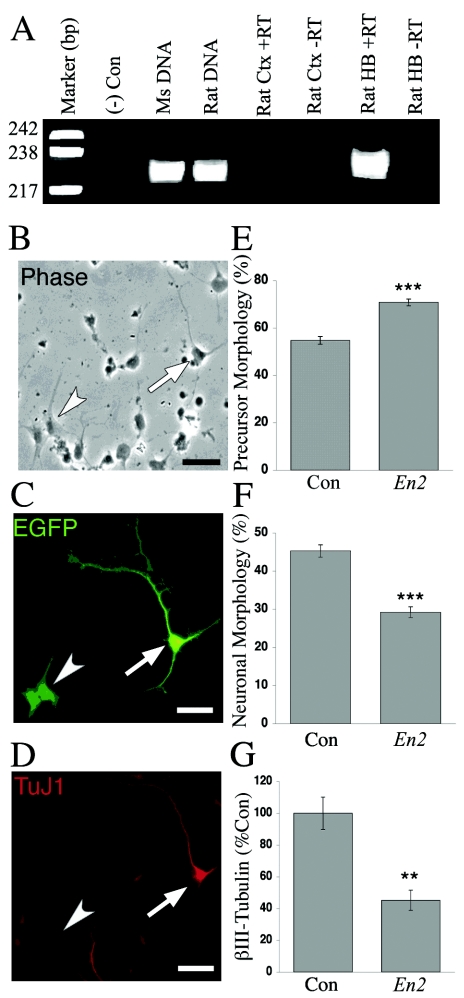
Figure 3.
Effects of En2 misexpression on neurogenesis. A, No En2 expression in rat cerebral cortex. En2 expression was detected by RT-PCR of RNAs obtained from freshly dissected E14.5 rat cortex (Ctx) and hindbrain (HB). The primers amplified the same 220-bp region of the En2 3′ UTR from both genomic mouse (Ms) and rat DNAs, consistent with the high degree of nucleotide homology. No expression was found in rat cortex, whereas a high level was detected in the hindbrain, recapitulating the known expression pattern of En2. +/− RT = with/without reverse transcriptase; Con = control. B–D, Transfected cortical cells exhibiting morphologies of undifferentiated precursors and differentiated neurons. The arrowheads denote cells with precursor morphology, whereas the arrows identify cells with neuronal morphology. Magnification 40×; scale bar = 30 μm. B, Phase photomicrograph demonstrating cells with both precursor and neuronal morphologies. Note the large, flat cell bodies and the thick, tapering, and short processes of the precursors (arrowhead). In contrast, neuron-like cells exhibit round cell bodies with long, thin, and uniform-diameter processes (arrow). C, Examples of both precursors and neurons transfected with the En2 vectors and identified for gene expression by immunostaining for EGFP marker protein. D, Cultures that were double labeled to detect EGFP and neuron-specific cytoskeletal protein βIII-tubulin (TuJ1), an early marker of neuronal differentiation. TuJ1 expression was only detected in cells exhibiting neuronal morphologies, a finding consistent with our previous studies (Nicot and DiCicco-Bloom 2001). E, Increase in the proportion of GFP+ precursor cells, noted 24 h after En2 transfection. F, Corresponding decrease in the proportion of _En2_-transfected cells exhibiting neuronal morphology. G, En2 transfection elicited a >50% reduction in TuJ1-immunoreactive cells, a change that parallels the decrease in neuronal morphology. **P<.0005, _N_=9; ***P<4×10-8, _N_=15.
We initially examined the effects of vector expression on cortical cell morphology, assessing undifferentiated precursors and mature neurons (fig. 3_B_–3D). Undifferentiated neural precursors appear as flat cells that sometimes extend processes of variable diameter and length, with distal filopodia. Precursors do not express the early marker of neuronal differentiation: cytoskeletal protein βIII-tubulin (fig. 3D). Differentiated neurons exhibit a round or pyramidal cell body, extend thin uniform processes, and express βIII-tubulin (fig. 3_B_–3D). In cultures transfected with control vectors, approximately equal proportions of cells exhibited morphologies of precursors and neurons (fig. 3E and 3F), reproducing ratios obtained previously with this model (Nicot and DiCicco-Bloom 2001). In contrast, after En2 transfection, the proportion of undifferentiated precursors increased from 55% in controls to 71% in _En2_-transfected cells (fig. 3E). Since overall numbers of cells were similar across conditions, the increase in precursors occurred at the expense of cells exhibiting neuronal morphology—neurons decreased from 45% in controls to only 29% after En2 transfection (fig. 3F). The reduction in neuronal differentiation was further verified by assessment of βIII-tubulin expression, which was decreased by 55% in _En2_-transfected cells (fig. 3G), raising the possibility that altered cytoskeletal protein expression may underlie changes observed in cellular morphology elicited by En2. These data demonstrate that En2 ectopic expression disrupts neuronal differentiation, indicating that the gene is an important regulator of neuronal development.
Discussion
In this study, we provide further genetic support that EN2 is involved in ASD susceptibility. Evidence for association is presented for rs1861972 and rs1861973 in the AGRE II and NIMH data sets. PAR estimations calculated using the entire sample of 518 families indicate that the risk allele responsible for _rs1861972_-rs1861973 association contributes to ∼40% of ASD cases in the general population. In addition, LD mapping with 18 SNPs distributed across the gene has currently localized the associated genomic region to the intron (fig. 2B). Analysis of this region has identified associated intronic alleles that will be characterized, in the future, for functional differences. Future LD studies will also examine the possibility of more-complex LD patterns across the gene, extending to 5′ and 3′ genomic regions. Finally, we also demonstrate that ectopic misexpression of En2 disrupts neuronal development. Together, these data suggest that variation within the EN2 gene plays an important role in ASD etiology and that one or more risk alleles that cause altered expression of EN2 could perturb neuronal development and contribute to the pathology associated with ASD.
A primary objective of this study was to examine whether our previous association of rs1861972 and rs1861973 with ASD could be replicated in other data sets. Many factors may contribute to a lack of replication in association studies of complex genetic traits (Terwilliger 2000; Riley 2004; Bartlett et al. 2005). Consequently, the significant replication of rs1861972 and rs1861973 association in the AGRE II and NIMH data sets strongly supports EN2 as a contributing factor in ASD etiology. When the transmissions from all three data sets were combined and analyzed, highly significant evidence of association was obtained. Although a P value is not a measure of effect size, these data demonstrate that EN2 association with ASD is maintained in this larger sample. The combined data set includes 518 families, or 2,336 individuals, and is one of the largest samples in which an association study for ASD has been conducted. The fact that very significant association is observed in this large sample recruited from multiple sources supports the possibility that EN2 is a risk factor for ASD and that EN2 contributes to ASD susceptibility in the general population.
Given that our replication data provided genetic evidence for a risk allele situated within EN2, LD mapping was used to localize the genomic position of the risk allele(s). The rs1861972-rs1861973 A-C haplotype has consistently demonstrated similar or more-significant association than either SNP individually. In addition, three- and four-SNP haplotypes that contain a core A-C haplotype are not consistently overtransmitted (Gharani et al. 2004) (table 5). These data have been interpreted as an indication that rs1861972 and rs1861973 are not risk alleles but, instead, are nonfunctional polymorphisms in LD with an unidentified risk allele. Eighteen polymorphisms, including the new intronic SNP (ss38341503) identified in our samples, have now been tested for association. According to dbSNP (build 124), 35 EN2 polymorphisms have been identified in the EN2 gene and 2.5 kb of promoter sequence. dbSNP reports 13 polymorphisms in the promoter, 2 in the protein-coding sequence, 11 in the intron, and 9 in the 3′ UTR. We have tested 3 of the 13 promoter polymorphisms, 11 of the 11 intronic SNPs, both protein-coding SNPs, and 1 polymorphism in the 3′ UTR. Thus, a significant proportion of the polymorphisms annotated for EN2 have now been tested for association.
Of the 14 newly genotyped SNPs, only 2 intronic SNPs (rs2361688 and rs3824068) individually exhibit minimal association with ASD. However, given that, for both these SNPs, association is only detected under a single diagnostic definition and that the significance level of this association is less than that obtained for the _rs1861972_-rs1861973 A-C haplotype, these variants are not thought to function as risk alleles under a single-locus model. Detection of LD between a disease locus and a marker is dependent on various factors, including disease and marker allele frequencies, recombination rates, and the population history. In particular, it has been shown that the greatest power to detect LD is obtained if the frequency of the marker allele is at least as large as the frequency of the causative mutation (Garner and Slatkin 2003). Given that the greatest association with ASD has been obtained for the common A-C haplotype of _rs1861872_-rs1861973 (with a frequency of >67%), this suggests that the risk allele is also likely to have a high frequency. The MAF of all intronic SNPs ranges from 1% to 40% (fig. 1), and, in all cases except rs2361688 (MAF of 27%), it is the minor allele that is in LD with the common alleles of rs1861872 or rs1861973 (data not shown). This may partly explain the observed association for rs2361688 and the lack of detectable association with ASD for most of the other intronic SNPs. Alternatively, the association of rs2361688 individually and in three-marker haplotype analysis may indicate that the associated allele of rs2361688 could be functioning in a multilocus manner. Minimal association is also detected for rs3824068 individually and in a three-SNP haplotype analysis under the narrow diagnosis only. Examination of the transmission data revealed that the rare T allele of rs3824068 displayed strong LD with the A-C haplotype, whereas the common C allele of rs3824068 is equally distributed between the A-C and G-T haplotypes of _rs1861872_-rs1861973 (data not shown). Thus, the T allele is almost always transmitted with a subset of the A-C haplotypes, and this may explain the weak association with ASD detected with this marker. These data suggest that rs3824068 is unlikely to be functional and is most likely a polymorphism in which the rare T allele is in stronger LD with the risk allele than are the other, nonassociated intronic SNPs.
Finally, haplotype analysis has also shown that the common _rs1861872_-rs1861973 A-C–containing haplotypes for markers rs6460013, rs7794177, ss38341503, and rs3808329 displayed statistically significant association, similar to that of the A-C haplotype alone, under at least one diagnostic definition. However, these SNPs were not associated individually with ASD and therefore are not likely to function as risk alleles in a single-locus model. Furthermore, none of the three-SNP haplotype analyses resulted in a dramatic reduction of the P value, which indicates that these polymorphisms are likely not functioning in a multilocus manner and that the minor fluctuations in P values reflect the strong LD that exists between these haplotypes and the risk allele.
In summary, our analysis thus far has demonstrated that only rs1861972 and rs1861973 are consistently and significantly associated with ASD alone and as a haplotype under both narrow and broad diagnostic criteria. We propose two alternate hypotheses to explain our current LD and association data. The first hypothesis is that the ASD risk allele(s) responsible for the rs1861972 and rs1861973 association is located in the intron of EN2. Resequencing of the intron identified only one additional rare SNP, which indicated that further common SNPs of similar frequency as that of the _rs1861972_-rs1861973 A-C haplotype are unlikely. Moreover, single- and multi-SNP association data indicate that rs1861972 and rs1861973, and possibly rs2361688, are candidate risk alleles that function together in cis to increase risk for ASD. The lack of association for some core A-C haplotypes in our three- and four-marker haplotype analysis could then be the result of unidentified epistatic genetic or environmental factors (Gharani et al. 2004). The prediction that the nonassociated G-T haplotype could potentially disrupt the binding of the LvC and CBP transcription factors supports this hypothesis. The Ets family of transcription factors has been shown to bind to the LvC consensus sequence, which has been implicated in neuronal differentiation (Gunther and Graves 1994; Hippenmeyer et al. 2005). CBP acts as a coactivator that promotes the interaction between tissue-specific transcription factors and the basal transcriptional machinery (Song et al. 2002). The nonassociated alleles of rs186172 and rs1861973 may then influence the expression of EN2, but it has yet to be demonstrated that the intron acts as a _cis_-regulatory sequence. Ongoing expression analysis is being done to investigate whether the intron acts as a _cis_-regulatory element and, if so, whether these putative transcription factor binding sites contribute to its activity.
The second, equally possible hypothesis is that the risk allele(s) maps outside the tested region or to some segment of the promoter or 3′ UTR that is in LD with rs1861972 and rs1861973. Future LD and association analyses of all markers mapped within the entire EN2 locus will investigate the possibility of more-complex LD patterns across the gene, such as the LD interdigitations across haplotype blocks that have been observed for a number of genomic regions (Carlson et al. 2004; Hinds et al. 2005).
Although the identity of the risk allele(s) is yet to be defined, we used this large combined sample of 518 families in an attempt to quantify the effect of this locus on ASD susceptibility. We estimated a modest relative risk for the predisposing A-C haplotype, but, because the risk haplotype occurs at such a high frequency, this translates into a large PAR—an influence on as many as 40% of autism cases in the general population. These data suggest a highly significant role for variation at this locus in the etiology of ASD.
Since the relative risk and PAR are calculated from sample populations ascertained for multiple affected siblings, the actual risk estimates may be different in the general ASD population. Estimates of relative risk in multiplex sibships, compared with singleton families, are thought to be distorted depending on the background heritability (a multilocus model of the disorder). In the case of multiplex families, the relative risk is, in fact, expected to be deflated at a rate that is proportional to the increase in background heritability and to be more accurately estimated in situations of low background heritability (Risch 2001). Given the complex polygenic model of inheritance, with anywhere between 3 and 15 interacting loci, that has been proposed for ASD (Risch et al. 1999), it is not clear how the true mode of inheritance in our sample may have affected our risk estimates. Further studies in other populations or single-ascertainment family-based data sets should confirm the true impact of this locus.
EN2 maps to chromosome 7q36, and suggestive linkage has been observed for this region in two different studies that used the AGRE data set (Liu et al. 2001; Alarcon et al. 2002). In the initial linkage analysis using 110 AGRE families, fine mapping on distal chromosome 7 demonstrated that D7S483, located 5.5 Mb proximal to EN2, displayed a LOD score of 2.13 (Liu et al. 2001). QTL analysis using the same set of microsatellite markers and 152 AGRE families demonstrated suggestive linkage of QTLs implicated in general language performance to 7q36 (_P_=.001) (Alarcon et al. 2002). Recently, Yonan et al. (2003) performed a genome scan on a set of 345 AGRE families (which includes most of the AGRE I and AGRE II samples in our present study) and observed only minimal linkage at distal chromosome 7 (LOD<1.3). However, since none of these studies used markers that are distal to _EN2,_ it is difficult to determine whether the contribution of _EN2_ to these linkage findings has been properly assessed. Genome-scan analysis of the NIMH data set also provided a nominal positive LOD score for chromosome 7q; however, this was at marker _D7S1804_ (maximum LOD score 0.93), which is ∼23 Mb proximal to _EN2_ (Risch et al. 1999). Given the modest relative risk and high haplotype frequency of the A-C predisposing haplotype, it is likely that the risk allele(s) at this locus will typically be transmitted from both parents, making the contribution of this locus difficult to detect by conventional linkage analysis (Risch and Merikangas 1996). Despite the high population impact of this common risk variant, very large samples may be required to obtain a significant LOD score. Considering our association results and the availability of this large sample set of >500 families, it will now be worthwhile to further investigate linkage of 7q36 to ASD by use of a set of genetic markers that span the EN2 locus.
Previously, a case-control study in a French population reported significant association of a _Pvu_II RFLP located in the 5′ region of EN2 with autism (Petit et al. 1995). We have now mapped the position of this polymorphism to the promoter region between rs6150410 and rs1345514 and have demonstrated that, within the original AGRE I data set, this polymorphism is not associated with ASD. This difference in results could either indicate a false-positive result in the case-control study as a result of population stratification or reflect population-specific variation in LD between markers and different causal variants within this gene. This last possibility is consistent with the _Pvu_II RFLP being in linkage equilibrium with both rs1861972 and rs1861973 in the AGRE I sample.
Our association data, which provide strong genetic evidence that EN2 is involved in ASD susceptibility, are further supported by previous mouse functional data (Millen et al. 1994, 1995; Kuemerle et al. 1997; Baader et al. 1998, 1999). Two mouse mutants already exist for En2: a knockout and a transgenic that misexpresses the gene in a subset of Purkinje cells. Interestingly, both mouse mutants cause an autistic-like cerebellar phenotype, including hypoplasia and a decrease in the number of Purkinje cells. Thus, both the activity and the spatial/temporal regulation of En2 are critical for normal cerebellar development. Neither mouse mutant has undergone an extensive behavioral analysis, but ongoing experiments are investigating whether the En2 knockout mouse displays characteristic behavioral phenotypes.
Our current studies now provide mechanistic insight into how risk alleles that affect EN2 expression might perturb neuronal development. Misexpression of mouse En2 in primary cortical cultures elicited a reduction in neuronal differentiation, as reflected by the number of process-bearing neurons that express βIII-tubulin. This finding is consistent, at the molecular level, with studies in which the Drosophila Engrailed protein was shown to directly repress the expression of βIII-tubulin (Serrano et al. 1997). These results suggest that an ASD risk allele that alters EN2 mRNA or protein levels or its spatial/temporal regulation could significantly affect neuronal differentiation.
In conclusion, the data presented in this article provide further genetic and functional evidence that EN2 may be acting as an ASD susceptibility locus. In light of the previous studies demonstrating En2 function during mouse cerebellar development, the similarities between mouse En2 mutants and autistic cerebellar phenotypes, and the activation of the cerebellum during cognitive tasks that are abnormal in ASD (Kim et al. 1994; Millen et al. 1994, 1995; Raichle et al. 1994; Gao et al. 1996; Akshoomoff et al. 1997; Allen et al. 1997; Courchesne and Allen 1997; Kuemerle et al. 1997; Baader et al. 1998, 1999; Allen and Courchesne 2003; Corina et al. 2003; McDermott et al. 2003), our studies further support the hypothesis that developmental abnormalities of the cerebellum contribute to the behavioral deficits observed in ASD.
Acknowledgments
We thank Cure Autism Now and the Autism Genetic Resource Exchange (AGRE) for supplying the resources necessary for this study. We also thank Jay Tischfield and the Rutgers Cell and DNA Repository for providing the AGRE and National Institute of Mental Health (NIMH) samples. The NIMH samples were provided by NIMH Center for Collaborative Genetic Studies on Mental Disorders grant MH068457 (to Jay Tischfield). This work was supported in part by research grants from the NIMH (R01 MH70366 [to L.M.B.] and R01 MH70366 [to N.G.]), the March of Dimes Birth Defects Foundation (12-FY01-110 [to L.M.B.]), the National Alliance for Autism Research (to L.M.B., J.H.M., and E.D.-B.), the New Jersey Governor’s Council on Autism (to L.M.B., J.H.M., and E.D.-B.), the Whitehall Foundation (2001-12-54-APL [to J.H.M.]), and the National Institutes of Health (P01 ES11256, P30 ES05022, and USEPA-R829391 [to E.D.-B.]); a National Research Service Award Individual MD/PhD Fellowship from the National Institute of Neurological Disorders and Stroke (1 F30 NS48649-01 [to I.R.]); and a Molecular and Developmental Basis of Mental Health and Aging training grant (MH/AG-19957 [to R.B.]). Most importantly, we thank the families who have participated in and contributed to these studies.
The collection of data and biomaterials in one project that participated in the NIMH Autism Genetics Initiative has been supported by National Institutes of Health grants MH52708, MH39437, MH00219, and MH00980; by National Health Medical Research Council grant 0034328; by grants from the Scottish Rite, the Spunk Fund, the Rebecca and Solomon Baker Fund, the APEX Foundation, the National Alliance for Research in Schizophrenia and Affective Disorders, and the endowment fund of the Nancy Pritzker Laboratory (Stanford); and by gifts from the Autism Society of America, the Janet M. Grace Pervasive Developmental Disorders Fund, and families and friends of individuals with autism. The Principal Investigators and Coinvestigators were Neil Risch, Richard M. Myers, Donna Spiker, Linda J. Lotspeich, Joachim Hallmayer, Helena C. Kraemer, Roland D. Ciaranello, and Luca L. Cavalli-Sforza (Stanford University, Stanford) and William M. McMahon and P. Brent Petersen (University of Utah, Salt Lake City). The Stanford team is indebted to the parent groups and the clinician colleagues who referred families. The Stanford team extends our gratitude to the families with individuals with autism who were our partners in this research.
Appendix A
Members of the AGRE Consortium
Daniel H. Geschwind (University of California at Los Angeles), Maya Bucan (University of Pennsylvania, Philadelphia), W. Ted Brown (New York State Institute for Basic Research in Developmental Disabilities, Staten Island), Joseph Buxbaum (Mount Sinai School of Medicine, New York), Edwin H. Cook, Jr. (University of Chicago), T. Conrad Gilliam (Columbia Genome Center, New York), David A. Greenberg (Mount Sinai Medical Center, New York), David H. Ledbetter (University of Chicago), Bruce Miller (University of California at San Francisco), Stanley F. Nelson (University of California at Los Angeles School of Medicine), Jonathon Pevsner (Kennedy Kreiger Institute, Baltimore), Jerome I. Rotter (Cedar-Sinai Medical Center, Los Angeles), Gerald D. Schellenberg (University of Washington, Seattle), Carol A. Sprouse (Children’s National Medical Center, Baltimore), Rudolph E. Tanzi (Massachusetts General Hospital, Boston), Kirk C. Wilhelmsen (University of California at San Francisco), and Jeremy M. Silverman (Mount Sinai Medical School, New York).
Appendix B: Supplemental Material
DNA Analysis
SNPs rs1345514, rs3824068, rs1861972, and rs1861973 were genotyped by simplex Pyrosequencing assays with use of the automated PSQ HS 96A platform as described elsewhere (Ronaghi et al. 1998; Ahmadian et al. 2000). The insertion/deletion polymorphism rs6150410 was genotyped by PCR amplification followed by allele separation on a 10% polyacrylamide gel. Genotypes were called on the basis of band-size differences of 263 bp (insertion allele) and 254 bp (deletion allele). Primers were designed using publicly available software. For rs6460013, rs7794177, rs1264067, rs3808332, rs3808331, and rs4717034, a tetra-primer ARMS-PCR strategy was used to genotype individuals (Ye et al. 2001; Gharani et al. 2004). For _Pvu_II and rs2361688, an RFLP assay with _Pvu_II and _Hinf_I, respectively, was used. For rs3735652, ss38341503, and rs3808329, a ligase-detection reaction and the Luminex 100 flow cytometry platform was used (Iannone et al. 2000).
The primers used were for rs6150410 (forward [F], ctagagggaaaacggggttc; reverse [R], aactccgcaaggtgtttcag), _Pvu_II (F, tggcagatgtgtgcctag; R, ccagaccggtcatctcgttttc), rs1345514 (F, agagctgccctatcggatgtt; R, aaactaattttgccggagagc; Pyrosequencing primer, cccaccaaacaccc), rs3735652 (F, ctgtcggtgagctcggact; R, tggaagacagagaggggaga; Luminex primers: G, gcgacattgtgtgaagctgacg; C, gcgacattgtgtgaagctgacc; common, ccggcccgggcagcggc), rs6460013 (forward outer [Fo], cgcatctcttcccagcccctagc; reverse outer [Ro], tgcatcctcctgagtcccaccg; forward inner [Fi], ccttccctacgatcttccaactcggg; reverse inner [Ri], gcatgcgtccccggcctaga), rs7794177 (Fo, cacagggaaggaggaaaataaa; Ro, tcatcagaaatatgcacgcata; Fi, agatctgcgattttaaaaaactaact; Ri, ttgatgatttctacaaggacaagg), rs3824068 (F, cattaacaagagccccagga; R, ccatgagagcacacacccta; Pyrosequencing primer, cagtgcctgtcttgc), rs2361688 (F, tgcacctacccctaccaaagcca; R, tgtggatctccttggaggccct), rs3824067 (Fo, ctccaaggagatccacattcctctt; Ro, gggtcgctgtaaggcttctaggac; Fi, cgagatgctccctaaagcccaa; Ri, ggtttcaatttgtgcggtgattcaa), rs1861972 (F, catacaccgcacaaattgaaac; R, gattcagacttatgaacctgacctg; Pyrosequencing primer, caccactccctgcca), rs1861973 (F, catacaccgcacaaattgaaac; R, gattcagacttatgaacctgacctg; Pyrosequencing primer, ccttacagcgaccct), ss38341503 (F, ccttctgctctcctccctct; R, ggcctggtttttcctagtcc; Luminex primers: C, ccctcctgtcctcagggcc; T, ccctcctgtcctcagggct; common, cacctgcccctgattcccac), rs3808332 (Fo, gcccttggctgggagtcataga; Ro, gggactatggggcaggcctagt; Fi, tttcccagtcttctctcctccacc; Ri, gcggtaggtgctgagagcga), rs3808331 (Fo, agtcttctctcctcccctctct; Ro, gaggactgcgtgtgatgtaagt; Fi, gaaagtgtggggagttttgatt; Ri, tctagataaaagtaaaactcctggat), rs4717034 (Fo, ccgccatccctgttcctgaaca; Ro, gtgtgccacccaataggcaccg; Fi, ccctcaccaagtggtggaggtcagt; Ri, gactgggcatgggctcaccg), and rs3808329 (F, gtttgtgttggcttggtgag; R, ccctctacagagccttctgc; Luminex primers: G, cctctcctcaccctcctgcg; A, cctctcctcaccctcctgca; common, ctaactccctcctccttctcc).
For rs6460013, rs7794177, rs1264067, rs3808332, rs3808331, and rs4717034, tetra-primer ARMS-PCR was conducted in a 10-μl reaction containing 1 pmol of each of the inner primers and 0.1 pmol of each of the outer primers, 0.25 mM dNTP, 1.5 mM MgCl2, 25 mM KCl, and 10 mM Tris-HCl (pH 8.3 for rs6460013, rs7794177, and rs1264067; pH 8.8 for rs3808332 and rs4717034; and pH 9.2 for rs3808331). For rs1264067, the same conditions were used, except for 3.5 mM MgCl2. For rs6150410, the same conditions were used as for rs1264067, with the following exceptions: 0.4 μM of each primer and 10 mM Tris-HCl (pH 8.8). Standard cycling conditions were used: an initial cycle at 94°C for 4 min; 35 cycles at 94°C for 30 s, _T_m for 30 s, and 74°C for 30 s; and a final step at 74°C for 10 min (_T_m = 50°C for rs1264067, 55°C for rs6460013 and rs7794177, 56°C for rs3808331, 59°C for rs6150410, and 62°C for rs3808332 and rs4717034).
SNPs rs1345514, rs3824068, rs1861972, and rs1861973 were genotyped using a Pyrosequencing assay. PCR was conducted in a 20-μl reaction containing 0.25 mM dNTP, 1.875 mM MgCl2, 6.25 mM KCl, 1.25 mM Tris-HCl (pH 9.0), 0.1% Triton X-100, and 0.05 μM of each primer for rs1861972 and rs1861973 and 0.075 μM of each primer for rs3824068. For rs1345514, the same PCR conditions as for rs1861972 were used, except for the use of 0.025 μM of each primer; 0.01 mM of dATP, dCTP, and dTTP; 2.5 μM dGTP; 7.5 μM 7-deaza-2′-deoxyguanosine triphosphate; and 1.25 mM MgCl2. Standard cycling conditions were used: an initial cycle at 94°C for 4 min; 40 cycles at 94°C for 30 s, _T_m for 30 s, and 74°C for 30 s; and a final step at 74°C for 10 min (_T_m = 60°C for rs3824068, rs1861972, and rs1861973 and 62°C for rs3808332 and rs4717034).
SNPs rs3808329, ss38341503, and rs3735652 were genotyped using Luminex 100 flow cytometry platform. For rs3808329 and ss38341503, PCR was conducted in a 20-μl reaction containing 0.4 μM of each primer, 0.125 mM dNTP, 1.875 mM MgCl2, 31.25 mM KCl, and 12.5 mM Tris-HCl (pH 8.8 for rs3808329 and pH 8.3 for ss38341503). For rs3735652, the reaction contained 1.25 mM MgCl2; 1.25 mM Tris-HCl (pH 9.0); 6.25 mM KCl; 0.1% Triton X-100; 0.01 mM dATP, dCTP, and dTTP; 2.5 μM dGTP; and 7.5 μM 7-deaza-2′-deoxyguanosine triphosphate. Standard cycling conditions were used: an initial cycle at 94°C for 4 min; 35 cycles at 94°C for 30 s, _T_m for 30 s, and 74°C for 40 s; and a final step at 74°C for 10 min (_T_m = 66.3°C for rs3808329, 58°C for rs3735652, and 57°C for ss38341503).
rs2361688 and _Pvu_II were genotyped using an RFLP assay. PCR was conducted in a 10-μl reaction containing 0.1 μM of each primer and 0.25 mM dNTP. For rs2361688, 10 mM Tris-HCl (pH 9.2), 1.5 mM MgCl2, and 25 mM KCl were used. For _Pvu_II, 1 mM Tris-HCl (pH 9.2), 5 mM KCl, 0.1% Triton X-100, and 1.25 mM MgCl2 were used. Cycling conditions were as follows: an initial cycle at 94°C for 4 min; 35 cycles at 94°C for 30 s, _T_m for 30 s, and 74°C for 30 s; and a final step at 74°C for 10 min (_T_m = 64°C for rs2361688 and 61°C for _Pvu_II).
Sequencing Analysis
Four sets of primers were used to sequence the EN2 intron, as follows. PCR product 1: F, ctgtcggtgagctcggactcgg; R, gccctgcagagatgctggatatat. PCR product 2: F, tagaaaggaccttctctcaggg; R, gtggttggaaacccagacagagat. PCR product 3: F, cattaacaagagccccaggaccagaag; R, gacaaggtcagctgggctac. PCR product 4: F, ttccccatggatagcaggtcctag; R, ggtctcgaaaaccaaagaagaagaacccga. For product 1, PCR was conducted in a 50-μl reaction containing 2 mM MgCl2; 5 mM KCl; 1 mM Tris-HCl (pH 9.2); 0.1% Triton X-100; 2 μl of GC melt (BD Biosciences); 5% dimethyl sulfoxide (Sigma); 0.4 μM of each primer; 8 μM dATP, dCTP, and dTTP; 2 μM dGTP; and 6 μM 7-deaza-2′-deoxyguanosine triphosphate. Standard cycling conditions were used: an initial cycle at 94°C for 1 min; 35 cycles at 94°C for 40 s, 59°C for 30 s, and 68°C for 3 min 30 s; and a final step at 68°C for 3 min. For products 2, 3, and 4, PCR was conducted in a 20-μl reaction containing 0.625 mM MgCl2, 5 mM KCl, 1 mM Tris-HCl (pH 9.2), 0.1% Triton X-100, 0.25 mM dNTPs, and 0.5 ng/μl of each primer. Standard cycling conditions for PCR products 2, 3, and 4 were used: an initial cycle at 94°C for 5 min; 35 cycles at 94°C for 30 s, _T_m for 30 s, and 74°C for 60 s; and a final step at 74°C for 10 min (_T_m = 60°C for PCR products 2 and 4 and 62°C for PCR product 3).
Web Resources
The URLs for data presented herein are as follows:
- Autism Genetic Resource Exchange (AGRE), http://www.agre.org/
- dbSNP, http://www.ncbi.nlm.nih.gov/SNP/
- National Institute of Mental Health (NIMH) Data Set, http://zork.wustl.edu/nimh/NIMH_initiative/NIMH_initiative_link.html
- NIMH Center for Collaborative Genetic Studies, http://zork.wustl.edu/nimh/home/d_autism.html
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for ASD and EN2) [PubMed]
- Pyrosequencing Technical Support, http://techsupport.pyrosequencing.com/
- Tetra-primer ARMS-PCR, http://cedar.genetics.soton.ac.uk/public_html/primer1.html
- TRANSFAC, http://www.gene-regulation.com/pub/databases.html#transfac
- Primer3, http://www.genome.wi.mit.edu/genome-software/other/primer3.html
References
- Abecasis GR, Cookson WO (2000) GOLD—graphical overview of linkage disequilibrium. Bioinformatics 16:182–183 [DOI] [PubMed] [Google Scholar]
- Ahmadian A, Gharizadeh B, Gustafsson AC, Sterky F, Nyren P, Uhlen M, Lundeberg J (2000) Single nucleotide polymorphism analysis by pyrosequencing. Anal Biochem 280:103–110 [DOI] [PubMed] [Google Scholar]
- Akshoomoff NA, Courchesne E, Townsend J (1997) Attention coordination and anticipatory control. Int Rev Neurobiol 41:575–598 [DOI] [PubMed] [Google Scholar]
- Alarcon M, Cantor RM, Liu J, Gilliam TC, the Autism Genetic Research Exchange Consortium, Geschwind DH (2002) Evidence for a language quantitative trait locus on chromosome 7q in multiplex autism families. Am J Hum Genet 70:60–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen G, Buxton RB, Wong EC, Courchesne E (1997) Attentional activation of the cerebellum independent of motor involvement. Science 275:1940–1943 [DOI] [PubMed] [Google Scholar]
- Allen G, Courchesne E (2003) Differential effects of developmental cerebellar abnormality on cognitive and motor functions in the cerebellum: an fMRI study of autism. Am J Psychiatry 160:262–273 [DOI] [PubMed] [Google Scholar]
- Altshuler D, Hirschhorn JN, Klannemark M, Lindgren CM, Vohl MC, Nemesh J, Lane CR, Schaffner SF, Bolk S, Brewer C, Tuomi T, Gaudet D, Hudson TJ, Daly M, Groop L, Lander ES (2000) The common PPARγ Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat Genet 26:76–80 [DOI] [PubMed] [Google Scholar]
- Auranen M, Vanhala R, Varilo T, Ayers K, Kempas E, Ylisaukko-Oja T, Sinsheimer JS, Peltonen L, Jarvela I (2002) A genomewide screen for autism-spectrum disorders: evidence for a major susceptibility locus on chromosome 3q25-27. Am J Hum Genet 71:777–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baader SL, Sanlioglu S, Berrebi AS, Parker-Thornburg J, Oberdick J (1998) Ectopic overexpression of Engrailed-2 in cerebellar Purkinje cells causes restricted cell loss and retarded external germinal layer development at lobule junctions. J Neurosci 18:1763–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baader SL, Vogel MW, Sanlioglu S, Zhang X, Oberdick J (1999) Selective disruption of “late onset” sagittal banding patterns by ectopic expression of Engrailed-2 in cerebellar Purkinje cells. J Neurosci 19:5370–5379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey A, Luthert P, Dean A, Harding B, Janota I, Montgomery M, Rutter M, Lantos P (1998) A clinicopathological study of autism. Brain 121:889–905 [DOI] [PubMed] [Google Scholar]
- Bartlett CW, Gharani N, Millonig JH, Brzustowicz LM (2005) Three autism candidate genes: a synthesis of human genetic analysis with other disciplines. Int J Dev Neurosci 23:221–234 [DOI] [PubMed] [Google Scholar]
- Bauman ML, Kemper TL (1985) Histoanatomic observations of the brain in early infantile autism. Neurology 35:866–874 [DOI] [PubMed] [Google Scholar]
- ——— (1986) Developmental cerebellar abnormalities: a consistent finding in early infantile autism. Neurology Suppl 1 36:190 [Google Scholar]
- ——— (1994) Neuroanatomic observations of the brain in autism. In: The neurobiology of autism. Johns Hopkins University Press, Baltimore, pp 119–145 [Google Scholar]
- Carey RG, Li B, DiCicco-Bloom E (2002) Pituitary adenylate cyclase activating polypeptide anti-mitogenic signaling in cerebral cortical progenitors is regulated by p57Kip2-dependent CDK2 activity. J Neurosci 22:1583–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA (2004) Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet 74:106–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corina DP, San Jose-Robertson L, Guillemin A, High J, Braun AR (2003) Language lateralization in a bimanual language. J Cogn Neurosci 15:718–730 [DOI] [PubMed] [Google Scholar]
- Courchesne E (1997) Brainstem, cerebellar and limbic neuroanatomical abnormalities in autism. Curr Opin Neurobiol 7:269–278 [DOI] [PubMed] [Google Scholar]
- Courchesne E, Allen G (1997) Prediction and preparation, fundamental functions of the cerebellum. Learn Mem 4:1–35 [DOI] [PubMed] [Google Scholar]
- Courchesne E, Carper R, Akshoomoff N (2003) Evidence of brain overgrowth in the first year of life in autism. JAMA 290:337–344 [DOI] [PubMed] [Google Scholar]
- Courchesne E, Karns CM, Davis HR, Ziccardi R, Carper RA, Tigue ZD, Chisum HJ, Moses P, Pierce K, Lord C, Lincoln AJ, Pizzo S, Schreibman L, Haas RH, Akshoomoff NA, Courchesne RY (2001) Unusual brain growth patterns in early life in patients with autistic disorder: an MRI study. Neurology 57:245–254 [DOI] [PubMed] [Google Scholar]
- Courchesne E, Yeung-Courchesne R, Press GA, Hesselink JR, Jernigan TL (1988) Hypoplasia of cerebellar vermal lobules VI and VII in autism. N Engl J Med 318:1349–1354 [DOI] [PubMed] [Google Scholar]
- Dudbridge F (2003) Pedigree disequilibrium tests for multilocus haplotypes. Genet Epidemiol 25:115–121 [DOI] [PubMed] [Google Scholar]
- Folstein S, Rutter M (1977) Infantile autism: a genetic study of 21 twin pairs. J Child Psychol Psychiatry 18:297–321 [DOI] [PubMed] [Google Scholar]
- Folstein SE, Rosen-Sheidley B (2001) Genetics of autism: complex aetiology for a heterogeneous disorder. Nat Rev Genet 2:943–955 [DOI] [PubMed] [Google Scholar]
- Gaffney GR, Kuperman S, Tsai LY, Minchin S, Hassanein KM (1987) Midsagittal magnetic resonance imaging of autism. Br J Psychiatry 151:831–833 [DOI] [PubMed] [Google Scholar]
- Gao JH, Parsons LM, Bower JM, Xiong J, Li J, Fox PT (1996) Cerebellum implicated in sensory acquisition and discrimination rather than motor control. Science 272:545–547 [DOI] [PubMed] [Google Scholar]
- Garner C, Slatkin M (2003) On selecting markers for association studies: patterns of linkage disequilibrium between two and three diallelic loci. Genet Epidemiol 24:57–67 [DOI] [PubMed] [Google Scholar]
- Geschwind DH, Sowinski J, Lord C, Iversen P, Shestack J, Jones P, Ducat L, Spence SJ, the AGRE Steering Committee (2001) The Autism Genetic Resource Exchange: a resource for the study of autism and related neuropsychiatric conditions. Am J Hum Genet 69:463–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharani N, Benayed R, Mancuso V, Brzustowicz LM, Millonig JH (2004) Association of the homeobox transcription factor, ENGRAILED 2, 3, with autism spectrum disorder. Mol Psychiatry 9:474–484 [DOI] [PubMed] [Google Scholar]
- Gunther CV, Graves BJ (1994) Identification of ETS domain proteins in murine T lymphocytes that interact with the Moloney murine leukemia virus enhancer. Mol Cell Biol 14:7569–7580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto T, Tayama M, Murakawa K, Yoshimoto T, Miyazali M, Harada M, Kuroda Y (1995) Development of the brainstem and cerebellum in autistic patients. J Autism Dev Disord 25:1–18 [DOI] [PubMed] [Google Scholar]
- Hinds DA, Stuve LL, Nilsen GB, Halperin E, Eskin E, Ballinger DG, Frazer KA, Cox DR (2005) Whole-genome patterns of common DNA variation in three human populations. Science 307:1072–1079 [DOI] [PubMed] [Google Scholar]
- Hippenmeyer S, Vrieseling E, Sigrist M, Portmann T, Laengle C, Ladle DR, Arber S (2005) A developmental switch in the response of DRG neurons to ETS transcription factor signaling. PLoS Biol 3:e159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iannone MA, Taylor JD, Chen J, Li MS, Rivers P, Slentz-Kesler KA, Weiner MP (2000) Multiplexed single nucleotide polymorphism genotyping by oligonucleotide ligation and flow cytometry. Cytometry 39:131–140 [PubMed] [Google Scholar]
- Kemper TL, Bauman ML (1993) The contribution of neuropathologic studies to the understanding of autism. Behav Neurol 11:175–187 [PubMed] [Google Scholar]
- Kim SG, Ugurbil K, Strick PL (1994) Activation of a cerebellar output nucleus during cognitive processing. Science 265:949–951 [DOI] [PubMed] [Google Scholar]
- Kleiman MD, Neff S, Rosman NP (1992) The brain in infantile autism: are posterior fossa structures abnormal? Neurology 42:753–760 [DOI] [PubMed] [Google Scholar]
- Kuemerle B, Zanjani H, Joyner A, Herrup K (1997) Pattern deformities and cell loss in Engrailed-2 mutant mice suggest two separate patterning events during cerebellar development. J Neurosci 17:7881–7889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Nyholt DR, Magnussen P, Parano E, Pavone P, Geschwind D, Lord C, Iversen P, Hoh J, the Autism Genetic Resource Exchange Consortium, Ott J, Gilliam TC (2001) A genomewide screen for autism susceptibility loci. Am J Hum Genet 69:327–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan C, Joyner AL (1989) PvuII and RsaI RFLPs for the human homeo box-containing gene EN2. Nucleic Acids Res 17:2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu N, DiCicco-Bloom E (1997) Pituitary adenylate cyclase-activating polypeptide is an autocrine inhibitor of mitosis in cultured cortical precursor cells. Proc Natl Acad Sci USA 94:3357–3362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin ER, Monks SA, Warren LL, Kaplan NL (2000) A test for linkage and association in general pedigrees: the pedigree disequilibrium test. Am J Hum Genet 67:146–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott KB, Petersen SE, Watson JM, Ojemann JG (2003) A procedure for identifying regions preferentially activated by attention to semantic and phonological relations using functional magnetic resonance imaging. Neuropsychologia 41:293–303 [DOI] [PubMed] [Google Scholar]
- Millen KJ, Hui CC, Joyner AL (1995) A role for En-2 and other murine homologues of Drosophila segment polarity genes in regulating positional information in the developing cerebellum. Development 121:3935–3945 [DOI] [PubMed] [Google Scholar]
- Millen KJ, Wurst W, Herrup K, Joyner AL (1994) Abnormal embryonic cerebellar development and patterning of postnatal foliation in two mouse Engrailed-2 mutants. Development 120:695–706 [DOI] [PubMed] [Google Scholar]
- Murakami JW, Courchesne E, Press GA, Yeung-Courchesne R, Hesselink JR (1989) Reduced cerebellar hemisphere size and its relationship to vermal hypoplasia in autism. Arch Neurol 46:689–694 [DOI] [PubMed] [Google Scholar]
- Nicot A, DiCicco-Bloom E (2001) Regulation of neuroblast mitosis is determined by PACAP receptor isoform expression. Proc Natl Acad Sci USA 98:4758–4763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell JR, Weeks DE (1998) PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmen SJ, van Engeland H, Hof PR, Schmitz C (2004) Neuropathological findings in autism. Brain 127:2572–2583 [DOI] [PubMed] [Google Scholar]
- Petit E, Herault J, Martineau J, Perrot A, Barthelemy C, Hameury L, Sauvage D, Lelord G, Muh JP (1995) Association study with two markers of a human homeogene in infantile autism. J Med Genet 32:269–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raichle ME, Fiez JA, Videen TO, MacLeod AM, Pardo JV, Fox PT, Petersen SE (1994) Practice-related changes in human brain functional anatomy during nonmotor learning. Cereb Cortex 4:8–26 [DOI] [PubMed] [Google Scholar]
- Riley B (2004) Linkage studies of schizophrenia. Neurotox Res 6:17–34 [DOI] [PubMed] [Google Scholar]
- Risch N (2001) Implications of multilocus inheritance for gene-disease association studies. Theor Popul Biol 60:215–220 [DOI] [PubMed] [Google Scholar]
- Risch N, Merikangas K (1996) The future of genetic studies of complex human diseases. Science 273:1516–1517 [DOI] [PubMed] [Google Scholar]
- Risch N, Spiker D, Lotspeich L, Nouri N, Hinds D, Hallmayer J, Kalaydjieva L, et al (1999) A genomic screen of autism: evidence for a multilocus etiology. Am J Hum Genet 65:493–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritvo ER, Freeman BJ, Mason-Brothers AM, Mo A, Ritvo AM (1985) Concordance for the syndrome of autism in 40 pairs of affected twins. Am J Psychiatry 142:74–77 [DOI] [PubMed] [Google Scholar]
- Ritvo ER, Freeman BJ, Scheibel AB, Duong T, Robinson H, Guthrie D, Ritvo A (1986) Lower Purkinje cell count in the cerebella of four autistic subjects: initial findings of the UCLA-NSAC Autopsy Research report. Am J Psychiatry 143:862–866 [DOI] [PubMed] [Google Scholar]
- Ronaghi M, Uhlen M, Nyren P (1998) A sequencing method based on real-time pyrophosphate. Science 281:363–365 [DOI] [PubMed] [Google Scholar]
- Serrano N, Brock HW, Maschat F (1997) β3-tubulin is directly repressed by the Engrailed protein in Drosophila. Development 124:2527–2536 [DOI] [PubMed] [Google Scholar]
- Song CZ, Keller K, Murata K, Asano H, Stamatoyannopoulos G (2002) Functional interaction between coactivators CBP/p300, PCAF, and transcription factor FKLF2. J Biol Chem 277:7029–7036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger JD (2000) In: Rao DC, Province MA (eds) Genetic dissection of complex traits. Vol. 42. Academic Press, New York, pp 351–391 [Google Scholar]
- Weeks DE, Lathrop M (1995) Polygenic disease: methods for mapping complex disease traits. Trends Genet 11:513–519 [DOI] [PubMed] [Google Scholar]
- Ye S, Dhillon S, Ke X, Collins AR, Day IN (2001) An efficient procedure for genotyping single nucleotide polymorphisms. Nucleic Acids Res 29:E88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonan AL, Alarcon M, Cheng R, Magnossun PKE, Spence SJ, Palmer AA, Grunn A, Juo SH, Terwilliger JD, Liu J, Cantor RM, Geschwind DH, Gilliam TC (2003) A genomewide screen of 345 families for autism-susceptibility loci. Am J Hum Genet 73:886–897 [DOI] [PMC free article] [PubMed] [Google Scholar]