Regulation of an intergenic transcript controls adjacent gene transcription in Saccharomyces cerevisiae (original) (raw)
Abstract
Recent studies have revealed that transcription of noncoding, intergenic DNA is abundant among eukaryotes. However, the functions of this transcription are poorly understood. We have previously shown that in Saccharomyces cerevisiae, expression of an intergenic transcript, SRG1, represses the transcription of the adjacent gene, SER3, by transcription interference. We now show that SRG1 transcription is regulated by serine, thereby conferring regulation of SER3, a serine biosynthetic gene. This regulation requires Cha4, a serine-dependent activator that binds to the SRG1 promoter and is required for SRG1 induction in the presence of serine. Furthermore, two coactivator complexes, SAGA and Swi/Snf, are also directly required for activation of SRG1 and transcription interference of SER3. Taken together, our results elucidate a physiological role for intergenic transcription in the regulation of SER3. Moreover, our results demonstrate a mechanism by which intergenic transcription allows activators to act indirectly as repressors.
Keywords: Intergenic transcription, noncoding RNA, transcription interference, transcription
The analysis of genome-wide transcription in many organisms, including bacteria, yeast, Drosophila, Arabidopsis, mouse, and human, has yielded a common, yet unexpected feature. In addition to the transcription of protein-coding sequences, there is also widespread transcription across non-protein-coding regions (for reviews, see Mattick 2003; Morey and Avner 2004; Huttenhofer et al. 2005; Johnson et al. 2005). In humans, such noncoding transcripts have been detected from intergenic DNA, from introns, and from antisense transcription of exons (Chen et al. 2002; Kapranov et al. 2002; Saha et al. 2002; Bertone et al. 2004; Kampa et al. 2004; Cheng et al. 2005). Additional analysis suggests that much of this transcription is regulated and, thus, may itself play regulatory roles (Cawley et al. 2004; Kim et al. 2005). Recent experiments in the yeast Saccharomyces cerevisiae have also provided evidence for extensive transcription of noncoding regions (Hurowitz and Brown 2003; Havilio et al. 2005; Wyers et al. 2005).
One large and broad class of noncoding RNAs that has been studied in prokaryotes and eukaryotes plays direct roles in the regulation of gene expression (for reviews, see Bernstein and Allis 2005; Storz et al. 2005). These regulatory noncoding RNAs have been shown to function at many different levels of gene expression. In Escherichia coli, >50 small noncoding RNA regulators have been identified (for reviews, see Gottesman 2004; Storz et al. 2005). In eukaryotes, one intensively studied group, short interfering RNAs (siRNAs), has been shown to control both chromatin structure and mRNA stability (for reviews, see Meister and Tuschl 2004; Bernstein and Allis 2005). Other well studied eukaryotic categories include the Xist and Tsix RNAs that are involved in X-inactivation in mammals, and the roX1 and roX2 RNAs that are required for dosage compensation in Drosophila (for reviews, see Andersen and Panning 2003; Bernstein and Allis 2005). In addition to these examples, noncoding RNAs have been shown to regulate virtually all known steps of gene expression from transcription initiation to mRNA translation.
Several studies have suggested a second class of noncoding transcription that plays an important role in transcription regulation. In this second class, it is the act of transcription that confers activation, rather than the RNA product itself (for review, see Morey and Avner 2004). One example of this type of regulation occurs in the Drosophila bithorax complex (BX-C). Several early studies demonstrated that transcription occurs across noncoding regions at BX-C, suggesting a possible role in mediating repression by the Polycomb group (PcG) complexes (Lipshitz et al. 1987; Sanchez-Herrero and Akam 1989; Cumberledge et al. 1990). More recent studies have shown a correlation between intergenic transcription across PcG response elements (PREs) at the BX-C locus and the relief of silencing by PcG complexes (Bender and Fitzgerald 2002; Hogga and Karch 2002; Rank et al. 2002). Most recently, studies using a transgenic reporter system have demonstrated that transcription in either direction across a PRE blocks repression by PcG complexes and that this activated state is inheritable (Schmitt et al. 2005). A second example occurs at the 50-kb human β-globin locus, where developmentally regulated intergenic transcription occurs in three large chromatin subdomains and is required for the proper developmental regulation of the five globin genes within this locus (Gribnau et al. 2000). The domains of intergenic transcription correlate with the regions of DNase I sensitivity, again strongly suggesting that transcription confers chromatin changes.
In contrast to cases in which intergenic transcription activates gene expression, we recently reported a case in S. cerevisiae in which intergenic transcription represses gene expression (Martens et al. 2004). Our work identified a noncoding RNA, SRG1, which is transcribed from intergenic DNA, and showed that transcription of SRG1 across the promoter of the adjacent SER3 gene represses SER3 transcription. We provided evidence for a model in which transcription of SRG1 represses SER3 expression by transcription interference. However, the role of SRG1 in normal SER3 regulation was not addressed.
In this work, we use the S. cerevisiae SRG1/SER3 system to provide new insights into the physiological role in regulation by an intergenic transcript. First, we show that SRG1 transcription is induced by high serine levels, resulting in repression of SER3. Second, we identify the serine-responsive activator Cha4 as acting directly in SRG1 induction, thereby indirectly repressing SER3. Third, we show that Cha4 recruits the SAGA and Swi/Snf coactivator complexes to the SRG1 promoter in a serine-dependent manner. Finally, we show that both SAGA and Swi/Snf are required for SER3 repression by facilitating Cha4-dependent activation of SRG1. Taken together, our results show that a physiological response to serine, repression of SER3 transcription, occurs by activating SRG1 intergenic transcription. Based on these results and previous work that implicates Cha4 as a direct activator of the serine catabolism CHA1 gene (Holmberg and Schjerling 1996; Sabet et al. 2003), intergenic transcription provides a mechanism for a single protein to simultaneously activate and repress opposing pathways.
Results
Serine-dependent regulation of SER3 requires the expression of SRG1 from intergenic DNA
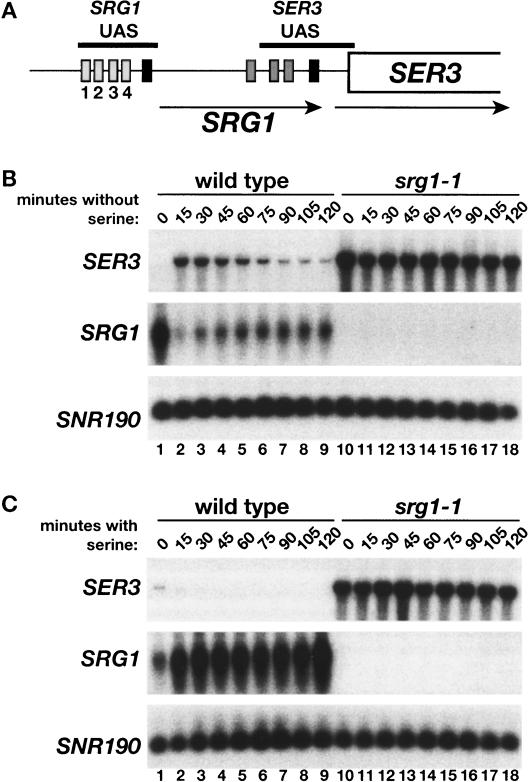
We have previously shown that transcription of SER3 is strongly repressed in YPD, a serine-containing medium, by transcription of SRG1 from intergenic DNA (Fig. 1A; Martens et al. 2004) To investigate a physiological role for this repression mechanism, we performed Northern analysis to assay the response of SER3 and SRG1 RNA levels to changes in serine levels. First, when wild-type cells were grown in minimal medium with serine (SD + ser) and then shifted to minimal medium without serine (SD), there was a rapid but transient increase in SER3 mRNA levels (Fig. 1B, lanes 1–9). Conversely, SRG1 RNA levels, initially high in the presence of serine, underwent a rapid but transient decrease when the cells were shifted from SD + ser to SD medium. When the complementary shift was performed, from SD to SD + ser medium, the opposite effect was observed, as SER3 mRNA levels decreased, while SRG1 RNA levels were induced to a significantly greater level (Fig. 1C, lanes 1–9). These results demonstrate that expression of SER3 and SRG1 are tightly but oppositely regulated by the availability of serine.
Figure 1.
Effect of serine on SER3 and SRG1 expression. (A) A schematic of SRG1 and SER3, showing TATA and putative UAS sequences conserved between S. cerevisiae and four related yeast strains. TATA elements are represented by black boxes, the putative SRG1 UAS elements by light-gray boxes 5′ of the SRG1 TATA (labeled 1–4), and the putative SER3 UAS elements by dark-gray boxes 5′ of SER3 TATA. The horizontal black bars mark the SRG1 and SER3 UAS regions that were amplified by PCR in ChIP experiments described later. The arrows indicate the orientation and positions of the SRG1 and SER3 transcripts. (B) Northern analysis of SER3 and SRG1 was performed on wild-type (FY2472) and srg1-1 (FY2471; contains a mutation of the SRG1 TATA) strains after a shift from SD + ser to SD medium. SNR190 served as a loading control. Total RNA was isolated at 15-min intervals. These data are representative of three independent experiments. (C) Northern analysis was performed on the strains described in B after the opposite shift, from SD to SD + ser. These data are representative of three independent experiments.
Next we tested whether transcription of SRG1 is required for the serine-dependent regulation of SER3. To do this we examined RNA levels in an srg1-1 mutant in which mutation of the SRG1 TATA sequence greatly reduces SRG1 RNA levels. In the srg1-1 mutant, SER3 was expressed at high levels independently of the presence or absence of serine (Fig. 1B [lanes 10–18], C [lanes 10–18]). Therefore, serine-dependent regulation of SER3 occurs indirectly through the serine-dependent control of SRG1 transcription.
Cha4 binds to the SRG1 promoter to control serine-dependent transcription of SRG1
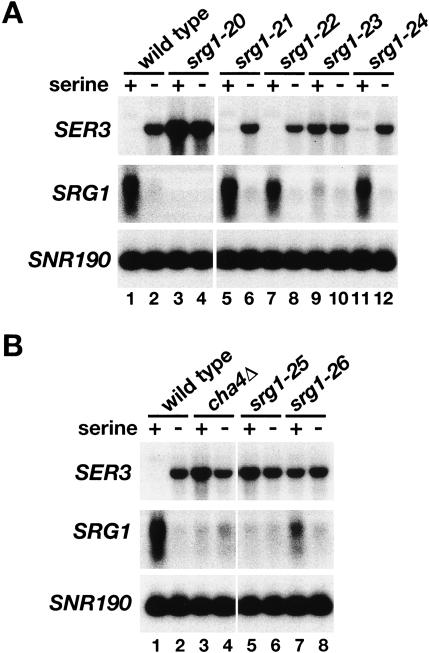
To test for a serine-dependent response element within the SRG1 promoter, we constructed mutations that delete four SRG1 promoter sequences that are highly conserved between S. cerevisiae and closely-related yeasts (labeled 1–4 in Fig. 1A; Martens et al. 2004). These mutants were then tested for SRG1 and SER3 mRNA levels in the presence or absence of serine. A deletion that encompasses all four regions (srg1-20) abolished SRG1 expression and caused SER3 derepression in SD + ser medium (Fig. 2A, cf. lanes 1 and 3). Deletion of three of the four individual conserved sequences (UAS1, UAS2, and UAS4) caused no effect on serine-dependent expression of SRG1 or SER3 (Fig. 2A). In contrast, deletion of UAS3 (srg1-23) resulted in significant decreases in the induction of SRG1 and repression of SER3 in SD + ser medium (Fig. 2B, cf. lanes 1 and 9).
Figure 2.
Identification of a serine response element in the SRG1 promoter. (A) Northern analysis of SER3 and SRG1 on a wild-type strain (FY2460) and on a series of srg1 promoter mutants. The srg1-20 mutant (FY2467) has the entire SRG1 UAS region deleted (Fig. 1A, regions 1–4). The srg1-21 (FY2464), srg1-22 (FY2465), srg1-23 (FY2466), and srg1-24 (FY2461) mutants are deletions of SRG1 UAS sequences 1, 2, 3, and 4, respectively (Fig. 1A). Total RNA was isolated from cells grown in SD + ser medium and from cells that had then been shifted to SD medium for 25 min. SNR190 served as a loading control. These data are representative of three independent experiments. (B) Northern analysis of SER3 and SRG1 was performed on wild-type (FY2460), _cha4_Δ (FY2459), srg1-25 (FY2463), and srg1-26 (FY2462) strains that were grown in SD + ser and SD medium as described in A. The srg1-25 mutant is a deletion of a putative Cha4-binding site in the SRG1 promoter and the srg1-26 mutant has a triple-point mutation within this putative Cha4-binding site. These data are representative of three independent experiments.
Further sequence inspection revealed that the UAS3 region overlaps a putative binding site (TGGAGATA CATCTCCA) for Cha4, an S. cerevisiae activator. Cha4 has been previously shown to be a serine-responsive activator of the CHA1 gene, which encodes an enzyme involved in serine catabolism (Holmberg and Schjerling 1996; Sabet et al. 2003). To determine whether Cha4 plays a role in activating SRG1 transcription we constructed and tested three mutations: a deletion of CHA4 (_cha4_Δ), and both a deletion (srg1-25) and a multiple-point mutant (srg1-26) of the putative Cha4-binding site. By Northern analysis, we observed greatly decreased levels of SRG1 RNA in all three mutants as compared with a wild-type strain when these strains were grown in SD + ser (Fig. 2B, cf. lanes 3,5,7 and 1). As expected, SER3 repression is also abolished in these three mutants (Fig. 2B).
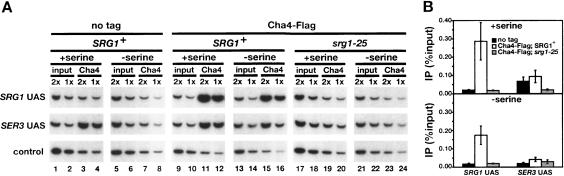
To confirm a direct role for Cha4 in serine-dependent activation of SRG1, we assayed Cha4 physical association with both the SRG1 and SER3 promoters by chromatin immunoprecipitation (ChIP). Our results show that when cells are grown in SD + ser, Cha4 strongly associates with the UAS region of the SRG1 promoter (Fig. 3A [lanes 9–12], B, top panel); this association is lost in srg1-25, in which the putative Cha4-binding site in the SRG1 promoter is deleted (Fig. 3A [lanes 17–20], B). We also observed a weak association of Cha4 with the SER3 UAS region (Fig. 3A [lanes 9–12], B). However, this signal is likely caused by the close proximity of the SRG1 and SER3 UAS regions (221 base pairs (bp) between probes), as the SER3 ChIP signal is lost in the srg1-25 mutant (Fig. 3A [lanes 17–20], B). Taken together, our results show that Cha4 binds to a single site within the SRG1 UAS to activate SRG1, thereby repressing SER3 transcription in serine-rich medium.
Figure 3.
ChIP analysis of Cha4 association with the SRG1 promoter. (A) ChIP analysis of Cha4 was performed on wild-type (FY2470) and srg1-25 (FY2468) strains expressing Cha4-Flag and on an untagged control strain (FY1350). Cha4-Flag was immunoprecipitated with anti-Flag antibody from cells grown in SD + ser medium (+serine) and from cells that had been shifted from SD + ser to SD medium for 25 min (–serine). A representative set of PCR reactions that amplify the SRG1 UAS and SER3 UAS regions (see diagram in Fig. 1A) from twofold dilutions of chromatin is shown. The control primer set amplifies a region of chromosome V that lacks open reading frames (Komarnitsky et al. 2000). (B) Quantitation of ChIP analysis. The amount of SRG1 UAS or SER3 UAS that was amplified from immunoprecipitated DNA is expressed as a percentage of the amount of input DNA. Each bar represents the average and standard error from three independent experiments.
To test if Cha4 binding to SRG1 is serine dependent, we compared Cha4 binding in minimal media with or without serine. Cha4 binding was only mildly decreased after shifting cells to SD medium for 25 min (Fig. 3A [lanes 13–16], B [bottom panel]), a condition in which SRG1 transcription is at a low level (Fig. 1B). This result suggests that Cha4 binding is not strongly controlled by serine levels; rather, induction of SRG1 in response to serine involves an event subsequent to Cha4 binding. This finding is consistent with previous studies that have characterized Cha4 activation at CHA1 (Sabet et al. 2003).
The Spt3 and Spt8 subunits of SAGA are required to repress SER3
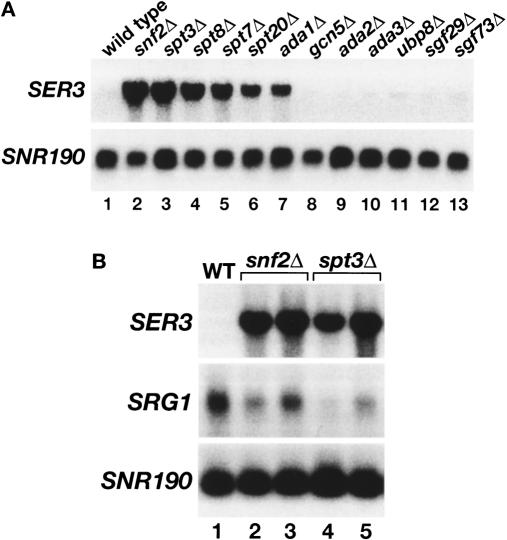
Whole-genome expression analyses originally highlighted SER3 as a gene with interesting regulation, as those studies suggested that both Swi/Snf (Holstege et al. 1998; Sudarsanam et al. 2000) and SAGA (Holstege et al. 1998) serve as repressors of SER3. Our more recent studies have examined the role of Swi/Snf in SER3 repression (Martens and Winston 2002; Martens et al. 2004). Now, to further investigate the role of SAGA in SER3 repression, we measured SER3 mRNA levels in eleven mutants, each lacking a different SAGA subunit and impairing different SAGA actitivies. Our results show that only particular classes of SAGA components are required for SER3 repression. Two SAGA mutants believed to impair TBP recruitment, _spt3_Δ and _spt8_Δ, both had dramatically increased levels of SER3 mRNA, similar to that previously observed for a deletion of SNF2, which encodes the catalytic subunit of the Swi/Snf chromatin remodeling complex (Fig. 4A, lanes 1–4). SER3 mRNA levels were also greatly derepressed in three other SAGA mutants, _ada1_Δ, _spt20_Δ, and _spt7_Δ although not quite to the extent observed for _spt3_Δ and _spt8_Δ (Fig. 4A, lanes 5–7). Ada1, Spt20, and Spt7 are required for the structural integrity of SAGA (Grant et al. 1997; Sterner et al. 1999). In contrast, none of the other SAGA mutants tested, including _gcn5_Δ and _ubp8_Δ, had any effect on SER3 repression (Fig. 4A, lanes 8–11). These results strongly suggest that the Spt3/Spt8 TBP recruitment activity of SAGA (Dudley et al. 1999b; Warfield et al. 2004) is required for SER3 repression, while the Gcn5 histone acetyltransferase activity (Grant et al. 1997) and the Ubp8 deubiquitylation activity (Henry et al. 2003; Daniel et al. 2004) of SAGA play little, if any, role.
Figure 4.
Repression of SER3 is dependent on the Spt3 and Spt8 subunits of SAGA. (A) Northern analysis of SER3. Total RNA was isolated from wild-type (FY3), _snf2_Δ (FY1360), _spt3_Δ (FY294), _spt8_Δ (FY50), _spt7_Δ (FY963), _spt20_Δ (FY1098), _ada1_Δ (FY1560), _gcn5_Δ (FY1600), _ada2_Δ (FY1548), _ada3_Δ (FY1596), _ubp8_Δ (FY2473), _sgf29_Δ (FY2474), and _sgf73_Δ (FY2475) strains that were grown in YPD. SNR190 served as a loading control. These data are representative of three independent experiments. (B) Northern analysis of SER3 and SRG1. Total RNA was isolated from wild-type (FY4), two _snf2_Δ (FY2150 and FY2151), and two _spt3_Δ (FY930 and FY2142) strains that were grown in SD + ser medium. RNA levels were averaged for at least three independent experiments. SER3 mRNA levels are 31.4 ± 2.7 and 27.5 ± 2.3 in _snf2_Δ and _spt3_Δ strains, respectively, as compared with wild type. SRG1 RNA levels are 0.59 ± 0.06 and 0.37 ± 0.07 in _snf2_Δ and _spt3_Δ strains, respectively, as compared with wild type.
To address the roles of both SAGA and Swi/Snf in activating SRG1 transcription, we assayed SRG1 RNA levels in both _spt3_Δ and _snf2_Δ mutants. To look under conditions maximally inducing for SRG1, cells were grown in SD + ser medium. Northern analysis shows that the serine-induced levels of SRG1 were decreased, albeit modestly, in both mutants compared with wild type. In the _snf2_Δ mutant, induction of SRG1 was reduced 1.5-fold to twofold (Fig. 4B, lanes 2,3) consistent with both our previous Northern analysis (Martens et al. 2004) and with ChIP experiments in which we found a twofold decrease in the association of the Rpb3 subunit of RNA polymerase II to SRG1 in a _snf2_Δ strain (data not shown). In the _spt3_Δ mutant, induction was reduced two- to threefold (Fig. 4B, lanes 4,5). Therefore, our results show that both SAGA and Swi/Snf are required for full induction of SRG1 in serine-rich medium.
Association of SAGA and Swi/Snf to the SRG1 promoter is dependent on Cha4 and serine
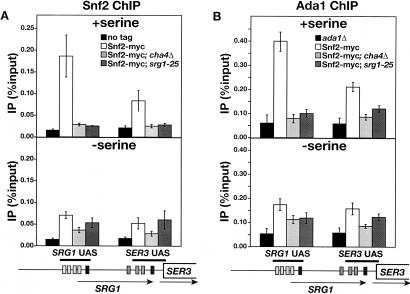
Our results show that Cha4, Spt3, and Snf2 are required for full induction of SRG1 in medium with serine, suggesting that Cha4 recruits SAGA and Swi/Snf to the SRG1 UAS. To test this possibility, we performed ChIP experiments on Swi2 of Swi/Snf (Fig. 5A) and Ada1 of SAGA (Fig. 5B). In wild-type strains, we detected a strong association of both proteins with the SRG1 UAS, with a lower level of association over the adjacent SER3 UAS (Fig. 5, top panels). Significantly, the association of both Snf2 and Ada1 with these regions was reduced to background levels when either CHA4 (_cha4_Δ) or the Cha4-binding site in the SRG1 UAS (srg1-25) was deleted. These results strongly suggest that both Swi/Snf and SAGA are recruited to the SRG1 UAS by Cha4.
Figure 5.
ChIP analysis of Snf2 and Ada1 association to the SRG1 promoter. (A) ChIP analysis of Snf2 was performed on wild-type (FY2470), _cha4_Δ (FY2469), and srg1-25 (FY2468) strains expressing Snf2-myc and an untagged control strain (FY1350). Snf2-myc was immunoprecipitated with anti-myc A14 antibody from chromatin isolated from cells that were grown in SD + ser medium (+serine) and from cells that were shifted from SD + ser to SD medium for 25 min (–serine). The amounts of SRG1 UAS and SER3 UAS that were amplified from immunoprecipitated DNA are expressed as percentages of the amounts of input DNA. Each bar represents the average and standard error from three independent experiments. (B) ChIP analysis of Ada1. Ada1 was immunoprecipitated with anti-Ada1 antibody from the same chromatin that is described in A with the exception of the untagged control strain, which was replaced by an _ada1_Δ control strain (FY1560). Each bar represents the average amount and standard error of three independent experiments.
Serine-dependent activation by Cha4 appears to involve a step that occurs subsequent to Cha4 binding (Fig. 3B; Sabet et al. 2003; our results). To test whether recruitment of SAGA and Swi/Snf by Cha4 are affected by serine levels, we examined the association of Snf2 and Ada1 to the SRG1 UAS after shifting cells from SD + ser to SD medium. In contrast to Cha4, which remained associated with the SRG1 UAS in the absence of serine, there was no significant recruitment of either Snf2 or Ada1 when cells were grown in SD medium (Fig. 5A,B; bottom panels). Therefore, Cha4 recruits SAGA and Swi/Snf to the SRG1 UAS in a serine-dependent manner.
Both SAGA and Swi/Snf are required for SRG1_-mediated transcription interference_
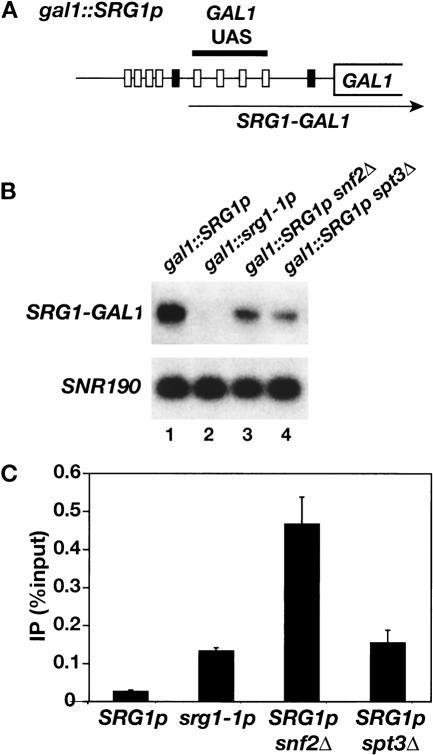
Because _snf2_Δ and _spt3_Δ have only modest defects in SRG1 transcription we performed experiments to determine the effects of these mutations on transcription interference by SRG1. Since the direct activators of SER3 have not been identified, we integrated the SRG1 promoter at the GAL1 locus, 5′ of the GAL1 UAS (Fig. 6A), in wild-type, _snf2_Δ, and _spt3_Δ strains. These strains were analyzed for both transcription from the SRG1 promoter and Gal4 binding. (Note that in this experiment GAL1 will not be transcribed as the cells are grown in glucose, a condition permissive for Gal4 binding, but repressive for GAL1 transcription. These conditions were necessary to avoid secondary effects of _snf2_Δ and _spt3_Δ mutations on galactose-grown cells.) Using a probe to the GAL1 UAS region we detected transcription from the SRG1 promoter that appears to extend to the end of GAL1. Consistent with our previous results, transcription from the SRG1 promoter is decreased two- to threefold in _snf2_Δ, and _spt3_Δ mutants and is undetectable in an srg1-1 mutant, containing a mutated SRG1 TATA (Fig. 6B; Martens et al. 2004). ChIP analysis demonstrates that, like the srg1-1 mutant, _snf2_Δ and _spt3_Δ mutations relieve the inhibition of Gal4 binding observed with wild type SRG1 (Fig. 6C). Unexpectedly, Gal4 binding to the GAL1 UAS is three- to fourfold higher in the _snf2_Δ strain than in either the _spt3_Δ or gal1::srg1-1p strains (Fig. 6C). However, in a control experiment, we found that Gal4 binding to wild-type GAL1 was similarly increased in a _snf2_Δ strain (Supplementary Fig. S2), which would account for this difference. The requirement for Swi/Snf and SAGA in transcription interference with Gal4 binding in this construct strongly suggests that Swi/Snf and SAGA are required for transcription interference of SER3. Although we cannot rule out the possibility that Swi/Snf and/or SAGA have additional functions in SER3 repression, our results suggest that one function of SAGA and Swi/Snf in SER3 repression is to facilitate transcription interference by activating transcription of SRG1.
Figure 6.
Effect of _snf2_Δ and _spt3_Δ on transcription interference by SRG1.(A) A schematic of gal1::SRG1p. The SRG1 UAS (gray boxes) and TATA (left-most black box) sequences were integrated into the GAL1 promoter, 5′ of the four Gal4-binding sites (white boxes). The arrow indicates the srg1-GAL1 RNA that is transcribed as a result of the SRG1 promoter insertion. The gal1::srg1-1p allele contains a similar insertion of the srg1-1 TATA mutant promoter. Under the conditions of the experiment, with cells grown on glucose, there is no transcription from the normal GAL1 initiation site; however, Gal4 is still bound under these growth conditions (Dudley et al. 1999b; Ren et al. 2000). (B) Northern analysis was performed on gal1::SRG1p (FY2476), gal1::srg1-1p (FY2477), _snf2_Δ gal1::SRG1p (FY2478), and _spt3_Δ gal1::SRG1p (FY2479) strains that were grown in YPD medium. Transcription from the SRG1 promoter was detected using a probe to the GAL1 UAS (SRG1-GAL1). SNR190 RNA was measured as a loading control. (C) ChIP analysis of Gal4 was performed on the same strains described in B. The amount of GAL1 UAS that was PCR-amplified from immunoprecipitated DNA was calculated relative to the amount amplified from input DNA. Each bar represents the average and standard error from three experiments.
Discussion
In this work we have elucidated a physiological role for an intergenic transcript in gene regulation. Our results have shown that the repression of the S. cerevisiae SER3 gene in response to high levels of serine occurs by the induction of intergenic transcription of SRG1. Our results also show that the serine-dependent induction of SRG1 depends upon Cha4, which binds to the SRG1 promoter, and that activation by Cha4 requires the SAGA and Swi/Snf coactivator complexes.
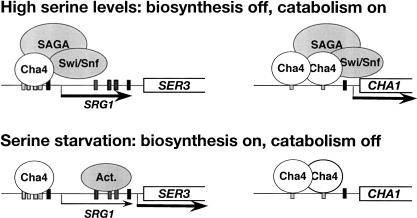
Taken together with previous studies (Holmberg and Schjerling 1996; Sabet et al. 2003), our results suggest a model in which the control of SRG1 intergenic transcription is the primary mechanism that regulates SER3 in response to changes in serine. In this model, SRG1 transcription also allows Cha4 to serve as both an activator and repressor of transcription. When cells are exposed to high levels of serine, Cha4 directly activates the CHA1 gene, required for serine catabolism, and indirectly represses the SER3 gene via activation of SRG1 (Fig. 7, top panel). The requirements for CHA1 activation may be similar to those for SRG1, as CHA1 activation is impaired in both _snf2_Δ and _spt3_Δ mutants, albeit only modestly in _snf2_Δ, and both Swi/Snf and SAGA are recruited to the CHA1 promoter in a serine-dependent fashion (Supplementary Fig. S1; R. Morse, pers. comm.). In serine-starvation conditions, the opposite regulation occurs, as Cha4 no longer recruits SAGA and Swi/Snf, resulting in a reversal of the transcription states of CHA1 and SER3 (Fig. 7, bottom panel; Supplementary Fig. S1). Thus, S. cerevisiae can quickly adapt to changes in intracellular serine by coordinately regulating serine catabolism and biosynthesis using a single activator. This model also illustrates a mechanism by which intergenic transcription increases the potential functions of regulatory proteins as it allows activators to act indirectly as repressors.
Figure 7.
A model for the coordinated regulation of serine biosynthesis and catabolism by Cha4. (Top) In the presence of high serine levels, Cha4 indirectly represses the serine biosynthetic gene SER3 via activation of SRG1 and directly activates the serine catabolic gene CHA1. (Bottome) Under serine-starvation conditions, when Cha4 is no longer able to recruit SAGA and Swi/Snf, the expression states of SER3 and CHA1 are reversed. In this model, the expression of SER3 also requires putative activators (Act.) that bind to the previously identified SER3 UAS (Martens et al. 2004). Thus, Cha4 can act as both an activator and as a repressor in response to serine.
In addition to the factors identified in this work that control SRG1 transcription in response to serine levels, other physiological conditions and factors may play important roles in this regulatory system. Sequence conservation suggests that, in addition to the Cha4-binding site, other SRG1 promoter sequences may be required for SRG1 transcription under some conditions (Fig. 2A; Cliften et al. 2003; Kellis et al. 2003; Martens et al. 2004). These sequences may respond to other physiological signals that control serine levels, such as general nitrogen metabolism, one-carbon metabolism, and fatty acid biosynthesis. In addition, we have previously identified a region of the SER3 promoter that is required for SER3 activation (Martens et al. 2004); however, the factors that bind to this putative SER3 UAS remain unknown and we have not found any evidence for a role for this region in response to serine. We also note that histone H3 mutants that cause defects in SER3 repression have been identified (Sabet et al. 2003; Duina and Winston 2004). The analysis of these mutants, as well as the identification of other factors required for SRG1 and SER3 regulation, will provide further insight into the physiological relevance for regulation of SER3 by intergenic transcription.
Another aspect of the SER3/SRG1 regulatory system that remains to be determined is the role of serine in the Cha4-dependent recruitment of SAGA and Swi/Snf. Like many S. cerevisiae activators that respond to intracellular metabolites (for review, see Sellick and Reece 2005), Cha4 binding to DNA is not significantly affected by serine levels (our results; Sabet et al. 2003). Possibly, serine or an intermediate in the serine biosynthetic pathway directly modulates Cha4 activity by inducing a conformational change that facilitates recruitment of SAGA and Swi/Snf. A similar mechanism has been suggested for Put3, an activator of the proline-utilization pathway, as proline directly interacts with Put3 to cause conformational changes that induce the transcription activity of Put3 (Axelrod et al. 1991; Sellick and Reece 2003). Alternatively, serine may indirectly control Cha4 activity by controlling the interaction of Cha4 with an as-yet-unidentified second protein, similar to the galactose-mediated regulation of Gal4 by interaction with Gal80 (Sellick and Reece 2005).
A number of different mechanisms have been proposed for transcription interference (for review, see Shearwin et al. 2005) and other naturally occurring cases of transcription interference have been identified (for examples, see Hausler and Somerville 1979; Cullen et al. 1984; Proudfoot 1986; Corbin and Maniatis 1989; Boussadia et al. 1997; Moseley et al. 2002). Our data show that loss of either SAGA or Swi/Snf causes only a two- to threefold decrease in SRG1 RNA levels, yet this modest decrease appears to be sufficient to abolish interference as it is accompanied by a 50-fold increase in SER3 mRNA levels. One possibility is that there is a threshold level of SRG1 transcription that is sufficient to confer transcription interference and that a modest decrease in that level allows activators full access to the SER3 promoter. However, given the modest effects on SRG1 RNA levels observed in the _snf2_Δ and _spt3_Δ mutants, SAGA and Swi/Snf may play additional roles to repress SER3 besides SRG1 activation. Consistent with this possibility, Snf2-dependent changes in chromatin structure extend to the SER3 TATA sequence (Martens and Winston 2002). Therefore, there may be two roles for Swi/Snf in SER3 repression, through activation of SRG1 and by inhibition of factor binding to the SER3 promoter.
In conclusion, our work has demonstrated a mechanism by which the regulation of noncoding, intergenic transcription provides a physiologically important response in a metabolic signaling pathway. Studies in other systems, including the Drosophila BX-C complex and the human β-globin locus, have demonstrated other cases where regulation of noncoding, intergenic transcription plays important regulatory roles (Gribnau et al. 2000; Schmitt et al. 2005). Additional recent studies have shown that noncoding transcription is widespread (Mattick 2003; Morey and Avner 2004; Huttenhofer et al. 2005; Johnson et al. 2005) and that a significant fraction is regulated (Cawley et al. 2004; Kim et al. 2005). Taken together, these studies suggest that the control of noncoding, intergenic transcription may represent a broadly used mechanism for transcriptional regulation.
Materials and methods
S. cerevisiae strains and media
All S. cerevisiae strains used (Table 1) are isogenic with a GAL2+ derivative of S288C (Winston et al. 1995). Strains were constructed by standard methods, either by crosses or by transformation (Ausubel et al. 1991). Details are available upon request. The spt8_Δ_302::LEU2 (Eisenmann et al. 1994), spt3-202 (Winston and Minehart 1986), spt3_Δ_203::TRP1 (Happel and Winston 1992), spt7_Δ_402::LEU2 (Gansheroff et al. 1995), spt20_Δ_100::URA3 (Roberts and Winston 1996), _snf2_Δ::LEU2 (Cairns et al. 1996), snf2_Δ_1::HIS3 (Abrams et al. 1986), _gcn5_Δ::HIS3 (Roberts and Winston 1997), ser33_Δ_0::kanMX (Martens and Winston 2002), srg1-1 (Martens et al. 2004), RPB3-HA::LEU2 (Kolodziej et al. 1990), SNF2-C18myc::TRP1 (Martens and Winston 2002), and cha4_Δ_0::kanMX (Open Biosystems) alleles have been previously described. The _ada2_Δ::_HIS3, ada1_Δ::HIS3, and _ada3_Δ::HIS3 mutations were constructed by replacing their open reading frames with HIS3 (S. Roberts and F. Winston, unpubl.). The ubp8_Δ_0::kanMX, sgf29_Δ_0::kanMX, and sgf73_Δ_0::kanMX mutations were constructed by replacing their open reading frames with the kanMX marker (Brachmann et al. 1998). The srg1-20 mutation replaces SRG1 promoter sequences from –660 to –560 (relative to the SER3 ATG) with three copies of the myc epitope. The srg1-21, srg1-22, srg1-23, srg1-24, and srg1-25 mutations replace SRG1 promoter sequences –659 to –646, –644 to –636, –611 to –604, –590 to –582, and –612 to –592 each with an AvrII site (CCTAGG). The srg1-26 mutation contains three point mutations in the Cha4 consensus binding site within the SRG1 promoter (TGGAGATACATCTCCA to ctGAGATACATCTCaA). This mutant sequence has been shown to be defective for Cha4 binding (Holmberg and Schjerling 1996). The CHA4-Flag::kanMX allele encodes one copy of the Flag epitope at the 3′ end of CHA4, which was constructed by PCR-mediated integration using plasmid pDM714 (kind gift from D. Moazed, Harvard Medical School, Boston, MA). The gal1::SRG1p allele contains the SRG1 promoter and transcription initiation sites (base pairs –713 to –445 relative to the SER3 ATG) at position –556 of GAL1 (relative to GAL1 ATG). The gal1::srg1-1p allele has an insertion of the same DNA except with the srg1-1 promoter containing the TATA mutation. As indicated, strains were grown in either YPD (1% yeast extract, 2% peptone, and 2% glucose) or minimal media (0.145% yeast nitrogen base, 0.5% ammonium sulfate, 2% glucose, and supplemented with required amino acids) with 1 mM serine (SD + ser) or without serine (SD).
Table 1.
Saccharomyces cerevisiae strains
| Name | Genotype |
|---|---|
| FY3 | MATaura3-52 |
| FY4 | MATa (prototroph) |
| FY50 | MATaura3-52 leu2Δ1 spt8Δ302::LEU2 trp1Δ63 his4-917δ |
| FY294 | MATaura3-52 lys2-173R2 leu2Δ1 trp1Δ63 his4-917δ spt3-202 |
| FY930 | MATaura3-52 lys2-173R2 his3Δ200 spt3-202 |
| FY963 | MATaura3-52 leu2Δ1 spt7Δ402::LEU2 his4-917δ |
| FY1098 | MATaura3-52 spt20Δ100::URA3 leu2Δ1 his3Δ200 |
| FY1350 | MATα ura3Δ0 lys2Δ0 leu2Δ0 |
| FY1360 | MATaura3-52 lys2-173R2 leu2Δ1 snf2Δ::LEU2 his3Δ200 |
| FY1548 | MATaura3-52 lys2-128δ leu2Δ1 his3Δ200 ada2Δ::HIS3 |
| FY1560 | MATaura3-52 lys2-173R2 his3Δ200 ada1Δ::HIS3 |
| FY1596 | MATaura3-52 lys2-173R2 his3Δ200 ada3Δ::HIS3 his4-912δ |
| FY1600 | MATaura3-52 leu2Δ1 his3Δ200 gcn5Δ::HIS3 his4-917δ |
| FY2142 | MATα spt3Δ0::kanMX |
| FY2150 | MATasnf2Δ0::kanMX |
| FY2151 | MATasnf2Δ0::kanMX |
| FY2245 | MATaura3-52 lys2-128δ leu2Δ1 RPB3-HA::LEU2 his3Δ200 trp1Δ63 SNF2-C18myc::TRP1 his4-912δ ser33Δ0::kanMX |
| FY2459 | MATaura3Δ0 lys2Δ0 leu2Δ0 cha4Δ0::kanMX |
| FY2460 | MATaura3Δ0 lys2Δ0 leu2Δ0 CHA4-Flag::kanMX |
| FY2461 | MATaura3Δ0 lys2Δ0 leu2Δ0 CHA4-Flag::kanMX srg1-24 |
| FY2462 | MATaura3Δ0 lys2Δ0 leu2Δ0 his3Δ200 CHA4-Flag::kanMX srg1-26 |
| FY2463 | MATaura3Δ0 lys2Δ0 leu2Δ0 his3Δ200 CHA4-Flag::kanMX srg1-25 |
| FY2464 | MATaura3Δ0 lys2Δ0 leu2Δ0 his3Δ200 CHA4-Flag::kanMX srg1-21 |
| FY2465 | MATaura3Δ0 lys2Δ0 leu2Δ0 his3Δ200 CHA4-Flag::kanMX srg1-22 |
| FY2466 | MATaura3Δ0 lys2Δ0 leu2Δ0 his3Δ200 CHA4-Flag::kanMX srg1-23 |
| FY2467 | MATaura3Δ0 lys2Δ0 leu2Δ0 CHA4-Flag::kanMX srg1-20 |
| FY2468 | MATα ura3Δ0 lys2Δ0 leu2Δ0 his3Δ200 met15Δ0 SNF2-C18myc::TRP1 CHA4-Flag::kanMX srg1-25 |
| FY2469 | MATα ura3Δ0 lys2Δ0 leu2Δ0 SNF2-C18myc::TRP1 cha4Δ0::kanMX |
| FY2470 | MATα ura3Δ0 lys2Δ0 leu2Δ0 his3Δ200 trp1Δ63 SNF2-C18myc::TRP1 CHA4-Flag::kanMX |
| FY2471 | MATaura3Δ0 lys2Δ0 his3Δ200 leu2Δ0 srg1-1 |
| FY2472 | MATaura3Δ0 lys2Δ0 his3Δ200 leu2Δ0 |
| FY2473 | MATα ura3-52 lys2-173R2 leu2Δ1 arg4-12 ubp8Δ0::kanMX |
| FY2474 | MATaura3-52 lys2-173R2 leu2Δ1 arg4-12 sgf29Δ0::kanMX his4-917δ |
| FY2475 | MATα ura3-52 lys2-173R2 leu2Δ1 arg4-12 sgf73Δ0::kanMX his4-917δ |
| FY2476 | MATaura3Δ0 lys2Δ0 leu2Δ0 his3Δ200 ser33Δ0::kanMX gal1::SRG1p |
| FY2477 | MATaura3Δ0 lys2Δ0 leu2Δ0 his3Δ200 ser33Δ0::kanMX gal1::srg1-1p |
| FY2478 | MATα ura3-52 lys2Δ0 leu2Δ1 snf2Δ::LEU2 his3Δ200 gal1::SRG1p |
| FY2479 | MATα ura3Δ0 lys2Δ0 leu2Δ1 his3Δ200 his4-917δtrp1Δ63 spt3Δ203::TRP1 gal1::SRG1p |
| FY2502 | MATaura3-52 lys2-128δ leu2Δ1 RPB3-HA::LEU2 snf2Δ1::HIS3 his4-912δ ser33Δ0::kanMX |
Northern analysis
Northern hybridization analysis was performed as previously described (Ausubel et al. 1991). Probes for SER3, SRG1, and SNR190 were previously described (Martens and Winston 2002; Duina and Winston 2004; Martens et al. 2004). A probe specific to the GAL1 5′ UTR was generated by random labeling a PCR product, containing the GAL1 sequence from –443 to –157 (relative to the GAL1 ATG), amplified from genomic DNA. RNA levels were quantitated using a PhosphorImager (Molecular Dynamics) and normalized to SNR190, whose levels are unaffected by the mutations and growth conditions studied here.
ChIP analysis
ChIP analysis was performed as previously described (Dudley et al. 1999b; Martens and Winston 2002) with the following modifications: For the Cha4, Snf2, and Ada1 ChIPs, 200-mL cultures were grown to a density of 0.8 × 107 to 1.0 × 107 cells/mL in SD + ser medium and then split into two equal volumes. One half was treated with buffered formaldehyde (11% formaldehyde, 0.1M NaCl, 1 mM EDTA, and 50 mM Hepes-KOH at pH 7.5) to a final concentration of 1%. Cells from the other half of the culture were harvested by centrifugation, resuspended in SD medium, incubated for 25 min at 30°C, and then treated with buffered formaldehyde to a final concentration of 1%. The following antibodies were used for ChIP from one-fifth of the total amount of cross-linked chromatin: 4 μL of mouse A14 anti-myc ascites (Santa Cruz Biotechnology), 2 μL of rabbit anti-Flag M2 serum (Sigma-Aldrich, Inc), and 1 μL of rabbit anti-Ada1 serum (a generous gift from L. Guarente, Massachusetts Institute of Technology, Cambridge, MA). Input DNA (0.002% and 0.001%) and immunoprecipitated DNA (4% and 2% for Cha4-Flag and Snf2-myc, 2% and 1% for Ada1) were subjected to quantitative radioactive PCR and the products were separated on an 8% nondenaturing polyacrylamide gel. For the Gal4 ChIPs, 200-mL cultures were grown to a density of 1 × 107 to 2 × 107 cells/mL in YPD and then 37% formaldehyde was added to a final concentration of 1%. Gal4 was immunoprecipitated from one-tenth of the total amount of cross-linked chromatin with 1 μL; of anti-Gal4 serum (Santa Cruz Biotechnology, Inc.). Input DNA (0.002% and 0.001%) and immunoprecipitated DNA (1.33% and 0.67%) were analyzed as described for the Cha4 ChIPs. The PCR primers amplify the following regions whose coordinates are given relative to the SER3 ATG: SRG1 UAS primers amplify a 259-bp product from –714 to –456 and SER3 UAS primers amplify a 298-bp product from –234 to +65. Primers that amplify the GAL1 UAS and the control region from chromosome V that lacks open reading frames have been previously described (Dudley et al. 1999a; Komarnitsky et al. 2000). All ChIP experiments were quantitated by PhosphorImager analysis (Molecular Dynamics). Association of factors to specific DNA sequences was calculated as a percentage of the amount of coimmunoprecipitated DNA relative to the input DNA.
Acknowledgments
We thank Dominique Helmlinger and Natalie Kuldell for valuable comments on the manuscript and Amine Nourani and Randy Morse for helpful discussions. This work was supported by NIH grant GM32967 to F.W., by a postdoctoral fellowship from the Canadian Institute for Health Research to J.A.M, and by a predoctoral fellowship from the Howard Hughes Medical Institute to P.-Y.J.W.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1367605.
References
- Abrams E., Neigeborn, L., and Carlson, M. 1986. Molecular analysis of SNF2 and SNF5, genes required for expression of glucose-repressible genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 6**:** 3643–3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen A.A. and Panning, B. 2003. Epigenetic gene regulation by noncoding RNAs. Curr. Opin. Cell Biol. 15**:** 504. [DOI] [PubMed] [Google Scholar]
- Ausubel F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K.E. 1991. Current protocols in molecular biology. Greene Publishing Associates and Wiley-Interscience, New York.
- Axelrod J.D., Majors, J., and Brandriss, M.C. 1991. Proline-independent binding of PUT3 transcriptional activator protein detected by footprinting in vivo. Mol. Cell. Biol. 11**:** 564–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender W. and Fitzgerald, D.P. 2002. Transcription activates repressed domains in the Drosophila bithorax complex. Development 129**:** 4923–4930. [DOI] [PubMed] [Google Scholar]
- Bernstein E. and Allis, C.D. 2005. RNA meets chromatin. Genes & Dev. 19**:** 1635–1655. [DOI] [PubMed] [Google Scholar]
- Bertone P., Stolc, V., Royce, T.E., Rozowsky, J.S., Urban, A.E., Zhu, X., Rinn, J.L., Tongprasit, W., Samanta, M., Weissman, S., et al. 2004. Global identification of human transcribed sequences with genome tiling arrays. Science 306**:** 2242–2246. [DOI] [PubMed] [Google Scholar]
- Boussadia O., Amiot, F., Cases, S., Triqueneaux, G., Jacquemin-Sablon, H., and Dautry, F. 1997. Transcription of unr (upstream of N-ras) down-modulates N-ras expression in vivo. FEBS Lett. 420**:** 20–24. [DOI] [PubMed] [Google Scholar]
- Brachmann C.B., Davies, A., Cost, G.J., Caputo, E., Li, J., Hieter, P., and Boeke, J.D. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14**:** 115–132. [DOI] [PubMed] [Google Scholar]
- Cairns B.R., Levinson, R.S., Yamamoto, K.R., and Kornberg, R.D. 1996. Essential role of Swp73p in the function of yeast Swi/Snf complex. Genes & Dev. 10**:** 2131–2144. [DOI] [PubMed] [Google Scholar]
- Cawley S., Bekiranov, S., Ng, H.H., Kapranov, P., Sekinger, E.A., Kampa, D., Piccolboni, A., Sementchenko, V., Cheng, J., Williams, A.J., et al. 2004. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell 116**:** 499–509. [DOI] [PubMed] [Google Scholar]
- Chen J., Sun, M., Lee, S., Zhou, G., Rowley, J.D., and Wang, S.M. 2002. Identifying novel transcripts and novel genes in the human genome by using novel SAGE tags. Proc. Natl. Acad. Sci. 99**:** 12257–12262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J., Kapranov, P., Drenkow, J., Dike, S., Brubaker, S., Patel, S., Long, J., Stern, D., Tammana, H., Helt, G., et al. 2005. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science 308**:** 1149–1154. [DOI] [PubMed] [Google Scholar]
- Cliften P., Sudarsanam, P., Desikan, A., Fulton, L., Fulton, B., Majors, J., Waterston, R., Cohen, B.A., and Johnston, M. 2003. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science 301**:** 71–76. [DOI] [PubMed] [Google Scholar]
- Corbin V. and Maniatis, T. 1989. Role of transcriptional interference in the Drosophila melanogaster Adh promoter switch. Nature 337**:** 279–282. [DOI] [PubMed] [Google Scholar]
- Cullen B.R., Lomedico, P.T., and Ju, G. 1984. Transcriptional interference in avian retroviruses—Implications for the promoter insertion model of leukaemogenesis. Nature 307**:** 241–245. [DOI] [PubMed] [Google Scholar]
- Cumberledge S., Zaratzian, A., and Sakonju, S. 1990. Characterization of two RNAs transcribed from the cis-regulatory region of the abd-A domain within the Drosophila bithorax complex. Proc. Natl. Acad. Sci. 87**:** 3259–3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel J.A., Torok, M.S., Sun, Z.W., Schieltz, D., Allis, C.D., Yates III, J.R., and Grant, P.A. 2004. Deubiquitination of histone H2B by a yeast acetyltransferase complex regulates transcription. J. Biol. Chem. 279**:** 1867–1871. [DOI] [PubMed] [Google Scholar]
- Dudley A.M., Gansheroff, L.J., and Winston, F. 1999a. Specific components of the SAGA complex are required for Gcn4- and Gcr1-mediated activation of the his4–912δ promoter in Saccharomyces cerevisiae. Genetics 151**:** 1365–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley A.M., Rougeulle, C., and Winston, F. 1999b. The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes & Dev. 13**:** 2940–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duina A.A. and Winston, F. 2004. Analysis of a mutant histone H3 that perturbs the association of Swi/Snf with chromatin. Mol. Cell Biol. 24**:** 561–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenmann D.M., Chapon, C., Roberts, S.M., Dollard, C., and Winston, F. 1994. The Saccharomyces cerevisiae SPT8 gene encodes a very acidic protein that is functionally related to SPT3 and TATA-binding protein. Genetics 137**:** 647–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gansheroff L.J., Dollard, C., Tan, P., and Winston, F. 1995. The Saccharomyces cerevisiae SPT7 gene encodes a very acidic protein important for transcription in vivo. Genetics 139**:** 523–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S. 2004. The small RNA regulators of Escherichia coli: Roles and mechanisms. Annu. Rev. Microbiol. 58**:** 303–328. [DOI] [PubMed] [Google Scholar]
- Grant P.A., Duggan, L., Cote, J., Roberts, S.M., Brownell, J.E., Candau, R., Ohba, R., Owen-Hughes, T., Allis, C.D., Winston, F., et al. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: Characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes & Dev. 11**:** 1640–1650. [DOI] [PubMed] [Google Scholar]
- Gribnau J., Diderich, K., Pruzina, S., Calzolari, R., and Fraser, P. 2000. Intergenic transcription and developmental remodeling of chromatin subdomains in the human β-globin locus. Mol. Cell 5**:** 377–386. [DOI] [PubMed] [Google Scholar]
- Happel A.M. and Winston, F. 1992. A mutant tRNA affects δ-mediated transcription in Saccharomyces cerevisiae. Genetics 132**:** 361–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausler B. and Somerville, R.L. 1979. Interaction in vivo between strong closely spaced constitutive promoters. J. Mol. Biol. 127**:** 353–356. [DOI] [PubMed] [Google Scholar]
- Havilio M., Levanon, E.Y., Lerman, G., Kupiec, M., and Eisenberg, E. 2005. Evidence for abundant transcription of noncoding regions in the Saccharomyces cerevisiae genome. BMC Genomics 6**:** 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry K.W., Wyce, A., Lo, W.S., Duggan, L.J., Emre, N.C., Kao, C.F., Pillus, L., Shilatifard, A., Osley, M.A., and Berger, S.L. 2003. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes & Dev. 17**:** 2648–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogga I. and Karch, F. 2002. Transcription through the iab-7 cis-regulatory domain of the bithorax complex interferes with maintenance of Polycomb-mediated silencing. Development 129**:** 4915–4922. [DOI] [PubMed] [Google Scholar]
- Holmberg S. and Schjerling, P. 1996. Cha4p of Saccharomyces cerevisiae activates transcription via serine/threonine response elements. Genetics 144**:** 467–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege F.C., Jennings, E.G., Wyrick, J.J., Lee, T.I., Hengartner, C.J., Green, M.R., Golub, T.R., Lander, E.S., and Young, R.A. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95**:** 717–728. [DOI] [PubMed] [Google Scholar]
- Hurowitz E.H. and Brown, P.O. 2003. Genome-wide analysis of mRNA lengths in Saccharomyces cerevisiae. Genome Biol. 5**:** R2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenhofer A., Schattner, P., and Polacek, N. 2005. Non-coding RNAs: Hope or hype? Trends Genet. 21**:** 289–297. [DOI] [PubMed] [Google Scholar]
- Johnson J.M., Edwards, S., Shoemaker, D., and Schadt, E.E. 2005. Dark matter in the genome: Evidence of widespread transcription detected by microarray tiling experiments. Trends Genet. 21**:** 93–102. [DOI] [PubMed] [Google Scholar]
- Kampa D., Cheng, J., Kapranov, P., Yamanaka, M., Brubaker, S., Cawley, S., Drenkow, J., Piccolboni, A., Bekiranov, S., Helt, G., et al. 2004. Novel RNAs identified from an in-depth analysis of the transcriptome of human chromosomes 21 and 22. Genome Res. 14**:** 331–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P., Cawley, S.E., Drenkow, J., Bekiranov, S., Strausberg, R.L., Fodor, S.P., and Gingeras, T.R. 2002. Large-scale transcriptional activity in chromosomes 21 and 22. Science 296**:** 916–919. [DOI] [PubMed] [Google Scholar]
- Kellis M., Patterson, N., Endrizzi, M., Birren, B., and Lander, E.S. 2003. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423**:** 241–254. [DOI] [PubMed] [Google Scholar]
- Kim T.H., Barrera, L.O., Zheng, M., Qu, C., Singer, M.A., Richmond, T.A., Wu, Y., Green, R.D., and Ren, B. 2005. A high-resolution map of active promoters in the human genome. Nature 436**:** 876–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodziej P.A., Woychik, N., Liao, S.M., and Young, R.A. 1990. RNA polymerase II subunit composition, stoichiometry, and phosphorylation. Mol. Cell. Biol. 10**:** 1915–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarnitsky P., Cho, E.J., and Buratowski, S. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes & Dev. 14**:** 2452–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipshitz H.D., Peattie, D.A., and Hogness, D.S. 1987. Novel transcripts from the Ultrabithorax domain of the bithorax complex. Genes & Dev. 1**:** 307–322. [DOI] [PubMed] [Google Scholar]
- Martens J.A. and Winston, F. 2002. Evidence that Swi/Snf directly represses transcription in S. cerevisiae. Genes & Dev. 16**:** 2231–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens J.A., Laprade, L., and Winston, F. 2004. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature 429**:** 571–574. [DOI] [PubMed] [Google Scholar]
- Mattick J.S. 2003. Challenging the dogma: The hidden layer of non-protein-coding RNAs in complex organisms. Bioessays 25**:** 930–939. [DOI] [PubMed] [Google Scholar]
- Meister G. and Tuschl, T. 2004. Mechanisms of gene silencing by double-stranded RNA. Nature 431**:** 343–349. [DOI] [PubMed] [Google Scholar]
- Morey C. and Avner, P. 2004. Employment opportunities for non-coding RNAs. FEBS Lett. 567**:** 27–34. [DOI] [PubMed] [Google Scholar]
- Moseley J.L., Page, M.D., Alder, N.P., Eriksson, M., Quinn, J., Soto, F., Theg, S.M., Hippler, M., and Merchant, S. 2002. Reciprocal expression of two candidate di-iron enzymes affecting photosystem I and light-harvesting complex accumulation. Plant Cell 14**:** 673–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N.J. 1986. Transcriptional interference and termination between duplicated α-globin gene constructs suggests a novel mechanism for gene regulation. Nature 322**:** 562–565. [DOI] [PubMed] [Google Scholar]
- Rank G., Prestel, M., and Paro, R. 2002. Transcription through intergenic chromosomal memory elements of the Drosophila bithorax complex correlates with an epigenetic switch. Mol. Cell. Biol. 22**:** 8026–8034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B., Robert, F., Wyrick, J.J., Aparicio, O., Jennings, E.G., Simon, I., Zeitlinger, J., Schreiber, J., Hannett, N., Kanin, E., et al. 2000. Genome-wide location and function of DNA binding proteins. Science 290**:** 2306–2309. [DOI] [PubMed] [Google Scholar]
- Roberts S.M. and Winston, F. 1996. SPT20/ADA5 encodes a novel protein functionally related to the TATA-binding protein and important for transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 16**:** 3206–3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 1997. Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcn5 proteins, with the Snf/Swi and Srb/mediator complexes. Genetics 147**:** 451–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabet N., Tong, F., Madigan, J.P., Volo, S., Smith, M.M., and Morse, R.H. 2003. Global and specific transcriptional repression by the histone H3 amino terminus in yeast. Proc. Natl. Acad. Sci. 100**:** 4084–4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S., Sparks, A.B., Rago, C., Akmaev, V., Wang, C.J., Vogelstein, B., Kinzler, K.W., and Velculescu, V.E. 2002. Using the transcriptome to annotate the genome. Nat. Biotechnol. 20**:** 508–512. [DOI] [PubMed] [Google Scholar]
- Sanchez-Herrero E. and Akam, M. 1989. Spatially ordered transcription of regulatory DNA in the bithorax complex of Drosophila. Development 107**:** 321–329. [DOI] [PubMed] [Google Scholar]
- Schmitt S., Prestel, M., and Paro, R. 2005. Intergenic transcription through a Polycomb group response element counteracts silencing. Genes & Dev. 19**:** 697–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellick C.A. and Reece, R.J. 2003. Modulation of transcription factor function by an amino acid: Activation of Put3p by proline. EMBO J. 22**:** 5147–5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2005. Eukaryotic transcription factors as direct nutrient sensors. Trends Biochem. Sci. 30**:** 405–412. [DOI] [PubMed] [Google Scholar]
- Shearwin K.E., Callen, B.P., and Egan, J.B. 2005. Transcriptional interference—A crash course. Trends Genet. 21**:** 339–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner D.E., Grant, P.A., Roberts, S.M., Duggan, L.J., Belotserkovskaya, R., Pacella, L.A., Winston, F., Workman, J.L., and Berger, S.L. 1999. Functional organization of the yeast SAGA complex: Distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol. Cell. Biol. 19**:** 86–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz G., Altuvia, S., and Wassarman, K.M. 2005. An Abundance of RNA Regulators. Annu. Rev. Biochem. 74**:** 199–217. [DOI] [PubMed] [Google Scholar]
- Sudarsanam P., Iyer, V.R., Brown, P.O., and Winston, F. 2000. Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 97**:** 3364–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warfield L., Ranish, J.A., and Hahn, S. 2004. Positive and negative functions of the SAGA complex mediated through interaction of Spt8 with TBP and the N-terminal domain of TFIIA. Genes & Dev. 18**:** 1022–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F. and Minehart, P.L. 1986. Analysis of the yeast SPT3 gene and identification of its product, a positive regulator of Ty transcription. Nucleic Acids Res. 14**:** 6885–6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F. Dollard, C., and Ricupero-Hovasse, S.L. 1995. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11**:** 53–55. [DOI] [PubMed] [Google Scholar]
- Wyers F., Rougemaille, M., Badis, G., Rousselle, J.C., Dufour, M.E., Boulay, J., Regnault, B., Devaux, F., Namane, A., Seraphin, B., et al. 2005. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121**:** 725–737. [DOI] [PubMed] [Google Scholar]