The Forkhead Transcription Factor FoxI1 Remains Bound to Condensed Mitotic Chromosomes and Stably Remodels Chromatin Structure (original) (raw)
Abstract
All forkhead (Fox) proteins contain a highly conserved DNA binding domain whose structure is remarkably similar to the winged-helix structures of histones H1 and H5. Little is known about Fox protein binding in the context of higher-order chromatin structure in living cells. We created a stable cell line expressing FoxI1-green fluorescent protein (GFP) or FoxI1-V5 fusion proteins under control of the reverse tetracycline-controlled transactivator doxycycline inducible system and found that unlike most transcription factors, FoxI1 remains bound to the condensed chromosomes during mitosis. To isolate DNA fragments directly bound by the FoxI1 protein within living cells, we performed chromatin immunoprecipitation assays (ChIPs) with antibodies to either enhanced GFP or the V5 epitope and subcloned the FoxI1-enriched DNA fragments. Sequence analyses indicated that 88% (106/121) of ChIP sequences contain the consensus binding sites for all Fox proteins. Testing ChIP sequences with a quantitative DNase I hypersensitivity assay showed that FoxI1 created stable DNase I sensitivity changes in condensed chromosomes. The majority of ChIP targets and random targets increased in resistance to DNase I in FoxI1-expressing cells, but a small number of targets became more accessible to DNase I. Consistently, the accessibility of micrococcal nuclease to chromatin was generally inhibited. Micrococcal nuclease partial digestion generated a ladder in which all oligonucleosomes were slightly longer than those observed with the controls. On the basis of these findings, we propose that FoxI1 is capable of remodeling chromatin higher-order structure and can stably create site-specific changes in chromatin to either stably create or remove DNase I hypersensitive sites.
In eukaryotes, gene expression is directly regulated by DNA-binding transcription factors and their associated cofactors. Most DNA-binding transcription factors can be grouped into large families of related proteins that have similar DNA-binding domains. One such motif is called the forkhead (Fkh) DNA binding domain. It folds into a variant of the helix-turn-helix motif and is made up of three helices and two characteristic large loops, or “wings,” Therefore, this DNA-binding motif is also named the “winged-helix” DNA-binding domain (FKH/WH) (29). Since the first forkhead gene was described in Drosophila melanogaster (67), over 100 members of the forkhead gene family have been identified in species ranging from Saccharomyces cerevisiae to humans (29). They have been implicated in such diverse roles as cell cycle regulation (75), early embryonic lineage decisions (22), and tumorigenesis (38). The nomenclature of the chordate forkhead transcription factors has recently been revised, and these genes, now termed Fox (after Forkhead box), are divided into 17 subclasses, or clades (A to Q), according to the amino acid sequence of their conserved forkhead domains (http://www.biology.pomona.edu/fox.html). Comparative genome analyses have shown that the number of forkhead transcription factors appears to have increased during evolution, with a greater number identified in vertebrates than in invertebrates (32). Among the organisms for which the genome sequences are completed or nearly so, there is indeed a correlation between anatomical complexity and forkhead gene number: 4 in S. cerevisiae, 15 in Caenorhabditis elegans, 20 in Drosophila melanogaster, and 39 in humans (9). These data suggest that the origin and expansion of forkhead genes is positively correlated with eukaryotic complexity.
A clue for understanding the expansion of forkhead genes is the discovery that the structure of forkhead transcription factor HNF3-γ (FoxA3) is remarkably similar to that of the central globular domain of avian erythrocyte linker histone H5 and its homologue, the ubiquitous histone H1, except that the linker histones lack the W2 loop (8, 14). They are thus classified into the same winged-helix class of DNA-binding proteins (19). Despite the structural similarity, the forkhead proteins have been clearly shown to be tissue-specific transcriptional regulators with sequence-specific DNA binding, whereas the linker histones do not have specific sequences to which they bind. In addition, genes coding for H1 linker histones with an evolutionarily conserved winged helix motif can be found in diverse protists (30), but forkhead genes have so far only been found in animals and fungi, not in protists (9). These data suggest that the roles for linker histones and the forkhead proteins have diverged over the course of evolution. However, the similarity in structure does suggest that there are overlapping mechanisms as to how the two types of proteins interact with the chromatin. For example, the winged-helix structure of H1 is sufficient to bind to linker DNA adjacent to the nucleosome core (2) and a murine HNF-3 could functionally substitute for the linker histone H1 in the mouse prealbumin gene enhancer (12). Therefore, a possibility is raised that Fox proteins are not only typical transcription factors but could also be involved in remodeling of the chromatin structure. Interestingly, in vitro studies by Cirillo and coworkers suggest that binding by wild-type HNF-3 could not compact chromatin as linker histones do, but site-directed mutants of HNF3 could compact nucleosomal DNA (12). Thus, the mode of interaction of forkhead transcription factors with chromatin is clearly an interesting issue that may provide new insights into gene regulation.
Recently, through genetic screens, we and other groups have independently identified a member of the Fox family, the zebra fish protein FoxI1 (31, 42, 58). By amino acid sequence, this protein is highly similar to the other members of the FoxI1 subfamily, human FREAC6 (FKHL10, HFH3) (44), mouse Fkh10 (27), and Xenopus FoxI1c (50). FoxI1-null mutants have highly specific phenotypes in both zebra fish and mouse ears (27, 42). As a method to study the effects FoxI1 has on transcription in a controlled system, we integrated the FoxI1 gene into the genome of the zebra fish embryonic cell line Pac2. Two FoxI1 stable cell lines were generated, a FoxI1-green fluorescent protein (GFP) fusion and a FoxI1-V5 epitope fusion, both regulated by a doxycycline-inducible system (65). Direct visualizations of FoxI1-GFP and FoxI1-V5 immunofluorescence indicated that association of FoxI1 with chromatin persisted through all stages of the cell cycle. Of particular note, FoxI1 remained bound even to mitotically condensed chromosomes, which is not typical of most transcription factors. Chromatin immunoprecipitation (ChIP) and sequencing of clones obtained from the precipitated DNA indicated that FoxI1 predominately recognized the consensus sequence of all Fox proteins. Using sequences identified from chromatin immunoprecipitations to test DNase I hypersensitivity, we found that FoxI1 created stable alterations in DNase I sensitivity that became more pronounced in condensed chromosomes. DNase I sensitivity assays and nucleosomal structural analysis suggested that FoxI1 could be a potential component for remodeling and maintaining nucleosome structure. It could modify the chromatin globally while having more specific effects on target locations. As FoxI1 is dynamically expressed in dividing cells of otic placode and pharyngeal pouches (42), we propose that the FoxI1 protein remodels chromatin structure during the rapid nuclear division of these cells during development. This would create a stable transcriptional “ground state” for the cells that would allow them to respond rapidly and appropriately to external developmental cues.
MATERIALS AND METHODS
Constructs for prokaryotic expression.
For FoxI1-prokaryotic expression, the coding region of the FoxI1 cDNA was subcloned in frame into the pBAD/Thio-TOPO expression vector (Invitrogen), which contains an 11.7-kDa HP-thioredoxin leader sequence and the V5 epitope. The resulting plasmid was called pBAD-foxi1-v5. For in vitro translation, the sequences corresponding to bases 141 to 1800 of the zebra fish FoxI1 cDNA, which included 36 bases of the 5′ untranslated region, the entire coding region, and the 3′ untranslated region, were amplified (forward primer, 5′-TTGGTACCTAGATCTTTGAGAAGAAGTGTGACAG-3′; reverse primer, 5′-GTTGGAATTCTAGAGTCGCGGCCGC-3′). The resulting BglII-XbaI insert was subcloned into the BamHI-XbaI sites of the pCS2+ vector, yielding plasmid pCS2+foxi1. To construct plasmid pCS2+foxi1-v5, the FoxI1-V5 sequence from pBAD-foxi1-v5 was subcloned into pCS2+foxi1 as a BamHI-XbaII insert. Similarly, pCS2+foxi1-GFP was generated by replacing the V5 tag with an enhanced GFP (eGFP) fusion. The resultant plasmids were used for in vitro syntheses of FoxI1 protein and FoxI1 fusion proteins using the TNT quick-coupled transcription/translation system (Promega).
Expression of FoxI1 fusion proteins in zebra fish PAC2 cells.
The entire open reading frame for zebra fish FoxI1 was amplified by PCR (forward primer 5-AGCTGCAAGCTTATGAGTCCATTCACTGCATGC-3; reverse primer 5-ATCTCGAGATGCATAACTTCTGATCCGTCCCGGTTGTATATG-3) and cloned into vector pFRT163-GFP (Fig. 1) as a HindIII-NsiI insert, generating pSP163-FoxI1-GFP. To construct the 163-FoxI1-V5 plasmid, the FoxI1-V5 fragment from pCS2+FoxI1-v5 was subcloned into pSP163-FoxI1-GFP after BamHI/XhoII digestion. The constructs were then transfected into the zebra fish fibroblast cell line PAC2 (33) using a modified version of the Invitrogen T-Rex system. Two stable cell lines were generated: FoxI1-GFP and FoxI1-V5. The expression of fusion proteins was induced by adding 1 μg/ml doxycycline (Invitrogen). For immunofluorescent detection, cells were fixed with 4% formaldehyde-0.1% Triton X-100 in phosphate-buffered saline at room temperature for 30 min and incubated with relevant antibodies (anti-GFP monoclonal antibody [Clontech] and anti-V5 antibody [Invitrogen]) to detect expression of the fusion proteins using confocal microscopy.
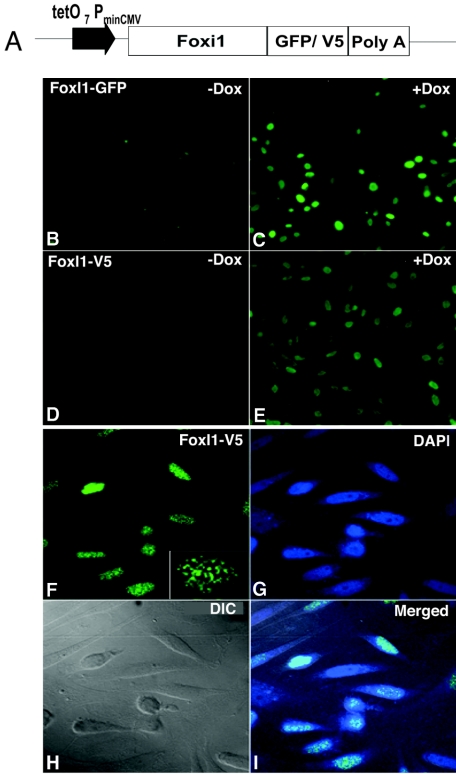
FIG. 1.
Generation of doxycycline (Dox)-inducible FoxI1-GFP and FoxI1-V5 stable cell lines. (A) Constructs for Dox-inducible FoxI1-GFP or FoxI1-V5 used for generation of FoxI1-GFP and FoxI1-V5 PAC2 cell lines. (B) FoxI1-GFP Pac2 cell line in the absence of Dox (−Dox). (C) FoxI1-GFP Pac2 cell line in the presence of Dox (+Dox) visualized by GFP fluorescence. (D) FoxI1-V5 cell line in the absence of Dox. (E) FoxI1-V5 cell line in the presence of Dox, visualized by anti-V5 antibodies. (F) Confocal images of FoxI1-V5 staining, results were similar for the GFP fusion (not shown). The lower right corner has a higher-magnification inset showing the punctate staining in more detail. (G) DAPI staining of the same cells. (H) Differential interference contrast (DIC) image of the same cells. (I) Merged image from panels F, G, and H.
ChIP analysis.
The chromatin immunoprecipitation was performed using the chromatin immunoprecipitation assay kit (Upstate Cell Signaling Solutions, Lake Placid, NY) and modified based on the Wells and Farnham protocol (70, 71). The detailed protocol is available upon request. In brief, Dox-induced or noninduced FoxI1-GFP or FoxI1-V5 cells (108) were treated with 1% formaldehyde on a rotating platform at room temperature for 15 min. The cross-linking reaction was stopped by the addition of glycine. Cells were treated with 0.25% trypsin at 32°C for 5 min and scraped. Trypsin was inactivated with 0.5 ml of sera. Following cold phosphate-buffered saline washes, the scraped cell pellet was resuspended in sodium dodecyl sulfate (SDS) lysis buffer containing protease inhibitor and sonicated. Soluble cross-linked chromatin was precleared by incubation with a preblocked salmon sperm DNA-protein A-agarose 50% slurry for 2 h at 4°C. Precleared chromatin was incubated with 3 μg of anti-V5 monoclonal or anti-GFP monoclonal antibody overnight at 4°C and precipitated by incubation with fresh blocked protein A-Sepharose for 2 h at 4°C. After a series of washes, immunoprecipitated FoxI1-DNA complexes were eluted with elution buffer (1% SDS, 0.1 M NaHCO3) and formaldehyde cross-linking was reversed by treatment with heat at 65°C for 6 h. After purification with proteinase K digestion and phenol-chloroform extraction, either the DNA was used directly in PCRs or immunoprecipitated DNA was ligated to linkers, amplified by PCR, and cloned into the PCR2.1-TOPO vector (Invitrogen). Random clones were sequenced. For PCR primer sequences, see the supplemental material.
DNase I sensitivity assays.
Quantification of DNase I sensitivity by real-time PCR was performed based on the methods previously described (15, 36). Nocodazole treatments were as follows. Cells were cultured to confluence, maintained for 2 to 3 days, and then split from 1 to 4 flasks. We added nocodazole (400 ng/ml) for 16 to 20 h. Ninety percent of the shake-off cells showed condensed nuclei. To test the viability of treated cells, the shake-off cells were transferred to fresh medium and cultured for 1 week. The cells grew a bit slower than those untreated but were otherwise normal.
Culture cells (107) were harvested and lysed in the presence of 0.2% NP-40-RSB buffer (10 mM Tris, pH 7.5, 10 mM NaCl, 3 mM MgCL2). The collected nuclei were resuspended in RSB buffer, aliquoted into 6 tubes, and digested at 37°C with 0.25 to 4 units of DNase I (Roche) for 5 min before being stopped by the addition of an equal volume of stop buffer (1% SDS, 600 mM NaCl, 10 mM EDTA, 20 mM Tris-HCl, pH 8.0). Isolation of nuclei was visually confirmed under a microscope with 4′,6′-diamidimo-2-phenylindole (DAPI) staining. After proteinase K digestion, phenol-chloroform extraction, and ethanol precipitation, the purified DNA was dissolved in Tris-EDTA, pH 8.0, quantified twice using the PicoGreen double-stranded DNA quantification reagent (Molecular Probes), and aliquoted into 96-well plates at 20 ng/well. Real-time quantitative PCR was performed using the QuantiTect SYBR green PCR kit (QIAGEN) and analyzed using the ABI PRISM 7900 (Applied Biosystems). ChIP primers to 20 individual genomic regions were designed based on the sequences identified in the ChIP experiments. Each amplicon (referred to as ChIP targets in the text) was approximately 200 to 300 bp in size and surrounded at least one putative FoxI1 binding site. Sixteen of the ChIP primer sets gave single bands when used to amplify zebra fish genomic DNA and were used for a quantitative PCR-based DNase I hypersensitivity assay, as previously described (15, 36). Each sample was repeated in duplicate at least twice (minimum of 4 data points; standard deviation, <0.32). Random genomic DNA sequences were selected manually by hand picking STS markers from the zebra fish genomic assembly. For sequences, see the supplemental material.
Analyses of MNase digests.
Digestion of nuclei with micrococcal nuclease (MNase) was carried out as previously described (20). Parallel aliquots of nuclei (∼300 _A_260 units) were resuspended in 500 μl of MNase digestion buffer (60 mM KCl, 15 mM NaCl, 15 mM Tris-HCl, pH 7.5, 2 mM MgCl2, 1 mM CaCl2, 1 mM dithiothreitol, and 0.25 M sucrose) and incubated at 37°C for 10 min at various concentrations (final concentration, 0.25, 1, and 2 units/ml). For DNA analyses, MNase-digested chromatin samples were subjected to organic extraction with proteinase K and DNase-free RNase treatment. For chromatin fractionation, MNase-digested nuclei were fractionated by the method of Zhao et al. (74). Nuclei (100 _A_260 units) were resuspended in 500 μl of MNase digestion buffer and incubated with 2 units of micrococcal nuclease for 10 min at 37°C. Three fractions (S1, S2, and P, respectively) were collected and analyzed by standard SDS-polyacrylamide gel electrophoresis (PAGE) and immunoblotting as described in the figure legends.
The preparation of nuclear matrix was modified as described by Berezney (5). Briefly, the above chromatin fractionation pellet (P fraction) was resuspended to 1 mg DNA/ml in STM buffer (0.25 M sucrose, 20 mM Tris, pH 7.5, 5 mM MgCl2) and digested with 50 units of DNase I (Roche) per 100 μl for 10 min on ice, followed by three high-salt extractions (2 M NaCl, 0.2 mM MgCl2, 10 mM Tris, pH 7.5) and one 1% Triton X-100 extraction. The soluble extractions, including the DNase I extraction, high-salt extraction, and Triton X-100 extraction, were collected and pooled as fraction P1. The insoluble pellet was washed twice in low-magnesium buffer (0.2 mM MgCl2, 10 mM Tris, pH 7.5). The remaining material was the isolated nuclear matrices (P2 insoluble chromatin).
Immunoblot and Southwestern analysis.
Protein extracts were separated on a 4 to 20% SDS-PAGE gel and immunoblotted with commercial anti-GFP (Clontech) and anti-V5 (Invitrogen) monoclonal antibodies. Southwestern analysis followed Papavassiliou's protocol (45), with the exception of using digoxigenin (DIG) labeling probes with DIG PCR labeling according to the manufacturer's suggestions (Roche). Briefly, _Escherichia coli_-expressed thioredoxin-FoxI1-V5 fusion protein was separated on a 4 to 20% SDS-PAGE gel and transferred onto nitrocellulose membranes (Protran BA85; Schleicher & Schuell). After renaturation, the bound proteins were hybridized with DIG-labeled DNA probe with a sheared herring sperm DNA fragment in the absence or presence of a 10-fold excess of unlabeled PCR products. The binding signal was detected using alkaline phosphatase-conjugated anti-DIG antibody and visualized with nitroblue tetrazolium-5-bromo-4-chloro-3-indolylphosphate (Roche).
Inverse one-hybrid analyses.
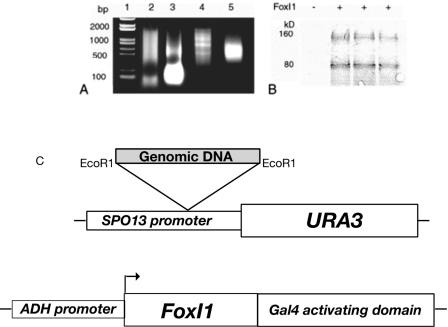
The procedure for the in vitro binding assay was performed using immunoprecipitation modified based on the method described by el-Deiry et al. (18). The URA3 reporter plasmid, pHQ366 (51), was modified by replacing the pstI-p53 binding site with a PstI-EcoRI linker (5′-GTATCTCGAGG-3′ and 5′-AATTCCTCGAGATACTGCA-3′), yielding the new plasmid pYoh366. An ADE2 and hemagglutinin (HA)-FoxI1 expression vector was constructed by inserting the FoxI1 coding region as an NcoI/XhoI fragment into pACT2 (Clontech) and replacing LEU2 with ADE2. The ADE2 gene was amplified from yeast genomic DNA using primers 5′-AATGCAATCGATTAACGCCGTATCGTGATTAAC-3′ and 5′-ACGTAAGCGGCCGCCGCTATCCTCGGTTCTGC-3′. FoxI1 expression was confirmed by Western blotting using an anti-HA tag antibody. FoxI1 expression was monitored by ADE2 gene expression determined by white cell growth on low-concentration adenine plates. Library screening was performed according to the strategy described by Tokino et al. (64). Yeast transformation was carried out using the alkali-cation yeast transformation kit (QBiogene). The yeast strains used were W303 MATa and _MAT_α, kindly provided by Carl Wu's laboratory. The final selected FoxI1-dependent clones were sequenced with two primers adjacent to the cloning site, 5′-GCGCTTTAAGAGAAAATATTTGTCCTG-3′ and 5′-CGGCTATTTCTCAATATACTCCTAATTAATAC-3′.
Sequence analyses.
ChIP sequences were mapped in the zebra fish genome using SSAHA on the Sanger Institute website (http://www.ensembl.org/Multi/blastview?species=Danio_rerio).
cDNA microarray analyses.
The detailed cDNA microarray analysis protocol is available upon request. In brief, total RNAs were isolated from Dox-induced and noninduced FoxI1-V5 cells (107). First-strand cDNA probes were generated by incorporation of Cy5-dCTP for Dox-induced and Cy3-dCTP for noninduced cDNA, respectively (Amersham Bioscience) during reverse transcription with SuperScript II (Invitrogen). The resulting cDNA probes were purified and concentrated. Equal amounts of Dox-induced and noninduced probes were hybridized to three zebra fish oligonucleotide arrays. Each chip contains 31,200 oligonucleotides (Compugen and MWG) designed from zebra fish expressed sequence tag assemblies and representing approximately 20,000 genes. After hybridization, the slides were washed, dried, and scanned using a DNA microarray scanner (Agilent Technologies) at 635 nm (Cy5) and then at 532 nm (Cy3). Fluorescence intensities were quantified using feature extraction software (Agilent Technologies). The experiment was repeated with the fluorescent dyes switched. The fluorescence intensities of the 6 data points were averaged, and the ratio of the intensity of the Dox-induced sample to that of the noninduced sample was then determined. The significant cutoffs for up- or down-regulated genes in FoxI1-expressing cells were determined as described previously (3). We chose cutoff values of ≥2.0 for up-regulated and ≤0.6 for down-regulated genes in FoxI1-expressing cells.
RESULTS
FoxI1 remains bound to chromatin through all cell cycle stages.
To test the translation of our cloned FoxI1 cDNA, we expressed FoxI1-V5 fusion proteins in E. coli and also in vitro-translated FoxI1-GFP and FoxI1-V5 fusion proteins. Immunoblotting analyses showed that anti-GFP and anti-V5 antibodies specifically recognized the predicted size bands (data not shown). These results confirmed that our cDNA sequence could be efficiently expressed. Based on our cDNA sequence, zebra fish FoxI1 can be divided into three regions: (i) an N-terminal domain containing a histidine-rich region (amino acids 90 to 104); (ii) a core forkhead DNA-binding domain (amino acids 183 to 278); and (iii) a C-terminal domain containing a small serine-rich region (amino acids 299 to 355). Inspection of the FoxI1 amino acid sequence also showed three potential nuclear localization signals: PPLKRTRT (amino acids 15 to 22), PFYKKSK (amino acids 218 to 224), and RRKRKRR (amino acids 273 to 279), two of which are in the DNA binding domain. Homology searches did not reveal significant homology with any reported transcriptional activation domains within other transcription factors (40).
Because some Fox proteins have demonstrated subcellular localizations (cytoplasmic versus nuclear) that changed depending on external cellular signals (7, 28), we examined FoxI1 distribution within living cells. We created two stable cell lines derived from the zebra fish embryonic cell line PAC2 (33); one cell line carried a doxycycline-inducible FoxI1-GFP fusion and the other carried an inducible FoxI1 protein tagged with the V5 epitope (Invitrogen). The expression profiles showed that both FoxI1 fusion proteins target to the nuclei and display a punctate staining pattern in the nuclei. A weak signal was detectable in the uninduced cells that was significantly increased with the addition of doxycycline (Fig. 1A to F). Cell cycle monitoring by flow cytometry showed that the initial expression of FoxI1 caused a transient delay in S phase which returned to normal after 24 h of expression (not shown). In contrast, a nearly identical cell line expressing eGFP alone expressed the protein throughout the cytoplasm (not shown). These results suggest that the FoxI1 protein has the expected subcellular nuclear localization and is potentially bound to certain regions of the chromatin in higher concentrations, causing the punctate pattern.
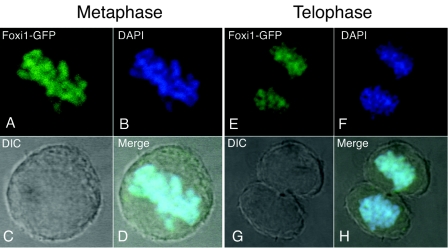
By comparing GFP fluorescence with DAPI staining in confocal images, we observed that the FoxI1 remained bound to the chromosomes even during mitosis (Fig. 2). When cells entered prophase, the fluorescence pattern of FoxI1-GFP was matched to the DAPI staining pattern in the same cells from prometaphase, metaphase, anaphase, and telophase, to cytokinesis (data not shown), indicating that the chromatin association of FoxI1 persists throughout mitosis. Little to no fluorescence was detected in other subcellular compartments, suggesting that most if not all of the FoxI1 remained bound to the chromatin (Fig. 1 and 2).
FIG. 2.
FoxI1-GFP associates with mitotic chromatin at metaphase (left) and telophase (right). FoxI1-GFP cells were counterstained with DAPI and imaged with a confocal microscope. (A) FoxI1-GFP signal at metaphase. (B) DAPI staining of the same cell as shown in panel A. (C) DIC image of the cell in panel A. (D) Merged image of panels A, B, and C. Note that the GFP signal colocalizes with the DAPI signal in the condensed chromosomes. (E) FoxI1-GFP signal in a dividing cell in telophase. (F) DAPI staining of the telophase cell in panel E. (G) DIC image of the cell in panel E. (H) Merged image of panels E, F, and G. Similar results were seen with the FoxI1-V5 fusion (not shown).
FoxI1 affects a subset of gene expression but not global transcription.
To assess how FoxI1 ectopic expression alters transcription, we used microarray analysis based on slides printed with oligonucleotides designed to zebra fish expressed sequence tags (Compugen, MWG). The analysis showed that FoxI1 transcript levels increased 14-fold when the cells were induced by doxycycline. However, of 31,200 zebra fish oligonucleotides covering approximately 20,000 protein-encoding genes, only 15 oligonucleotides, representing 12 genes, showed significant responses to FoxI1 expression, with an average calibrated ratio of 4.02 (2.3 to 6) for activated genes (8 elements) and 0.49 (0.15 to 0.6) for repressed genes (7 elements) (Table 1). In addition to 5 unknown genes, we identified 7 known genes that are changed on the array (Table 1). Several genes are consistent with the developmental role of FoxI1 in zebra fish, and four have known roles in forming the ear and jaw (the structures affected in the zebra fish FoxI1 mutations). For example, Sox9a has known roles in ear and jaw development in zebra fish (34, 72). Additionally, Pro alpha 1(I) collagen (COL1A1) and gelatinase b (matrix metalloproteinase 9 [MMP-9]) have been shown to be coregulated in bone formation (37). The targeted knockout of FoxI1 in mice has kidney defects with a loss of appropriate anion transporters, proton pumps, and anion-exchange proteins (6). The monocarboxylate transporter 4 (MCT4) shows differential expression with FoxI1 on the arrays and has been shown to be expressed in kidney-derived cells (41). MMP-9, Pax6b, and MCT4 changes were confirmed by quantitative real-time PCR, and results were consistent with the microarray data (data not shown).
TABLE 1.
FoxI1-responsive genesa
| Gene no. | Dox+ Cy5/Dox− Cy3 | Dox+ Cy3/Dox− Cy5 | Avg calibrated ratio | Clone title |
|---|---|---|---|---|
| 1 | 13.28, 12.63, 12.82 | 15.38, 15.54, 13.91 | 13.93 | FoxI1 |
| 2 | 3.13, 2.91, 3.41 | 8.25, 8.70, 6.83 | 5.54 | Unknown |
| 3 | 3.64, 2.52, 3.41 | 7.63, 7.43, 8.24 | 5.48 | Unknown |
| 4 | 4.02, 6.12, 4.98 | 6.40, 7.63, 7.01 | 6.03 | MCT4 |
| 5 | 5.74, 6.02, 6.02 | 3.60, 2.02, 3.28 | 4.45 | Es1 |
| 6 | 2.98, 2.92, 3.73 | 2.03, 2.62, 2.47 | 2.79 | Es1 |
| 7 | 2.90, 2.57, 2.79 | 2.64, 2.83, 2.82 | 2.76 | MMP-9 |
| 8 | 1.92, 2.05, 2.45 | 2.48, 2.63, 2.35 | 2.31 | MMP-9 |
| 9 | 2.99, 2.24, 2.53 | 2.85, 3.27, 3.06 | 2.82 | Unknown |
| 10 | 0.56, 0.60, 0.58 | 0.55, 0.51, 0.49 | 0.55 | Unknown |
| 11 | 0.57, 0.52, 0.58 | 0.51, 0.68, 0.54 | 0.57 | Unknown |
| 12 | 0.49, 0.47, 0.55 | 0.53, 0.51, 0.38 | 0.49 | Pro alpha 1(I) collagen, COL1A1 |
| 13 | 0.68, 0.79, 0.78 | 0.42, 0.42, 0.37 | 0.58 | Pax6b |
| 14 | 0.72, 0.83, 0.77 | 0.43, 0.40, 0.47 | 0.60 | Pax6b |
| 15 | 0.16, 0.15, 0.15 | 0.03, 0.03, 0.37 | 0.15 | Nuclear matrix protein p84 |
| 16 | 0.34, 0.32, 0.39 | 0.60, 0.52, 0.74 | 0.49 | Sox9a |
In contrast to its global chromatin binding pattern, FoxI1 alone does not have large effects on global transcription. Given the small number of genes that changed with expression of FoxI1 protein, we argue that FoxI1 generally requires other transcription factors to directly activate or repress genes. It is neither a potent activator nor repressor of gene expression on its own but more likely acts as a gene-specific coregulator of transcription.
Identifying FoxI1 chromatin-binding targets by ChIP.
Like many transcription factors, the binding sites for forkhead transcription factors have been experimentally determined, with the core recognition sequence being TRTTTR. This core region is necessary for forkhead binding, and bases immediately flanking the core contribute to the binding specificity of the different family members (44, 48). However, there is increasing evidence that Fox proteins recognize multiple related binding sites. For example, FHX (forkhead homologous X) was able to bind to two different types of sequences (46). The type A binding site contained a core element, (A/G)(T/C)AAA(C/T)A, whereas type B differed significantly from the consensus sequence and could not be determined with confidence. Although gel mobility shift assays are typically used to identify the short length and degenerative nature of binding site sequences under in vitro conditions, in vivo binding conditions are often difficult to recreate in vitro and the in vivo environment may impact binding specificity.
To isolate DNA fragments directly bound by the FoxI1 protein within living cells, we performed ChIP assays with antibodies to either eGFP or the V5 epitope. Compared to a nonspecific antibody control or a no-FoxI1 control, FoxI1-V5 immunoprecipitated DNA was approximately 27-fold higher, indicating that the ChIPs were, in fact, enriching for DNA sequences bound by FoxI1 (Table 2). After removal of small fragment DNAs (containing the preblocked herring DNA), the immunoprecipitated DNA was subcloned and sequenced. Of the 192 sequences tested (96 each from eGFP and V5 fusions), 164 clones contained inserts longer than 100 bp. Forty-three sequences were identical to or overlapping other clones (some isolated in both cell lines), representing 121 unique DNA fragments. Eighty-eight percent (106/121) of the DNA sequences contained the consensus core sequence of all Fox genes (TRTTTR), typically in multiple copies (95/106) (see the supplemental material). Not all target sequences contained the consensus site, suggesting that FoxI1 may interact with DNA targets through nonconsensus sequences or by binding structural features of the chromatin. Also, some ChIP sequences may represent the background noise of the technique. Therefore, we limited additional analysis to the 106 ChIP sequences that contained consensus binding sites.
TABLE 2.
ChIP assays
| Parameter | Result for cell line with doxycycline/nocodazole treatmentb | ||||||
|---|---|---|---|---|---|---|---|
| Foxi1-V5 | Pac2-Tet | Foxi1-V5, +/+ | Pac2-Tet, +/+ | Blocked beads, −/− | |||
| +/− | +/− | +/− | +/− | ||||
| Antibody | Anti-V5 | Anti-GFP | Anti-V5 | Anti-GFP | Anti-V5 | Anti-V5 | |
| Amt of input DNA (μg) | 170.7 | 170.7 | 229.6 | 229.6 | 208 | 265.8 | 0 |
| Amt of immunoprecipitated DNA (μg) | 0.424 | 0.186 | 0.165 | 0.178 | 0.304 | 0.184 | 0.177 |
| Amt of enriched DNA (μg)a | 0.247 | 0.009 | 0 | 0.001 | 0.127 | 0.007 | 0 |
Genomic locations of ChIP sequences.
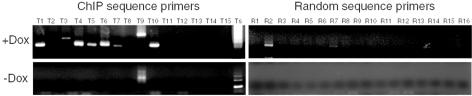
None of the isolated sequences bound to regions near the 12 genes identified on the array based on BLAT or BLAST comparisons. Sixty-eight of the 106 ChIP sequences were unambiguously mappable to 20 of 25 zebra fish chromosomes with aligned fragments longer than 90 bp in size and with equal or greater than 95% identity (as of June 28, 2005) (http://www.ensembl.org/Danio_rerio). The ChIP sequences included 14 tandemly repetitive DNAs. One example of the repeat sequences recognized by FoxI1 is ChIP sequence 32 (identical to sequences 106 and 168), which represents the type I satellite repetitive sequence (17). It has been reported that this type I satellite-like DNA accounts for 8% of the zebra fish genome (17). Fluorescence in situ hybridization analysis on zebra fish metaphase chromosomes detected localization of the identified DNA on all chromosomes, mostly at the centromeric and pericentromeric regions (57). To demonstrate that the cloned genomic fragments were genuine targets of FoxI1 binding, we chose 20 of the sequences and designed primers to approximately 300-bp amplicons for each target region. In parallel, we designed 20 sets of primers to random genomic regions. Sixteen of the 20 primer pairs amplified specific targets and were used for further analysis. We performed ChIP on nocodazole-treated, mitotically arrested cells induced with FoxI1 and cells not induced with FoxI1 using equal amounts of input DNA. The precipitated DNA was then tested with the 16 PCR pairs to ChIP targets or 16 primer sets to the random sequences. Nine of the 16 sequences were positive in cells induced with FoxI1, while only two of these targets (including the satellite DNA target Ts) were amplified in the absence of FoxI1, and the signals to these positives were significantly weaker (Fig. 3). Only 1 of the 16 random genomic targets was amplified. This represented a statistically significant difference in the two populations (P = 0.008, χ2 with Yates' correction). Thus, at least one-half of the sequences identified by the sequencing of ChIP clones were verifiable by PCR and required FoxI1 expression. To determine if FoxI1 sites could be computationally identified based on our microarray studies, we used 3 of the genes that had high-quality associated genomic sequence (Pax6, MMP-9, and MCT4) and examined the 2-kb upstream promoter region for putative FoxI1 binding sites. We identified 9 such sites and designed 4 primer sets to test for FoxI1 binding in the ChIP-enriched DNA. None of the primer sets were able to amplify fragments from the ChIP DNA, but 3 of the 4 sets successfully amplified fragments from the input DNA (not shown). This suggests that FoxI1 activation of these genes may not be through the traditional route of binding in the promoter.
FIG. 3.
PCR-coupled ChIP analyses of mitotic chromatin. Doxycycline (Dox)-induced (+Dox) and uninduced (−Dox) FoxI1-V5 cells were treated with nocodazole as described in Materials and Methods. Equal amounts of input DNA were subject to ChIP, and the enriched DNA was amplified using primers designed to amplify sequences identified in the subcloned ChIP fragments (T1 to T15, Ts). Data were compared to amplifications by primers designed to random genomic sequences (R1 to R16). The Ts primers were selected from type I satellite DNA, which flank the 186-bp repeat unit which creates a ladder when amplified. Nine of the 16 ChIP fragments amplified in the presence of FoxI1, while only 1 of 16 amplified in the random control, and this fragment only amplified in the presence of FoxI1, suggesting it may be a bona fide target of FoxI1 binding. Two of the ChIP-identified sequences amplified in the uninduced state, one of which was the satellite DNA (T9, Ts). These may represent high-affinity targets that are bound by a low level of FoxI1 that is detectable even in the uninduced state.
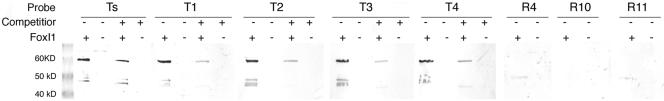
To further confirm the binding of some of these targets, we performed Southwestern blotting using _E. coli_-expressed FoxI1 and either labeled ChIP targets or labeled random targets (Fig. 4). We tested 5 of the PCR-positive fragments (Ts and T1 to T4) and 3 of the random targets. All of the target fragments were bound by a protein of the predicted size for FoxI1 (60 kDa) and could be competed by an excess of unlabeled probe, while none of the random targets showed any binding to the blot. This confirmed that the ChIP-identified sequences were genuinely being bound by FoxI1protein.
FIG. 4.
Confirmation of FoxI1-DNA binding by southwestern analysis of FoxI1 protein. _E. coli_-expressed FoxI1-V5, FoxI1+, and a FoxI1-null vector, FoxI1- (FoxI1 coding region was inserted into the opposite orientation), were induced with 0.02% arabinose. Protein extract (100 μg) was subject to SDS-PAGE and transferred to a nitrocellulose membrane. Probes were DIG labeled by PCR, amplifying the inserts from ChIP-cloned targets (T1, T2, T3, T4, and Ts) and random targets (R4, R10, and R11). DIG-labeled probes (10 μg) were hybridized with the bound proteins in the presence of 100 μg of sonicated herring sperm DNA and a 10-fold excess of competitor DNA (unlabeled probe DNA, 100 μg). The ChIP probes specifically recognized the thioredoxin-FoxI1-V5 fusion protein (predicted size of 60 kDa) and could be competed with unlabeled probe, while random probes (R4, R10, and R11) did not bind to FoxI1.
FoxI1 binds to type I satellite DNA sequence.
As the type I satellite DNA was identifiable in both induced and uninduced cells, we were interested in establishing whether FoxI1 could really bind these sequences, as it is possible that the satellite DNA is so highly represented in the genome that some contamination might be expected. We cannot determine whether the FoxI1 protein levels in our cell culture system are comparable to physiological levels, thus there is the additional concern that overexpression may cause FoxI1 to bind to chromatin by indirect association instead of sequence-specific binding. To further confirm identified binding sites of FoxI1 protein, we combined in vitro immunoprecipitation and a modification of the yeast one-hybrid system (4, 51, 66), which we termed the yeast “inverse one-hybrid” analysis, to demonstrate that FoxI1 can bind to the same sequences in an entirely different cellular context (Fig. 5). The in vitro-translated FoxI1-V5 fusion protein was incubated with sheared zebra fish genomic DNA, immunoprecipitated with anti-V5 antibodies, and purified with protein A-agarose beads. After four rounds of repeated FoxI1-DNA binding selection (Fig. 6A), the FoxI1-enriched DNA fragments were shotgunned upstream of a promoterless URA3 reporter vector (pYoh366) (Fig. 5C) containing the TRP1 gene, yielding a library of genomic DNA fragments containing a total of 1.2 × 106 clones (average size, 500 bp). The library was then transformed into Saccharomyces cerevisiae strain W303, which carried a plasmid expressing a FoxI1-Gal4 activating domain fusion. A total of 2 × 104 transformants was obtained, of which 210 colonies showed growth on media lacking uracil. Plasmids from these yeast colonies were transformed into E. coli strain KC8, and the library plasmid was isolated by growth of the KC8 cells on bacterial minimal media (64) without tryptophan. The recovered plasmids were then transfected into yeast strain W303 containing an empty vector control, selected on SD-Trp-Ura3 medium. Of 200 clones tested, 17 (8.5%) were found to grow on media lacking uracil only when FoxI1 was also expressed in the cells. These inserts were sequenced. A BLAST search revealed that three clones contained the type I satellite DNA, identical to the three ChIP-identified sequences: 32, 106, and 168. We therefore have 3 independent techniques that demonstrate that FoxI1 can bind to type I satellite DNA (ChIP, yeast inverse one-hybrid assay, and Southwestern blotting). From these results, we concluded that FoxI1 can bind to this repeat satellite DNA both in vitro and in vivo. Despite the fact that satellite DNA comprises approximately 8% of the genomic DNA (17), this binding cannot account for all the association to the chromatin, as satellite DNA has been shown to be primarily pericentric (57), yet the FoxI1-GFP signal can be detected throughout the entire chromatin (Fig. 1).
FIG. 5.
Strategy for SELEX immunoprecipitation coupled with a yeast inverse one-hybrid test system. (A) Enrichment of genomic DNA by FoxI1-V5 using SELEX immunoprecipitation. Lane 1, DNA marker; lane 2, sonicated genomic DNA; lane 3, PCR amplification of the first round of precipitated DNA; lane 4, PCR amplification of the third round of enriched DNA; lane 5, PCR amplification of the final selected DNA fragments. (B) Immunoblot detection of Gal4AD-HA-FoxI1 fusion proteins in yeast (+), with an empty vector used as a control (−). FoxI1 expression was detected by Western blotting using anti-HA-tagged antibody. (C) Construction of the URA3 reporter plasmid and the FoxI1 fusion protein. The EcoRI site was used for shotgunned insertion of test DNA fragments. The FoxI1 protein was fused to the transcription-activating domain of the yeast GAL4 protein to ensure transcriptional activation in yeast. The protein was constitutively expressed in the test strain. Three unique sequences were identified using this technique including type 1 satellite DNA.
FIG. 6.
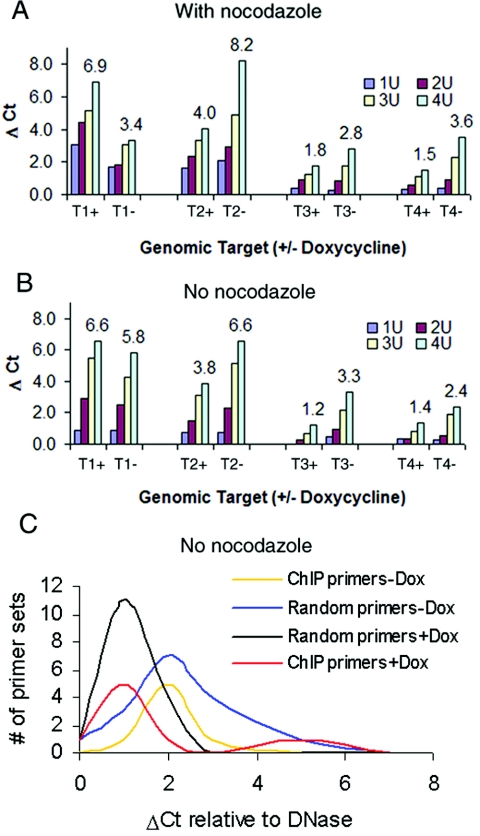
Quantitative analyses of DNase I sensitivity by real-time PCR. (A and B) FoxI1-V5 cells were induced for 24 h or grown without induction. The intact nuclei were digested with increasing concentrations of DNase I (0.25 to 4 U). Primer sets were either designed using ChIP-isolated DNA sequences (ChIP primers) or primers randomly selected from zebra fish STS sequences (random primers). ΔCt indicates the number of additional cycles necessary to amplify the target to the CT compared to a 0.25-U DNase I control. The number at the top of each target indicates the Δ_CT_ derived by subtracting the CT at 0.25 U DNase I from the CT at 4 U of DNase I. A Δ_CT_ of 1 is equivalent to a twofold change in sensitivity, for example, T1 changes from 3.4 to 6.9 in the presence of FoxI1, this is equal to an ∼11-fold increase in sensitivity. (A) Expression of FoxI1 alters DNase I hypersensitivity in condensed chromatin treated with nocodazole. Mitotically arrested cells were separated by shake off of the culture after 16 h in nocodazole (400 ng/ml). The x axis indicates different specific genomic targets in the presence (+) or absence (−) of FoxI1 and ranges of DNase I concentration from 1 to 4 U. (B) Alterations in DNase I hypersensitivity in unsynchronized cells. Δ_CT_s are not as large under these conditions compared to condensed chromatin. (C) Δ_CT_s for all ChIP targets compared to the random targets in the absence and presence of FoxI1 expression. Note the shift to the left for both ChIP and random targets and the bimodal distribution of ChIP targets in the presence of FoxI1 (red line). Dox, doxycycline.
FoxI1 expression alters chromatin accessibility to DNase I.
Because FoxI1 remained bound to chromatin and heterochromatin even during mitosis and Fox proteins have a structure similar to the linker histone H5 (12, 14), we decided to address the effect of FoxI1 expression on condensed chromatin architecture. DNase I sensitivity assays have been used to demonstrate structural changes in chromatin architecture related to the transcriptional activity of the chromatin regions (21, 56). More interestingly in regard to our observation of FoxI1 binding the condensed chromatin, some hypersensitive sites have been shown to persist during mitotic chromatin condensation (39). Accordingly, we examined the DNase I sensitivity of our ChIP-isolated sequences by using a quantitative PCR assay (36). FoxI1-GFP and FoxI1-V5 cells were cultured in either the presence or absence of doxycycline. Nocodazole, a microtubule-destabilizing molecule, was used to arrest cells in mitosis (39). The nuclei of mitotic cells in the presence or absence of FoxI1 were treated with various concentrations of DNase I, and the purified DNA was subjected to quantitative PCR using primers to 8 positive PCR samples (the satellite DNA is not amenable to this type of analysis). Many of the ChIP targets in mitotically condensed chromatin did not show a significant difference in hypersensitivity from the unsynchronized population in the presence or absence of FoxI1. However, 4 of the 8 targets (50%) did show significant differences in DNase I sensitivity when FoxI1 was induced. Three of the genomic targets, T2, T3, and T4, displayed a reduction in DNase I hypersensitivity, while one genomic target, T1, had increased DNase I sensitivity (Fig. 6A and B). These sensitivity differences can be quite large in the nocodazole-treated condensed chromatin. Each cycle change in the cycle threshold (CT) represents a twofold change in DNase I sensitivity. Thus, for target 1 (T1) the region is 11.3-fold more sensitive to DNase I digestion in the presence of FoxI1, while target 2 (T2) is greater than 16-fold more resistant to DNase I digestion in the presence of FoxI1. Of interest is that the differences in DNase I sensitivity between the presence and absence of FoxI1 are typically much greater in the condensed chromatin than they are in the unsynchronized population (compare Fig. 6A and B), suggesting that other transcription factors are modifying the effects of FoxI1 when the chromatin is in a more relaxed state, allowing those other factors access to the DNA.
To further analyze the effects on chromatin DNA not specifically bound by FoxI1, we examined the 16 random genomic targets used in Fig. 3 and compared them to the ChIP-identified targets by real-time PCR. With no Dox induction, there is a similar distribution of Δ_CT_s for both ChIP targets and randomly selected targets (random targets), with an average Δ_CT_ value of approximately 2 (Fig. 6C). After induction of the FoxI1 protein, random targets significantly reduced their Δ_CT_ values (the Δ_CT_ mode) for random targets dropped from 2 to 1 (Fig. 6C). The 8 ChIP targets showed a bimodal distribution. Many of the sequences showed a reduction of the Δ_CT_, but other targets maintained or increased in value (Fig. 6C), suggesting FoxI1 can have both global and site-specific effects on chromatin structure.
FoxI1 plays a role in altering the nucleosomal array.
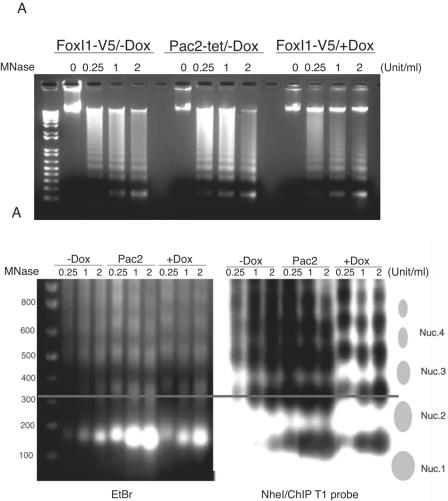
The DNase sensitivity assays suggested that FoxI1 protein might reduce or inhibit accessibility of chromatin DNA to DNase I. However, we do not know whether FoxI1 modified the higher-order structure of nucleosomes. Nuclear digestions with MNase have been used to analyze the nucleosomal array structure of chromatin (20, 23, 61, 74). This enzyme cuts the DNA threads at the junctions between nucleosomes and releases mono- and oligonucleosomes. In the cells expressing prothymosin α, a chromatin-remodeling protein, MNase partial digestion generated a ladder in which all DNA fragments were slightly shorter than those observed with controls (20). We tested the MNase sensitivity of Pac2 cells under three conditions: (i) in Pac2 cells induced by doxycycline but containing no inducible FoxI1 transgene, (ii) in Pac2 cells with the inducible FoxI1 transgene but not doxycycline and (iii) in Pac2 cells where FoxI1 is induced by doxycycline (Fig. 7A). In contrast to prothymosin α, which facilitates accessibility to chromatin of MNase, we found that the cells expressing FoxI1 showed a significantly increased general resistance to MNase digestion. Most of the chromosomal DNA remained in the largest sized DNA fragments, even at the highest concentrations of MNase, while in the absence of FoxI1, the genomic DNA was clearly digested into mono- and oligonucleosomes. We then went on to more carefully analyze the size of the nucleosome oligomers in the presence or absence of FoxI1. We ran the MNase-digested DNA and imaged the gel either using ethidium bromide or by Southern blot using the T1 target as a probe (T1 contains a SINE element so it hybridizes to many sites in the genome) (Fig. 7B). At every MNase concentration, FoxI1 shifted the size of the oligomers slightly larger. The line on the figure passes through the center of a trinucleosome (0.25U/ml MNase) in the absence of FoxI1. The matching trinucleosome in the presence of FoxI1 is approximately 50 bp larger. This suggests that FoxI1 is partially protecting the MNase-sensitive linker regions of the nucleosomal array. This then raises the question of whether FoxI1 is directly competing with the linker histones in packaging chromatin or having an additive effect to the linker histones.
FIG. 7.
Nucleosome structural analysis in FoxI1-expressing cells. Nuclei were isolated from induced non-FoxI1 integration Pac2-tet-on cells (Pac2/+Dox), uninduced FoxI1-V5 cells (FoxI1/-Dox), and induced FoxI1-V5 cells (FoxI1/+Dox). (A) Isolated nuclei were incubated with the indicated concentrations (0.25, 1, or 2 units/ml) of MNase; 0 indicates incubation without nuclease. Purified DNA fragments were electrophoresed on a 1.4% agarose gel. Marker is the 1-kb Plus DNA ladder (Invitrogen). (B) MNase digests (2 μg each) were digested with NdeI and separated on a 1.4% agarose gel at 16°C, 70 V for 6 h, stained with ethidium bromide (EtBr), and after photography (shown in panel EtBr, on the left), blotted to nylon membranes and probed with 32P-labeled ChIP target T1 (right panel). The T1 target is 279 bp in length and contains 72-bp SINE repeat elements. The line passes through the center of the trioligonucleosome at 0.25 U/ml MNase in the absence of FoxI1 to facilitate visualization of the nucleosome size change.
FoxI1 is strongly enriched in the insoluble chromatin fraction.
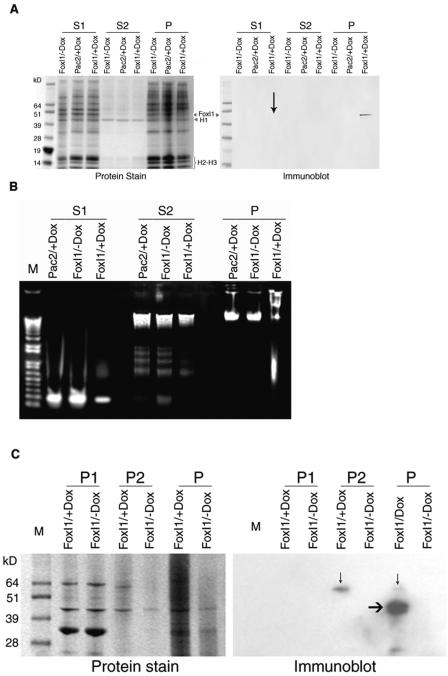
To establish whether FoxI1 is bound to active or inactive chromatin, we fractionated chromatin according to the method described by Zhao and colleagues (74). Three fractions were obtained: an active chromatin fraction (S1), an H1-enriched inactive chromatin fraction (S2), and nuclear matrix proteins containing insoluble pellet (P) (24, 53, 74). Satellite DNA is markedly depleted in S1, but is present in other chromatin fractions (24, 25). If FoxI1 is involved in associations with heterochromatin, it would be expected to be enriched in the S2 and/or P fraction. We demonstrated that the P fraction was strongly enriched in FoxI1 protein relative to the S1 fraction (approximately 99% to 1% by calculating the intensity). This result indicated that FoxI1 is different from most chromatin remodeling proteins that have been shown to facilitate chromatin accessibility to MNase and are enriched in the active chromatin fraction (S1), such as HMG1 (74), prothymosin α (20), and hSWI/SNF proteins (52).
Comparing protein staining with the immunodetection of FoxI1 in Fig. 8A, we can infer that (i) linker histone is enriched in the S2 fraction where FoxI1 is not detectable and (ii) FoxI1 expression is much lower than linker histone H1 because no significant band of FoxI1 could be seen on the protein staining gel corresponding to the FoxI1 detected on an immunoblot (FoxI1-V5 expected size, 48 kDa). Taken together, while structurally similar to the linker histones, FoxI1 is not globally competing with linker histones to alter MNase resistance. It appeared that FoxI1 is effecting change through interactions with the nuclear matrix.
FIG. 8.
Chromatin fractionation analysis. Isolated nuclei were digested with 2 units/ml of MNase for 10 min. Three fractions were obtained: S1, soluble first fraction; S2, soluble second fraction; P, the insoluble pellet fraction. Aliquots from each cell line were separated on a NuPAGE 12% Bis-Tris gel. M, SeeBlue Plus2 prestained standard (Invitrogen). (A) Gels were stained with SimplyBlue Safe Satin (Invitrogen), and the duplicate gel was immunoblotted with anti-V5 monoclonal antibody. FoxI1-labeled lanes have the induced FoxI1 protein. Linker H1 protein is clearly seen as a 40-kDa protein enriched in the S2 fraction in all three cell lines. FoxI1 was detectable in the S1 fraction (marked with black arrow), but the majority was present in the pellet (P). (B) An ethidium bromide-stained agarose gel showed that isolated DNA in the S2 fraction was low, relative to the S1 and P fractions. (C) The initial pellet fraction (P) was subjected to DNase I digestion and high-salt extractions, as described in Materials and Methods. The soluble extraction (P1) and insoluble fraction (P2) were examined using the ECL Western blot analysis system (Amersham Bioscience). A band detected by anti-V5 antibody in insoluble chromatin (P2) (marked with black arrow) was 62 kDa in size, and the untreated pellet fraction was 48 to 62 kDa (P). The mobility anomaly may be associated with the high-salt treatment of the chromatin.
It is possible that FoxI1 is expressed at much higher than physiological levels in this artificial system and that this overexpression could simply be affecting the ability to release DNA into the S2 fraction, which would incorrectly identify FoxI1 as being associated with the nuclear matrix. To further establish that FoxI1 is enriched in the insoluble pellet (P fraction), the initial P fraction was subsequently fractionated according to the method for preparation of nuclear matrix described by Berezney (5). DNase I digestion was followed with consecutive high-salt and 1% Triton X-100 extractions. The P fraction was subfractionated into P1 (soluble) and P2 (insoluble chromatin, nuclear matrices). FoxI1 was detected in insoluble chromatin (P2), suggesting that FoxI1 was tightly bound to the nuclear matrix. The alterations in micrococcal nuclease sensitivity and FoxI1's close association with the nuclear matrix suggests that FoxI1 is altering chromatin compaction by increasing association with the nuclear matrix.
DISCUSSION
We have previously cloned and characterized a new gene: zebra fish foxi one. It encoded a nuclear protein with 419 amino acids and was expressed specifically in a small number of tissues in the developing zebra fish embryo. FoxI1 has been shown to possess complex signaling connections with other genes, such as fibroblast growth factors, pax8, ngn, phox2a, pendrin, band3 anion exchangers (AE1), and possibly, the ATP6B subunit of the vacuolar H+-ATPase (1, 6, 26, 31, 42, 58-60). The mechanism of these multi-interactions is not clear. We speculate that FoxI1 may play a central role in a complex signaling network necessary for ear and jaw development.
We created two zebra fish cell lines where we could regulate FoxI1 expression under the control of a doxycycline responsive system. Unlike most transcription factors that are actively excluded from the condensed chromatin during mitosis (35), we demonstrated that the zebra fish forkhead transcription factor FoxI1 remains bound to condensed chromatin throughout the cell cycle. Only two examples of transcription factors have been shown to remain bound to the mitotic chromosomes: the general transcription factor TATA binding protein (11) and the ubiquitous Drosophila proteins GAGA factor and Prod (49). Thus, FoxI1 is the first example of a transcription factor that is expressed tissue specifically yet is able to stably bind to condensed chromatin. The developmental defects demonstrated in both zebra fish and mouse when FoxI1 is mutated suggest that this stable chromatin binding is an essential aspect of the early developmental program. The punctate nuclear staining of FoxI1 is reminiscent of the subcellular localizations of GAGA and Prod in Drosophila simulans and Drosophila manuritiana brain interphase nuclei (49). Platero interpreted this granular pattern as the protein predominately binding to euchromatic sites, with the intense spots representing interphase binding of the proteins to satellite DNA targets. We believe that the forkhead transcription factors could be part of a class of factors including GAGA and prod that bind chromatin stably and modulate developmental responses of the cells by altering the genomic transcriptional template in a stable manner, in contrast to other types of transcription factors and nuclear proteins which have rapid DNA association dynamics (47, 62). While the cell lines we generated create a carefully controlled environment for studying FoxI1, there is the caveat that this artificial system, because of overexpression or inappropriate cellular context, may not represent the function of FoxI1 in vivo. What we can conclude is that FoxI1 demonstrates unusual characteristics for a transcription factor in that it is able to maintain its DNA interactions in condensed chromatin, and we feel strongly that this ability to bind cannot be explained by an overexpression of the protein. The logical conclusion is that this binding to condensed chromatin is related to the actual function of FoxI1 in vivo.
Remarkably, the induction of FoxI1 expression has a very modest effect on gene expression in these cells. Only 12 genes of the approximately 20,000 (0.06%) sampled by microarray changed in levels by any significant amount. Given that FoxI1 can be shown to widely bind to the chromatin (Fig. 2), it can be concluded that FoxI1 is not likely to be a potent activator or repressor on its own and more likely requires a combinatorial effect with other transcription factors to cause significant changes in expression. This is consistent with a role in global chromatin remodeling. Similarly, linker histone H1 does not have a major effect on global transcription but can act as either a positive or negative gene-specific regulator of transcription in vivo (55). There is also the possibility that by making a fusion of FoxI1 to GFP or the V5 epitope, the transcriptional regulation activity of FoxI1 is disrupted or altered. Because this is an artificial system and gene regulation is a complex interaction from multiple transcription factors, it is not clear whether any of the genes changed in the array analysis are bona fide targets of FoxI1 regulation. We do not argue that the direction of the gene changes are relevant to the normal developmental context, as many modifying factors may be present in the developing organism that are not present in the tissue culture system, but we do believe that many of these genes will prove to be relevant targets of regulation by FoxI1. The genes that are affected do appear to be enriched for genes known to be involved in ear and jaw development or kidney function (the precise locations of FoxI1 expression). Sox9a is involved in both ear and jaw development (72, 73). MMP-9 is a key enzyme in cartilage formation (43), and MCT4 is expressed specifically in the mouse kidney (41). The establishment of these genes as genuine targets of regulation in zebra fish embryos is ongoing, although a clear genetic relationship between FoxI1 and Sox9a has already been established (34).
The nature of association of FoxI1 with the heterochromatic satellite DNA is not clear. It may be a simple coincidence that these repetitive DNAs contain multiple copies of the consensus binding site of Fox proteins but are not functionally relevant sites in vivo. Alternatively, it may be a consequence of the coevolution of forkhead genes with satellite DNAs. This concept follows from the theory of mitotic borrowing proposed by Csink and Henikoff (16). The authors proposed that expansion of a new satellite DNA repeat would borrow an appropriate DNA-binding protein, which ensures packaging of this satellite DNA array during mitosis. Only those repeat motifs that are able to borrow appropriate mitotic proteins can be expanded into large blocks of satellite DNA arrays. In interphase, these DNA-binding proteins would have other functions unrelated to satellite DNAs (16). In that case, forkhead gene expansion may be responding or correlating with satellite DNA evolution. Fitting with that hypothesis, the forkhead class of transcription factor has an overall structure similar to those of the linker histones H1 and H5. This similarity of structure may allow the forkhead transcription factors to bind a DNA context (i.e., condensed chromatin) from which most transcription factors are excluded. Additionally, Cirillo and Zaret showed that the forkhead protein HNF3 bound to DNA more stably in the context of nucleosomes than when bound to naked DNA (13), again suggesting that the similarity of three-dimensional structure implies a preferred chromatin context for the forkhead transcription factors and providing for a possible “crossover” role as described by Csink and Heinkoff. This would not necessarily imply that FoxI1 is directly competing with H1 for DNA binding, merely that the general three-dimensional structure of the forkhead proteins conveys an ability to bind condensed chromatin, in sharp contrast to most transcription factors.
To examine the influence of FoxI1 expression on chromatin structure, we performed DNase I hypersensitivity assays using the sequences we isolated from ChIP enrichment. We found that most of the confirmed genomic targets bound by FoxI1 maintained or decreased DNase I sensitivity in the presence of FoxI1. A smaller number of sites bound by FoxI1 displayed increases in DNase I sensitivity that were maintained even in mitotically condensed chromatin. In fact, these differences in DNA structure were magnified in condensed chromatin compared to unsynchronized cells. In some cases, this sensitivity to DNase I could change more than 10-fold, with some locations becoming more sensitive and others becoming less sensitive. Importantly, some of the changes in DNase I sensitivity were not correlated with FoxI1-binding sites, suggesting that FoxI1 was also having longer-range effects on chromatin structure. The DNase I studies suggest that FoxI1 has two effects on chromatin: direct effects on specifically bound targets and indirect effects on the global organization of chromatin. In support of this argument, Carroll et al. demonstrated that the estrogen receptor bound to many sites on chromosomes 21 and 22, many of which were far from regulated genes (10). They were then able to demonstrate that the binding of the estrogen receptor required the presence of FoxA1 binding in close proximity. This is consistent with the permissive role we hypothesized for FoxI1.
Additional chromatin structural analyses demonstrated that FoxI1 was strongly enriched in insoluble chromatin, with a much smaller amount of FoxI1 associated with the active chromatin S1 fraction. No FoxI1 was detected in the S2 fraction, which is enriched for the linker histones H1 and H5 (74). The association of FoxI1 with both the transcriptionally active chromatin (S1) and competent chromatin (P) again demonstrates the two modes of FoxI1 function, with the S1 fraction having effects on specific transcripts and the P fraction of FoxI1 involved in overall chromatin architecture. Studies have shown that repetitive elements could influence chromatin structure in two ways: (i) dispersed repeated copies containing binding sites for chromatin-organizing proteins can form a basis for local chromatin structure (54) and (ii) tandemly repetitive sequences can nucleate the highly compacted structure called “heterochromatin” and negatively affect the expression of genetic loci at distances of many kilobase pairs. Both types of position effect variegation are well documented in fruit flies (68, 69). Consistent with a FoxI1 role in the association with satellite DNA, the Domina (Dom) protein, another member of the FKH/WH transcription factor gene family in Drosophila, was shown to be accumulated in the chromocenter and function as a suppressor of position-effect variegation (PEV) (63). In addition, the GAGA protein which has a similar staining pattern to FoxI1 does not have structural similarity to the FKH/WH family but, like Dom, was also originally identified as being involved in PEV in Drosophila. It may not, therefore, be a coincidence that both the zebra fish and mouse mutations in FoxI1 displayed somewhat variable phenotypes (27, 42), which could suggest PEV effects in a vertebrate context. It is widely believed that living cells can use repetitive DNA sequences in various ways to affect the expression of coding sequences. Our results provide evidence that FoxI1-expressing cells can modulate chromatin structures, both local effects of FoxI1 binding and global or long distance changes in nucleosome organization. The model for regulation by forkhead transcription factors becomes one where the protein is bound stably to the chromatin with both isolated and global effects on chromatin structure, essentially establishing a genomic “template.” This template will allow cells expressing the forkhead protein to rapidly and appropriately respond to the external induction factors in the context of the developing embryo. We demonstrated previously that FoxI1 was necessary for an appropriate response to fibroblast growth factor signaling in the zebra fish embryo, and the present study gives a framework for how that response could take place.
Supplementary Material
[Supplemental material]
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, at the National Human Genome Research Institute.
We thank Weihua Wu for discussion during preparation of the manuscript and L. Brody and R. Nissen for critical reading of the manuscript. We thank A. Dultra, E. Pak, and P. Leo for assistance in confocal microscopy and fluorescence in situ hybridization, S. Anderson for flow cytometry examination, Guojian Jiang for microarray hybridizations, Tao Tao and M. Portnoy for instruction in Standalone Blastall and UCSC BLAT searches. Thanks go to R. Brachmann for providing the pHQ366 plasmid.
There are no competing financial interests for any author.
Footnotes
REFERENCES
- 1.Al-Awqati, Q., and G. J. Schwartz. 2004. A fork in the road of cell differentiation in the kidney tubule. J. Clin. Investig. 113**:**1528-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allan, J., P. G. Hartman, C. Crane-Robinson, and F. X. Aviles. 1980. The structure of histone H1 and its location in chromatin. Nature 288**:**675-679. [DOI] [PubMed] [Google Scholar]
- 3.Azumi, K., M. Fujie, T. Usami, Y. Miki, and N. Satoh. 2004. A cDNA microarray technique applied for analysis of global gene expression profiles in tributyltin-exposed ascidians. Mar. Environ. Res. 58**:**543-546. [DOI] [PubMed] [Google Scholar]
- 4.Bell, P. J., I. W. Davies, and P. V. Attfield. 1999. Facilitating functional analysis of the Saccharomyces cerevisiae genome using an EGFP-based promoter library and flow cytometry. Yeast 15**:**1747-1759. [DOI] [PubMed] [Google Scholar]
- 5.Berezney, R. 1980. Fractionation of the nuclear matrix. I. Partial separation into matrix protein fibrils and a residual ribonucleoprotein fraction. J. Cell Biol. 85**:**641-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blomqvist, S. R., H. Vidarsson, S. Fitzgerald, B. R. Johansson, A. Ollerstam, R. Brown, A. E. Persson, G. G. Bergstrom, and S. Enerback. 2004. Distal renal tubular acidosis in mice that lack the forkhead transcription factor Foxi1. J. Clin. Investig. 113**:**1560-1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brownawell, A. M., G. J. Kops, I. G. Macara, and B. M. Burgering. 2001. Inhibition of nuclear import by protein kinase B (Akt) regulates the subcellular distribution and activity of the forkhead transcription factor AFX. Mol. Cell. Biol. 21**:**3534-3546.11313479 [Google Scholar]
- 8.Burley, S. K., X. Xie, K. L. Clark, and F. Shu. 1997. Histone-like transcription factors in eukaryotes. Curr. Opin. Struct. Biol. 7**:**94-102. [DOI] [PubMed] [Google Scholar]
- 9.Carlsson, P., and M. Mahlapuu. 2002. Forkhead transcription factors: key players in development and metabolism. Dev. Biol. 250**:**1-23. [DOI] [PubMed] [Google Scholar]
- 10.Carroll, J. S., X. S. Liu, A. S. Brodsky, W. Li, C. A. Meyer, A. J. Szary, J. Eeckhoute, W. Shao, E. V. Hestermann, T. R. Geistlinger, E. A. Fox, P. A. Silver, and M. Brown. 2005. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122**:**33-43. [DOI] [PubMed] [Google Scholar]
- 11.Chen, D., C. S. Hinkley, R. W. Henry, and S. Huang. 2002. TBP dynamics in living human cells: constitutive association of TBP with mitotic chromosomes. Mol. Biol. Cell 13**:**276-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cirillo, L. A., C. E. McPherson, P. Bossard, K. Stevens, S. Cherian, E. Y. Shim, K. L. Clark, S. K. Burley, and K. S. Zaret. 1998. Binding of the winged-helix transcription factor HNF3 to a linker histone site on the nucleosome. EMBO J. 17**:**244-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cirillo, L. A., and K. S. Zaret. 1999. An early developmental transcription factor complex that is more stable on nucleosome core particles than on free DNA. Mol. Cell 4**:**961-969. [DOI] [PubMed] [Google Scholar]
- 14.Clark, K. L., E. D. Halay, E. Lai, and S. K. Burley. 1993. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature 364**:**412-420. [DOI] [PubMed] [Google Scholar]
- 15.Crawford, G. E., I. E. Holt, J. C. Mullikin, D. Tai, R. Blakesley, G. Bouffard, A. Young, C. Masiello, E. D. Green, T. G. Wolfsberg, F. S. Collins, and National Institutes Of Health Intramural Sequencing Center. 2004. Identifying gene regulatory elements by genome-wide recovery of DNase hypersensitive sites. Proc. Natl. Acad. Sci. USA 101**:**992-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Csink, A. K., and S. Henikoff. 1998. Something from nothing: the evolution and utility of satellite repeats. Trends Genet. 14**:**200-204. [DOI] [PubMed] [Google Scholar]
- 17.Ekker, M., A. Fritz, and M. Westerfield. 1992. Identification of two families of satellite-like repetitive DNA sequences from the zebrafish (Brachydanio rerio). Genomics 13**:**1169-1173. [DOI] [PubMed] [Google Scholar]
- 18.el-Deiry, W. S., S. E. Kern, J. A. Pietenpol, K. W. Kinzler, and B. Vogelstein. 1992. Definition of a consensus binding site for p53. Nat. Genet. 1**:**45-49. [DOI] [PubMed] [Google Scholar]
- 19.Gajiwala, K. S., H. Chen, F. Cornille, B. P. Roques, W. Reith, B. Mach, and S. K. Burley. 2000. Structure of the winged-helix protein hRFX1 reveals a new mode of DNA binding. Nature 403**:**916-921. [DOI] [PubMed] [Google Scholar]
- 20.Gomez-Marquez, J., and P. Rodriguez. 1998. Prothymosin alpha is a chromatin-remodelling protein in mammalian cells. Biochem. J. 333(Pt 1)**:**1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gregory, R. I., S. Khosla, and R. Feil. 2001. Probing chromatin structure with nuclease sensitivity assays. Methods Mol. Biol. 181**:**269-284. [DOI] [PubMed] [Google Scholar]
- 22.Guo, Y., R. Costa, H. Ramsey, T. Starnes, G. Vance, K. Robertson, M. Kelley, R. Reinbold, H. Scholer, and R. Hromas. 2002. The embryonic stem cell transcription factors Oct-4 and FoxD3 interact to regulate endodermal-specific promoter expression. Proc. Natl. Acad. Sci. USA 99**:**3663-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hatzis, P., and I. Talianidis. 2002. Dynamics of enhancer-promoter communication during differentiation-induced gene activation. Mol. Cell 10**:**1467-1477. [DOI] [PubMed] [Google Scholar]
- 24.Huang, S. Y., and W. T. Garrard. 1989. Electrophoretic analyses of nucleosomes and other protein-DNA complexes. Methods Enzymol. 170**:**116-142. [DOI] [PubMed] [Google Scholar]
- 25.Huang, S. Y., and W. T. Garrard. 1986. Temperature-dependent cleavage of chromatin by micrococcal nuclease near the nucleosome center. FEBS Lett. 199**:**89-91. [DOI] [PubMed] [Google Scholar]
- 26.Hulander, M., A. E. Kiernan, S. R. Blomqvist, P. Carlsson, E. J. Samuelsson, B. R. Johansson, K. P. Steel, and S. Enerback. 2003. Lack of pendrin expression leads to deafness and expansion of the endolymphatic compartment in inner ears of Foxi1 null mutant mice. Development 130**:**2013-2025. [DOI] [PubMed] [Google Scholar]
- 27.Hulander, M., W. Wurst, P. Carlsson, and S. Enerback. 1998. The winged helix transcription factor Fkh10 is required for normal development of the inner ear. Nat. Genet. 20**:**374-376. [DOI] [PubMed] [Google Scholar]
- 28.Jacobs, F. M., L. P. van der Heide, P. J. Wijchers, J. P. Burbach, M. F. Hoekman, and M. P. Smidt. 2003. FoxO6, a novel member of the FoxO class of transcription factors with distinct shuttling dynamics. J. Biol. Chem. 278**:**35959-35967. [DOI] [PubMed] [Google Scholar]
- 29.Kaestner, K. H., W. Knochel, and D. E. Martinez. 2000. Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev. 14**:**142-146. [PubMed] [Google Scholar]
- 30.Kasinsky, H. E., J. D. Lewis, J. B. Dacks, and J. Ausio. 2001. Origin of H1 linker histones. FASEB J. 15**:**34-42. [DOI] [PubMed] [Google Scholar]
- 31.Lee, S. A., E. L. Shen, A. Fiser, A. Sali, and S. Guo. 2003. The zebrafish forkhead transcription factor Foxi1 specifies epibranchial placode-derived sensory neurons. Development 130**:**2669-2679. [DOI] [PubMed] [Google Scholar]
- 32.Lehmann, O. J., J. C. Sowden, P. Carlsson, T. Jordan, and S. S. Bhattacharya. 2003. Fox's in development and disease. Trends Genet. 19**:**339-344. [DOI] [PubMed] [Google Scholar]
- 33.Lin, S., N. Gaiano, P. Culp, J. C. Burns, T. Friedmann, J. K. Yee, and N. Hopkins. 1994. Integration and germ-line transmission of a pseudotyped retroviral vector in zebrafish. Science 265**:**666-669. [DOI] [PubMed] [Google Scholar]
- 34.Liu, D., H. Chu, L. Maves, Y. L. Yan, P. A. Morcos, J. H. Postlethwait, and M. Westerfield. 2003. Fgf3 and Fgf8 dependent and independent transcription factors are required for otic placode specification. Development 130**:**2213-2224. [DOI] [PubMed] [Google Scholar]
- 35.Martinez-Balbas, M. A., A. Dey, S. K. Rabindran, K. Ozato, and C. Wu. 1995. Displacement of sequence-specific transcription factors from mitotic chromatin. Cell 83**:**29-38. [DOI] [PubMed] [Google Scholar]
- 36.McArthur, M., S. Gerum, and G. Stamatoyannopoulos. 2001. Quantification of DNaseI-sensitivity by real-time PCR: quantitative analysis of DNase I- hypersensitivity of the mouse beta-globin LCR. J. Mol. Biol. 313**:**27-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McClelland, P., J. E. Onyia, R. R. Miles, Y. Tu, J. Liang, A. K. Harvey, S. Chandrasekhar, J. M. Hock, and J. P. Bidwell. 1998. Intermittent administration of parathyroid hormone (1-34) stimulates matrix metalloproteinase-9 (MMP-9) expression in rat long bone. J. Cell. Biochem. 70**:**391-401. [PubMed] [Google Scholar]
- 38.Medema, R. H., G. J. Kops, J. L. Bos, and B. M. Burgering. 2000. AFX-like Forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27kip1. Nature 404**:**782-787. [DOI] [PubMed] [Google Scholar]
- 39.Michelotti, E. F., S. Sanford, and D. Levens. 1997. Marking of active genes on mitotic chromosomes. Nature 388**:**895-899. [DOI] [PubMed] [Google Scholar]
- 40.Mitchell, P. J., and R. Tjian. 1989. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science 245**:**371-378. [DOI] [PubMed] [Google Scholar]
- 41.Nagasawa, K., K. Nagai, A. Ishimoto, and S. Fujimoto. 2003. Transport mechanism for lovastatin acid in bovine kidney NBL-1 cells: kinetic evidences imply involvement of monocarboxylate transporter 4. Int. J. Pharm. 262**:**63-73. [DOI] [PubMed] [Google Scholar]
- 42.Nissen, R. M., J. Yan, A. Amsterdam, N. Hopkins, and S. M. Burgess. 2003. Zebrafish foxi one modulates cellular responses to Fgf signaling required for the integrity of ear and jaw patterning. Development 130**:**2543-2554. [DOI] [PubMed] [Google Scholar]
- 43.Ortega, N., D. Behonick, D. Stickens, and Z. Werb. 2003. How proteases regulate bone morphogenesis. Ann. N. Y. Acad. Sci. 995**:**109-116. [DOI] [PubMed] [Google Scholar]
- 44.Overdier, D. G., H. Ye, R. S. Peterson, D. E. Clevidence, and R. H. Costa. 1997. The winged helix transcriptional activator HFH-3 is expressed in the distal tubules of embryonic and adult mouse kidney. J. Biol. Chem. 272**:**13725-13730. [DOI] [PubMed] [Google Scholar]
- 45.Papavassiliou, A. G. 2001. Determination of a transcription-factor-binding site by nuclease protection footprinting onto southwestern blots. Methods Mol. Biol. 148**:**135-149. [DOI] [PubMed] [Google Scholar]
- 46.Perez-Sanchez, C., M. A. Gomez-Ferreria, C. A. de La Fuente, B. Granadino, G. Velasco, A. Esteban-Gamboa, and J. Rey-Campos. 2000. FHX, a novel fork head factor with a dual DNA binding specificity. J. Biol. Chem. 275**:**12909-12916. [DOI] [PubMed] [Google Scholar]
- 47.Phair, R. D., and T. Misteli. 2000. High mobility of proteins in the mammalian cell nucleus. Nature 404**:**604-609. [DOI] [PubMed] [Google Scholar]
- 48.Pierrou, S., M. Hellqvist, L. Samuelsson, S. Enerback, and P. Carlsson. 1994. Cloning and characterization of seven human forkhead proteins: binding site specificity and DNA bending. EMBO J. 13**:**5002-5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Platero, J. S., A. K. Csink, A. Quintanilla, and S. Henikoff. 1998. Changes in chromosomal localization of heterochromatin-binding proteins during the cell cycle in Drosophila. J. Cell Biol. 140**:**1297-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pohl, B. S., S. Knochel, K. Dillinger, and W. Knochel. 2002. Sequence and expression of FoxB2 (XFD-5) and FoxI1c (XFD-10) in Xenopus embryogenesis. Mech Dev. 117**:**283-287. [DOI] [PubMed] [Google Scholar]
- 51.Qian, H., T. Wang, L. Naumovski, C. D. Lopez, and R. K. Brachmann. 2002. Groups of p53 target genes involved in specific p53 downstream effects cluster into different classes of DNA binding sites. Oncogene 21**:**7901-7911. [DOI] [PubMed] [Google Scholar]
- 52.Reyes, J. C., C. Muchardt, and M. Yaniv. 1997. Components of the human SWI/SNF complex are enriched in active chromatin and are associated with the nuclear matrix. J. Cell Biol. 137**:**263-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rose, S. M., and W. T. Garrard. 1984. Differentiation-dependent chromatin alterations precede and accompany transcription of immunoglobulin light chain genes. J. Biol. Chem. 259**:**8534-8544. [PubMed] [Google Scholar]
- 54.Shapiro, J. A. 2002. Repetitive DNA, genome system architecture and genome reorganization. Res. Microbiol. 153**:**447-453. [DOI] [PubMed] [Google Scholar]
- 55.Shen, X., and M. A. Gorovsky. 1996. Linker histone H1 regulates specific gene expression but not global transcription in vivo. Cell 86**:**475-483. [DOI] [PubMed] [Google Scholar]
- 56.Shim, E. Y., C. Woodcock, and K. S. Zaret. 1998. Nucleosome positioning by the winged helix transcription factor HNF3. Genes Dev. 12**:**5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sola, L., and E. Gornung. 2001. Classical and molecular cytogenetics of the zebrafish, Danio rerio (Cyprinidae, Cypriniformes): an overview. Genetica 111**:**397-412. [DOI] [PubMed] [Google Scholar]
- 58.Solomon, K. S., T. Kudoh, I. B. Dawid, and A. Fritz. 2003. Zebrafish foxi1 mediates otic placode formation and jaw development. Development 130**:**929-940. [DOI] [PubMed] [Google Scholar]
- 59.Solomon, K. S., S. J. Kwak, and A. Fritz. 2004. Genetic interactions underlying otic placode induction and formation. Dev. Dyn. 230**:**419-433. [DOI] [PubMed] [Google Scholar]
- 60.Solomon, K. S., J. M. Logsdon, Jr., and A. Fritz. 2003. Expression and phylogenetic analyses of three zebrafish FoxI class genes. Dev. Dyn. 228**:**301-307. [DOI] [PubMed] [Google Scholar]
- 61.Soutoglou, E., and I. Talianidis. 2002. Coordination of PIC assembly and chromatin remodeling during differentiation-induced gene activation. Science 295**:**1901-1904. [DOI] [PubMed] [Google Scholar]
- 62.Stavreva, D. A., W. G. Muller, G. L. Hager, C. L. Smith, and J. G. McNally. 2004. Rapid glucocorticoid receptor exchange at a promoter is coupled to transcription and regulated by chaperones and proteasomes. Mol. Cell. Biol. 24**:**2682-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Strodicke, M., S. Karberg, and G. Korge. 2000. Domina (Dom), a new Drosophila member of the FKH/WH gene family, affects morphogenesis and is a suppressor of position-effect variegation. Mech. Dev. 96**:**67-78. [DOI] [PubMed] [Google Scholar]
- 64.Tokino, T., S. Thiagalingam, W. S. el-Deiry, T. Waldman, K. W. Kinzler, and B. Vogelstein. 1994. p53 tagged sites from human genomic DNA. Hum. Mol. Genet. 3**:**1537-1542. [DOI] [PubMed] [Google Scholar]
- 65.Urlinger, S., U. Baron, M. Thellmann, M. T. Hasan, H. Bujard, and W. Hillen. 2000. Exploring the sequence space for tetracycline-dependent transcriptional activators: novel mutations yield expanded range and sensitivity. Proc. Natl. Acad. Sci. USA 97**:**7963-7968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vidal, M., R. K. Brachmann, A. Fattaey, E. Harlow, and J. D. Boeke. 1996. Reverse two-hybrid and one-hybrid systems to detect dissociation of protein-protein and DNA-protein interactions. Proc. Natl. Acad. Sci. USA 93**:**10315-10320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weigel, D., G. Jurgens, F. Kuttner, E. Seifert, and H. Jackle. 1989. The homeotic gene fork head encodes a nuclear protein and is expressed in the terminal regions of the Drosophila embryo. Cell 57**:**645-658. [DOI] [PubMed] [Google Scholar]
- 68.Weiler, K. S., and B. T. Wakimoto. 1995. Heterochromatin and gene expression in Drosophila. Annu. Rev. Genet. 29**:**577-605. [DOI] [PubMed] [Google Scholar]
- 69.Weiler, K. S., and B. T. Wakimoto. 2002. Suppression of heterochromatic gene variegation can be used to distinguish and characterize E(var) genes potentially important for chromosome structure in Drosophila melanogaster. Mol. Genet. Genomics 266**:**922-932. [DOI] [PubMed] [Google Scholar]
- 70.Weinmann, A. S., S. M. Bartley, T. Zhang, M. Q. Zhang, and P. J. Farnham. 2001. Use of chromatin immunoprecipitation to clone novel E2F target promoters. Mol. Cell. Biol. 21**:**6820-6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wells, J., and P. J. Farnham. 2002. Characterizing transcription factor binding sites using formaldehyde crosslinking and immunoprecipitation. Methods 26**:**48-56. [DOI] [PubMed] [Google Scholar]
- 72.Yan, Y. L., C. T. Miller, R. M. Nissen, A. Singer, D. Liu, A. Kirn, B. Draper, J. Willoughby, P. A. Morcos, A. Amsterdam, B. C. Chung, M. Westerfield, P. Haffter, N. Hopkins, C. Kimmel, J. H. Postlethwait, and R. Nissen. 2002. A zebrafish sox9 gene required for cartilage morphogenesis. Development 129**:**5065-5079. [DOI] [PubMed] [Google Scholar]
- 73.Yan, Y. L., J. Willoughby, D. Liu, J. G. Crump, C. Wilson, C. T. Miller, A. Singer, C. Kimmel, M. Westerfield, and J. H. Postlethwait. 2005. A pair of Sox: distinct and overlapping functions of zebrafish sox9 co-orthologs in craniofacial and pectoral fin development. Development 132**:**1069-1083. [DOI] [PubMed] [Google Scholar]
- 74.Zhao, K., E. Kas, E. Gonzalez, and U. K. Laemmli. 1993. SAR-dependent mobilization of histone H1 by HMG-I/Y in vitro: HMG-I/Y is enriched in H1-depleted chromatin. EMBO J. 12**:**3237-3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhu, G., P. T. Spellman, T. Volpe, P. O. Brown, D. Botstein, T. N. Davis, and B. Futcher. 2000. Two yeast forkhead genes regulate the cell cycle and pseudohyphal growth. Nature 406**:**90-94. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental material]