Contributions of Trf4p- and Trf5p-dependent polyadenylation to the processing and degradative functions of the yeast nuclear exosome (original) (raw)
Abstract
The nuclear exosome is involved in a large number of RNA processing and surveillance pathways. RNase III cleavage intermediates destined to be 3′-processed or degraded can be detected when the Rrp6p subunit of the nuclear exosome is absent. Here we show that these processing and degradation intermediates are polyadenylated, and that their polyadenylation is dependent on the activity of Trf4p and Trf5p, two variant poly(A) polymerases. Polyadenylation of cleavage intermediates was inhibited when Trf4p was absent, and reduced to various extents in the absence of Trf5p, suggesting that these two poly(A) polymerases play functionally distinct roles in the polyadenylation of these RNA species. Finally, in the absence of Trf4p, we observed 3′-extended forms of the U4 snRNA that are similar to those observed in the absence of Rrp6p. These results suggest that polyadenylation of RNA processing intermediates plays a functional role in RNA processing pathways and is not limited to RNA surveillance functions.
Keywords: Rrp6p, Rnt1p, RNase III, polyadenylation, processing, exosome
INTRODUCTION
The exosome is a complex of 3′→5′ exonucleases (Mitchell et al. 1997), which plays important roles in the processing, surveillance, and degradation of many eukaryotic RNAs (Allmang et al. 1999; Mitchell and Tollervey 2003; LaCava et al. 2005; Wyers et al. 2005). Production of the mature 3′-end of virtually all noncoding RNAs occurs by exonucleolytic digestion of precursor RNA species or of cleavage intermediates, many of which are generated in yeast by the double-stranded RNA endonucleolytic activity of the RNase III enzyme Rnt1p (Abou Elela et al. 1996; Chanfreau et al. 1997; Abou Elela and Ares 1998; Allmang et al. 1999; Kufel et al. 1999; Seipelt et al. 1999). The exosome activity also contributes to several RNA surveillance pathways that prevent the accumulation of aberrant species by degrading incorrect or incompletely processed RNAs (Bousquet-Antonelli et al. 2000; Hilleren et al. 2001; Kadaba et al. 2004). These RNAs can be degraded exclusively through the exonucleolytic activities of the nuclear exosome or by RNase III cleavage followed by exonucleolytic digestion (Danin-Kreiselman et al. 2003). A canonical view of the processing or degradation pathways carried out by the eukaryotic exosome is that a free RNA 3′-end provides an entry site for exonucleolytic digestion, and that degradation will proceed to the mature 3′-end for processing pathways or to the end of the transcript in the case of degradative pathways. However, recent results have shown that RNA degradation by the exosome is stimulated by polyadenylation of the 3′-end of the RNA substrates. Initial genetic studies have shown that surveillance of hypomodified tRNAiMet by the exosome is enhanced by in vivo polyadenylation of the hypomodified tRNA precursor (Kadaba et al. 2004). Strikingly, polyadenylation of theses species is dependent on a variant poly(A) polymerase, Trf4p, rather than on the classical poly(A) polymerase Pap1p. Further biochemical studies have shown that a complex containing Trf4p, the RNA binding proteins Air1/2p, and the putative helicase Mtr4p exhibits polyadenylation activity in vitro, and that Trf4p-mediated polyadenylation stimulates the degradative activities of the exosome in vitro (LaCava et al. 2005; Vanacova et al. 2005; Wyers et al. 2005). More generally, Trf4p-mediated polyadenylation has been involved in the degradation of cryptic transcripts generated by RNA polymerase II (Wyers et al. 2005) and has been involved in the degradation of some noncoding Pol.II and Pol.III transcripts (LaCava et al. 2005; Wyers et al. 2005). From these studies, it was concluded that Trf4p-mediated polyadenylation mostly plays a role in the surveillance of some noncoding RNA species.
In this study, we show that RNA processing or degradation intermediates that accumulate in a strain lacking the nuclear exosome component Rrp6p are polyadenylated and that the polyadenylation of these intermediates requires Trf4p. Interestingly the level of polyadenylation of some of these intermediates is also reduced in the absence of Trf5p, another variant poly(A) polymerase, suggesting a partially redundant and specialized role for these enzymes in the polyadenylation of these species. These results suggest that polyadenylation by Trf4p and Trf5p stimulates the processing and degradative activities of the nuclear exosome.
RESULTS AND DISCUSSION
3′-Processing and degradation intermediates of snRNAs, snoRNAs, and mRNAs generated by RNase III cleavage are polyadenylated
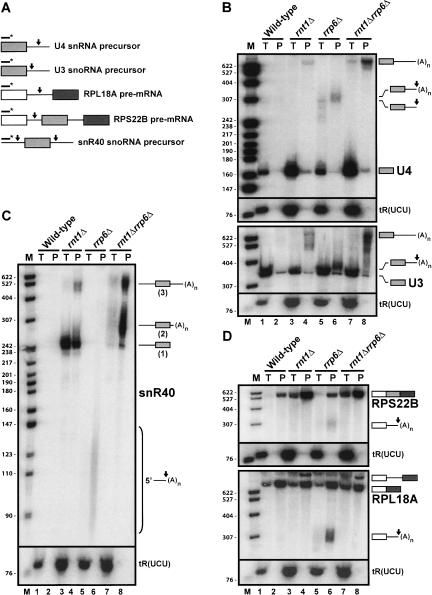
Recent results have shown that hypomodified tRNAiMet precursors are degraded by the nuclear exosome (Kadaba et al. 2004). Interestingly, polyadenylated forms of these precursors accumulate in a _rrp6_-deficient strain, suggesting that efficient degradation of this particular substrate by the nuclear exosome requires polyadenylation of the 3′-end of the hypomodified tRNA precursor. This result led us to revisit the mechanism of processing or degradation of RNase III cleavage products. Previous studies have shown that the upstream cleavage products generated by the yeast RNase III Rnt1p are stabilized in the absence of the nuclear exosome component Rrp6p (Allmang et al. 1999; van Hoof et al. 2000; Danin-Kreiselman et al. 2003). We thus examined the level of polyadenylation of 3′-processing or degradation intermediates that accumulate in the _rrp6_Δ strain, by oligo-(dT) affinity purification and Northern blot. We analyzed the level of polyadenylation of two substrates that are normally processed at their 3′-end by Rnt1p cleavage followed by 3′→5′ exosomal digestion: the U3 snoRNA and the U4 snRNA (Allmang et al. 1999; Kufel et al. 2000; van Hoof et al. 2000; Fig. 1A ▶). In addition, we also analyzed the level of polyadenylation of several RNA species generated by Rnt1p cleavage that undergo exosome-mediated degradation rather than processing. Cleavage by Rnt1p in the 5′-extension present in the snR40 box C/D snoRNA precursor (Fig. 1A ▶) releases a downstream intermediate that is further processed to the mature 5′ end, presumably by the 5′→3′ exonucleases Xrn1p and Rat1p (Chanfreau et al. 1998a). The upstream cleavage product is normally degraded, presumably by the nuclear exosome. Thus, we analyzed the fate of this upstream cleavage product and whether or not polyadenylated forms of this product also accumulate in _rrp6_-deficient cells. Finally, Rnt1p contributes to the degradation of some unspliced pre-mRNA precursors, by cleaving intronic stem-loops present in unspliced precursors and lariat introns (Danin-Kreiselman et al. 2003; Fig. 1A ▶). The cleaved intermediates are degraded by the nuclear exosome and in the 5′→3′ direction by the exonucleases Xrn1p or Rat1p but can be detected in vivo when these exonucleases are inactivated (Danin-Kreiselman et al. 2003). We thus analyzed the levels of polyadenylation of cleaved mRNA species in the _rrp6_Δ strain. To this end, RNAs were prepared from various mutant strains and analyzed by Northern blot using oligonucleotide probes hybridizing to different regions of these various RNA substrates (Fig. 1A ▶).
FIGURE 1.
Analysis of the in vivo polyadenylation status of Rnt1p cleavage products and primary precursors. (A) Schematic representation of the primary precursors of the U4 snRNA, the U3 and snR40 snoRNAs, and the RPL18A and RPS22B mRNAs. Boxes represent mature or exonic sequences. Arrows represent the Rnt1p cleavage sites. Black lines with asterisks represent the location of the oligonucleotide probes used. (B) 3′-Processing intermediates and unprocessed precursors of the U3 snoRNA and of the U4 snRNA are polyadenylated. Total RNAs (T) or polyadenylated RNAs selected by oligo-dT affinity (P) extracted from the indicated strains were loaded on acrylamide gels, transferred to nylon membranes, and probed with oligonucleotides hybridizing to the mature sequences of the U3 snoRNA or U4 snRNA. Species with arrowheads represent the species that result from Rnt1p cleavage. The Arginine tRNA tR(UCU) was used as a negative control to test for contamination of purified polyadenylated RNAs by total RNAs. (C) The upstream degradation intermediate of the snR40 box C/D snoRNA is polyadenylated. Legends as in B. The probe used hybridizes against the 5′-extension of snR40 (see A); therefore, the mature snR40 snoRNA is not detected. (D) Degradation intermediates of the RPL18A and RPS22B unspliced pre-mRNAs are polyadenylated. Legends as in B. Probes used hybridize against the 5′-exon sequences of RPL18A or RPS22B. The same cleaved intermediate was detected by using an intronic oligonucleotide probe in the case of RPS22B (data not shown). For RPS22B, the unspliced precursor is not included in the gel.
This analysis revealed that all Rnt1p cleavage products that accumulate in the _rrp6_Δ strain are polyadenylated (Fig. 1B–D ▶). 3′-Extended processing intermediates of the U3 snoRNA (~360 nt) and of the U4 snRNA (~240–300nt) were detected in total RNAs extracted from the _rrp6_Δ strain (Fig. 1B ▶, lane 5). Analysis of oligo(dT) purified RNAs on the same membrane revealed the presence of higher molecular mass species of ~380 nt for U3 and of ~310 nt for U4 (Fig. 1B ▶, lane 6). Based on their slower mobility and on their accumulation in oligo(dT) purified RNAs, these correspond to polyadenylated processing intermediates that accumulate in the _rrp6_Δ strain. These results are consistent with previous studies showing that the U4 processing intermediates are polyadenylated (van Hoof et al. 2000). Analysis of species that undergo 3′→5′ degradation following Rnt1p cleavage showed that polyadenylated forms of these species also accumulate in the _rrp6_Δ strain (Fig. 1C,D ▶, lane 6). The upstream cleavage product of the snR40 5′-extension was found to accumulate in the _rrp6_Δ strain as a smear of 85–130 nt–long polyadenylated species in the oligo(dT) fractions prepared from this strain (Fig. 1C ▶, lane 6). Estimation of the length of the polyadenylated species of the snR40 5′-cleavage product revealed that the average length of the poly(A) tail of these species is 30–45 nt. This number is in agreement with the length of the poly(A) tails detected in the case of the hypomodifed tRNAiMet (Kadaba et al. 2004), significantly shorter than the classical poly(A) tails observed in yeast mRNAs (~80 nt). In the _rnt1_Δ strain, we also detected a significant amount of 250-nt unprocessed snR40 snoRNA precursor in the oligo(dT)-selected RNA preparations (Fig. 1C ▶, lane 4, species labeled “1”). Strikingly these species did not exhibit a size difference between the total and oligo-(dT) lanes, and did not show the smeared pattern typical of poly(A) tails, suggesting that their presence in the oligo-(dT) fraction was not due to polyadenylation per se. Upon inspection of the snR40 5′-extension sequence, we found that it contains several stretches of continuous adenosines. Therefore the presence of these unprocessed precursors in the oligo(dT) fractions is very likely due to their high adenosine content, rather than to a genuine polyadenylation. Cleavage products (~300 nt long) of the unspliced precursors of the RPS22B and RPL18A mRNAs were also found to be polyadenylated (Fig. 1D ▶, lane 6), showing that polyadenylation also affects cleaved species that are generated from unspliced pre-mRNAs. For all species examined, the identity of the bands accumulating in the _rrp6_Δ strain as Rnt1p cleavage products was confirmed based on their migration and on the fact that these species are not detected in a _rnt1_Δ _rrp6_Δ double-mutant strain where Rnt1p activity is lacking (Fig. 1B–D ▶, lanes 7,8). Thus, these species do not correspond to exonucleolytic pauses of the exosome but rather to genuine Rnt1p cleavage products.
Analysis of products that accumulate in the _rnt1_Δ mutant and in the double-mutant _rnt1_Δ _rrp6_Δ also revealed the presence of long 3′-extended forms of U3 (500–650 nt) and U4 (>600 nt) that are enriched in the polyadenylated fractions (Fig. 1B ▶, lanes 4,8). The presence of these long species was reported previously (van Hoof et al. 2000), but it was not shown that they are polyadenylated. These species were readily detectable in poly(A) RNAs from the _rnt1_Δ strain but were further stabilized by the depletion of Rrp6p (Fig. 1B ▶, lane 8). Similarly, 500–600 nt–long polyadenylated 3′-extended species of the snR40 precursor also accumulated in a strain lacking Rnt1p (Fig. 1C ▶, lane 4, species labeled “3”), and these species were further stabilized in the absence of Rrp6p (lane 8). Another extended species, 250–310 nt long and labeled as “2” in Figure 1C ▶, was detected only in the _rnt1_Δ _rrp6_Δ strain (Fig. 1C ▶, lanes 7,8). These observations show that polyadenylated RNA precursors accumulate in the absence of Rnt1p and are further stabilized by inactivation of the nuclear exosome component Rrp6p. Thus, two types of poly-adenylated noncoding RNAs accumulate when various RNA processing enzymes are inactivated: long 3′-extended precursor RNAs that are found in the absence of Rnt1p, and cleavage intermediates that accumulate in the absence of Rrp6p. We decided to focus our study on the mechanism of polyadenylation of RNase III cleavage intermediates.
Polyadenylation of processing and degradation intermediates generated by Rnt1p cleavage requires the Trf4p and Trf5p poly(A) polymerases
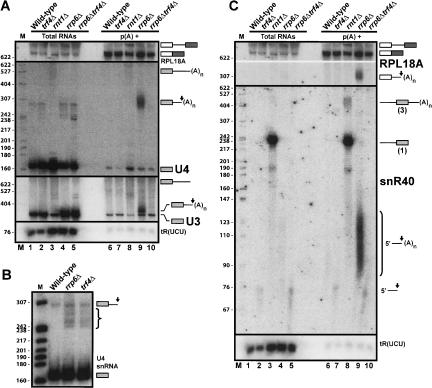
To understand the mechanism of polyadenylation of Rnt1p cleavage products, we next searched for the poly(A) polymerase activities responsible for the polyadenylation of processing and degradation intermediates detected in the _rrp6_Δ strain. The classical poly(A) polymerase Pap1p was suggested in an earlier study to contribute to this polyadenylation activity (van Hoof et al. 2000). However, another candidate was Trf4p, based on the observation that Trf4p-mediated polyadenylation is required for degradation of a number of RNA species by the exosome (Kadaba et al. 2004; LaCava et al. 2005; Vanacova et al. 2005; Wyers et al. 2005). Finally, Trf5p, another protein with poly(A) polymerase motifs similar to those of Trf4p, could be involved as well, since it was shown that overexpression of Trf5p compensates for the polyadenylation defects of a strain lacking Trf4p (LaCava et al. 2005). To investigate the putative roles of Trf4p and Trf5p, we constructed the _rrp6_Δ _trf4_Δ and _rrp6_Δ _trf5_Δ double-mutant strains, and we analyzed the status of the polyadenylated species in these strains by oligo-(dT) affinity purification and Northern analysis. The _trf4_Δ and _trf5_Δ single-mutant strains were also analyzed in parallel.
Strikingly, polyadenylated species that were previously shown to accumulate in the _rrp6_Δ strain were no more detectable in the _rrp6_Δ _trf4_Δ double-mutant strain (Fig. 2A,C ▶, lanes 10). This result was observed for both RNA species that correspond to processing intermediates (U3, U4) (Fig. 2A ▶, lane 10) or degradation intermediates (snR40 5′-extension and RPL18A) (Fig. 2C ▶, lane 10). This result suggests that Trf4p plays a major role in the polyadenylation of these species. In addition, we noticed the presence in total RNA samples extracted from the _trf4_Δ strain of 3′-extended forms of the U4 snRNA (240–300 nt long), which migrate between the mature form of U4 and the Rnt1p cleavage product (Fig. 2B ▶, species labeled “}”). The pattern of these species was remarkably similar to the one observed in the _rrp6_Δ strain, suggesting that the absence of Trf4p significantly impairs the efficiency of U4 3′-processing, as it is the case for Rrp6p. These species were even further stabilized in the _rrp6_Δ _trf4_Δ double-mutant strain (Fig. 2A ▶, lane 5), showing that Trf4p and Rrp6p cooperate functionally in the processing of 3′-extended forms of U4. In addition, we observed in total RNA samples extracted from the _rrp6_Δ _trf4_Δ strain two 3′-extended forms of the U3 snoRNA (~450–470 nt long) that comigrate with the precursor species accumulating in the _rnt1_Δ strain (Fig. 2A ▶, cf. lanes 3 and 5). These observations suggest that inactivation of both Rrp6p and Trf4p has a synergistic effect on U3 and U4 3′-processing and results in a phenotype similar to that observed in the absence of Rnt1p, strengthening the idea that Trf4p-mediated polyadenylation is required for efficient processing. Similarly, we observed an accumulation of the 76 nt–long nonpolyadenylated snR40 5′-cleavage product in total RNAs extracted from the _trf4_Δ strain (Fig. 2C ▶, lane 2), suggesting that Trf4p activity is required for its degradation. Note that the same nonpolyadenylated upstream cleavage product was also found in poly(A)+ RNAs (Fig. 2C ▶, lanes 7,10) because of the previously mentioned stretch of adenosines present in the 5′-extension of snR40.
FIGURE 2.
Polyadenylation of Rnt1p cleavage intermediates requires Trf4p. (A) Analysis of the polyadenylation status of the U3 and U4 cleavage intermediates in _trf4_Δ-derived strains. The membrane was also hybridized with the RPL18A probe to test for equal loading in the poly(A)+RNA lanes. (B) 3′-Extended forms of the U4 snRNA accumulate in the absence of Trf4p. A membrane with total RNAs extracted from wild-type, _trf4_Δ, or _rrp6_Δ strains was hybridized with the U4 probe. The brace indicates the 3′-extended forms that accumulate in the absence of Trf4p or Rrp6p. (C) Analysis of the polyadenylation status of the RPL18A and snR40 cleavage intermediates in _trf4_Δ-derived strains. Legends as in Figure 1 ▶.
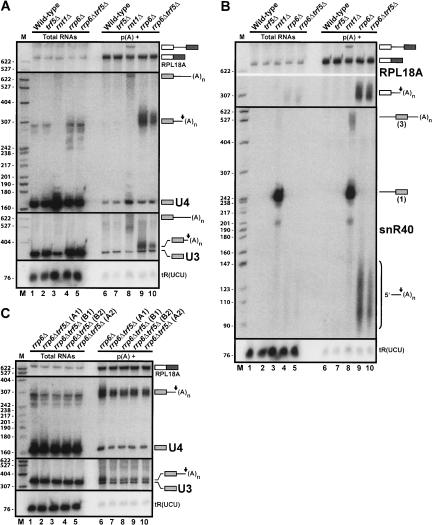
Similarly to Trf4p, we investigated the function of Trf5p in the polyadenylation of Rnt1p cleavage and degradation intermediates (Fig. 3 ▶). In contrast to the result observed with Trf4p, depletion of Trf5p in the context of the absence of Rrp6p had a weaker effect on the accumulation of polyadenylated intermediates. These species were still detected, but to a lesser extent in the _rrp6_Δ _trf5_Δ double-mutant strain than in the _rrp6_Δ strain (Fig. 3A,B ▶, cf. lanes 9 and 10), showing that Trf5p contributes to some extent to the polyadenylation status of the Rnt1p cleavage intermediates. Interestingly, the effect of Trf5p depletion was variable depending on the substrates analyzed. The levels of polyadenylated U3 cleavage intermediates in the _rrp6_Δ _trf5_Δ double-mutant strain were reduced to 30% of the levels observed in the _rrp6_Δ strain (Fig. 3A ▶). The polyadenylated intermediates were reduced to 40% for U4 (Fig. 3A ▶), and to 50% for snR40 (Fig. 3B ▶), while the effect on the polyadenylation of the RPL18A cleavage products was marginal (72%) (Fig. 3B ▶). Thus, it appears that Trf5p depletion affects polyadenylation of various RNA targets to different extents. The effects observed on the U3 and U4 cleavage intermediates were reproducible, as they were observed on two independent clones and two independent RNA preparations of the double deletion strain (Fig. 3C ▶).
FIGURE 3.
Polyadenylation of Rnt1p cleavage intermediates is influenced by Trf5p. (A) Analysis of the polyadenylation status of the U3 and U4 cleavage intermediates in _trf5_Δ-derived strains. The membrane was also hybridized with the RPL18A probe to test for equal loading in the poly(A)+RNA lanes. (B) Analysis of the polyadenylation status of the RPL18A and snR40 cleavage intermediates in _trf5_Δ-derived strains. Legends as in Figure 1 ▶. (C) The reduction of polyadenylated cleavage intermediates observed in the _rrp6_Δ _trf5_Δ double deletion strain for the U3 and U4 RNAs is reproducible. “A” and “B” represent RNAs extracted from two independent clones carrying the _rrp6_Δ _trf5_Δ double deletion; “1” and “2” represent two independent RNA preparations.
Overall, these results show that Trf4p plays a major role in the polyadenylation of Rnt1p cleavage products that correspond to intermediates in the processing and degradative pathways carried out by the nuclear exosome. Given the previous results showing that the Trf4p-associated TRAMP complex exhibits poly(A) polymerase activity (LaCava et al. 2005; Vanacova et al. 2005; Wyers et al. 2005), our genetic results suggest that Trf4p carries the major poly(A) polymerase activity responsible for the polyadenylation of Rnt1p cleavage products that correspond to nuclear exosome degradation or processing intermediates. This result is retrospectively not surprising, as it is likely that the 3′-hydroxyl groups generated by Rnt1p cleavage may not be good substrates for the classical poly(A) polymerase Pap1p in vivo. Pap1p poly(A) polymerase activity occurs in vivo in the context of cleavage and polyadenylation complexes, and Pap1p activity in vivo might normally be limited to the 3′-hydroxyl groups generated by the cleavage subunits of these complexes.
One puzzling result is that Trf4p depletion abolishes polyadenylation of all Rnt1p cleavage intermediates, while Trf5p depletion only reduces polyadenylation of some intermediates to different extents. If the two enzymes were truly redundant in their polyadenylation activities of processing intermediates, one might not expect a complete absence of the polyadenylated species in the absence of Trf4p, but only a reduction. One possibility is that Trf5p may not be directly responsible for the polyadenylation of these intermediates but may influence directly or indirectly the activity of Trf4p for some specific RNA species. In this scenario, the reduction in the level of polyadenylated intermediates observed in the _rrp6_Δ _trf5_Δ strain might result from a perturbation of Trf4p activity for some particular substrates. Alternatively, it is possible that the complete loss of poly(A) tails observed in the _rrp6_Δ _trf4_Δ strain might be partially due to the severe growth defect observed in this particular strain (Wyers et al. 2005). While the _rrp6_Δ _trf5_Δ strain grows relatively normally, the _rrp6_Δ _trf4_Δ strain is extremely sick (Wyers et al. 2005; data not shown). Thus the complete loss of poly(A) tails of cleavage intermediates observed in the _rrp6_Δ _trf4_Δ strain might be due to a combination of the direct loss of Trf4p poly(A) polymerase activity and to some other indirect effects linked to a slower RNA metabolism in this strain. In conclusion, we favor a model where polyadenylation of these processing intermediates involves both Trf4p and Trf5p, and where the quantitative contribution of Trf5p might vary depending on the RNA substrates.
While previous studies have shown that surveillance or degradative functions of the exosome are stimulated by polyadenylation, our results suggest that polyadenylation of Rnt1p cleavage products stimulates their subsequent processing or degradation by the exosome. We cannot strictly rule out that the cleaved polyadenylated intermediates detected in the _rrp6_Δ strain correspond to nonproductive polyadenylated species that are destined to be degraded rather than undergoing 3′-processing. However, the 3′extended species of U3 and U4 detected in total RNAs extracted from the _trf4_Δ and/or _rrp6_Δ _trf4_Δ strain suggests that Trf4p plays a significant role in 3′-processing of these noncoding RNAs. Overall, these results show that variant poly(A) polymerases such as Trf4p and Trf5p are not solely-involved in degradation or surveillance mechanisms but also in processing activities.
MATERIALS AND METHODS
Yeast strains
The wild-type yeast strains used in this study are either strain BMA64 (_ura3-1; trp1_Δ; ade2-1; leu2-3,112; his3-11,15) (Baudin et al. 1993) or strain BY4742 (MAT α; _his3_Δ1; _leu2_Δ0; _lys2_Δ0; ura3_Δ_0) purchased from Open Biosystems. Two _rnt1_Δ strains have been used depending on the figures with identical results. They were obtained by direct disruption using a PCR-based method (Baudin et al. 1993; Longtine et al. 1998) of the RNT1 open reading frame either in the BMA64 genetic background (using the TRP1 selection marker) or in the BY4742 genetic background (using the his5 + selection marker). Two _rrp6_Δ strains have been used (MAT α; _his3_Δ1; _leu2_Δ0; _lys2_Δ0; _ura3_Δ0; rrp6::kan r and _MAT a; his3_Δ1; _leu2_Δ0; _met15_Δ0; _ura3_Δ0; rrp6::kan r), both purchased from Open Biosystems. The _rnt1_Δ _rrp6_Δ double-mutant strain was created by mating a _rnt1_Δ strain generated in the BMA64 genetic background (MAT α; _ura3_-1; _trp1_Δ; ade2-leu2-3,112; his3-11,15; rnt1::his5 +) with the _rrp6_Δ strain of opposite mating type (MAT a) described above (Open Biosystems), dissecting tetrads after sporulation and selecting the histidine prototroph, G418 resistant spores. The _trf4_Δ (MAT α; _his3_Δ1; _leu2_Δ0; _lys2_Δ0; _ura3_Δ0; trf4::kan r) and _trf5_Δ (MAT α; _his3_Δ1; _leu2_Δ0; _lys2_Δ0; _ura3_Δ0; trf5::kan r) single-mutant strains were purchased from Open Biosystems. The _trf4_Δ _rrp6_Δ (MAT α; his3_Δ_1; leu2_Δ_0; lys2_Δ_0; ura3_Δ_0; rrp6::kan r; trf4::his5 +) double-mutant strain was obtained by direct disruption (Baudin et al. 1993; Longtine et al. 1998) of the TRF4 open reading frame in the _rrp6_Δ strain purchased from Open Biosystems. The _trf5_Δ _rrp6_Δ (MAT α; his3_Δ_1; leu2_Δ_0; lys2_Δ_0; ura3_Δ_0; trf5::kan r; rrp6::his5 +) double-mutant strain was obtained by direct disruption of the RRP6 open reading frame in the _trf5_Δ strain purchased from Open Biosystems.
RNA analysis
Northern blot analysis on 5% or 6% polyacrylamide gels was performed as described (Chanfreau et al. 1998b). Five micrograms of total RNAs or 2 μg of oligo-(dT) purified RNAs were used for each membrane. Poly(A) purification from total yeast RNAs was performed by using Ambion Poly(A) Purist Mag kit following the manufacturer’s instructions. Oligonucleotides probes used for Northern analysis were (5′-3′) U3: CCAAGTTGGATTCAGTGGCTC; U4: ggTATTCCAAAAATTCCCTACATAgTC; RPS22B exon1: TTGTTAATGTGGTATCTTATAATTTATTTGTTCACCTTAAATGTGTGGAAATTGACTTTGAACGTCTCCATATTTCAAAGG; RPL18A exon1: GGAGCAGTTCTGTGACCAGATCT; snR40-upstream: CAAAGTTACGAGACCGGTTCGCAC; and tR(UCU): AACCCATAATCTTCTGATTAGAAGTC.
Acknowledgments
We thank D. Tollervey for communication of unpublished results. This work was supported by a Human Frontier Science Program Organization Postdoctoral Fellowship (A.K.H) and by NIGMS grant R01 GM61518 from the NIH to G.C.
NOTE ADDED IN PROOF The in vitro poly(A) polymerase activity of Trf5p was demonstrated after the acceptance of this paper for publication (Haracska et al. 2005).
REFERENCES
- Abou Elela, S. and Ares Jr., M. 1998. Depletion of yeast RNase III blocks correct U2 3′ end formation and results in polyadenylated but functional U2 snRNA. EMBO J. 17**:** 3738–3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abou Elela, S., Igel, H., and Ares Jr., M. 1996. RNase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell 85**:** 115–124. [DOI] [PubMed] [Google Scholar]
- Allmang, C., Kufel, J., Chanfreau, G., Mitchell, P., Petfalski, E., and Tollervey, D. 1999. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 18**:** 5399–5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin, A., Ozier-Kalogeropoulos, O., Denouel, A., Lacroute, F., and Cullin, C. 1993. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 21**:** 3329–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousquet-Antonelli, C., Presutti, C., and Tollervey, D. 2000. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell 102**:** 765–775. [DOI] [PubMed] [Google Scholar]
- Chanfreau, G., Elela, S.A., Ares Jr., M., and Guthrie, C. 1997. Alternative 3′-end processing of U5 snRNA by RNase III. Genes & Dev. 11**:** 2741–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanfreau, G., Legrain, P., and Jacquier, A. 1998a. Yeast RNase III as a key processing enzyme in small nucleolar RNAs metabolism. J. Mol. Biol. 284**:** 975–988. [DOI] [PubMed] [Google Scholar]
- Chanfreau, G., Rotondo, G., Legrain, P., and Jacquier, A. 1998b. Processing of a dicistronic small nucleolar RNA precursor by the RNA endonuclease Rnt1. EMBO J. 17**:** 3726–3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danin-Kreiselman, M., Lee, C.Y., and Chanfreau, G. 2003. RNAse III–mediated degradation of unspliced pre-mRNAs and lariat introns. Mol. Cell 11**:** 1279–1289. [DOI] [PubMed] [Google Scholar]
- Haracska, L., Johnson, R.E., Prakash, L., and Satya Prakash. 2005. Trf4 and Trf5 proteins of Saccharomyces cerevisiae exhibit poly(A) RNA polymerase activity but no DNA polymerase activity. Mol. Cell. Biol. 25**:** 10183–10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleren, P., McCarthy, T., Rosbash, M., Parker, R., and Jensen, T.H. 2001. Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature 413**:** 538–542. [DOI] [PubMed] [Google Scholar]
- Kadaba, S., Krueger, A., Trice, T., Krecic, A.M., Hinnebusch, A.G., and Anderson, J. 2004. Nuclear surveillance and degradation of hypo-modified initiator tRNAMet in S. cerevisiae. Genes & Dev. 18**:** 1227–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufel, J., Dichtl, B., and Tollervey, D. 1999. Yeast Rnt1p is required for cleavage of the pre-ribosomal RNA in the 3′ ETS but not the 5′ ETS. RNA 5**:** 909–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufel, J., Allmang, C., Chanfreau, G., Petfalski, E., Lafontaine, D.L., and Tollervey, D. 2000. Precursors to the U3 small nucleolar RNA lack small nucleolar RNP proteins but are stabilized by La binding. Mol. Cell. Biol. 20**:** 5415–5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCava, J., Houseley, J., Saveanu, C., Petfalski, E., Thompson, E., Jacquier, A., and Tollervey, D. 2005. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121**:** 713–724. [DOI] [PubMed] [Google Scholar]
- Longtine, M.S., McKenzie III, A., Demarini, D.J., Shah, N.G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J.R. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14**:** 953–961. [DOI] [PubMed] [Google Scholar]
- Mitchell, P. and Tollervey, D. 2003. An NMD pathway in yeast involving accelerated deadenylation and exosome-mediated 3′→5′ degradation. Mol. Cell 11**:** 1405–1413. [DOI] [PubMed] [Google Scholar]
- Mitchell, P., Petfalski, E., Shevchenko, A., Mann, M., and Tollervey, D. 1997. The exosome: A conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell 91**:** 457–466. [DOI] [PubMed] [Google Scholar]
- Seipelt, R.L., Zheng, B., Asuru, A., and Rymond, B.C. 1999. U1 snRNA is cleaved by RNase III and processed through an Sm site-dependent pathway. Nucleic Acids Res. 27**:** 587–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacova, S., Wolf, J., Martin, G., Blank, D., Dettwiler, S., Friedlein, A., Langen, H., Keith, G., and Keller, W. 2005. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 3**:** e189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof, A., Lennertz, P., and Parker, R. 2000. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol. Cell. Biol. 20**:** 441–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyers, F., Rougemaille, M., Badis, G., Rousselle, J.C., Dufour, M.E., Boulay, J., Regnault, B., Devaux, F., Namane, A., Seraphin, B., et al. 2005. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121**:** 725–737. [DOI] [PubMed] [Google Scholar]