Internalization and phagosome escape required for Francisella to induce human monocyte IL-1β processing and release (original) (raw)
Abstract
Macrophage responses to Francisella infection have been characterized previously by subdued proinflammatory responses; however, these studies have generally focused on macrophage cell lines or monocyte-derived macrophages. Therefore, we studied the ability of fresh human blood monocytes to engulf and respond to Francisella by using the live vaccine strain variant and Francisella novicida. Because Francisella organisms have been reported to escape from the phagolysosome into the cytosol, we hypothesized that this escape may trigger the activation of caspase-1. Francisella tularensis variants were readily taken up by fresh human CD14+ monocytes, inducing the release of IL-1β, as well as IL-8, in a time- and dose-dependent fashion. Importantly, whereas live and dead Escherichia coli, F. novicida, and live vaccine strain, as well as the LPS of E. coli, were able to induce abundant IL-1β mRNA synthesis and intracellular pro-IL-1β production, only live Francisella induced enhanced IL-1β processing and release (51 ± 10 vs. 7.1 ± 2.1 ng/ml, for F. novicida vs. E. coli LPS; P = 0.0032). Cytochalasin D blocked the Francisella internalization and the _Francisella_-induced monocyte IL-1β processing and release but not that induced by the exogenous stimulus E. coli LPS. Also, killing bacteria did not block uptake but significantly diminished the IL-1β processing and release that was induced by Francisella. Blocking bacterial escape from the phagosome into the cytosol also decreased IL-1β but not IL-8 release. These findings demonstrate that Francisella organisms efficiently induce IL-1β processing and release in fresh monocytes by means of a sensing system that requires the uptake of live bacteria capable of phagosome escape.
Keywords: bacteria, caspase-1, cytokine, phagocytosis
Monocytes and macrophages, which are the first-line guards of the innate host-defense system, recognize and phagocytose pathogens. The innate immune system senses the invasion of pathogenic microorganisms through the recognition of pathogen-associated molecular patterns by Toll-like receptors (TLRs), of which there are 10 that are known in humans, as well as non-TLRs (1–3). Pathogen recognition induces the release of many proinflammatory and antiinflammatory cytokines, which provide a balanced inflammatory response. At the same time, prototypical intracellular pathogens like Francisella tularensis depend on the intracellular milieu of mononuclear cells for growth and replication (4, 5). In this context, a class of intracellular innate host-defense molecules, which recognize and respond to internalized pathogens, has been identified (3, 6). These molecules [historically called CATERPILLERs (3) and nucleotide-binding oligomerization domain–leucine-rich repeats (NOD-LRRs) (7), and, most recently, NACHT leucine-rich repeats (NLRs) (8)] are homologues of plant-disease-resistance genes and contain leucine-rich repeats, much like TLRs. The human family of NLR consists of 22 (8) or 23 (7) proteins. These NLRs, similar to TLRs, function to regulate pathogen recognition and response, thus inducing an inflammatory reaction.
IL-1β has a key role in initiating and maintaining the inflammatory response. It is synthesized as biologically inactive 31-kDa precursor (pro-IL-1β). To be active, pro-IL-1β must first be processed to a 17-kDa mature molecule and released. This process is controlled by the IL-1β-converting enzyme (caspase-1) (9). A specific configuration of the intracellular host-defense molecules that is composed predominately of members of the NLR family is called the inflammasome (10). The inflammasome directly or indirectly senses extracellular and intracellular signals from pathogens to induce the activation and release of IL-1β (11). Hence, NLRs may function as intracellular equivalents of TLRs (i.e., intracellular sensors of pathogen-associated molecular patterns).
In this article, we present evidence that mononuclear cells respond to Francisella most efficiently by means of an intracellular innate defense system. An intracellular response is relevant because Francisella escapes into the cytoplasm after being taken up into macrophage phagosomes (12). Although both live and dead Francisella induced pro-IL-1β synthesis, intracellular engulfment of live Francisella was required for effective IL-1β processing and release. Because IL-1β processing is tightly regulated and depends on inflammasome-induced caspase-1 activation, we hypothesize that the effect of Francisella on IL-1β processing depends on its interaction with the inflammasome of the cytosol and the related intracellular response elements.
Materials and Methods
Bacterial Strains and LPS. Escherichia coli (BL21DE3) and three strains of Francisella [F. tularensis live vaccine strain (LVS) (strain JSG2225; ATCC29684) and Francisella novicida (strain JSG2401; U112), both expressing GFP; and mglA mutant of F. novicida (provided by Fran Nano, University of Victoria, Victoria, BC, Canada)] were used for bacterial stimuli. Bacteria were grown on chocolate II agar (Becton Dickinson) at 37°C, harvested, and resuspended in RPMI medium 1640 before being added to cell cultures. The number of bacterial colony-forming units (CFU) was determined by counting the colonies on chocolate II agar. Bacteria were killed by heat at 95°C for 10 min or 56°C for 40 min or by resuspending in 4% paraformaldehyde (PFM) in PBS for 40 min, followed by five washes in PBS and two washes in RPMI medium 1640 to quench residual aldehydes. In some experiments, bacteria were killed by incubating with 50 μg of gentamicin for 1 h. Effective killing by heat, PFM, or gentamicin was confirmed by plating bacteria on chocolate II agar to count CFU. LPS was obtained from E. coli strain 0127:B8 (Westphal preparation, phenol extraction; Difco) or a commercially available highly purified LPS (phenol plus chromatography) from strain 0111:B4 (Alexis Biochemicals, San Diego). Highly purified LPS from _F. tularensis_–LVS and F. novicida were used in selected experiments (13).
Monocyte Isolation. Human monocytes were isolated from fresh blood by Histopaque-1077 (Sigma-Aldrich) density-gradient centrifugation at 600 × g for 20 min at room temperature. The mononuclear layer was removed and washed twice in RPMI medium 1640. Monocytes were isolated by positive selection with anti-CD14-coated magnetic beads (Miltenyi Biotec, Auburn, CA) according to the manufacturer's instructions. This procedure consistently leads to a ≥98% pure population of CD14+ cells, confirmed by flow cytometry analysis. Purified monocytes were used in experiments immediately after isolation.
Monocyte Infection. Monocytes were resuspended in 12 × 75-mm polypropylene tubes (Fisher Scientific) in RPMI medium 1640 supplemented with 10% FBS (endotoxin-free; HyClone), at 1 or 2 × 106 cells per tube. Live or heat-killed bacteria (95°C for 10 min) were added to cells at a multiplicity of infection (moi) of 100. LPS was added at 1 μg/ml concentration. In selected experiments, monocytes were pretreated with 5 μg/ml actin polymerization inhibitor cytochalasin D (cD) (Sigma-Aldrich) for 30 min to block phagocytosis of bacteria. In experiments designed to prevent bacteria from escaping the phagosome, a lysosomotropic agent (NH4Cl; endotoxin-free; Hospira, Lake Forest, IL) that blocks endosome acidification, was added to monocytes at a concentration of 20 mM. Cells were harvested 2, 8, and 24 h after infection; separated from bacteria by low-speed centrifugation at 1,000 × g for 5 min; and lysed in TRIzol (Invitrogen, Life Technologies) or hypotonic lysis buffer for RNA or protein isolation, respectively. After low-speed centrifugation, cell culture media was cleared from bacteria by high-speed centrifugation at 16,000 × g for 5 min and used for cytokine determination.
Cytokine Determination. Human IL-1β and IL-8 were measured by using the Immulite automated chemiluminometer (Diagnostic Products, Los Angeles). Cytokine standards were run before each batched group of samples to verify accuracy. The mature IL-1β level was verified by a sandwich ELISA that was developed in our laboratory (14). Cell extracts were analyzed by immunoblotting with antibodies that were developed by our laboratory to selectively detect pro-IL-1β or mature IL-1β, respectively.
Real-Time PCR. RNA was extracted by TRIzol reagent and 1–2 μg of total RNA was reverse transcribed to cDNA by ThermoScript RNase H– reverse transcriptase (Invitrogen, Life Technologies) and diluted to 100 μl. We used 20–60 ng of the converted cDNA for quantitative PCR with SYBR green I PCR master mixture in the PRISM 7700 apparatus (Applied Biosystems). The PCR occurred in 20 μl, with 2 μl of cDNA template and 0.25 μM primers. We used relative quantification to evaluate the expression of selected genes linked to endotoxin signaling, cytokines, receptors, and the recently described NLR family. All primers were designed with the following similar criteria: to give an 80- to 200-nt amplicon, GC range of 30–70%, melting temperature of 59–60°C, free of secondary structure, and a free-energy change (Δ_G_) of less than or equal to –7 kcal/mol. When selected, primer pairs were validated by PCR and high-resolution gel electrophoresis to have a single band of desired size that was free of primer dimers. For selected genes, PCR products were sequenced. After optimization, only primer pairs that satisfied our criteria were used. Amplification of genomic DNA was used to verify that the PCR conditions did not amplify any contaminating genomic DNA. Relative copy numbers (RCN) and expression ratios of selected genes were normalized to the expression of two housekeeping genes [GAPDH and CAP-1 (cAMP-accessory protein)] and calculated with the following equation: RCN = _E_–ΔCt × 100, where E is the efficiency of PCR and ΔCt is the Ct(target) – Ct(reference) (average of two housekeeping genes). PCR efficiency was calculated with the following equation: E = 10(–1/slope). Based on our preliminary experiments, we observed that E(target) = E(reference) ≅ 2; therefore, we substituted 2 for E in the equation, which represents an efficiency of nearly 100% for all selected genes in the PCR.
Microscopy. Human cells infected with GFP expressing Francisella were visualized at ×40 magnification by fluorescent microscopy (Olympus BX40). For transmission electron microscopy, monocytes were fixed overnight in 2.5% glutaraldehyde, washed, and resuspended in 2% agarose to produce a solid matrix containing the cells. The cells in agarose were postfixed in 1% osmium tetraoxide and embedded in Spurr's resin. Thin (90 nm) sections were cut and stained with uranylacetate and Reynold's lead citrate. Samples were examined by using a Philips CM12 transmission electron microscope operating at 60 kV at ×8,000 magnification.
Statistical Analysis. All data were expressed as mean ± SEM. Comparisons of groups for statistical difference were done by using Student's two-tailed t test. Significance was set at P < 0.05.
Results
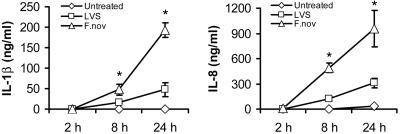
Mononuclear Cells Respond Variably to Different Strains of Francisella. _F. tularensis_–LVS and F. novicida were engulfed by monocytes and macrophages within 1–2 h in the absence of serum opsonization. GFP expression by bacteria allowed for real-time monitoring and quantification of intracellular bacteria. Infected mononuclear cells responded to bacterial invasion by secretion of inflammatory cytokines. We observed a time- and dose-dependent release of IL-1β by monocytes after infection with each of two Francisella strains. For _F. tularensis_–LVS, IL-1β release was induced over time in proportion to the bacterial dose (moi). For F. novicida, all moi (1–1,000) converged to produce similar IL-1β release by 24 h. Also, the cytokine levels released by human monocytes infected with Francisella were extremely high (i.e., 1–2 logs above conventional stimuli, e.g., E. coli endotoxin), suggesting that bacterial invasion triggers a unique immune response. As shown in Fig. 1, F. novicida is a more potent activator of human monocytes as compared with _F. tularensis_–LVS.
Fig. 1.
Comparison of F. novicida with _F. tularensis_–LVS for IL-1β and IL-8 release. Monocytes at 106 cells per ml in RPMI medium 1640 were cultured either alone or with F. novicida or _F. tularensis_–LVS at an moi of 100. Supernatants were harvested at 2, 8, and 24 h and analyzed for cytokines by immunoassay. Data for IL-1β and IL-8 are given as mean ± SEM for n = 9 independent donors. *, P < 0.01, for differences between the two organisms.
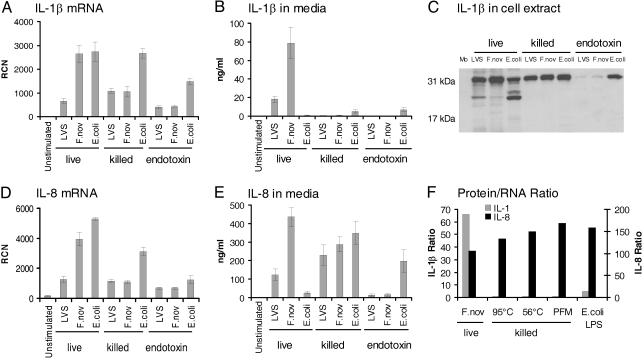
Live Bacteria Are Necessary for IL-1β Maturation and Release. To determine whether the effect of F. novicida on IL-1β processing and release depended on an active bacterial process, monocytes were cultured with live or killed bacteria. Live Francisella induced significantly more monocyte IL-1β release than did live E. coli or the conventional stimulus, E. coli endotoxin (1 μg/ml; Fig. 2). Also, highly purified endotoxin of _F. tularensis_–LVS and F. novicida did not induce IL-1β release. Consistent with the endotoxin data, heat-killed Francisella did not induce detectable IL-1β release, but heat-killed E. coli were more effective than live E. coli (Fig. 2_B_). We speculate that the thermal stress may release additional E. coli endotoxins. All methods of killing Francisella prevented the robust IL-1β release.
Fig. 2.
Requirement of live Francisella for induction of mature IL-1β release by human monocytes. (A) Monocytes were stimulated with live or killed _F. tularensis_–LVS (LVS), F. novicida (F.nov), or E. coli or the purified endotoxins of these organisms at 1 μg/ml and harvested at 8 h for mRNA. IL-1β mRNA was determined by real-time PCR expressed as RCN. (B) Supernatants of the samples from A were analyzed for IL-1β by immunoassay. (C) Monocyte lysates from the experiment were also analyzed for intracellular pro-IL-1β synthesis by immunoblotting using rabbit polyclonal antibody to IL-1β as detected by chemiluminescence. (D) Samples from A were also analyzed for IL-8 mRNA. (E) Supernatants of the samples from A were analyzed by immunoassay for IL-8. (F) To demonstrate the relative ability of mRNA to be translated and then subsequently released, the protein to mRNA ratio (ng/ml protein/RCN) is plotted for IL-1β and IL-8 for live F. novicida and for F. novicida killed by heating (95°C and 56°C) vs. PFM as compared with E. coli LPS. Experiments reflect an n = 4 for killed bacteria and bacterial endotoxin and n = 9 separate experiments for live F. tularensis.
To determine at what level in IL-1β production, processing or release heat-killed Francisella differed from live Francisella, monocytes were evaluated for IL-1β by mRNA as well as for intracellular protein. Pro-IL-1β mRNA was measured in monocytes that were stimulated with live or killed bacteria or with bacterial endotoxin. All three stimuli induced monocyte IL-1β mRNA (Fig. 2 A) and pro-IL-1β synthesis (Fig. 2_C_). Whereas monocytes responded to live and killed bacteria at both the message and protein precursor levels, only live Francisella significantly activated pro-IL-1β cleavage and release (Fig. 2_B_). As a specificity control for intracellular processing of IL-1β, IL-8 mRNA synthesis and protein release was also studied. Monocyte IL-8 mRNA levels correlated with IL-1β message levels for stimulation with live and killed bacteria, and bacterial endotoxin (Fig. 2_D_). However, in contrast to their requirement for live Francisella to induce IL-1β release, monocytes released IL-8 well after contact with either live or killed Francisella, as well as with E. coli endotoxin (Fig. 2_E_). A comparison of cytokine protein release per mRNA copy number demonstrates that live F. novicida and, to a lesser extent, E. coli endotoxin induce release of IL-1β by human monocytes, whereas killed forms of F. novicida do not. In contrast, IL-8 release did not depend on bacterial viability (Fig. 2_F_). Note that Francisella endotoxin was relatively inefficient in its ability to activate either IL-1β or IL-8 production in monocytes (Fig. 2). Together, these results suggest that the ability of Francisella to activate IL-1β processing and release by human monocytes is restricted to live organisms and is independent of surface endotoxin content.
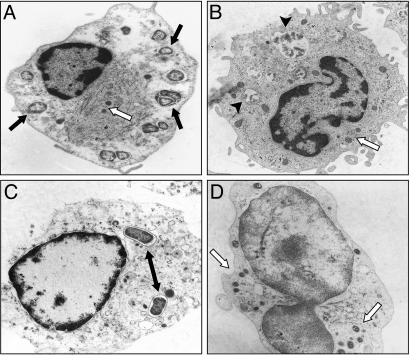
Francisella Internalization Required for IL-1β Maturation and Release. Transmission EM imaging of monocytes infected with live and heat-killed bacteria were analyzed at 8 h after infection to confirm that F. novicida is internalized (Fig. 3). As shown, both live (Fig. 3_A_) and heat-killed (Fig. 3_B_) Francisella were engulfed by monocytes. PFM and gentamicin-killed Francisella were also observed inside of monocytes (data not shown). Although E. coli was also found inside the monocytes (Fig. 3_C_), in contrast to Francisella, E. coli organisms were typically surrounded by a well defined phagosome membrane.
Fig. 3.
Live and dead bacteria engulfed by monocytes. Transmission EM images of human monocytes infected with live (A) and heat-killed (B) F. novicida, both showing engulfment of the organisms. (C) E. coli can be seen within more well defined phagosomes. (D) cD inhibits F. novicida engulfment. The black arrow indicates F. novicida, the white arrow indicates mitochondria, the double arrow indicates E. coli, and the arrowhead indicates killed F. novicida.
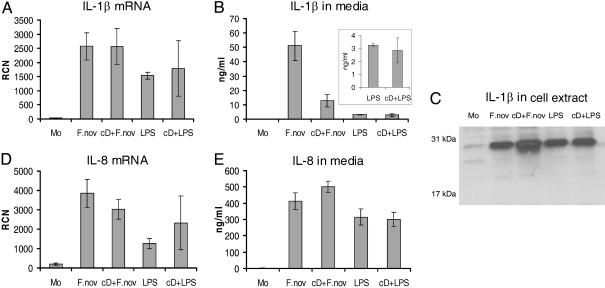
To determine whether phagocytosis was required for the Francisella effect, monocytes were pretreated for 30 min with 5 μg/ml actin polymerization inhibitor cD. Fig. 3_D_ shows that the cellular architecture was significantly affected by cD and only rarely were bacteria internalized. Pretreatment of cells with cD reduced mature IL-1β release by monocytes infected with live F. novicida by 4-fold but did not significantly affect E. coli endotoxin-induced IL-1β release (Fig. 4_B_). This difference in the cD response further supports the hypothesis that live intracellular Francisella organisms are required to optimally activate caspase-1. Note that inhibition of actin polymerization did not reduce IL-1β mRNA message (Fig. 4_A_) nor the synthesis of intracellular IL-1β precursor (Fig. 4_C_). Also, IL-8 mRNA transcription and protein release in response to live F. novicida and E. coli endotoxin were not affected by inhibition of actin polymerization (Fig. 4 D and E).
Fig. 4.
Effect of Francisella internalization on IL-1β processing and release by monocytes. (A) IL-1β mRNA detection in monocytes by real-time PCR after stimulation with either F. novicida (F.nov) or E. coli LPS as affected by pretreatment with cD. (B) Effect of cD pretreatment on monocyte mature IL-1β release induced by either live F.nov or LPS. (Inset) LPS data enlarged. (C) Effect of cD on intracellular pro-IL-1β production as detected by immunoblotting of monocyte cell lysates prepared as described above. (D) Effect of cD on IL-8 mRNA. (E) Effect of cD on IL-8 release into media from monocytes stimulate as above. Results are the average of three experiments.
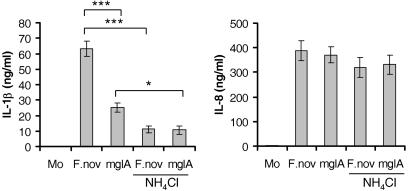
Francisella Escape from the Phagosome Is Necessary for IL-1β Release. To test whether escape from the phagosome is critical for effective stimulation of IL-1β processing and release by human monocytes, we used the F. novicida mglA mutant. MglA is a transcriptional protein that regulates Francisella pathogenicity island protein IglC (15, 16), and virulence factors IglA and PdpA (17). F. novicida mglA mutant demonstrates defective replication within mononuclear cells (18). The defect in intracellular replication of the mglA mutant is associated with an inability to disrupt the phagosome and escape into the cytoplasm (18). As seen from Fig. 5, monocytes infected with mglA mutant showed significantly (P < 0.0001) decreased IL-1β release, as compared with monocytes infected with WT F. novicida at the same moi. As expected, the mglA mutant induced normal amount of IL-8 release (Fig. 5). Plating monocyte lysates on chocolate II agar showed similar bacterial counts for WT and mutant F. novicida 8 h after infection (data not shown). However, these counts of intracellular bacteria cannot differentiate between bacteria in the phagosome and free cytosolic Francisella. Therefore, to further demonstrate that the escape of Francisella from the phagosome is important for IL-1β processing, we used ammonium chloride, a chemical agent that blocks endosome acidification and thus abrogates endosomal escape and intracellular bacterial growth (19). Monocytes were incubated for 8 h with WT or mglA mutant in the presence of 20 mM NH4Cl. IL-1β release was significantly decreased for both the WT (P < 0.0001) and mglA mutant (P = 0.003) (Fig. 5). However there was no reduction in IL-8 release (P = 0.2 and P = 0.4 for F. novicida and mglA, respectively) by NH4Cl (Fig. 5_B_).
Fig. 5.
Preventing escape of Francisella from phagosome impairs IL-1β release. Monocytes (Mo) were incubated with F. novicida (F.nov) or mglA mutant in the presence or absence of 20 mM ammonium chloride (NH4Cl), which is a lysosomotropic agent that prevents bacteria from escaping the phagosome. At 8 h later, supernatant was collected and assayed for IL-1β and IL-8. *, P < 0.01; ***, P < 0.0001. (n = 5 for NH4Cl treatment and n = 11 for the rest of the experiments.)
Discussion
The innate immune system recognizes and defends against pathogenic microbial invasion. Pathogen recognition utilizes transmembrane TLRs and intracellular NLRs. Most Gram-positive and negative bacteria, viral, fungal, and parasitic infection can be recognized by the repertoire of 10 TLRs (20). In general, TLRs trigger multiple signaling pathways that result in the activation of immune defenses that help to clear the organisms (21). For example, Gram-negative bacteria endotoxin is recognized by CD14 in conjunction with MD2 and TLR4 (22) and results in the synthesis and secretion of many inflammatory cytokines including IL-1β (23, 24). IL-1β generates a proinflammatory local and systemic response to infection and injury (9). IL-1β requires posttranslational processing by activated caspase-1 to create a biologically active, mature 17-kDa IL-1β (9, 25). The machinery that activates caspase-1 is complex and involves proteins collectively termed the inflammasome (10). In this context, it was recently reported that an intracellular pathogen Salmonella typhimurium activates caspase-1 by means of inflammasome-related molecules, such as the adaptor proteins ASC and IPAF (CLAN) (26). Another intracellular pathogen, Chlamydia trachomatis, is also able to induce production of pro-IL-1β in monocyte-derived dendritic cells, but only viable bacteria could activate caspase-1 and mature IL-1β release (27). Similarly, Yersinia enterocolitica by means of bacterially derived proteins trigger caspase-1 mediated posttranslational processing of pro-IL-1β in macrophages, although Yersinia is not an intracellular pathogen (28).
F. tularensis, a Gram-negative intracellular bacteria, is the causative agent of the disease tularemia (5) and a potential biological weapon (29). Francisella is highly infectious. Inhalation of 10–25 organisms can cause tularemia (30), accompanied by a pronounced inflammatory response. However, the bacterial component that causes such a response is unknown. Francisella requires macrophages for growth. After phagocytosis, Francisella has been shown to escape from phagosomal degradation (4). In this context, we found that engulfment of Francisella by monocytes induces a remarkably efficient activation of inflammatory cytokines that is significantly greater than that seen after challenge with a high dose of E. coli LPS. Surprisingly, F. novicida, which has been isolated only twice from humans (31) and is considered to be less pathogenic than F. tularensis, was found to infect and efficiently activate human mononuclear cells.
Our data indicate that monocytes sense both live and dead bacteria and respond by induction of IL-1β and IL-8 messages. Cytokine synthesis depends on NF-κB activation, which may be induced by TLR or NLR signaling (6, 32). Many cytokines, including IL-8, are synthesized and released from the cell immediately. In contrast, IL-1β requires an additional step, posttranslation processing (25, 33). Although there is no difference between the ability of live and dead bacteria to induce IL-8 expression and release, nor in their ability to induce intracellular pro-IL-1β accumulation, there is a very clear difference in IL-1β processing and release. Note that since submission of this article, similar findings have been described in human and mouse mononuclear phagocytes (34, 35).
The relative efficiency of live vs. the killed Francisella, as well as vs. Francisella endotoxin, is of particular interest. The LPS of _F. tularensis_–LVS is atypical and does not induce cellular responses typical for Gram-negative bacteria LPS (36). Incubation of human monocytes with highly purified _F. tularensis_–LVS and F. novicida endotoxin did not induce significant cytokine synthesis. The lack of response to LPS supports the argument that the effect of Francisella on IL-1β processing and release is independent of TLR4 (36, 37). The finding that TLR4 has a relatively minor role in murine defense against intradermal infection with _F. tularensis_–LVS (38) supports this concept. However, although it has not been shown, Francisella may signal through TLR2. It was shown that the IL-8 gene may be activated by TLR2 (39). TLR2-deficient mice show decreased cytokine production, including IL-1β (40). However, the lack of available TLR2 and TLR4 blocking agents prevent us from testing this hypothesis directly in human monocytes.
Thus, intracellular NLR proteins may be able to sense bacteria that enter the host. To determine whether monocytes recognize Francisella with extracellular or intracellular sensors, we performed experiments to inhibit phagocytosis. cD effectively prevented live bacteria uptake by monocytes and blocked IL-1β processing and release. However, preventing bacterial internalization did not affect IL-1β gene transcription and protein precursor synthesis. Importantly, inhibition of phagocytosis by blocking actin polymerization with cD did not inhibit the monocyte response to E. coli LPS, which signals by means of TLR4 and had no significant effect on IL-8, which does not require intracellular processing.
Thus, our data demonstrate that the effect of Francisella on IL-1β processing requires active uptake of living bacteria. The requirement for living bacteria suggests that only live organisms are able to escape from the phagosome, or alternatively, that only living bacteria release caspase-1 “activating” factors. In an effort to further address the involved mechanisms, we used several approaches. First, we blocked phagosome escape with ammonium chloride (Fig. 5). Ammonium chloride markedly suppressed IL-1β processing but had little effect on IL-8 release. Second, we used the F. novicida mutant mglA, which lacks key pathogenic factors that are required for pathogen survival and replication in the host (17, 18). Consistent with the weak ability of mglA to escape into the cytosol (18), mglA mutants were also diminished in their ability to activate caspase-1 (Fig. 5). Together, these findings provide evidence to support the hypothesis that F. novicida activates the caspase-1 machinery (presumably by means of an inflammasome-like structure), an event that requires bacteria to be phagocytosed and then allowed to escape into the cytosol.
Based on the knowledge that caspase-1 is activated during assembly of the multiprotein complex called the inflammasome, we hypothesize that Francisella may activate at least one form of the inflammasome (11, 24, 26, 41, 42). In summary, F. novicida is a very effective inducer of IL-1β synthesis, processing, and release. Its ability to do so requires bacterial internalization and is most efficient when bacteria are alive and escape from the phagosome into cytosol. Intracellular Francisella represents a tool for studying the regulation and activation of IL-1β processing and release.
Acknowledgments
We thank Tim Eubank for assistance with figures. We acknowledge assistance from the Ohio State University Microscopy and Imaging Facility and the Davis Heart and Lung Research Institute's Microarray–Genetics Core Laboratory. This work was supported by National Heart, Lung, and Blood Institute Grants HL40871 and HL76278.
Author contributions: M.A.G., I.J.B., and M.D.W. designed research; M.A.G., N.L.K., M.D.D., and M.W.H. performed research; J.S.G. contributed new reagents/analytic tools; and M.A.G. and M.D.W. analyzed data and wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: moi, multiplicity of infection; LVS, live vaccine strain; RCN, relative copy number; PFM, paraformaldehyde; TLR, Toll-like receptor; NLR, NACHT leucine-rich repeats; cD, cytochalasin D.
References
- 1.Akira, S. & Takeda, K. (2004) Nat. Rev. Immunol. 4**,** 499–511. [DOI] [PubMed] [Google Scholar]
- 2.O'Connor, W., Jr., Harton, J. A., Zhu, X., Linhoff, M. W. & Ting, J. P. (2003) J. Immunol. 171**,** 6329–6333. [DOI] [PubMed] [Google Scholar]
- 3.Harton, J. A., Linhoff, M. W., Zhang, J. & Ting, J. P. (2002) J. Immunol. 169**,** 4088–4093. [DOI] [PubMed] [Google Scholar]
- 4.Anthony, L. D., Burke, R. D. & Nano, F. E. (1991) Infect. Immun. 59**,** 3291–3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ellis, J., Oyston, P. C., Green, M. & Titball, R. W. (2002) Clin. Microbiol. Rev. 15**,** 631–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Inohara, N. & Nunez, G. (2003) Nat. Rev. Immunol. 3**,** 371–382. [DOI] [PubMed] [Google Scholar]
- 7.Inohara, N., Chamaillard, M., McDonald, C. & Nunez, G. (2005) Annu. Rev. Biochem. 74**,** 355–383. [DOI] [PubMed] [Google Scholar]
- 8.Martinon, F. & Tschopp, J. (2005) Trends Immunol. 26**,** 447–454. [DOI] [PubMed] [Google Scholar]
- 9.Dinarello, C. A. (1998) Ann. N.Y. Acad. Sci. 856**,** 1–11. [DOI] [PubMed] [Google Scholar]
- 10.Martinon, F., Burns, K. & Tschopp, J. (2002) Mol. Cell 10**,** 417–426. [DOI] [PubMed] [Google Scholar]
- 11.Martinon, F. & Tschopp, J. (2004) Cell 117**,** 561–574. [DOI] [PubMed] [Google Scholar]
- 12.Golovliov, I., Baranov, V., Krocova, Z., Kovarova, H. & Sjostedt, A. (2003) Infect. Immun. 71**,** 5940–5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vinogradov, E., Conlan, W. J., Gunn, J. S. & Perry, M. B. (2004) Carbohydr. Res. 339**,** 649–654. [DOI] [PubMed] [Google Scholar]
- 14.Wewers, M. D., Dare, H. A., Winnard, A. V., Parker, J. M. & Miller, D. K. (1997) J. Immunol. 159**,** 5964–5972. [PubMed] [Google Scholar]
- 15.Baron, G. S. & Nano, F. E. (1998) Mol. Microbiol. 29**,** 247–259. [DOI] [PubMed] [Google Scholar]
- 16.Gray, C. G., Cowley, S. C., Cheung, K. K. & Nano, F. E. (2002) FEMS Microbiol. Lett. 215**,** 53–56. [DOI] [PubMed] [Google Scholar]
- 17.Lauriano, C. M., Barker, J. R., Yoon, S. S., Nano, F. E., Arulanandam, B. P., Hassett, D. J. & Klose, K. E. (2004) Proc. Natl. Acad. Sci. USA 101**,** 4246–4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Santic, M., Molmeret, M., Klose, K. E., Jones, S. & Kwaik, Y. A. (2005) Cell. Microbiol. 7**,** 969–979. [DOI] [PubMed] [Google Scholar]
- 19.Fortier, A. H., Leiby, D. A., Narayanan, R. B., Asafoadjei, E., Crawford, R. M., Nacy, C. A. & Meltzer, M. S. (1995) Infect. Immun. 63**,** 1478–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qureshi, S. T. & Medzhitov, R. (2003) Genes Immun. 4**,** 87–94. [DOI] [PubMed] [Google Scholar]
- 21.Iwasaki, A. & Medzhitov, R. (2004) Nat. Immunol. 5**,** 987–995. [DOI] [PubMed] [Google Scholar]
- 22.Aderem, A. & Ulevitch, R. J. (2000) Nature 406**,** 782–787. [DOI] [PubMed] [Google Scholar]
- 23.Barton, G. M. & Medzhitov, R. (2002) Curr. Top. Microbiol. Immunol. 270**,** 81–92. [DOI] [PubMed] [Google Scholar]
- 24.Tschopp, J., Martinon, F. & Burns, K. (2003) Nat. Rev. Mol. Cell Biol. 4**,** 95–104. [DOI] [PubMed] [Google Scholar]
- 25.Burns, K., Martinon, F. & Tschopp, J. (2003) Curr. Opin. Immunol. 15**,** 26–30. [DOI] [PubMed] [Google Scholar]
- 26.Mariathasan, S., Newton, K., Monack, D. M., Vucic, D., French, D. M., Lee, W. P., Roose-Girma, M., Erickson, S. & Dixit, V. M. (2004) Nature 430**,** 213–218. [DOI] [PubMed] [Google Scholar]
- 27.Gervassi, A., Alderson, M. R., Suchland, R., Maisonneuve, J. F., Grabstein, K. H. & Probst, P. (2004) Infect. Immun. 72**,** 7231–7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schotte, P., Denecker, G., Van Den Broeke, A., Vandenabeele, P., Cornelis, G. R. & Beyaert, R. (2004) J. Biol. Chem. 279**,** 25134–25142. [DOI] [PubMed] [Google Scholar]
- 29.Vogel, G. (2003) Science 302**,** 222–223. [DOI] [PubMed] [Google Scholar]
- 30.Tarnvik, A. (1989) Rev. Infect. Dis. 11**,** 440–451. [PubMed] [Google Scholar]
- 31.Kieffer, T. L., Cowley, S., Nano, F. E. & Elkins, K. L. (2003) Microbes Infect. 5**,** 397–403. [DOI] [PubMed] [Google Scholar]
- 32.Ting, J. P. & Davis, B. K. (2005) Annu. Rev. Immunol. 23**,** 387–414. [DOI] [PubMed] [Google Scholar]
- 33.Hazuda, D. J., Lee, J. C. & Young, P. R. (1988) J. Biol. Chem. 263**,** 8473–8479. [PubMed] [Google Scholar]
- 34.Bolger, C. E., Forestal, C. A., Italo, J. K., Benach, J. L. & Furie, M. B. (2005) J. Leukocyte Biol. 77**,** 893–897. [DOI] [PubMed] [Google Scholar]
- 35.Mariathasan, S., Weiss, D. S., Dixit, V. M. & Monack, D. M. (2005) J. Exp. Med. 202**,** 1043–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ancuta, P., Pedron, T., Girard, R., Sandstrom, G. & Chaby, R. (1996) Infect. Immun. 64**,** 2041–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Forestal, C. A., Benach, J. L., Carbonara, C., Italo, J. K., Lisinski, T. J. & Furie, M. B. (2003) J. Immunol. 171**,** 2563–2570. [DOI] [PubMed] [Google Scholar]
- 38.Chen, W., Kuolee, R., Shen, H., Busa, M. & Conlan, J. W. (2005) Immunol. Lett. 97**,** 151–154. [DOI] [PubMed] [Google Scholar]
- 39.Wang, Q., Dziarski, R., Kirschning, C. J., Muzio, M. & Gupta, D. (2001) Infect. Immun. 69**,** 2270–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bornstein, S. R., Zacharowski, P., Schumann, R. R., Barthel, A., Tran, N., Papewalis, C., Rettori, V., McCann, S. M., Schulze-Osthoff, K., Scherbaum, W. A., et al. (2004) Proc. Natl. Acad. Sci. USA 101**,** 16695–16700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Agostini, L., Martinon, F., Burns, K., McDermott, M. F., Hawkins, P. N. & Tschopp, J. (2004) Immunity 20**,** 319–325. [DOI] [PubMed] [Google Scholar]
- 42.Yu, J. W., Wu, J., Zhang, Z., Datta, P., Ibrahimi, I., Taniguchi, S., Sagara, J., Fernandes-Alnemri, T. & Alnemri, E. S. (July 22, 2005) Cell Death Differ., 10.1038/sj.cdd.4401734. [DOI] [PubMed]