Essential genes of a minimal bacterium (original) (raw)
Abstract
Mycoplasma genitalium has the smallest genome of any organism that can be grown in pure culture. It has a minimal metabolism and little genomic redundancy. Consequently, its genome is expected to be a close approximation to the minimal set of genes needed to sustain bacterial life. Using global transposon mutagenesis, we isolated and characterized gene disruption mutants for 100 different nonessential protein-coding genes. None of the 43 RNA-coding genes were disrupted. Herein, we identify 382 of the 482 M. genitalium protein-coding genes as essential, plus five sets of disrupted genes that encode proteins with potentially redundant essential functions, such as phosphate transport. Genes encoding proteins of unknown function constitute 28% of the essential protein-coding genes set. Disruption of some genes accelerated M. genitalium growth.
Keywords: minimal cell, Mycoplasma genitalium, synthetic biology, transposon mutagenesis
One consequence of progress in the new field of synthetic biology is an emerging view of cells as assemblages of parts that can be put together to produce an organism with a desired phenotype (1). That perspective begs the question: “How few parts would it take to construct a cell?” In an environment that is free from stress and provides all necessary nutrients, what would constitute the simplest free-living organism? This problem has been approached theoretically and experimentally in our laboratory and elsewhere.
In a comparison of the first two bacterial genomes sequenced, Mushegian and Koonin (2) projected that the 256 orthologous genes shared by the Gram-negative Haemophilus influenzae and the Gram-positive Mycoplasma genitalium genomes are a close approximation of a minimal gene set for bacterial life. More recently Gil et al. (3) proposed a 206-protein-coding gene core of a minimal bacterial gene set based on analysis of several free-living and endosymbiotic bacterial genomes.
In 1999 we reported the use of global transposon mutagenesis to experimentally determine the genes not essential for laboratory growth of M. genitalium (4). Since then, there have been numerous other experimental determinations of bacterial essential gene sets using our approach and other methods such as site-directed gene knockouts and antisense RNA (5–12). Most of these studies analyzed human pathogens, often with the aim of identifying essential genes that might be used as antibiotic targets. Almost all of these organisms contain relatively large genomes that include many paralogous gene families. Disruption or deletion of such genes shows that they are nonessential but does not determine whether their products perform essential biological functions. It is only through gene essentiality studies of bacteria that have near minimal genomes that we bring empirical verification to the compositions of hypothetical minimal gene sets.
We believe that the Mollicutes, generically known as the mycoplasmas, are the best experimental platform for experimentally defining a minimal gene set. These wall-less bacteria evolved from more conventional progenitors in the Firmicutes taxon by a process of massive genome reduction. Mycoplasmas are obligate parasites that live in relatively unchanging niches requiring little adaptive capability. M. genitalium, a human urogenital pathogen, is the extreme manifestation of this genomic parsimony, having only 482 protein-coding genes and the smallest genome, at ≈580 kb, of any known free-living organism capable of being grown in axenic culture (13). Although more conventional bacteria with larger genomes used in gene essentiality studies have on average 26% of their genes in paralogous gene families, M. genitalium has only 6% (Table 1, which is published as supporting information on the PNAS web site). Thus, with its lack of genomic redundancy and contingencies for different environmental conditions, M. genitalium is already close to being a minimal bacterial cell.
In our 1999 report (4) on the essential microbial gene for M. genitalium and its closest relative Mycoplasma pneumoniae, we mapped ≈2,200 transposon insertion sites in these two species, and identified 130 putatively nonessential M. genitalium protein-coding genes or M. pneumoniae orthologs of M. genitalium genes. We estimated that 265–350 of the protein-coding genes of M. genitalium are essential under laboratory growth conditions (4). However, proof of gene dispensability requires isolation and characterization of pure clonal populations, which we did not do. We grew Tn4001-transformed cells in mixed pools for several weeks and then isolated genomic DNA from those mixtures of mutants. Amplicons from inverse PCRs were sequenced by using that DNA as a template to identify the transposon insertion sites in the mycoplasma genomes. Most of the genes containing transposon insertions encoded either hypothetical proteins or other proteins not expected to be essential. Nonetheless, some of the putatively disrupted genes, such as isoleucyl and tyrosyl-tRNA synthetases (MG345 and MG455), DNA replication gene dnaA (MG469), and DNA polymerase III, subunit α (MG261) are thought to perform essential functions. We explained how genes generally thought to be essential might be disrupted and consequently scored inappropriately: a gene may be tolerant of the transposon insertion and not actually disrupted, cells could contain two copies of a gene, or the gene product may be supplied by other cells in the same mixed pool of mutants. Herein, we report an expanded study in which we have isolated and characterized M. genitalium Tn4001 insertion mutants that were present in individual colonies picked from agar plates. From this analysis, we made a more thorough estimate of the number of genes essential for growth of this minimalist bacterium.
Results
We sequenced across the transposon-genome junctions of mutants using a primer specific for Tn4001tet. Presence of a transposon in a gene indicated that that gene was disrupted and therefore nonessential. Because some genes might possibly tolerate transposon insertions and we did not confirm the absence of the gene products, the disruptions are technically only putative. To exclude the possibility that gene disruptions were the result of a transposon insertion in one copy of a duplicated gene, we used PCR to detect genes lacking the insertion. These PCRs showed us that almost all of our colonies contained both disrupted and WT versions of the genes identified as having the Tn4001. Further analysis using quantitative PCR showed that most colonies were mixtures of two or more mutants; thus, we operationally refer to them and any DNA isolated from them as colonies rather than clones. This cell clumping led us to isolate individual mutants using filter cloning. To break up clumps of cells possibly containing multiple different mutants, we forced cells through 0.22-μm filters before plating. We used these cells to produce subcolonies that we both sequenced and analyzed using quantitative PCR. Each disrupted gene was defined by subcloning at least one primary colony.
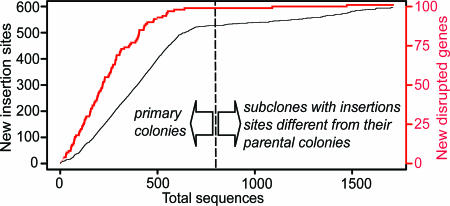
In total, we analyzed 3,321 M. genitalium transposon insertion mutant primary colonies and subcolonies to determine the locations of Tn4001tet inserts. For 74% of these, we generated sequence data that enabled us to map the transposon insertion sites. Colonies containing significant populations of multiple Tn4001tet insertions cannot be characterized using this approach. Only 62% of primary colonies generated useful sequence. This result was likely because of the tendency of mycoplasma cells to form persistent cell aggregates leading to colonies containing mixtures of multiple mutants that proved refractory to sequencing. For subcolonies, the success rate was 82%. Of the successfully sequenced subcolonies, 59% revealed a transposon insertion at a different site from that in the parental primary colony. The rate at which mutants accumulated in previously undetected sites of insertion on the genome was higher for the primary colonies than the subcolonies. However, the rate of accumulation of new insertion sites dropped after our first 600 sequences from independent colonies, indicating that we were approaching saturation mutagenesis of all nonlethal insertion sites (Fig. 1).
Fig. 1.
Accumulation of new disrupted M. genitalium genes (red) and new transposon insertion sites in the genome (black) as a function of the total number of analyzed primary colonies and subcolonies with insertion sites different from that of the parental primary clone.
We mapped a total of 2,462 different transposon insertion sites on the genome (Fig. 2). Eighty-four percent of the mutations were in protein-coding genes. No rRNA, tRNA, or structural RNA genes contained insertions. To address our central question of which M. genitalium genes were not essential for growth in SP4 (a rich laboratory medium) (14), we used the same criteria to designate a gene disruption as in our previous study. We considered transposon insertions disruptive if they were after the first three codons and before the 3′-most 20% of the coding sequence of a gene. Thus, nondisruptive mutations resulting from transposon-mediated duplication of short sequences at the insertion site (15, 16) and potentially inconsequential COOH-terminal insertions do not result in erroneous determination of gene expendability. Using these criteria, we identified a total of 100 dispensable M. genitalium genes (Table 2, which is published as supporting information on the PNAS web site). In Fig. 1, it can be seen that the rate of accumulation of new genes disrupted as a function of primary colonies and subcolonies plateaus, suggesting that we have or very nearly have disrupted all nonessential genes.
Fig. 2.
Global transposon mutagenesis of M. genitalium. The locations of transposon insertions from the current study are noted by a Δ below the insertion site on the map. Insertions mapped in our previous study (4) are noted with a ▿. Sites with 10 or more insertions are noted by a red filled triangle (▴).
We wanted to determine whether any of our disrupted genes were in cells bearing two copies of the gene. Unexpectedly, PCRs using primers flanking the transposon insertion sites produced amplicons of the size expected for WT templates from all five colonies initially tested (data not shown). End-stage analysis of PCRs could not tell us whether the WT sequences we amplified were the result of a low level of transposon jumping out of the target gene, or whether there was a gene duplication. Therefore, for at least one colony or subcolony for each disrupted gene, we used quantitative PCR to measure how many copies of contaminating WT versions of that gene there were in the sequenced DNA preps.
Analysis of the quantitative PCR results showed that most colonies were mixtures of multiple mutants. This result was likely a consequence of a high transformation efficiency, resulting in aggregates of M. genitalium cells containing more than one cell with a transposon. The direct genomic sequencing identified only the plurality member of the population. To address this issue, we adapted our mutant isolation protocol to include one or two rounds of filter cloning. Existing colonies of interest were filter-subcloned. We isolated 10 subcolonies, and the sites of their Tn4001 insertions were determined. We took both rapidly growing colonies and M. genitalium colonies that were delayed in their appearance. Often only a minority of the subcolonies had inserts in the same location as found with the parental colony. For the two colonies we isolated with mutations in conserved hypothetical gene MG116, we were unable to isolate subcolonies with mutations in that gene. Quantitative PCR showed that, in ≈30% of the cells derived from those colonies, the MG116 gene was not disrupted. This result was consistent with MG116 encoding an essential gene product that could be supplied in trans by other cells in the population. For our other 100 gene disruption mutants, even after filter cloning, we still found that almost every subcolony had some low level of a WT copy of the disrupted gene. This finding was likely the result of Tn4001 jumping (17). After subcloning, we were able to isolate gene disruption mutant colonies for 99 different disrupted M. genitalium genes that had <1% WT sequence, and one had between 1 and 2% WT DNA. These results indicated that none of those mutants contained a duplicate nondisrupted copy of their mutated gene.
Several mutants manifested remarkable phenotypes. Although some of the mutants grew slowly, mutants in lactate/malate dehydrogenase (MG460) and conserved hypothetical proteins MG414 and MG415 mutants had doubling times up to 20% faster than WT M. genitalium (data not shown). Mutants in MG185, which encodes a putative lipoprotein, floated rather than adhering to plastic as do WT cells. Cells with transposon insertions in the transketolase gene (MG066), which encodes a membrane protein and pentose phosphate pathway enzyme, grew in chains of clumped cells rather than in the monolayers characteristic of WT M. genitalium. Additionally, the MG066 mutants would lyse when washed with PBS and thus had to be processed in either SP4 medium or 100% serum.
We isolated mutants with transposon insertions at some sites much more frequently than others (Fig. 4, which is published as supporting information on the PNAS web site). We found colonies with mutations at hot spots in four genes: MG339 (recA), the fast growing MG414 and MG415, and MG428 (putative regulatory protein) constituted 31% of the total mutant pool. There was a striking difference in the most frequently found transposon insertion sites among primary colonies relative to the subcolonies having different insertions sites from their parental colonies (Fig. 4). We isolated 169 colonies and subcolonies having different insertions sites from their parental colonies with Tn4001tet inserted at base pair 517,751, which is in MG414. Only 5 (3%) of those were primary colonies. Conversely, we isolated 209 colonies with inserts in the 520,114–520,123 region, which is in MG415, and 56% of those were in primary colonies. The MG414 mutants were probably due both to rapid growth and to Tn4001 preferential jumping to that genome region, whereas the high frequency and near equal distribution of MG415 primary and subcolony transposon insertions may only be because those mutants grow more rapidly than others.
Discussion
We believe that we have identified nearly all of the nonessential genes in M. genitalium because, as shown in Fig. 1, the number of new gene disruptions reached a plateau as a function of the number of colonies analyzed. We found only 100 putatively nonessential genes as compared with our previous study conducted on batch-wise cultures (4). We tailored our protocols to minimize the chief shortcomings of the earlier study. First, we monitored plates for the appearance of transposon mutants up to 4 weeks to detect slow growing mutants. Second, as in our previous study, the rigorous criteria we used to classify a gene as disrupted minimized the likelihood that we overestimated the nonessential gene set because some genes might be tolerant of the transposon insertions.
The key procedural improvement in this project relative to our earlier report was the isolation and characterization of individual transposon mutants. Although we initially isolated many colonies that were mixtures of more than a single mutant, the filter cloning approach we applied to this problem proved to be effective for separating mixtures of mutants. The quantitative PCR characterization of clone purity revealed that Tn4001tet jumping also contributed to clone heterogeneity.
The current study gives a significantly different set of nonessential genes from our original study. In that study, we identified disruptions in 130 different M. genitalium protein-coding genes or M. pneumoniae orthologs of those genes. Our new study shows disruptions in only 67 of those 130 genes. Of the remaining 63 genes, 26 were disrupted only in M. pneumoniae in the earlier study. This finding suggests that some M. genitalium orthologs of nonessential M. pneumoniae genes are in fact essential. In the current study, transposon mutants both formed colonies on solid agar and grew as isolated clones in liquid culture, both under tetracycline selection. Previously, our transposon expressed a gentamicin resistance gene, and the mixed mutant pools grew only in liquid culture. These differences may be the reason some genes such as lipoprotein gene MG395 and lipase/esterase gene MG310 were hit frequently in the first study and not at all in the current study. Also, in our earlier work we did not reach a point where we mutagenized all or nearly all nonessential genes such as we have currently, which may account for many of the 34 genes only disrupted in our current study.
Another possible explanation for differences between the two studies is the opportunity for mutants to complement each other in our earlier work. Because our 1999 study analyzed Tn4001 insertions in mixed pools of mutants, we could have disrupted essential genes that can function in trans. This result would most likely be true for secreted proteins. In the current study, we disrupted 6 of the predicted 19 M. genitalium lipoproteins. Previously, we disrupted the same 6 lipoprotein genes plus 6 others. Among the genes we hit only in our first study was MG186, a homolog of an extracellular staphylococcal nuclease that might possibly catabolize DNA to provide the cell with nucleosides. Likewise, the other presumably essential lipoproteins disrupted previously might also be capable of acting in trans as enzymes, or possibly the gene products might be exchanged among cells in the mixed population of mutants used in our previous study. Our MG116 mutant may be such an example of a mutation that yields viable cells only when cultured with other cells WT for that gene, although the MG116 protein most likely is cytoplasmic.
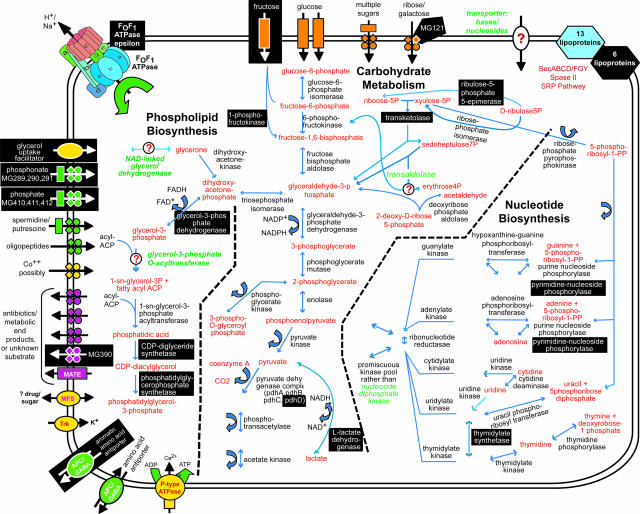
Mostly, we mutated the kinds of genes we expected to be nonessential: 48 of the 100 disrupted genes were either hypothetical proteins or proteins of unknown function, and there was an absence of disruptions in genes and regions expected to be universally essential. Still, many disrupted genes encoded proteins that one might think would be important because of the roles they play in metabolism or information storage and processing (Fig. 3).
Fig. 3.
Metabolic pathways and substrate transport mechanisms encoded by M. genitalium. Metabolic products are colored red, and mycoplasma proteins are black. White letters on black boxes mark nonessential functions or proteins based on our current gene disruption study. Question marks denote enzymes or transporters not identified that would be necessary to complete pathways, and those missing enzyme and transporter names are colored green. Transporters are colored according to their substrates: yellow, cations; green, anions and amino acids; orange, carbohydrates; purple, multidrug and metabolic end product efflux. The arrows indicate the predicted direction of substrate transport. The ABC type transporters are drawn as follows: rectangle, substrate-binding protein; diamonds, membrane-spanning permeases; circles, ATP-binding subunits.
None of the genes encoding the key enzymes of DNA replication were disrupted. However, we isolated mutants in other DNA metabolism genes that were expendable for the duration of our experiment, but might be necessary to maintain the M. genitalium genome over periods much longer than this experiment. These were six genes involved in recombination and DNA repair: recA (MG339), recU (MG352), Holliday junction DNA helicases ruvA (MG358) and ruvB (MG359), formamidopyrimidine-DNA glycosylase mutM (MG262.1), which excises oxidized purines from DNA, and a likely DNA damage inducible protein gene (MG360). Perhaps because of an accumulation of cell damage over time, mutants in chromosome segregation protein SMC (MG298) and hypothetical gene MG115, which is similar to the cinA gene of Streptococcus pneumoniae competence-inducible (cin) operon, grew more poorly after repeated passage.
Even with its near minimal gene set, M. genitalium has apparent enzymatic redundancy. We disrupted two complete ABC transporter gene cassettes for phosphate (MG410, MG411, and MG412) and putatively phosphonate (MG289, MG290, and MG291) import. The PhoU regulatory protein gene (MG409) was not disrupted, suggesting that it is needed for both cassettes. Phosphate is an essential metabolite that must be imported. Either phosphate might be imported by both transporters as a result of relaxed substrate specificity by the phosphonate system, or there is a metabolic capacity to interconvert phosphate and phosphonate. Although we disrupted both of these three gene cassettes, cells presumably need at least one phosphate transporter, so we added three ABC transporter genes for phosphate importation to our proposed minimal gene set. Relaxed substrate specificity is a recurring theme proposed and shown for several M. genitalium enzymes as a mechanism by which this bacterium meets its metabolic needs with fewer genes (18, 19).
M. genitalium generates ATP through glycolysis, and, although none of the genes encoding enzymes involved in the initial glycolytic reactions were disrupted, mutations in two energy generation genes suggested that there may be still more unexpected genomic redundancy in this essential pathway. We identified viable insertion mutants in genes encoding lactate/malate dehydrogenase (MG460) and the dihydrolipoamide dehydrogenase subunit of the pyruvate dehydrogenase complex (MG271). Mutations in either of these dehydrogenases would be expected to halve glycolytic ATP production, and unbalance NAD+ and NADH levels, which are the primary oxidizing and reducing agents in glycolysis. These mutations should have greatly reduced growth rate and accelerated acidification of the growth medium. Inexplicably, the lactate dehydrogenase mutants grow ≈20% faster than WT cells, whereas the MG271 mutants grew ≈20% slower. We also isolated a mutant in glycerol-3-phospate dehydrogenase (MG039), a phospholipid biosynthesis enzyme. The loss of functions in these mutants could have been compensated for by other M. genitalium dehydrogenases or reductases. This could be another case of mycoplasma enzymes having a relaxed substrate specificity, as has been reported for lactate/malate dehydrogenase (18) and nucleotide kinases (19).
We did not isolate any mutants in the M. genitalium cytoskeletal protein genes. That absence suggests that these genes are essential, which is at odds with the reported excision by Dhandayuthapani et al. (20) of the M. genitalium cytoskeletal protein HMW2 gene (MG218). They used homologous recombination using plasmids containing sequences flanking hmw2 (20). Because MG218 is a large gene (<5 Kb), it is very unlikely that we failed to find a disruption mutant simply because we did not analyze enough colonies. Our mutant isolation protocols were essentially the same. Possibly, MG218 mutants escaped detection because they were slow growing and were overshadowed by faster growing mutants in the mixed primary colonies.
Given the evolutionary path of extreme genome reduction taken by M. genitalium, it is likely that all its 482 protein-coding genes are in some way necessary for effective growth in its natural habitat, the human urogenital tract, either as an extracellular or intracellular parasite. More rapid growth of MG414 and MG415 mutants than WT M. genitalium suggests that these conserved hypothetical genes encode some sort of brake on cell growth. Slow growth in a human urogenital tract may help the bacterium avoid immunological surveillance.
Under our laboratory conditions, we identified 100 nonessential genes. Logically, the remaining 382 M. genitalium protein-coding genes, 3 phosphate transporter genes, and 43 RNA-coding genes presumably constitute the set of genes essential for growth of this minimal cell (Table 2). We disrupted genes in only 5 of the 12 M. genitalium paralogous gene families. Only for the two families comprised of lipoproteins MG185 and MG260 and glycerophosphoryl diester phosphodiesterases MG293 and MG385 did we disrupt all members. Accordingly, these families' functions may be essential, and we expanded our projection of the set of essential genes to 387 to include them. This is a significantly greater number of essential genes than the 265–350 predicted in our previous study of M. genitalium (4), or in the gene knockout/disruption study that identified 279 essential genes in Bacillus subtilis, which is a more conventional bacterium from the same Firmicutes taxon as M. genitalium (6). Similarly, our finding of 387 essential protein-coding genes greatly exceeds theoretical projections of how many genes comprise a minimal genome such as Mushegian and Koonin's 256 genes shared by both H. influenzae and M. genitalium (2), and the 206-gene core of a minimal bacterial gene set proposed by Gil et al. (3). One of the surprises about the essential gene set is its inclusion of 110 hypothetical proteins and proteins of unknown function. Some of these genes likely encode enzymes with activities reported in M. genitalium, such as transaldolase (21), but for which no gene has yet been annotated.
These data suggest that a genome constructed to encode 387 protein-coding and 43 structural RNA genes could sustain a viable synthetic cell, a Mycoplasma laboratorium (22). Rapid advances in gene synthesis technology and efforts at developing genome transplantation methods set the stage for experimental determination of how close this M. genitalium essential gene set based on disruption of individual genes comes to being the minimal set of genes needed to support cellular growth.
Materials and Methods
Cells and Plasmids. We obtained WT M. genitalium G37 (ATCC no. 33530**)** from the American Type Culture Collection. As part of this project, we resequenced and reannotated the genome of this bacterium. The new M. genitalium G37 sequence differed from the previous M. genitalium (13) genome sequence at 34 sites. The new sequence will replace the original M. genitalium genome sequence in GenBank as the reference sequence for the organism under the original accession number L43967. Notable consequences of the resequencing include merger of genes MG016, MG017, and MG018, which were reannotated as a DEAD helicase MG018. Similarly, DNA polymerase III γ/τ subunit genes MG419 and MG420 were merged to make MG419. Our transposon mutagenesis vector was the plasmid pIVT-1, which contains the Tn4001 transposon with a tetracycline resistance gene (tetM) (23), and was a gift from K. Dybvig (University of Alabama at Birmingham).
Transformation of M. genitalium with Tn4001 by Electroporation. Confluent flasks of M. genitalium cells were harvested by scraping into electroporation buffer (EB) comprised of 8 mM Hepes plus 272 mM sucrose at pH 7.4. We washed and then resuspended the cells in a total volume of 200–300 μl of EB. On ice, 100 μl of cells were mixed with 30 μg of pIVT-1 plasmid DNA and transferred to a 2-mm chilled electroporation cuvette (Bio-Rad). We electroporated using 2,500 V, 25 μF, and 100 Ω. After electroporation, we resuspended the cells in 1 ml of 37°C SP4 medium and allowed the cells to recover for 2 h at 37°C with 5% CO2. Aliquots of 200 μl of cells were spread onto SP4 agar plates containing 7 μg/ml tetracycline hydrochloride (VWR Scientific). The plates were incubated for 3–4 weeks at 37°C with 5% CO2 until colonies were visible. When colonies were 3–4 weeks old, we transferred individual M. genitalium colonies into SP4 medium plus 7 μg/ml tetracycline in 96-well plates. We incubated the plates at 37°C with 5% CO2 until the SP4 in most of the wells began to turn acidic and became yellow or orange (≈4 days). We froze those mutant stock cells at -80°C.
Amplification of Isolated Colonies for DNA Extraction. We inoculated 4 ml of SP4 containing 7 μg/ml tetracycline in six-well plates with 20 μl of transposon mutant stock cells and incubated the plates at 37°C with 5% CO2 until the cells reached 100% confluence. To extract genomic DNA from confluent cells, we scraped the cells and then transferred the cell suspension to a tube for pelleting by centrifugation. Thus, any nonadherent cells were not lost. We washed the cells in PBS (Mediatech, Herndon, VA) and then resuspended them in a mixture of 100 μl of PBS and 100 μl of the chaotropic MTL buffer from a Qiagen MagAttract DNA Mini M48 Kit (Qiagen, Valencia, CA). Tubes were stored at -20°C until the genomic DNA could be extracted using a Qiagen BioRobot M48 workstation.
Location of Tn4001tet Insertion Sites by DNA Sequencing from M. genitalium Genomic Templates. Our 20-μl sequencing reactions contained ≈0.5 μg of genomic DNA, 6.4 pmol of the 30-base oligonucleotide GTACTCAATGAATTAGGTGGAAGACCGAGG (Integrated DNA Technologies, Coralville, IA). The direct genomic sequencing protocol is described in Supporting Methods, which is published as supporting information on the PNAS web site. The primer anneals to the tetM gene103 base pairs from one of the transposon/genome junctions. Using blast, we located the insertion site on the M. genitalium genome.
Quantitative PCR to Determine Colony Homogeneity and Gene Duplication. We designed quantitative PCR primers (Integrated DNA Technologies) flanking transposon insertion sites using the default conditions for the primer design software primer express 1.5 (Applied Biosystems). Using quantitative PCR done on an Applied Biosystems 7700 Sequence Detection System, we determined the amounts of the target genes lacking a Tn4001 insertion in genomic DNA prepared from mutant colonies relative to the amount of those genes in WT M. genitalium. Reactions were done in Eurogentec (San Diego) qPCR Mastermix Plus SYBR Green. Genomic DNA concentrations were normalized after determining their relative amounts by using a TaqMan quantitative PCR specific for the 16S rRNA gene, which was done in Eurogentec qPCR Mastermix Plus. We calculated the number of target genes lacking the transposon in mutant genomic DNA preparations relative to the number of WT using the delta-delta Ct method (24).
Supplementary Material
Supporting Information
Acknowledgments
We thank Scott Peterson, Kim Wise, Barbara Culliton, and Jennifer Glass for helpful comments on the manuscript. We thank Bill Nelson at The Institute for Genomics Research for assistance with this project. This work was supported by grants from the U.S. Department of Energy (DE-FG02-02ER63453) and the J. Craig Venter Science Foundation.
Author contributions: J.I.G., N.A.-G., C.A.H., H.O.S., and J.C.V. designed research; J.I.G., N.A.-G., N.A., M.R.L., and M.M. performed research; J.I.G. contributed new reagents/analytic tools; J.I.G. and S.Y. analyzed data; and J.I.G., C.A.H., and H.O.S. wrote the paper.
Conflict of interest statement: No conflicts declared.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (Mycoplasma genitalium G37 ATCC 33530 genomic sequence; accession no. L43967).
References
- 1.Ferber, D. (2004) Science 303**,** 158-161. [DOI] [PubMed] [Google Scholar]
- 2.Mushegian, A. R. & Koonin, E. V. (1996) Proc. Natl. Acad. Sci. USA 93**,** 10268-10273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gil, R., Silva, F. J., Pereto, J. & Moya, A. (2004) Microbiol. Mol. Biol. Rev. 68**,** 518-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hutchison, C. A., Peterson, S. N., Gill, S. R., Cline, R. T., White, O., Fraser, C. M., Smith, H. O. & Venter, J. C. (1999) Science 286**,** 2165-2169. [DOI] [PubMed] [Google Scholar]
- 5.Forsyth, R. A., Haselbeck, R. J., Ohlsen, K. L., Yamamoto, R. T., Xu, H., Trawick, J. D., Wall, D., Wang, L., Brown-Driver, V., Froelich, J. M., et al. (2002) Mol. Microbiol. 43**,** 1387-1400. [DOI] [PubMed] [Google Scholar]
- 6.Kobayashi, K., Ehrlich, S. D., Albertini, A., Amati, G., Andersen, K. K., Arnaud, M., Asai, K., Ashikaga, S., Aymerich, S., Bessieres, P., et al. (2003) Proc. Natl. Acad. Sci. USA 100**,** 4678-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salama, N. R., Shepherd, B. & Falkow, S. (2004) J. Bacteriol. 186**,** 7926-7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herring, C. D., Glasner, J. D. & Blattner, F. R. (2003) Gene 311**,** 153-163. [DOI] [PubMed] [Google Scholar]
- 9.Mori, H., Isono, K., Horiuchi, T. & Miki, T. (2000) Res. Microbiol. 151**,** 121-128. [DOI] [PubMed] [Google Scholar]
- 10.Ji, Y., Zhang, B., Van Horn, S. F., Warren, P., Woodnutt, G., Burnham, M. K. & Rosenberg, M. (2001) Science 293**,** 2266-2269. [DOI] [PubMed] [Google Scholar]
- 11.Reich, K. A., Chovan, L. & Hessler, P. (1999) J. Bacteriol. 181**,** 4961-4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sassetti, C. M., Boyd, D. H. & Rubin, E. J. (2001) Proc. Natl. Acad. Sci. USA 98**,** 12712-12717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fraser, C. M., Gocayne, J. D., White, O., Adams, M. D., Clayton, R. A., Fleischmann, R. D., Bult, C. J., Kerlavage, A. R., Sutton, G., Kelley, J. M., et al. (1995) Science 270**,** 397-403. [DOI] [PubMed] [Google Scholar]
- 14.Tully, J. G., Rose, D. L., Whitcomb, R. F. & Wenzel, R. P. (1979) J. Infect. Dis. 139**,** 478-482. [DOI] [PubMed] [Google Scholar]
- 15.Dyke, K. G., Aubert, S. & el Solh, N. (1992) Plasmid 28**,** 235-246. [DOI] [PubMed] [Google Scholar]
- 16.Rice, L. B., Carias, L. L. & Marshall, S. H. (1995) Antimicrob. Agents Chemother. 39**,** 1147-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mahairas, G. G., Lyon, B. R., Skurray, R. A. & Pattee, P. A. (1989) J. Bacteriol. 171**,** 3968-3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cordwell, S. J., Basseal, D. J., Pollack, J. D. & Humphery-Smith, I. (1997) Gene 195**,** 113-120. [DOI] [PubMed] [Google Scholar]
- 19.Pollack, J. D., Myers, M. A., Dandekar, T. & Herrmann, R. (2002) OMICS 6**,** 247-258. [DOI] [PubMed] [Google Scholar]
- 20.Dhandayuthapani, S., Rasmussen, W. G. & Baseman, J. B. (1999) Proc. Natl. Acad. Sci. USA 96**,** 5227-5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pollack, J. D., Williams, M. V. & McElhaney, R. N. (1997) Crit. Rev. Microbiol. 23**,** 269-354. [DOI] [PubMed] [Google Scholar]
- 22.Reich, K. A. (2000) Res. Microbiol. 151**,** 319-324. [DOI] [PubMed] [Google Scholar]
- 23.Dybvig, K., French, C. T. & Voelker, L. L. (2000) J. Bacteriol. 182**,** 4343-4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Applied Biosystems (December 11, 1997) Relative Quantitation of Gene Expression (PerkinElmer, Foster City, CA), User Bulletin No. 2.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information