Agpat6—a Novel Lipid Biosynthetic Gene Required for Triacylglycerol Production in Mammary Epithelium (original) (raw)
. Author manuscript; available in PMC: 2011 Oct 19.
Published in final edited form as: J Lipid Res. 2006 Jan 31;47(4):734–744. doi: 10.1194/jlr.M500556-JLR200
Abstract
In analyzing the sequence tags for mutant mouse embryonic stem (ES) cell lines in BayGenomics (a mouse gene-trapping resource), we identified a novel gene, Agpat6, with sequence similarities to previously characterized glycerolipid acyltransferases. _Agpat6_’s closest family member is another novel gene that we have provisionally designated Agpat8. Both Agpat6 and Agpat8 are conserved from plants, nematodes, and flies to mammals. AGPAT6, which is predicted to contain multiple membrane-spanning helices, is found exclusively within the endoplasmic reticulum in mammalian cells. To gain insights into the in vivo importance of Agpat6, we used the Agpat6 ES cell line from BayGenomics to create _Agpat6_-deficient (_Agpat6_−/−) mice. _Agpat6_−/− mice lacked full-length Agpat6 transcripts, as judged by northern blots. One of the most striking phenotypes of _Agpat6_−/− mice was a defect in lactation. Pups nursed by _Agpat6_−/− mothers die perinatally. Normally, Agpat6 is expressed at high levels in the mammary epithelium of breast tissue, but not in the surrounding adipose tissue. Histological studies revealed that the aveoli and ducts of _Agpat6_−/− lactating mammary glands were underdeveloped, and there was a dramatic decrease in size and number of lipid droplets within mammary epithelial cells and ducts. Also, the milk from _Agpat6_−/− mice was markedly depleted in diacylglycerols and triacylglycerols. Thus, we identified a novel glycerolipid acyltransferase of the endoplasmic reticulum, AGPAT6, which is crucial for the production of milk fat by the mammary gland.
Supplementary key words: LPAAT, acyltransferase, transacylase, milk fat
INTRODUCTION
BayGenomics is a genomics program that uses gene-trapping vectors to produce mutant lines of mouse embryonic stem (ES) cells (http://baygenomics.ucsf.edu/) (1). The gene inactivated by the insertion of the gene-trapping vector can easily be identified with a unique DNA sequence tag. Thus far, BayGenomics has inactivated more than 3,000 unique genes in ES cells and has distributed more than 2,500 different cell lines to the research community, for the purpose of creating knockout mice. Aside from producing mutant ES cell clones, BayGenomics also produces a few knockout mice from the gene-trap ES cell lines, with the goal of identifying genes relevant to lipid metabolism and cardiopulmonary disease.
While analyzing the sequence tags for BayGenomics ES cell clones, we encountered a novel gene with sequence similarities to 1-acylglycerol-3-phosphate O-acyltransferases [AGPATs, which convert lysophosphatidic acid to phosphatidic acid (2, 3)] and other members of the glycerolipid acyltransferase family. Because of amino acid sequence similarities to AGPAT1, AGPAT2, and other putative AGPATs (AGPAT3, AGPAT4, and AGPAT5), the novel gene was provisionally designated Agpat6. Although this provisional name was based on similarities to other AGPATs, it should be noted that AGPAT6 also has sequence similarities to a variety of “non-AGPAT” glycerolipid acyltransferases. At about the same time, another report drew attention to the existence of Agpat6 in the human cDNA databases (4); however, there have been no published data on the intracellular localization of the enzyme or its in vivo importance.
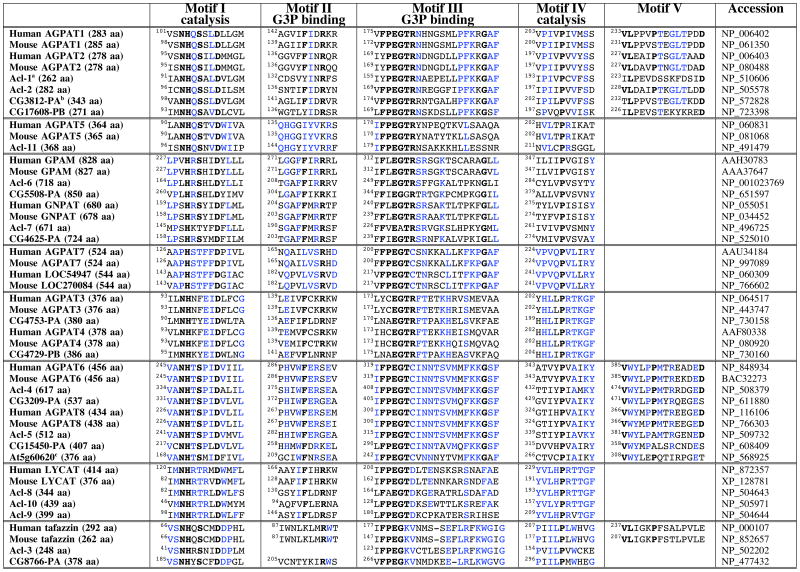
The glycerolipid acyltransferase protein family, which has been defined largely on the basis of amino acid sequence similarities, includes glycerol-3-phosphate acyltransferase (GPAM), glyceronephosphate O-acyltransferase (GNPAT), tafazzin, lysocardiolipin acyltransferase (LYCAT), 2-acylglycerophosphoethanolamine acyltransferase, the AGPATs, and a number of novel enzymes of unknown function. Amino acid sequence alignments of GPAM, AGPAT1, AGPAT2, and GNPAT have defined four regions of homology (motifs I–IV) that represent signatures for glycerolipid acyltransferases (5–7). Site-directed mutagenesis, followed by expression studies in Escherichia coli, have indicated that motifs I and IV are involved in catalysis, whereas motifs II and III are required for glycerol-3-phophate binding (5, 8, 9). Recent studies of naturally occurring mutations in AGPAT2 (10, 11) have uncovered a fifth important sequence motif (V), but its function has not yet been identified.
The biochemical properties, subcellular localization, and biological relevance of GPAM, GNPAT, tafazzin, LYCAT, AGPAT1, and AGPAT2, have been at least partially characterized either in humans or mice (2, 3, 12–15). GPAM is located in mitochondria and catalyzes the acylation of glycerol-3-phosphate at the sn-1 position to generate lysophosphatidic acid, and studies with _Gpam_-deficient mice have revealed that this enzyme plays a major role in triacylglycerol synthesis (12). GNPAT is located in peroxisomes and catalyses the acylation of glycerone phosphate to generate 1-acyl-glycerone-3-phosphate, a precursor in plasmalogen synthesis (16). Mutations in GNPAT cause rhizomelic chondrodysplasia punctata (RCDP) type 2, characterized by shortening of the upper extremities, mental retardation, and cataracts (13). The activity of tafazzin, which is located in mitochondria, is not known with certainty, but it could be involved in transferring acyl groups from phosphatidylcholine or phosphatidylethanolamine to monolysocardiolipin (17). Mutations in tafazzin cause Barth syndrome, an X-linked disease associated with dilated cardiomyopathy, skeletal myopathy, neutropenia, and growth retardation (14). Another cardiolipin biosynthetic enzyme, acyl-CoA:monolysocardiolipin acyltransferase (LYCAT), has been recently identified (15).
Biochemical roles for AGPAT1 and AGPAT2 in generating phosphatidic acid have been well documented (2, 3). Mutations in AGPAT2, which is expressed largely in adipose tissue, cause congenital generalized lipodystrophy (10), a disease characterized by a striking absence of subcutaneous and abdominal fat, hypertriglyceridemia, and severe insulin resistance. The other putative AGPATs (AGPAT3, AGPAT4, AGPAT5, and AGPAT7) (18, 19) were identified by sequence homology, and little is known about their biochemical properties or physiologic importance. AGPAT activity was attributed to AGPAT3, AGPAT4, and AGPAT5 in one study (18), but the activity levels were very low and in no way comparable to that observed for AGPAT2 (18). The physiological relevance of the novel BayGenomics gene, Agpat6, had never been previously investigated.
We sought to determine the intracellular localization of AGPAT6 and to define its relatedness, in terms of amino acid sequence, to known glycerolipid acyltransferases. In addition, we sought to ascertain the biological importance of AGPAT6 by examining the phenotypes of _Agpat6_-deficient mice. Here, we report that AGPAT6 is located exclusively in the endoplasmic reticulum (ER) and that its absence leads to underdeveloped mammary epithelium and the production of milk depleted in diacylglycerols and triacylglycerols. In addition, we report the identification of another novel gene, provisionally designated Agpat8, which is the closest homolog of Agpat6 within the glycerolipid acyltransferase family. Remarkably, AGPAT6 and AGPAT8 are conserved from plants, flies and worms to mammals.
MATERIALS AND METHODS
_Agpat6_-deficient mice
A mouse ES cell line (DTM030, strain 129/OlaHsd) containing an insertional mutation in Agpat6 was identified by BayGenomics, a gene-trapping resource (1). The gene-trap vector used (pGT1dTMpfs) contains a splice-acceptor sequence upstream of the reporter gene βgeo (a fusion of β-galactosidase and neomycin phosphotransferase II) (1). As judged by 5′ rapid amplification of cDNA ends (20), the insertional mutation in DTM030 was located in the second intron of Agpat6. Thus, the mutation results in the production of an in-frame fusion transcript consisting of exons 1 and 2 from Agpat6 and βgeo. Another BayGenomics ES cell line, RRF360, was used to create Agpat4 knockout mice.
We determined the exact site of insertion of the vector within intron 2, which allowed us to design a PCR strategy to genotype the mice. The following primers were used for genotyping: primer 1, 5′-ACAGGCTTTTGTGGTTTGGTTTGCT-3′; primer 2, 5′-AGAAATCCTCCCCAACAGTGGGACT-3′; primer 3 (from vector sequences), 3′-CGTGTCCTACAACACACACTCCAACC-5′. The wild-type allele was detected with primers 1 and 2 (located in sequences flanking the insertion, yielding a 458-bp product), while the mutant allele was detected with primers 1 and 3, yielding a 378-bp fragment.
ES cell line DTM030 was injected into C57BL/6 blastocysts to generate chimeric mice, which were bred to establish Agpat6 knockout mice. All mice had a mixed genetic background (C57BL/6 and 129/OlaHsd). The mice were weaned at 21 days of age, housed in a barrier facility with a 12-h light/12-h dark cycle, and fed a chow diet containing 4.5% fat (Ralston Purina, St. Louis, MO).
Northern blots
Total RNA was isolated from 50–150 mg of mouse tissue with Tri-Reagent (Sigma, St. Louis, MO). Total RNA (5 μg) was separated by electrophoresis on 1% agarose/formaldehyde gels and transferred to a Nytran SuPerCharge membrane (Schleicher & Schuell, Keene, NH). A mouse multiple-tissue poly(A)+ RNA blot and a mouse embryo poly(A)+ RNA blot (Clontech, Palo Alto, CA) were used to determine the tissue pattern of Agpat6 expression in adult mice and to examine the temporal expression of Agpat6 during embryogenesis. Bands on northern blots were visualized by autoradiography (Fuji Super RX films, Fujifilm, Tokyo, Japan) and quantified by densitometry (Molecular Imager FX, Bio-Rad, Hercules, CA). A 3′ UTR Agpat6 cDNA probe (a region not conserved in Agpat8) was amplified by RT-PCR with 5′-GTGGCAGGACAAGGTCAGAGCTACA-3′ and 5′-TCCCTCCTGACTCACCAGTTCTTCC-3′. The lacZ probe was prepared as described before (21). β-Actin and 18S cDNA probes were used as controls for RNA integrity and loading. [32P]dCTP-labeled cDNA probes were prepared with All-in-One random prime labeling mixture (Sigma). Standard prehybridization, hybridization, and washing procedures were used (21).
Western blots
The complete open reading frame of Agpat6 was amplified from a mouse embryo cDNA library by RT-PCR with primers 5′-ACCATGTTCCTGTTGCTACCT-3′ and 5′-GGACCGGCTGCGGTCCTCATGGTTTCC-3′. For expression in mammalian cells, the Appat6 coding sequence was cloned in-frame with a carboxyl-terminal V5-His tag into pcDNA3.1/V5-His TOPO TA expression vector (Invitrogen, Carlsbad, CA). For expression in insect cells, Agpat6 was amplified by PCR from pcDNA3.1/V5-His TOPO with the carboxyl-terminal V5-His tag with primers 5′-ATCGGAATTCATGTTCCTGTTGCTACCT-3′ and 5′-CGATGAATTCTCAATGGTGATGGTGATGATGACC-3′, subcloned into pCR2.1 (Invitrogen), and then subsequently cloned into the _Eco_RI site into pBacPAK8 (Clontech). The integrity of the Agpat6 sequence within each construct was verified by sequencing. High-Five insect cells infected with carboxyl-teminal-V5-tagged Agpat6, Agpat2, or _Gpam_-recombinant baculovirus, or COS-7 cells transfected with the _Agpat6_-pcDNA3.1/V5-His construct were collected in RIPA buffer containing a mixture of protease inhibitors (Complete Mini EDTA-free, Roche Applied Science, Indianapolis, IN). Samples were sonicated on ice and clarified by centrifugation at 500 × g for 10 min at 4°C. The protein content of the supernatant fluid (whole-cell extract) was determined with a Bradford assay (Bio-Rad). Denatured proteins (5 ng for insect cell extracts; 1 μg for COS-7 cell extracts) were size-fractionated on 4–12% Bis-Tris NuPAGE gels (Invitrogen). After electrotransfer to a sheet of nitrocellulose membrane (Invitrogen), the blots were blocked with phosphate-buffered saline containing 0.2% Tween and 5% nonfat powdered milk overnight at 4°C, and then incubated for 1 h at room temperature with a horseradish peroxidase–conjugated mouse monoclonal antibody against the V5 tag (Invitrogen) (1:5,000). Antibody binding was detected with the ECL Plus Western blotting kit (Amersham Biosciences, Piscataway, NJ) and Fuji Super RX x-ray film (Fujifilm).
Histology and immunohistochemistry
Mice were anesthetized intraperitoneally with a ketamine/xylazine cocktail and perfusion-fixed with normal saline followed by 4% paraformaldehyde. For β-galactosidase staining, tissues were fixed and stained as previously described (21). For hematoxylin/eosin staining, mammary glands were fixed in 10% buffered formalin for 24 h, dehydrated in ethanol, transitioned into xylene, embedded in paraffin, sectioned (1 μm-thick), and stained.
For fat staining, mice were deeply anesthetized intraperitoneally with a ketamine/xylazine cocktail, and perfusion-fixed with 0.1 M cacodylate followed by 2.5% glutaraldehyde in 0.1 M cacodylate. Mammary glands were dissected out and cleaved into thin slices to ensure thorough fixation. After fixation, tissues were stained in 1% osmium tetroxide, dehydrated in ethanol, transitioned into acetonitrile, and then embedded in Epon resin.
For some immunohistochemistry studies, the _Agpat6_-pcDNA3.1/V5-His construct described above was transfected into HeLa cells. AGPAT6 subcellular localization was compared to that of protein disulfide isomerase (PDI, an ER marker), and of manganese superoxide dismutase (MnSOD, a mitochondrial marker). The following antibodies were used: affinity purified rabbit anti-6xHistine (1:6,000; Immunology Consultants Laboratory, Newberg, OR); Alexa Fluor 568-labeled goat anti-rabbit IgG (1:800, Molecular Probes, Eugene, OR); mouse monoclonal anti-PDI (1:700, Abcam, Cambridge, MA); Alexa Fluor 488-labeled goat anti-mouse IgG (1:800, Molecular Probes); FITC-conjugated mouse anti-V5 tag (1:1,500, Invitrogen); rabbit polyclonal anti-MnSOD (1 μg/ml, Stressgen, Victoria, BC, Canada); Cy3-labeled goat anti-rabbit IgG (1:800, Abcam).
In addition, the complete coding sequences for Agpat2, Agpat6, and Gpam were inserted in-frame with a carboxyl-terminal enhanced green fluorescent protein marker in pECFP-N1 (Clontech). The ECFP-tagged constructs were expressed in COS-1 cells along with M1-YFP, a YFP-tagged ER marker (22). To assess mitochondrial localization, ECFP-tagged constructs were expressed in COS-1 cells labeled with a MitoTracker dye (Molecular Probes). Cells were imaged alive with an inverted Zeiss 510 laser scanning confocal microscope (63× lens).
Analysis of lipids in milk
Three hours after removing newborn pups, lactating females were given two successive intraperitoneal injections of oxytocin (5 μl/g of body weight of a 20 U/ml solution, injections separated by 20 min). At the time of the second oxytocin injection, mice were deeply anesthetized with a ketamine/xylazine cocktail, and milk was collected from the mammary glands and stored at −80°C for analysis. For all experiments, the milk was collected 24 h postpartum, when pups were still alive and suckling.
Milk lipids were extracted in the presence of authentic internal standards by the method of Folch et al. (23) with chloroform:methanol (2:1, v/v). Twenty microliters of milk were used for each analysis. In some experiments, neutral lipids were separated by thin layer chromatography in hexane:ethyl ether:acetic acid (80:20:2), subsequently visualized with iodine vapor, and lipids were identified by comigration with lipid standards. In other experiments, individual lipid classes within each extract were separated by preparative HPLC. Each isolated lipid class fraction was trans-esterified in 3N methanolic-HCl in a sealed vial under nitrogen at 100°C for 45 min. The fatty acid methyl esters were extracted from the mixture with hexane containing 0.05% butylated hydroxytoluene and prepared for gas chromatography by sealing the hexane extracts under nitrogen. Fatty acid methyl esters were then separated and quantified by capillary gas chromatography with a gas chromatograph (Hewlett-Packard model 6890, Wilmington, DE) equipped with a 30-m DB-225MS capillary column (J&W Scientific, Folsom, CA) and a flame-ionization detector, as described previously (24). Lipid metabolome data were expressed as nanomoles (nM) per gram and were assembled in tables as mean ± standard deviation for each group.
RESULTS
Agpat6 knockout mice
The BayGenomics library of mutant ES cells contained a mouse ES cell line with an insertional mutation in Agpat6, which was used to generate _Agpat6_-deficient mice. The site of insertion was identified within intron 2, making it possible to design a PCR strategy for distinguishing heterozygous (Agpat6+/−) from homozygous (_Agpat6_−/−) mice (Fig. 1A). As expected, full-length Agpat6 transcripts were absent in _Agpat6_−/− embryos (Fig. 1B).
Fig. 1.
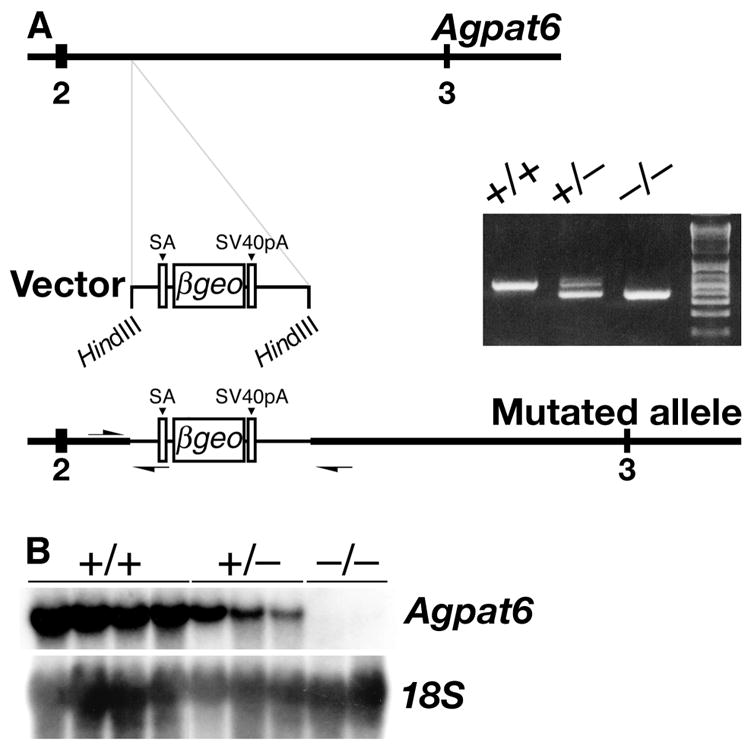
An insertional mutation in Agpat6. (A) Schematic of the insertion event, and the detection of the insertional mutation by PCR with genomic DNA from Agpat6+/+, Agpat6+/−, and _Agpat6_−/− mice. Numbers indicate exons. Primer orientation and location are indicated with arrows. SA, splice acceptor; SV40pA, poly(A) tail. (B) Northern blot with total RNA showing the expression of Agpat6 in Agpat6+/+, Agpat6+/−, and _Agpat6_−/−embryos. An 18S cDNA was used for normalization.
AGPAT6 structure
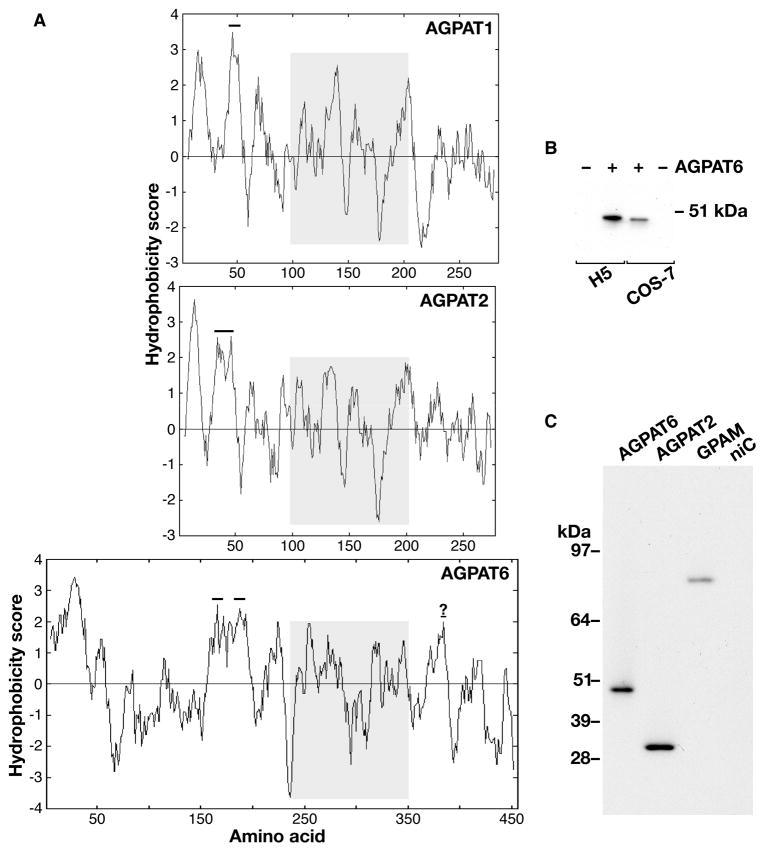
AGPAT6 is predicted to contain at least two transmembrane helices (Fig. 2A), as well as a peptide signal and a peptide cleavage site after residue 38 (http://www.cbs.dtu.dk/services/SignalP/). When expressed into COS-7 or insect cells, the C-terminal V5-tagged version of AGPAT6 migrates at 48 kDa, lower than the predicted size (52.2 kDa) for the full open reading frame (Fig. 2B, C). However, the predicted size of AGPAT6 after cleavage of the 38 amino acids signal peptide is 48 kDa, the size that we observed by western blotting. In contrast, the molecular weight of AGPAT2 by western blotting, 31 kDa, was the same as the predicted size of its entire open reading frame.
Fig. 2.
Structural features of AGPAT6. (A) Kyte-Doolittle hydrophobicity profiles for mouse AGPAT6, AGPAT1, and AGPAT2. Numbers on the X axis refer to amino acid residues. AGPAT6 (456 amino acids) is larger than AGPAT1 (285 amino acids) and AGPAT2 (278 amino acids), mainly because AGPAT6 contains 140 extra amino acid residues upstream to the region containing the signature glycerolipid acyltransferase sequence motifs (5–9) (highlighted in grey). Bold horizontal lines indicate predicted transmembrane domains (as judged by http://www.cbs.dtu.dk/services/TMHMM-2.0/, http://sosui.proteome.bio.tuat.ac.jp/sosuiframe0E.html, and http://smart.embl-heidelberg.de/). The question mark in the AGPAT6 sequence indicates a potential transmembrane domain that is predicted by one of the three sequence-analysis programs (http://sosui.proteome.bio.tuat.ac.jp/sosuiframe0E.html). (B) Western blot, with an anti-V5 antibody, showing that the molecular weight of the tagged version of AGPAT6 is 48 kDa, both in extracts from COS-7 cells transfected with V5-tagged Agpat6 (+) and in High-Five cells infected with a V5-tagged Agpat6 recombinant baculovirus (+). The 48-kDa band was absent in extracts from nontransfected COS-7 cells (−) and noninfected High-Five cells (−). (C) Western blot of total membrane fractions (5 ng each) from High-Five cells infected with V5-tagged AGPAT6 (48 kDa), AGPAT2 (31 kDa), or GPAM (93.7 kDa) baculoviruses. The molecular weight of AGPAT2 was identical to that predicted for the full-length open reading frame. Membranes from noninfected cells (niC) were included as a control.
AGPAT6 is located in the ER
To assess the subcellular localization of AGPAT6, we constructed a green fluorescent protein–tagged AGPAT6 and transfected it into COS-1 cells. These studies revealed that AGPAT6 is located in the ER, and none was in the mitochondria (Fig. 3A). As expected, GPAM was located entirely in the mitochondria, and AGPAT2 was located entirely in the ER (Fig. 3A). Immunofluorescence studies with a V5-His-tagged AGPAT6 in HeLa cells also indicated that AGPAT6 is an ER protein (Fig. 3B).
Fig. 3.
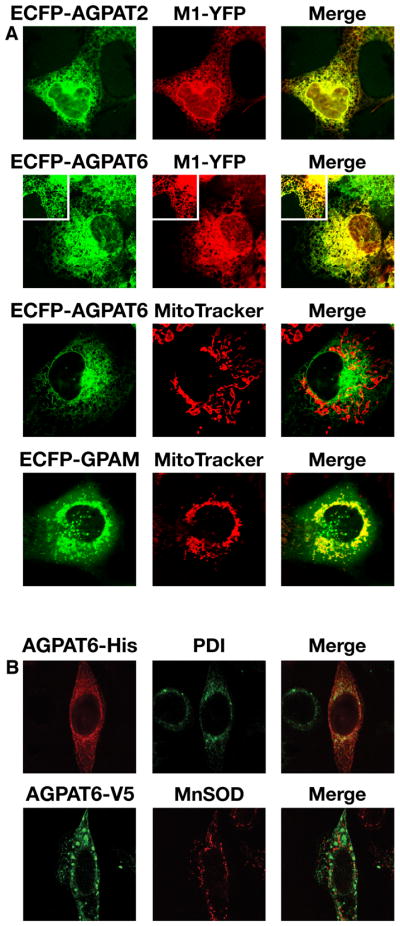
AGPAT6 localizes to the ER. (A) ECFP-tagged Agpat2, Agpat6, or Gpam constructs were cotransfected into COS-1 cells with M1-YFP (a YFP-tagged ER marker (22)). To assess mitochondrial localization, ECFP-tagged constructs were expressed in COS-1 cells labeled with a MitoTracker dye. ECFP–AGPAT6 colocalized with M1-YFP, but not with the MitoTracker dye, indicating an ER membrane localization for AGPAT6. A representative enlargement is shown as an insert in the panels for AGPAT6. AGPAT2 and GPAM were used as controls for ER and mitochondrial localization, respectively. (B) HeLa cells transfected with an expression vector for a V5-His-tagged Agpat6. The subcellular localization of AGPAT6 was compared with that of an ER marker, protein disulfide isomerase (PDI), and a mitochondrial marker, manganese superoxide dismutase (MnSOD). Merged images show colocalization of AGPAT6 and PDI but not MnSOD.
AGPAT6 expression patterns in mice
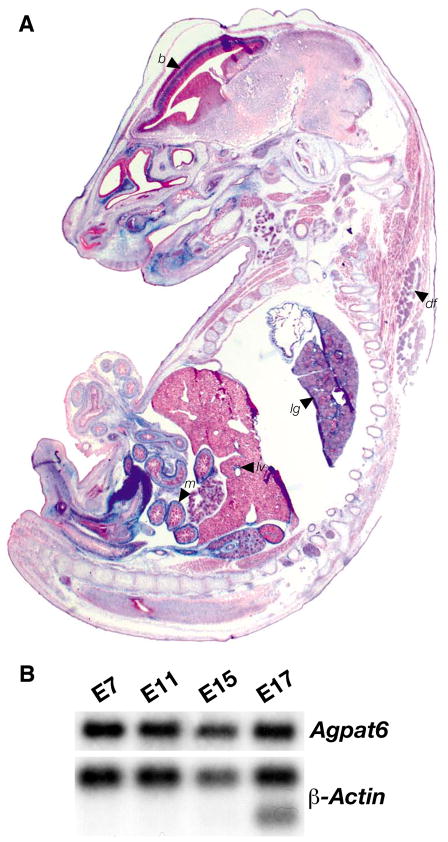
β-Galactosidase staining of tissues from _Agpat6_−/− embryos revealed that Agpat6 is expressed in the brain, dorsal fat pad, lung, liver, and a wide variety of mesenchymal tissues, including the mesenchyme of the gut and lung (Fig. 4A). Northern blots revealed that Agpat6 is expressed at high levels throughout embryogenesis (Fig. 4B).
Fig. 4.
Agpat6 expression during development. (A) β-Galactosidase staining of an Agpat6 knockout embryo (E15). Arrows point to expression in brain (b), dorsal fat pad (df), lung (lg), liver (lv) and mesenchyme (m) of the guts. (B) A mouse embryo poly(A)+ RNA blot showing the expression of Agpat6 throughout embryogenesis in wild-type mice. β-Actin was used for normalization.
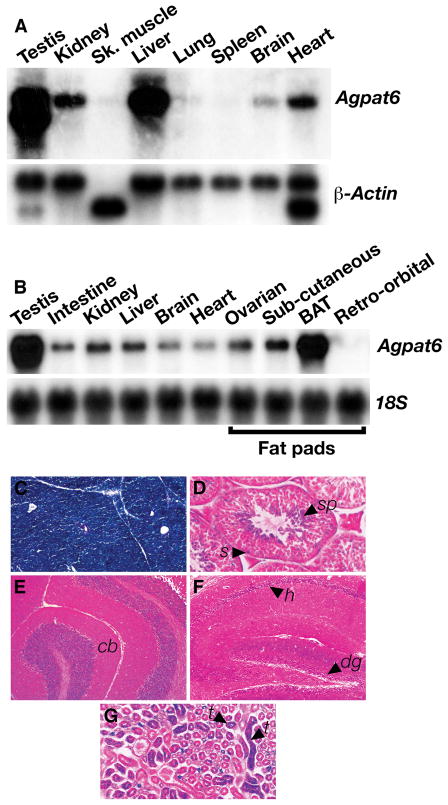
In adult wild-type mice, Agpat6 is expressed in a wide variety of tissues, including kidney, liver, brain, the ovarian fat pad, and testes, as judged by northern blots (Figs. 5A, B). Agpat6 is expressed at particularly high levels in brown adipose tissue (Fig. 5B). β-Galactosidase staining confirmed high-level of Agpat6 expression in brown adipose tissue (Fig. 5C) and showed that, in the testis, Agpat6 is expressed primarily in spermatids, with lower levels of expression in Sertoli cells (Fig. 5D). In the adult brain, Agpat6 is expressed predominantly in cerebellum (Fig. 5E) and hippocampus (Fig. 5F). In the kidney, Agpat6 is expressed in tubular cells (Fig. 5G).
Fig. 5.
Agpat6 expression in adult mice. (A) A mouse poly(A)+ RNA blot showing the expression of Agpat6 in adult tissues. A β-Actin cDNA was used for normalization. (B) A mouse total RNA blot showing prominent expression of Agpat6 in testis and brown adipose tissue in adult mice. A 18S cDNA was used for normalization. BAT, brown adipose tissue. (C–G) β-Galactosidase staining of brown adipose tissue (C), testis (D), cerebellum (E), hippocampus (F), and kidney (G) from a 6-week-old _Agpat6_−/− male. Arrows point to expression in spermatids (sp), Sertoli cells (s), cerebellar lobule (cb), hippocampus (h), dentate gyrus (dg), and tubular cells (t).
A role for AGPAT6 in milk production
_Agpat6_−/− mice were born at the expected mendelian frequency from crosses between heterozygous mice, indicating that, despite prominent expression in embryos, Agpat6 is not required for survival. However, offspring derived from _Agpat6_−/− females die within 48 h unless they are transferred to a foster mother. Very rarely, pups that are nursed by _Agpat6_−/− females survive the early postnatal period, but those mice are invariably runts and die by 3–4 weeks of age. These observations led us to suspect that Agpat6 could have a role in lactation.
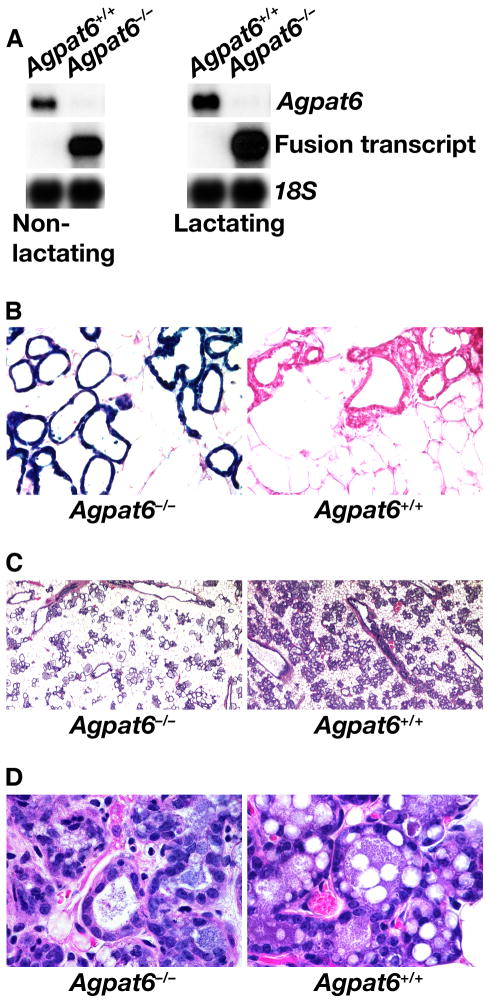
Agpat6 is expressed in nonlactating mammary gland, and the expression levels are upregulated during lactation, as judged by northern blot analysis. This upregulation was evident both for the full-length Agpat6 transcript in wild-type mice, as well as for the _Agpat6_–βgeo fusion transcript in _Agpat6_−/− mice (Fig. 6A). β-Galactosidase staining revealed that Agpat6 is expressed predominantly in epithelial cells of the mammary gland; no staining could be detected in the surrounding white adipose tissue (Fig. 6B). Hematoxylin and eosin–stained sections revealed that the alveoli and ducts of the mammary glands of _Agpat6_−/− lactating females were underdeveloped compared with those of wild-type mice (Fig. 6C). Furthermore, reduced numbers of fat droplets were evident in the epithelial cells of _Agpat6_−/− mammary glands when compared to wild-type controls (Fig. 6D).
Fig. 6.
Expression of Agpat6 in mammary gland. (A) Northern blot analysis of total RNA from Agpat6+/+ and _Agpat6_−/− mammary glands. An Agpat6 cDNA was used to detect the full-length Agpat6 mRNA in wild-type tissues, and a lacZ probe was used to detect the fusion transcript in the knockout mice. An 18S cDNA was used for normalization. (B) β-Galactosidase staining of mammary gland from _Agpat6_−/− and Agpat6+/+ lactating females, revealing a high level of Agpat6 expression in the epithelial cells of the mammary gland. (C, D) Hematoxylin and eosin staining of _Agpat6_−/− and Agpat6+/+ mammary glands, showing reduced size and number of alveoli in the mammary glands of lactating _Agpat6_−/− mice (C) as well as reduced numbers of fat droplets in _Agpat6_-deficient epithelial cells (D). In all experiments, the mammary glands were dissected 24 h postpartum, when pups were still alive and suckling.
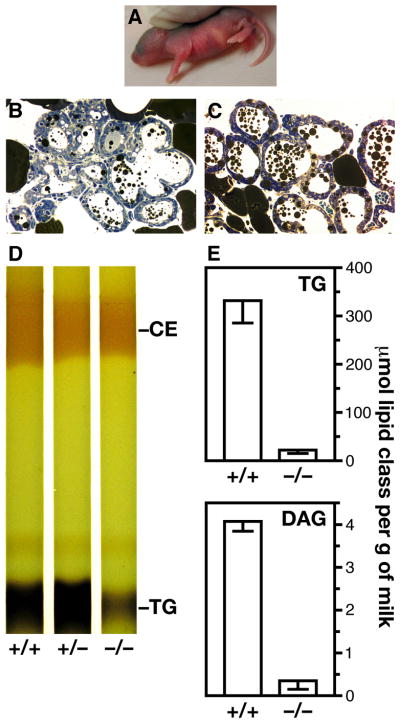
_Agpat6_−/− mothers had some capacity to make milk—evident by a milk stripe in newborn mouse pups (Fig. 7A). However, the amount of fat in the milk was reduced. Osmium tetroxide–stained sections of mammary gland revealed a decrease in the size and the amount of lipid droplets in the alveoli and ducts of _Agpat6_−/− mammary glands (Fig. 7B), compared with the alveoli of wild-type females (Fig. 7C). In addition, thin layer chromatography revealed that the milk from homozygous lactating females was depleted in triacylglycerols (Fig. 7D). Gas chromatography–based measurements revealed an ~90% reduction in diacylglycerols and triacylglycerols in the milk of _Agpat6_−/− mice (Fig. 7E).
Fig. 7.
Influence of Agpat6 deficiency on the composition of milk. (A) Milk streak in a pup nursed by an _Agpat6_−/− female. (B, C) Osmium tetroxide–stained sections of mammary glands from _Agpat6_−/− (B) and Agpat6+/+ (C) lactating females, showing decreased lipid droplets in the alveoli and ducts of the mammary glands from the _Agpat6_−/− female. (D) Reduced triacylglycerol (TG) content of milk from an _Agpat6_−/− female, compared to heterozygous and wild-type controls, as assessed by thin layer chromatography. The intensity of the cholesterol ester (CE) band was not significantly reduced in milk from _Agpat6_−/− females. (E) Reduced diacylglycerol (DAG) and triacylglycerol (TG) content of milk from _Agpat6_−/− females, as assessed by gas chromatography. In all experiments, the milk was collected (or the mammary glands were dissected) 24 h postpartum, when pups were still alive and suckling.
The fact that minimal amount of milk fat were evident in the mammary epithelium of _Agpat6_-deficient mice suggests the possibility that another glycerolipid acyltransferase might have a redundant role in producing triglycerides in mammary epithelium. Ultimately, understanding lipid synthesis in mammary epithelium will probably require the development of reporter alleles for all of the different glylcerolipid acyltransferase enzymes. Thus far, we have developed a reporter allele for Agpat4 (BayGenomics cell line RRF360); that gene is clearly not expressed in mammary epithelium or in the surrounding adipose tissue (Fig. 8A), while it is expressed strongly in the Sertoli cells of the testis (Fig. 8B).
Fig. 8.
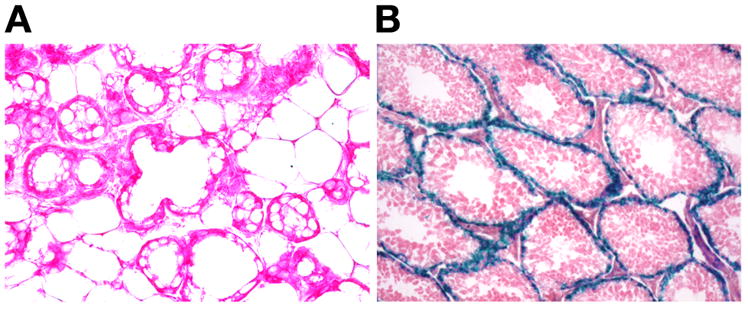
Agpat4 expression in mammary gland (A) and testis (B), as judged by β-galactosidase staining. Agpat4 expression in the mammary gland was undetectable, whereas it was robust in the Sertoli cells of the testis.
DISCUSSION
In this study, we report the discovery, within the BayGenomics gene-trapping resource, of Agpat6, a new member of the glycerolipid acyltransferase family. AGPAT6 is 48 kDa in size (with the V5-His tag) and is found exclusively within the ER. Agpat6 is predominantly expressed in brown adipose tissue and mammary epithelium. The milk from _Agpat6_−/− mice is depleted in diacylglycerols and triacylglycerols, and the mammary epithelium from _Agpat6_−/− mice is underdeveloped and depleted in intracellular fat droplets. In an accompanying article (25), we show that the triacylglycerol content of brown and white adipose tissue in _Agpat6_−/− mice is also significantly reduced. These experimental findings, together with sequence similarities between AGPAT6 and other glycerolipid acyltransferases (Table 1), suggest that AGPAT6 is a bona fide acyltransferase with an important role in the synthesis of triacylglycerols.
TABLE 1.
Proposed glycerolipid acyltransferase subgroups as assessed by alignment of amino acid sequences of putative orthologues from human, mouse, C. elegans, and D. melanogaster.
The mammary gland abnormalities in _Agpat6_−/− mice are reminiscent of those in mice lacking acylCoA:diacylglycerol acyltransferase 1 (DGAT1), an enzyme that adds an acyl group to diacylglycerol to generate triacylglycerols. _Dgat1_−/− nursing mothers have underdeveloped mammary glands and lack the ability to produce triacylglycerol-rich milk droplets (26). The fact that the mammary glands from Agpat6 and Dgat1 knockout females appeared underdeveloped suggests that defective lipid biosynthetic pathways interfere, directly or indirectly, with mammary gland development.
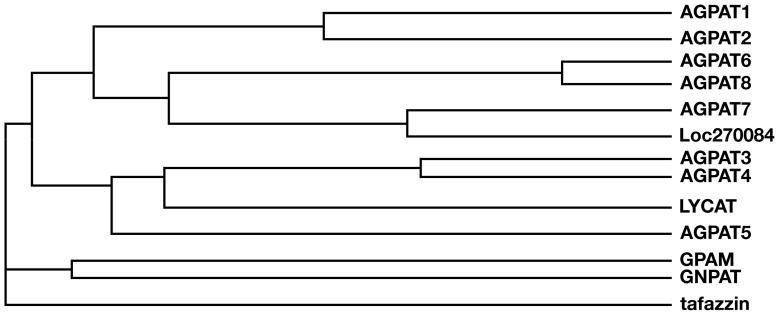
This identification of Agpat6 within BayGenomics illustrates the utility of gene trapping for inactivating a broad spectrum of genes, including novel genes that had escaped scientific scrutiny. Once the sequence tag for the Agpat6 ES cell line was in hand, we were able to classify Agpat6 as being a member of the glycerolipid acyltransferase family, and then move on to examine the mouse and human genomes for additional _Agpat6_-like sequences. By searching the DNA sequence databases, we quickly identified a novel, never-previously-reported gene resembling Agpat6, which we have provisionally designated Agpat8 (Table 1). Both AGPAT6 and AGPAT8 contain the classic sequence motifs (I–IV) characteristic of glycerolipid acyltransferases (5–7). When the sequences spanning motifs I–IV were analyzed for relatedness, it was apparent that AGPAT6 and AGPAT8 were most related to each other (Fig. 9) (66% identical at the amino acid level, but only ~2–15% identical with the other family members). More importantly, we found that both AGPAT6 and AGPAT8 contain sequences within motifs I–IV that distinguish them from other members of the family (Table 1). Domain III is strikingly conserved between AGPAT6 and AGPAT8, representing a signature sequence for these two proteins (Table 1). Also, the arginine in the VPEGTR consensus sequence within motif III is changed to a cysteine in AGPAT6 and AGPAT8, a feature shared only by AGPAT7 and the unknown protein at locus 270084. Remarkably, orthologues for Agpat6 and Agpat8 appear to exist in the genomes of plants, worms, and flies (Table 1), suggesting that, together, AGPAT6 and AGPAT8 probably play a unique, fundamental, and conserved function in lipid biosynthesis.
Fig. 9.
Dendrogram illustrating the amino acid sequence relatedness of mouse glycerolipid acyltransferases within the region of the proteins spanning functional domains I–IV (5–7). Alignments were performed with the Clustal W algorithm (http://www.ebi.ac.uk/clustalw/).
By analyzing conserved sequences (motifs I–IV) from multiple glycerolipid acyltransferases in four species (human, mouse, D. melanogaster, and C. elegans), we divided the family into eight subgroups (separated by double horizontal lines in Table 1). AGPAT1 and AGPAT2, which have been proven to carry out the AGPAT reaction (conversion of lysophosphatidic acid to phosphatidic acid) (2, 3), belong to the same subgroup. AGPAT3, AGPAT4, and AGPAT5, which have been reported to have very weak AGPAT activities (18), constitute two distinct subgroups. GPAM and GNPAT, two enzymes that add acyl groups to the sn-1 position (16, 27–29), are more closely related to each other than to any other member of the family. Lysocardiolipin acyltransferase (LYCAT) (15) falls into a fifth subgroup. Interestingly, we identified three putative C. elegans orthologues for mammalian LYCAT (15). Tafazzin falls into a sixth subgroup; tafazzin is involved in cardiolipin remodeling but its precise biochemical role remains to be established (17). AGPAT7 (19) and a closely related novel protein (locus 270084) form a seventh subgroup; their biological importance and biochemical function remain to be determined. AGPAT6 and AGPAT8 form the eighth subgroup.
For the entire glycerolipid acyltransferase family, we hypothesize that sequence relatedness within domains I–IV will ultimately be shown to correlate with biochemical function. For example, in the case of AGPAT6 and AGPAT8, we hypothesize that these two enzymes will ultimately be shown to have acyl acceptor and/or donor preferences that are similar to each other and distinct from those of other AGPATs, GPAMs, GNPATs, or LYCATs.
The identification of enzymatic activities for putative lipid biosynthetic enzymes can be straightforward (2, 3, 30, 31), but in some cases has been very challenging. For example, despite sequence motifs suggesting an acyltransferase activity (32) and despite years of biochemical studies by multiple groups, the biochemical role for tafazzin in cardiolipin remodeling has not yet been identified with certainty. This has certainly been the case for several of the AGPATs.
We expressed AGPAT2, GPAM, and AGPAT6 in insect cells, with sequence-verified plasmids, and then tested the membrane fractions for GPAM or AGPAT activities under a variety of reaction conditions. While the biochemical activities of our experimental controls, GPAM and AGPAT2, were invariably extremely robust (>5–10×background), we have not observed any AGPAT or GPAM activities in insect cell membranes overexpressing AGPAT6 (at least no activity above background) (A. Beigneux, S. G. Young, unpublished results). Similarly, we have not identified AGPAT or GPAM activities in E. coli membranes overexpressing AGPAT6.
One way to explain these results, of course, is to postulate that none of the reaction conditions were appropriate for AGPAT6, although they were perfectly suitable for the AGPAT2 and GPAM controls. This is definitely possible. For example, DGAT1 and DGAT2 carry out the same reaction but have different requirements for magnesium (31), so it would be a mistake to assume that AGPAT2 and AGPAT6 would share identical in vitro reaction conditions. Given the phenotypes of the mice, an AGPAT or GPAM activity for AGPAT6 would make a lot of sense. A reduced capacity for synthesizing lysophosphatidic acid or phosphatidic acid would probably explain reduced amounts of diacylglycerols and triacylglycerols in the milk and in the brown fat (25) of _Agpat6_−/− mice.
On the other hand, it is possible that AGPAT6 catalyzes a distinct biochemical activity. One reason for suspecting a different activity is that we have been unable, thus far, to detect even a small increase in AGPAT or GPAM activity in AGPAT6-enriched membranes, under a variety of assay conditions with various _sn_-1-acylglycerols, and acyl-CoA species as substrates. A second reason for suspecting a different enzymatic function is that it would be astonishing if mammals (and lower organisms such as worms and flies) truly require seven or more distinct AGPAT enzymes. Enzymes for crucial steps in lipid biosynthesis are frequently redundant (6), but the need for seven or more different AGPATs would be quite remarkable, particularly since the fatty acyl chain specificities of AGPAT1 and AGPAT2 are broad (2, 3) and since the loss of AGPAT2 causes such striking disease phenotypes (33).
In the end, biochemical studies, as well as the characterization of knockout mouse for each of the glycerolipid acyltransferases, will be critical for understanding lipid synthesis and lipid storage in different tissues, and for understanding the potential relevance of this family of enzymes to health and disease.
Acknowledgments
The authors are grateful to Drs. Tal Lewin and Diana Mehedint for biochemical protocols. This work was supported by BayGenomics, a Program for Genomics Applications from the National Heart, Lung, and Blood Institute (UO1 HL66621).
Abbreviations
AGPAT
1-acylglycerol-3-phosphate O-acyltransferase
GPAM
glycerol-3-phosphate acyltransferase
GNPAT
glyceronephosphate O-acyltransferase
LYCAT
lysocardiolipin acyltransferase
References
- 1.Stryke D, Kawamoto M, Huang CC, Johns SJ, King LA, Harper CA, Meng EC, Lee RE, Yee A, L’Italien L, Chuang PT, Young SG, Skarnes WC, Babbitt PC, Ferrin TE. BayGenomics: a resource of insertional mutations in mouse embryonic stem cells. Nucleic Acids Res. 2003;31:278–281. doi: 10.1093/nar/gkg064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aguado B, Campbell RD. Characterization of a human lysophosphatidic acid acyltransferase that is encoded by a gene located in the class III region of the human major histocompatibility complex. J Biol Chem. 1998;273:4096–4105. doi: 10.1074/jbc.273.7.4096. [DOI] [PubMed] [Google Scholar]
- 3.Eberhardt C, Gray PW, Tjoelker LW. Human lysophosphatidic acid acyltransferase. cDNA cloning, expression, and localization to chromosome 9q34.3. J Biol Chem. 1997;272:20299–20305. doi: 10.1074/jbc.272.32.20299. [DOI] [PubMed] [Google Scholar]
- 4.Li D, Yu L, Wu H, Shan Y, Guo J, Dang Y, Wei Y, Zhao S. Cloning and identification of the human LPAAT-zeta gene, a novel member of the lysophosphatidic acid acyltransferase family. J Hum Genet. 2003;48:438–442. doi: 10.1007/s10038-003-0045-z. [DOI] [PubMed] [Google Scholar]
- 5.Lewin TM, Wang P, Coleman RA. Analysis of amino acid motifs diagnostic for the sn-glycerol-3-phosphate acyltransferase reaction. Biochemistry. 1999;38:5764–5771. doi: 10.1021/bi982805d. [DOI] [PubMed] [Google Scholar]
- 6.Coleman RA, Lee DP. Enzymes of triacylglycerol synthesis and their regulation. Prog Lipid Res. 2004;43:134–176. doi: 10.1016/s0163-7827(03)00051-1. [DOI] [PubMed] [Google Scholar]
- 7.Dircks LK, Ke J, Sul HS. A conserved seven amino acid stretch important for murine mitochondrial glycerol-3-phosphate acyltransferase activity. Significance of arginine 318 in catalysis. J Biol Chem. 1999;274:34728–34734. doi: 10.1074/jbc.274.49.34728. [DOI] [PubMed] [Google Scholar]
- 8.Heath RJ, Rock CO. A conserved histidine is essential for glycerolipid acyltransferase catalysis. J Bacteriol. 1998;180:1425–1430. doi: 10.1128/jb.180.6.1425-1430.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heath RJ, Rock CO. A missense mutation accounts for the defect in the glycerol-3-phosphate acyltransferase expressed in the plsB26 mutant. J Bacteriol. 1999;181:1944–1946. doi: 10.1128/jb.181.6.1944-1946.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agarwal AK, Arioglu E, De Almeida S, Akkoc N, Taylor SI, Bowcock AM, Barnes RI, Garg A. AGPAT2 is mutated in congenital generalized lipodystrophy linked to chromosome 9q34. Nat Genet. 2002;31:21–23. doi: 10.1038/ng880. [DOI] [PubMed] [Google Scholar]
- 11.Magre J, Delepine M, Van Maldergem L, Robert JJ, Maassen JA, Meier M, Panz VR, Kim CA, Tubiana-Rufi N, Czernichow P, Seemanova E, Buchanan CR, Lacombe D, Vigouroux C, Lascols O, Kahn CR, Capeau J, Lathrop M. Prevalence of mutations in AGPAT2 among human lipodystrophies. Diabetes. 2003;52:1573–1578. doi: 10.2337/diabetes.52.6.1573. [DOI] [PubMed] [Google Scholar]
- 12.Hammond LE, Gallagher PA, Wang S, Hiller S, Kluckman KD, Posey-Marcos EL, Maeda N, Coleman RA. Mitochondrial glycerol-3-phosphate acyltransferase-deficient mice have reduced weight and liver triacylglycerol content and altered glycerolipid fatty acid composition. Mol Cell Biol. 2002;22:8204–8214. doi: 10.1128/MCB.22.23.8204-8214.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ofman R, Hettema EH, Hogenhout EM, Caruso U, Muijsers AO, Wanders RJ. Acyl-CoA:dihydroxyacetonephosphate acyltransferase: cloning of the human cDNA and resolution of the molecular basis in rhizomelic chondrodysplasia punctata type 2. Hum Mol Genet. 1998;7:847–853. doi: 10.1093/hmg/7.5.847. [DOI] [PubMed] [Google Scholar]
- 14.Bione S, D’Adamo P, Maestrini E, Gedeon AK, Bolhuis PA, Toniolo D. A novel X-linked gene, G4.5. is responsible for Barth syndrome. Nat Genet. 1996;12:385–389. doi: 10.1038/ng0496-385. [DOI] [PubMed] [Google Scholar]
- 15.Cao J, Liu Y, Lockwood J, Burn P, Shi Y. A novel cardiolipin-remodeling pathway revealed by a gene encoding an endoplasmic reticulum-associated acyl-CoA:lysocardiolipin acyltransferase (ALCAT1) in mouse. J Biol Chem. 2004;279:31727–31734. doi: 10.1074/jbc.M402930200. [DOI] [PubMed] [Google Scholar]
- 16.Webber KO, Hajra AK. Purification of dihydroxyacetone phosphate acyltransferase from guinea pig liver peroxisomes. Arch Biochem Biophys. 1993;300:88–97. doi: 10.1006/abbi.1993.1013. [DOI] [PubMed] [Google Scholar]
- 17.Xu Y, Kelley RI, Blanck TJJ, Schlame M. Remodeling of Cardiolipin by Phospholipid Transacylation. J Biol Chem. 2003;278:51380–51385. doi: 10.1074/jbc.M307382200. [DOI] [PubMed] [Google Scholar]
- 18.Lu B, Jiang YJ, Zhou Y, Xu FY, Hatch GM, Choy PC. Cloning and characterization of murine 1-acyl-sn-glycerol 3-phosphate acyltransferases and their regulation by PPARalpha in murine heart. Biochem J. 2005;385:469–477. doi: 10.1042/BJ20041348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ye GM, Chen C, Huang S, Han DD, Guo JH, Wan B, Yu L. Cloning and characterization a novel human 1-acyl-sn-glycerol-3-phosphate acyltransferase gene AGPAT7. DNA Seq. 2005;16:386–390. doi: 10.1080/10425170500213712. [DOI] [PubMed] [Google Scholar]
- 20.Townley DJ, Avery BJ, Rosen B, Skarnes WC. Rapid sequence analysis of gene trap integrations to generate a resource of insertional mutations in mice. Genome Res. 1997;7:293–298. doi: 10.1101/gr.7.3.293. [DOI] [PubMed] [Google Scholar]
- 21.Beigneux AP, Kosinski C, Gavino B, Horton JD, Skarnes WC, Young SG. ATP-Citrate Lyase Deficiency in the Mouse. J Biol Chem. 2004;279:9557–9564. doi: 10.1074/jbc.M310512200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chiu VK, Bivona T, Hach A, Sajous JB, Silletti J, Wiener H, Johnson RL, II, Cox AD, Philips MR. Ras signalling on the endoplasmic reticulum and the Golgi. Nat Cell Biol. 2002;4:343–350. doi: 10.1038/ncb783. [DOI] [PubMed] [Google Scholar]
- 23.Folch J, Lees M, Stanley GHS. A simple method for the isolation and purification of total lipids from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- 24.Watkins SM, Lin TY, Davis RM, Ching JR, DePeters EJ, Halpern GM, Walzem RL, German JB. Unique phospholipid metabolism in mouse heart in response to dietary docosahexaenoic or alpha-linolenic acids. Lipids. 2001;36:247–254. doi: 10.1007/s11745-001-0714-8. [DOI] [PubMed] [Google Scholar]
- 25.Vergnes L, Beigneux AP, Davis R, Watkins SM, Young SG, Reue K. Agpat6 deficiency causes subdermal lipodystrophy and resistance to obesity. Submitted to J Lipid Res. 2005 doi: 10.1194/jlr.M500553-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cases S, Zhou P, Shillingford JM, Wiseman BS, Fish JD, Angle CS, Hennighausen L, Werb Z, Farese RV., Jr Development of the mammary gland requires DGAT1 expression in stromal and epithelial tissues. Development. 2004;131:3047–3055. doi: 10.1242/dev.01158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Daae LN. The acylation of glycerol 3 -phosphate in different rat organs and in the liver of different species (including man) Biochim Biophys Acta. 1973;306:186–193. doi: 10.1016/0005-2760(73)90224-5. [DOI] [PubMed] [Google Scholar]
- 28.Stern W, Pullman ME. Acyl-CoA:sn-glycerol-3-phosphate acyltransferase and the positional distribution of fatty acids in phospholipids of cultured cells. J Biol Chem. 1978;253:8047–8055. [PubMed] [Google Scholar]
- 29.Haldar D, Tso WW, Pullman ME. The acylation of sn-glycerol 3-phosphate in mammalian organs and Ehrlich ascites tumor cells. J Biol Chem. 1979;254:4502–4509. [PubMed] [Google Scholar]
- 30.Cases S, Smith SJ, Zheng YW, Myers HM, Lear SR, Sande E, Novak S, Collins C, Welch CB, Lusis AJ, Erickson SK, Farese RV., Jr Identification of a gene encoding an acyl CoA:diacylglycerol acyltransferase, a key enzyme in triacylglycerol synthesis. Proc Natl Acad Sci U S A. 1998;95:13018–13023. doi: 10.1073/pnas.95.22.13018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cases S, Stone SJ, Zhou P, Yen E, Tow B, Lardizabal KD, Voelker T, Farese RV., Jr Cloning of DGAT2, a second mammalian diacylglycerol acyltransferase, and related family members. J Biol Chem. 2001;276:38870–38876. doi: 10.1074/jbc.M106219200. [DOI] [PubMed] [Google Scholar]
- 32.Neuwald AF. Barth syndrome may be due to an acyltransferase deficiency. Curr Biol. 1997;7:R465–R466. doi: 10.1016/s0960-9822(06)00237-5. [DOI] [PubMed] [Google Scholar]
- 33.Garg A. Acquired and inherited lipodystrophies. N Engl J Med. 2004;350:1220–1234. doi: 10.1056/NEJMra025261. [DOI] [PubMed] [Google Scholar]