Genome-wide Analysis of Re-replication Reveals Inhibitory Controls That Target Multiple Stages of Replication Initiation (original) (raw)
Abstract
DNA replication must be tightly controlled during each cell cycle to prevent unscheduled replication and ensure proper genome maintenance. The currently known controls that prevent re-replication act redundantly to inhibit pre-replicative complex (pre-RC) assembly outside of the G1-phase of the cell cycle. The yeast Saccharomyces cerevisiae has been a useful model organism to study how eukaryotic cells prevent replication origins from reinitiating during a single cell cycle. Using a re-replication-sensitive strain and DNA microarrays, we map sites across the S. cerevisiae genome that are re-replicated as well as sites of pre-RC formation during re-replication. Only a fraction of the genome is re-replicated by a subset of origins, some of which are capable of multiple reinitiation events. Translocation experiments demonstrate that origin-proximal sequences are sufficient to predispose an origin to re-replication. Origins that reinitiate are largely limited to those that can recruit Mcm2-7 under re-replicating conditions; however, the formation of a pre-RC is not sufficient for reinitiation. Our findings allow us to categorize origins with respect to their propensity to reinitiate and demonstrate that pre-RC formation is not the only target for the mechanisms that prevent genomic re-replication.
INTRODUCTION
Eukaryotic DNA replication is tightly controlled to ensure that the genome is copied exactly once before chromosome segregation and cytokinesis. Inappropriate replication after S-phase leads to severe DNA damage (Green and Li, 2005) and cell death (Yanow et al., 2001; Melixetian et al., 2004; Wilmes et al., 2004). To prevent these catastrophic effects, cells use multiple overlapping mechanisms to prevent unscheduled replication (Diffley, 2004; Blow and Dutta, 2005).
The initiation of eukaryotic DNA replication is divided into two stages: origin selection and origin activation. Origins of DNA replication are selected by the formation of the pre-replicative complex (pre-RC; Mendez and Stillman, 2003). The first event in pre-RC formation is the binding of the origin recognition complex (ORC) to origin DNA. During G1, ORC recruits other members of the pre-RC, including Cdc6 and Cdt1. Together these proteins load the six-subunit mini-chromosome maintenance complex (Mcm2-7), the putative replicative helicase (Takahashi et al., 2005), onto origin DNA. As cells enter S-phase, origins are activated by cyclin-dependent kinases (CDKs) and the Dbf4-dependent kinase (Bell and Dutta, 2002). These kinases target both pre-RC components and other replication factors to trigger the recruitment of replication proteins necessary for origin unwinding and DNA synthesis.
Eukaryotic chromosomes require multiple origins spread over their length to ensure that each chromosome is copied during S-phase. Although pre-RCs are assembled at all potential origins during G1, origins are not all activated at the same time. A temporal replication program leads to the activation of each origin at a characteristic time during S-phase with some origins initiating early in S-phase, others later, and still others not at all (Donaldson, 2005). The mechanisms controlling this program are poorly understood, but specific cyclins (Donaldson et al., 1998; Hu and Aparicio, 2005), checkpoint proteins (Santocanale and Diffley, 1998; Shirahige et al., 1998), and levels of chromosome acetylation (Vogelauer et al., 2002; Aparicio et al., 2004) have each been shown to affect this temporal program.
Once an origin has initiated, multiple mechanisms exist in all eukaryotes to prevent inappropriate reinitiation from occurring within the same cell cycle (Gopalakrishnan et al., 2001; Nguyen et al., 2001; Yanow et al., 2001). These mechanisms all inhibit the formation of pre-RCs outside of G1. For example, CDK-dependent phosphorylation targets pre-RC components to prevent new pre-RC formation (Machida et al., 2005). During each round of division, cells oscillate once between low (G1) and high (S, G2, M) CDK activity, meaning pre-RCs can only form and be activated once per cell cycle. Multicellular eukaryotes have at least one additional CDK-independent inhibitor of re-replication called geminin. This protein binds and inhibits Cdt1 (Wohlschlegel et al., 2000) outside of G1, thereby preventing new pre-RC formation (Mihaylov et al., 2002; Melixetian et al., 2004; Zhu et al., 2004).
In Saccharomyces cerevisiae B-type CDKs, which are composed of the Cdk1/Cdc28 kinase and one of six different B-type cyclins (Clb1-6), inhibit pre-RC formation by phosphorylating three components of the pre-RC (Nguyen et al., 2001). The resulting modifications have distinct consequences for each target. Phosphorylation of Cdc6 and Mcm2-7 leads to degradation (Elsasser et al., 1999; Drury et al., 2000) and export to the cytoplasm (Labib et al., 1999; Nguyen et al., 2000), respectively. Cdc28 also phosphorylates at least two of the six ORC subunits, Orc2 and Orc6 (Nguyen et al., 2001), but how these modifications inhibit ORC's role in pre-RC formation is unknown. All of the phosphorylation events described above prevent new pre-RC formation, which, in turn, prevents reinitiation. In addition to these mechanisms, direct interactions between ORC and cyclins prevent pre-RC formation in Schizosaccharomyces pombe (Wuarin et al., 2002) and S. cerevisiae (Wilmes et al., 2004).
In S. cerevisiae, the controls preventing reinitiation can be overcome by disrupting three of the CDK-dependent mechanisms described above. A strain modified to express nondegradable Cdc6, constitutively localize Mcm2-7 to the nucleus and inhibit phosphorylation of Orc2 and Orc6, can initiate a second round of initiation during a single cell cycle (Nguyen et al., 2001). Disrupting the interaction between the S-phase cyclin, Clb5, and the smallest ORC subunit Orc6 (in addition to the above mutations) results in further re-replication (Wilmes et al., 2004). Analysis of DNA content from re-replicating S. cerevisiae strains has shown that the majority of cells in the population do not fully re-replicate their genome. Interestingly, when a subset of origins was monitored for the ability to initiate during re-replication, only some of those tested showed reinitiation (Nguyen et al., 2001). These data suggest that not all origins are sensitive to reinitiation.
To gain insight into how the genome protects itself from re-replication, we bypassed all known re-replication control mechanisms in S. cerevisiae and identified origins across the genome that reinitiated. We show that re-replication initiates from a subset of origins used in S-phase, that the sensitivity to re-replication varies between origins, and that some origins are capable of reinitiating multiple times. Finally, we also show that Mcm2-7 loading is required, but not sufficient, for origins to reinitiate, indicating that there are layers of control beyond those inhibiting pre-RC formation that prevent re-replication.
MATERIALS AND METHODS
Plasmids
To integrate ARS418 at iYDR309c, plasmid pLys2-418-309CB was generated by first amplifying the intergenic region containing ARS418 using primers SB2558 and SB2559 (see Supplementary Table 1 for primer sequences) and putting the resulting DNA into the PstI and XbaI sites of pUC119-Lys2 to create pLys2-418. The intergenic region between YDR309C and YDR310C was then amplified using primers SB2664 and SB2665 and inserted into the SphI site in pLys2-418. To integrate the mutant ARS418 at iYDR309C, plasmid pLys2-418mut-309CB was generated by QuikChange XL (Stratagene, La Jolla, CA) mutagenesis, using primers SB2753 and SB2754. The mutant ARS418 was then amplified using primers SB2558 and SB3125 and inserted into the PstI/SacI sites of pLys2-418-309CB. ARS214 was amplified from the genome using primers SB3279 and SB3280 and inserted into the EcoRI/HinDIII sites of pARS1, replacing ARS1. The origin was then amplified using primers SB3182 and SB3183 and inserted into the PstI/SacI sites of pLys2-418-309CB, replacing ARS418.
Strains
All strains in this study are previously described except SB1808, SB1809, SB2023, SB2052, and SB2125 (see Supplementary Table 2 for genotypes). For density transfer, strains SB1808 and SB1809 were made ADE2 by transforming SB1507 or W303BLa with plasmid pASZ10 (Stotz and Linder, 1990) that had been linearized by BglII. To integrate ARS418 into the iYDR09c locus, strain SB1507 was transformed with BsmI-linearized pLys2-418-309CB (SB2023) or pLys2-mut418-309CB (SB2052). To integrate ARS214 into the iYDR309C locus, strain SB1507 was transformed with MluI-linearized pLys2-214-309C to create SB2125.
Re-replication Microarray Assays
Exponentially growing cells (OD600 of 0.4) were washed with sterile water and transferred into YP-raffinose + 15 μg/ml nocodazole. Once arrested, 2% galactose was added to induce cdc6_Δ_2-49 expression. After 3 h, cells were collected and genomic DNA was isolated by bead beating. Briefly, whole cells were mixed with 200 μl of gDNA buffer (10 mM Tris, pH 7.5, 1% SDS, 100 mM NaCl, 1 mM EDTA, 2% Triton X-100), 300 μl of glass beads, and 200 μl of phenol:chloroform:isoamyl-alcohol (25:24:1, GE Healthcare) and vortexed for 4 min. The DNA in the aqueous phase was precipitated and resuspended in 200 μl of TE (10 mM Tris, 1 mM EDTA). RNA was removed with RNAse (3 μg) treatment for 3 h 37°C. The DNA was then sheared to ∼1 kb (Branson Sonicator 250, Danbury, CT), phenol:chloroform extracted, and EtOH precipitated.
DNA from re-replicating cells and G1-arrested wild-type cells (10 μg each) were differentially labeled with 2 nmol of either Cy3-dUTP or Cy5-dUTP (Amersham Biosciences, Piscataway, NJ; GE Healthcare) using 4 μg random nonamer oligo (IDT) and 0.25 λ of high-concentration Klenow (New England Biolabs, Beverly, MA). Unincorporated dye was removed using a microcon column (30-kDa MW cutoff, Millipore, Bedford, MA) by washing the sample three times with TE. The labeled DNAs were then cohybridized onto either 11K or custom-made (Pokholok et al., 2005) 44K DNA microarrays from Agilent Technologies (Wilmington, DE) using Agilent Technologies' standard protocol for cDNA hybridization and washing (see http://www.ncbi.nlm.nih.gov/geo/ accession no. GPL3499 for a complete description of the arrays). For each set of triplicate experiment, one of the replicates was labeled as a dye swap.
Hydroxyurea-arrested Replication Profiles
W303 cells were arrested in 200 mM hydroxyurea (HU) for 90 min and then collected. Genomic DNA was then isolated, labeled, and hybridized to high-density DNA miroarrays as described above. DNA from G1-arrested cells was used as a reference population.
Density Transfer
Cells (SB1808 and SB1809) were grown for at least seven generations in 15N- and 13C-containing medium to an OD600 0.25. Alpha-factor was added and cells were grown until the population was ≥95% unbudded. Cells were then washed and resuspended in YP (14N 12C) + 2% raffinose + alpha-factor. After 1 h, cells were washed twice with water and released in to YP (14N 12C) + 2% raffinose + 0.1 mg/ml pronase + 15 μg/ml nocodazole. When the population was ≥95% large-budded, galactose was added to 2% to induce re-replication. After 3 h, 30-ml samples were collected.
DNA was isolated as described above and digested for 8 h with EcoRI at 37°C. The digested DNA was separated on a CsCl gradient (1.255g CsCl/g TE, refractive index = 1.4041). The resulting gradient was fractionated and each fraction was slot-blotted onto a nylon membrane (GeneScreen Plus, PerkinElmer, Boston, MA). The membrane was then probed using the indicated radio-labeled origin fragments (see Supplementary Table 1 for primers used to generate probes).
Genome-wide Location Analysis
Standard chromatin immunopreciptation (IP) assay was performed as previously described (Aparicio et al., 1997) at specific time points using a polyclonal antibody against Mcm2-7, UM185, (1:250 dilution) or ORC (1:250 dilution). The resulting IP DNA and one tenth of the input DNA were differentially labeled and cohybridized to custom-made 44K DNA microarrays from Agilent Technologies.
Data Analysis
Cy3 and Cy5 levels were quantitated using Agilent's Feature Extraction software. The resulting log ratios of experimental DNA/reference DNA for each spot on the array were then determined using the sma package [45] for R (v2.1.0, http://www.r-project.org), which is a computer language and environment for statistical computing. We also performed scale normalization across the slides for each set of triplicate experiments so each experiment had the same median absolute deviation.
For all replication profiles (HU and re-replication), the average log ratio of enrichment for each spot on the array was calculated for three independent experiments. The resulting average was used for all subsequent analysis. The averaged data were smoothed using the loess function in R to predict the average log ratio of experimental DNA/reference DNA every 50 bp.
Sites of absolute re-replication were defined as any spot having a log ratio re-replicated/unreplicated value above the bottom quarter percentile. The bottom quarter percentile represents the midpoint of the normal distribution of the re-replication data (Supplementary Figure 9A) and was used as a cutoff across the entire genome rather than determining the lowest site on each individual chromosome (Supplementary Figure 9B). The value of the log ratio replicated/unreplicated that defined the threshold of the bottom quarter percentile was then added to the log ratio replicated/unreplicated ratio for all spots on the array. All spots with a final value over zero were considered to represent re-replicated regions.
Genome-wide location analysis for both mitotic and re-replication Mcm2-7 was performed in triplicate. Data from the individual experiments were treated with the loess function to predict the log ratio IP DNA/input DNA (IP/IN) every 50 bp. Peaks on the smoothed and/or predicted data sets were determined using the turnpoints function in the pastecs package (v1.2-1) in R. True peaks in the genome-wide location analysis data sets were defined by three independent criteria: a confidence value >80 given by the turnpoints function, the log ratio IP/IN value at the peak p < 0.001, and that there was another point within 2 kb whose log ratio IP/IN had p < 0.05. The last criterion was to prevent identification of false peaks that arose because of gaps in the array data. True peaks in the replication data sets were defined as peaks that had the highest confidence values (infinite) and had a log ratio replicated/unreplicated value at the peak that was greater than 0. True peaks in the HU data set were defined as peaks that had the highest confidence value and had a log ratio replicated/unreplicated at the peak with p < 0.001.
Comparison of peaks between data sets was done by scanning each of the data sets for all true peaks on a chromosome and determining if a true peak in another data set was within 7.5 kb Averaged and raw data sets are available online in the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/, accession no. GSE4487).
RESULTS
Re-replication Initiates at Distinct Sites in the Genome
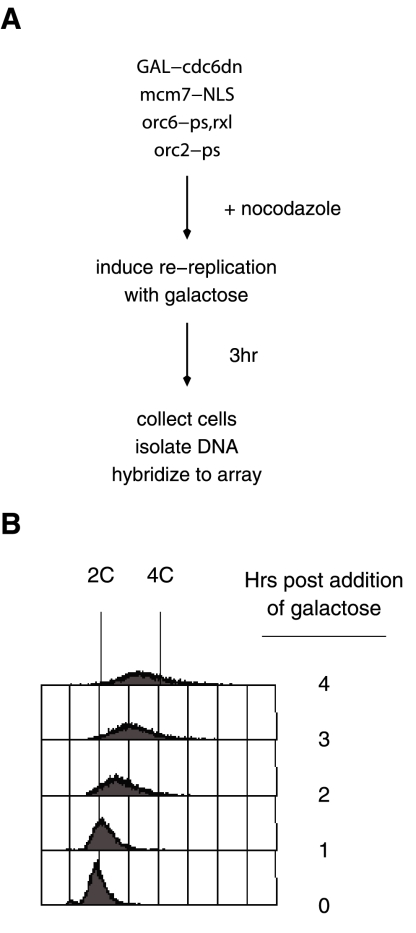
To assess the extent of re-replication across the S. cerevisiae genome, we used DNA microarrays to determine changes in DNA copy number as cells underwent re-replication. This technique has been used previously to identify sites of replication initiation by detecting newly synthesized DNA as cells pass through S-phase (Raghuraman et al., 2001; Yabuki et al., 2002). Our experiments were conducted using an S. cerevisiae strain with mutations that overcome all currently known mechanisms that prevent reinitiation (Wilmes et al., 2004). Re-replication in this strain is controlled using a galactose-inducible, nondegradable Cdc6. To ensure that all observed replication was due to re-replication, cells were arrested in G2/M before the induction of re-replication (Figure 1A). DNA was isolated from re-replicating cells at various time points after addition of galactose (Figure 1B). Unreplicated DNA isolated from G1-arrested cells served as a hybridization reference. The two populations of DNA were differentially labeled and cohybridized to a high-density DNA microarray with 44,000 features distributed throughout the genome (Pokholok et al., 2005). Initial experiments showed that cells after 3 h of Cdc6 induction had the most significant re-replication (Figure 1B), thus this time point was used in all subsequent experiments.
Figure 1.
Multiple pre-RC mutations result in induced re-replication. (A) An outline of the re-replication experiment. Re-replication-sensitive cells were grown to an OD600 of 0.4 in YPD and then arrested in nocodazole. After the cells were arrested, galactose was added to induce expression of Cdc6ΔN. After 3 h, cells were collected for further experiments. (B) FACS analysis of the re-replication-sensitive strain SB1507 (See Supplementary Table 2) several hours after induction of re-replication.
Analysis of three independent experiments showed that re-replication occurs at specific sites in the genome. To visualize the sites of re-replication, the log ratio of re-replicated/unreplicated DNA for each spot on the array was plotted as a function of its position along the chromosome (Figure 2A). The resulting profiles have distinct peaks, identifying sequences present in elevated copy number and that have re-replicated. Control experiments using strains lacking the genetic changes required for re-replication showed no significant variation in DNA copy number across the genome (Supplementary Figure 1).
Figure 2.
Analysis of genome-wide re-replication. (A) Re-replication is detected by copy number analysis using DNA microarrays. DNA from re-replicating cells and from G1-arrested cells was differentially labeled and cohybridized to a high-density DNA microarray. The log ratio re-replicated/unreplicated for each spot was plotted as a function of its position along the chromosome. Chromosome IV (see Supplementary Figure 2 for other chromosomes) is shown here as an example. (B) Sites of re-replication initiation are associated with G1 Mcm2-7 binding sites. A smoothing algorithm and a significance cutoff was applied to the re-replication data (see Materials and Methods) and plotted here for Chromosome IV (gray histogram). G1 Mcm2-7 binding sites (black histogram), determined by genome-wide location analysis, are superimposed on top of the re-replication data. Key sites discussed throughout the text, ARS1, ARS418, ARS428, and iYDR309C, are marked with a gray dashed line.
Re-replication Initiates from Sites of G1 pre-RC Formation
Peaks in the re-replication profile represent the most frequently re-replicated sequences, suggesting that they are sites of reinitiation. To determine if these sites are coincident with previously identified, potential origins, we compared the re-replication profile to sites of G1 pre-RC formation in wild-type cells as determined by genome-wide location analysis of Mcm2-7. This comparison allowed us to determine if peaks of re-replication colocalized with sites that have the capability to initiate replication during S-phase.
To compare the re-replication profile and G1 Mcm2-7 binding sites, we determined the midpoint of the peaks in each of the data sets. Before analysis, we applied a smoothing algorithm to the re-replication profile to help delineate the peaks by reducing random noise (Figure 2B, gray histogram, Supplementary Figure 2). Initial analysis showed that peaks on the re-replication profile substantially overlapped sites of Mcm2-7 binding (Figure 2B, black histogram, Supplementary Figure 3). To conduct a more quantitative analysis, a peak-finding algorithm was used to define the mid-points of the peaks along the chromosome in both data sets. We monitored the overlap between peaks on the re-replication profile and sites of Mcm2-7 binding using a range of window sizes and found that a 7.5-kb window was optimal. Using this window size, 82% of re-replication peaks overlapped with Mcm2-7 binding sites (Table 1). We noted that the peak-finding algorithm was not able to identify all sites of re-replication and thus possible reinitiation (e.g., peaks at chromosome ends; see Supplementary Figure 4). Accounting for these uncalled initiation sites in the re-replication data set, the final percent of re-replication peaks that are within 7.5 kb of an Mcm2-7 binding site is 91%. We conclude that re-replication largely occurred at sites that normally direct pre-RC formation during G1.
Table 1.
Comparison of overlap of peaks between all data sets
| Data set | Total no. of peaksa | % of peaks that overlap with G1 Mcm2-7 peaks | % of peaks that overlap with re-replication peaks | % of peaks that overlap with re-replication Mcm2-7 peaks | % of peaks that overlap with HU peaks | % of peaks that overlap with re-replication ORC peaks |
|---|---|---|---|---|---|---|
| G1 Mcm2-7 | 377 | 32 (120)c | 45 (169) | 33 (125) | 76 (285) | |
| Re-replication profile | 123 | 82 (101)b | 71 (87) | 48 (59) | 83 (102) | |
| Re-replication Mcm2-7 | 183 | 92 (169) | 51 (93) | 40 (74) | 93 (171) | |
| HU profile | 114 | 98 (112) | 52 (59) | 63 (72) | 96 (109) | |
| Re-replication ORC | 331 | 86 (284) | 35 (117) | 52 (171) | 34 (114) |
Although nearly all sites of re-replication overlapped with Mcm2-7 binding sites, the converse was not true. There were many sites of G1 pre-RC formation that did not align with peaks of re-replication. Using the same computational-based analysis as described above, we found that only 31% of all sites of pre-RC formation during G1 showed significant re-replication (Table 1). Together, our data show on a genome-wide level that not all potential sites of initiation can re-replicate and, therefore, that the extent of protection from re-replication is not uniform across the genome.
Origins Direct Re-replication
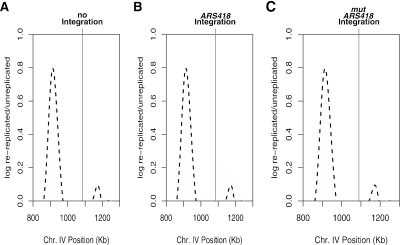
To directly address if origin sequences are required for re-replication, we asked if moving an origin sequence associated with a peak of re-replication was sufficient to establish a new site of re-replication in the genome. These experiments focused on the ARS418 locus, which is a site of G1 pre-RC formation (Figure 2B, black histogram), provides origin function on a plasmid (unpublished data), is a peak on an S-phase timing curve (Raghuraman et al., 2001; Yabuki et al., 2002; MacAlpine and Bell, 2005 and unpublished data), and is closely associated with a prominent peak on the re-replication profile (Figure 2B, gray histogram). Six hundred base pairs surrounding the ARS418 locus were integrated at an ectopic intergenic region (iYDR309C) that showed little, if any, re-replication (Figure 3A, gray histogram and see Figure 5B, closed circles). Using the re-replication-sensitive strain containing the ectopic ARS418, we performed the same re-replication experiment described above. The resulting re-replication profile showed that the insertion of ARS418 at the iYDR309C locus induced substantial re-replication (Figure 3B, gray histogram) compared with the strain without the ectopic ARS418 (Figure 3, A and B, dashed line). We also ectopically inserted 200 base pairs surrounding another re-replicating origin into iYDR309C: ARS214 (see dotted black line on Chromosome II in Supplementary Figure 2 for location). This second origin also induced re-replication at iYDR309C (Supplementary Figure 5). Thus, moving only origin-proximal DNA is sufficient to direct re-replication at a new locus.
Figure 3.
An origin sequence directs reinitiation (A) The re-replication profile surrounding iYDR309C, a segment that does not re-replicate, in the absence of an ectopic origin is depicted as both the gray histogram and the black dashed line. (B) ARS418, an origin that is associated with a peak on the re-replication profile, directs re-replication at an ectopic locus. The 600-base pair intergenic region containing ARS418 was moved to iYDR309C. Re-replication was induced and the resulting DNA was hybridized to a low-density DNA microarray (gray histogram, Chromosome IV, 800-1400 kb). Superimposed on top is the re-replication profile from the strain without the ectopic ARS418 (black dashed line). (C) An origin with a mutant ACS is not capable of directing re-replication. The essential ACS of ARS418 was mutated and integrated into the re-replicating strain. Re-replication was induced and the resulting DNA hybridized on to a low-density array (gray histogram, Chromosome IV, 800-1400 kb). The re-replication profile of the strain without the ectopic mutant ARS418 is superimposed on top (black dashed line).
Figure 5.
Origins are capable of reinitiating multiple times. (A) Diagram of density transfer experiment. A cartoon depicts what products will look like during the experiment with “heavy” DNA strands shown in black and “light” DNA strands shown in gray. The table briefly describes possible results. (B) ARS428 and ARS418 reinitiate multiple times. DNA from cells that underwent the density transfer protocol described in A was fractionated by CsCl gradient. The resulting fractions were probed for three different classes of DNA sequences as determined by DNA microarray analysis: two origins of reinitiation (ARS418, ▴; ARS428, □), two origins that are re-replicated, but are not sites of reinitiation (ARS1, ▪; ARS1413, ○) and an intergenic sequence that does not re-replicate (iYDR309C, •). The data were normalized by setting the peak of the HL density to a copy number of one.
To demonstrate that the origin was necessary for the new re-replication peak, we mutated the ectopically inserted ARS418 so that it was no longer functional. Our laboratory recently refined an algorithm (Breier et al., 2004) to identify functional ARS Consensus Sequences (ACS) across the S. cerevisiae genome (our unpublished results). Using this algorithm, we predicted the site of the essential ACS of ARS418 and mutated this sequence. This mutation eliminated the function of ARS418 on a plasmid (unpublished data). The mutant ARS418 was integrated at iYDR309C and the re-replication of this strain was analyzed by microarray (Figure 3C, gray histogram). Unlike the wild-type ARS418, the mutant ARS418 did not induce re-replication at iYDR309C, showing that the same sequence that is required for origin function in a plasmid context during normal S-phase is also required to direct re-replication. This observation is consistent with previously published data concerning ARS305 (Nguyen et al., 2001).
Timing of Initiation during S-phase Does Not Correlate with the Ability to Re-replicate
Having demonstrated that sites of reinitiation correspond to a subset of potential origins, we asked if sites of reinitiation represented a particular class of origins. We compared origins that reinitiate to the time of initiation of those same origins during S-phase. We used a previously described protocol (Yabuki et al., 2002) to identify origins that initiated in the presence of HU (Supplementary Figure 3). HU allows early origins to initiate but inhibits activation from later-initiating origins of replication (Santocanale and Diffley, 1998; Shirahige et al., 1998).
Comparing the profile generated in the presence of HU with the re-replication profile showed that some of the reinitiating origins are early, but not all. Similarly, there are early origins that do not re-replicate. Using a window of 7.5 kb and computationally based analysis, 48% of re-replication peaks are associated with HU-initiating origins (Table 1). Conversely, 52% of HU-initiating origins re-replicate. These data suggest that there is not a strong correlation between origins that re-replicate and when that origin initiates during S-phase. Thus, the factors that determine timing of initiation in S-phase are not the same as the factors that sensitize origins to re-replication during G2/M.
The telomeres and centromeres of the S. cerevisiae genome are specialized regions of the genome that replicate at specific times during S-phase (telomeres replicate late, whereas centromeres replicate early), so we also analyzed the ability of these regions to re-replicate. The subtelomeric chromosomal regions appeared overrepresented in the re-replicated fraction of the DNA (Supplementary Figure 2). To examine this feature further, we plotted the relative level of re-replication for each point on the array as a function of its distance from the telomere (Figure 4A, black plot). For comparison, we plotted the relative level of re-replication for a wild-type strain under re-replicating conditions (Figure 4A, gray plot). The resulting plot showed a positive correlation between the proximity of a sequence to the telomere and its extent of re-replication. We also plotted each point on the array as a function of its distance from the centromere (Figure 4B, black plot) and found that there was no correlation between distance from the centromere and sensitivity to re-replication.
Figure 4.
Subtelomeric regions have a high probability of re-replicating. (A) There is a positive correlation between the proximity of a sequence to the telomere and its probability of re-replicating. The relative enrichment for each spot on the microarray was plotted as a function of its distance to the closest telomere for both the re-replicating strain (black) and wild-type strain (gray) 3 h after addition of galactose. (B) There is no correlation between re-replication and proximity to centromeres. The relative enrichment for each spot on the microarray was plotted as a function of its distance to the centromere for both the re-replicating strain (black) and wild-type strain (gray) 3 h after addition of galactose.
Origins Can Reinitiate Multiple Times
FACS analysis 3 h after induction of re-replication shows that most cells in the population have ∼3C DNA content; however, some cells appear to have DNA content greater than 4C (Figure 1B). The existence of cells with >4C DNA content suggests that at least a subset of origins is capable of multiple rounds of reinitiation. To determine if origins can reinitiate more than once, we used a density transfer approach to more accurately determine the extent of re-replication at particular regions.
Cells were labeled with dense isotopes as outlined in Figure 5A. As illustrated, at the nocodazole arrest, cells will have passed through S-phase and therefore have one heavy and one light DNA strand. Induction of re-replication in the nocodazole-arrested cells will result in a third species of DNA composed of entirely light DNA strands. If a segment of DNA re-replicates exactly once then the ratio of Light-Light (LL) DNA to Heavy-Light (HL) DNA will be 1:1. If a segment has re-replicated more than once, the ratio will increase.
We examined several sites that represented different features of the re-replication profile to determine their extent of re-replication. We tested two origins that were prominent sites of reinitiation (ARS418 and ARS428; see Figure 2B), two origins that did not seem to be efficient sites of reinitiation (ARS1 and ARS1413; see Figure 2B and Supplementary Figure 2), and one sequence that was not substantially re-replicated, iYDR309C (Figure 2B). Consistent with their prominence in the re-replication profiles, both ARS418 and ARS428 have at least twice as much LL DNA as HL DNA (Figure 5B, closed triangles and open squares). Consistent with the re-replication profile, these data definitively demonstrate that some origins are capable of reinitiating multiple times. iYDR309C, however, showed no LL DNA, indicating that other regions of the genome do not re-replicate at all. Together, these data strongly support the model that re-replication is limited across the genome but that origins that reinitiate can do so more than once.
Pre-RC Formation Is Not the Only Determinant of the Ability to Re-replicate
We have shown that not all sites of G1 pre-RC formation reinitiate. Because previous data strongly suggest that Mcm2-7 loading onto origin DNA is required for reinitiation (Nguyen et al., 2001), there are two possible explanations for only a subset of these G1 pre-RC sites undergoing re-replication. First, it is possible that Mcm2-7 is only recruited to those origins that reinitiate. Alternatively, similar to the pre-RCs assembled in G1, Mcm2-7 could load at all potential origins, but only a subset is competent to reinitiate. To distinguish between these hypotheses, we asked where pre-RCs were formed during re-replication using Mcm2-7 genome-wide location analysis. To avoid confusing sites of pre-RC formation with fork movement, samples were taken 45 min after induction when re-replication is limited as determined by FACS (Figure 1B) and array analysis (unpublished data).
We first asked if Mcm2-7 binds to the same sites during re-replication as seen during G1. Because both genome-wide location analysis data sets have narrow peaks (compared with the re-replication profile), we could use a much smaller window when comparing G1 and re-replication Mcm2-7 binding sites. Using a 1-kb window, 92% of the re-replication Mcm2-7 binding sites overlap with G1 Mcm2-7 binding sites (Table 1). In contrast, only 45% of G1 Mcm2-7 binding sites overlap with re-replication Mcm2-7 binding sites (Figure 6A, Table 1, and Supplementary Figure 3), demonstrating that only a subset of sites that assemble pre-RCs in G1 also do so in re-replicating cells.
Figure 6.
Recruitment of Mcm2-7 is not sufficient for reinitiation. (A) Mcm2-7 binds only a fraction of possible origins during re-replication. Genome-wide location analysis of Mcm2-7 and ORC was performed 45 min after induction of re-replication. The binding sites of Mcm2-7 during re-replication (•) were compared with binding sites of ORC during re-replication (dark gray circles) and binding sites of Mcm2-7 during G1 (light gray circles). Plotted are only the points on the array that satisfied the significance cutoff (see Materials and Methods) for each of the data sets. (B) Mcm2-7 binds to origins that do not reinitiate. The binding sites of Mcm2-7 (black histogram) are overlaid on top of the re-replication profile for Chromosome IV (gray histogram). Each peak of re-replication is associated with an Mcm2-7 binding site, but the converse is not true.
We were concerned that Mcm2-7 associated with a subset of origins during re-replication because in the re-replication-sensitive strain, which has several ORC mutations, ORC only associated with the same subset of origins. To determine the location of ORC binding during re-replication, we performed ORC genome-wide location analysis as described above (Figure 6A and Supplementary Figure 3). We found that the majority of G1 Mcm2-7 binding sites overlap with sites of re-replication ORC binding sites (Table 1), suggesting that ORC containing two nonphosphorylatable subunits can bind to most potential origins. Therefore, ORC binding does not limit Mcm2-7 loading. Similar to G1 Mcm2-7 binding sites, only 52% of re-replication ORC binding sites are associated with a re-replication Mcm2-7 binding site. Thus the reduction in pre-RC formation during re-replication is not due to a reduced number of ORC binding sites.
We then determined how many sites of reinitiation overlap with re-replication Mcm2-7 binding sites. We used the same approach to compare these two data sets as when we compared the re-replication profile to G1 Mcm2-7 binding sites. We found that 71% of the re-replication profile peaks overlapped with a re-replication Mcm2-7 peak within a 7.5-kb window (Table 1). Taking into account the peaks that were not identified by the peak-finding algorithm (Supplementary Figure 4), the percentage increased to 80%. These comparisons show that Mcm2-7 is found at most sites of reinitiation, supporting the model that Mcm2-7 is required at origins that reinitiate.
We then asked what percentage of re-replication Mcm2-7 binding sites overlapped with sites of re-replication. Fiftyone percent of re-replication Mcm2-7 binding sites overlapped with re-replication peaks (Table 1), suggesting that only a subset of sites that exhibit Mcm2-7 association during induced re-replication go on to reinitiate (Figure 6B and Supplementary Figure 3). We also measured the interorigin distance between sites of re-replication as well as the distance between pre-RC binding during re-replication (Supplementary Figure 5). The median distance between origins that initiate during re-replication is 84 kb, but the median distance between Mcm2-7 binding sites during re-replication is only 57 kb. Thus, there are substantially more Mcm2-7 binding sites during induced re-replication than there are re-replication initiation sites.
These data support the first hypothesis presented above, which stated that reinitiation was limited to sites that load Mcm2-7 during re-replication. We found, however, that loading of Mcm2-7 was not sufficient to induce reinitiation because there were many origins throughout the genome that loaded Mcm2-7 but did not re-replicate (Figure 6B). With respect to the ability to re-replicate, sites of G1 pre-RC formation can be grouped into three classes: those that do not form pre-RCs during re-replication, those that form pre-RCs but do not reinitiate, and those that form pre-RCs and reinitiate. The recruitment of Mcm2-7, therefore, is not the only obstacle to re-replication and there must be other levels of control that act after pre-RC formation to prevent reinitiation.
DISCUSSION
Prevention of re-replication during a single cell cycle is critical for cell survival. Without such control, cells undergo gross chromosomal damage (Green and Li, 2005) and eventually death (Nguyen et al., 2001). Here, we have monitored the increase in DNA copy number and pre-RC formation during re-replication of the S. cerevisiae genome. We found that re-replication initiates from specific sites in the genome and that these sites are coincident with origins of replication. Our findings allow us to categorize origins with respect to their propensity to reinitiate and demonstrate that pre-RC formation is not the only target for mechanisms that prevent genomic re-replication.
In the course of these studies, we determined that at least 123 sites in the genome are capable of reinitiation. Concurrent with this study, Li and colleagues performed an analogous study (Green et al., 2006). A comparison between the results from each group show that 53% of our reinitiating sites (65 total) overlap with a reinitiating site in the Green et al. data set within 10 kb. There are a number of factors that might lead to a less than perfect correlation, including different strain backgrounds, a mutation that induces additional re-replication in the strain analyzed in this study, and different numbers of features on the DNA microarray slides (this study: ∼44,000; Green et al.: ∼13,000). We expect that the more sensitive reinitiation sites would be more likely to overlap. Indeed, of the 30 most-efficient reinitiation sites in our data set, 77% overlap with a peak in the Green et al. data set within 10 kb. This is more than a 25% increase over the entire data set, suggesting that many of the same origins reinitiate in both studies despite the differences in strain background, mutations, methodology, and analysis.
Limited Replication Fork Processivity Prevents Complete Genome Re-replication
The extent of re-replication varies widely over the genome, including substantial regions that show little or no re-replication. The differences in the amount of re-replication are likely to be due to a combination of asynchronous re-replication, inefficient re-replication, low replication fork processivity (see below), and the ability of some sequences to reinitiate more than once. The height of the peaks reflects two features of a re-replicating origin: 1) the percentage of cells in which each origin reinitiated, and 2) the number of rounds of reinitiation the associated origin(s) underwent (see Figure 5).
The lack of full-genome re-replication suggests that the replication forks derived from flanking origins stop before replicating the intervening DNA. These data are consistent with previously reported 2D-gel data (Nguyen et al., 2001), suggesting that replication forks have trouble reaching a site 30-35 kb from an origin. The inability of forks derived from adjacent origins to fully replicate intervening regions could be due to a reduced number of sites of initiation or from reduced processivity of forks. Although there is a notable increase in the interorigin distance during re-replication (84 kb compared with 43 kb in S-phase; see Supplementary Figure 5), this change cannot fully explain the incomplete re-replication. It is known that origins separated by 100 kb can direct the replication of the intervening DNA without affecting chromosome stability or cell viability (Dershowitz and Newlon, 1993). Thus, reduced fork processivity must play a role in the incomplete nature of re-replication.
Multiple factors could contribute to reduced fork processivity during re-replication. One possibility is that fork processivity could be affected by changes in chromatin that occur during G2/M. Alternatively, the “forks chasing forks” generated after multiple initiation events from the same origin could contribute to reduced processivity. Recent studies showed that the DNA damage response is elicited in S. cerevisiae when re-replication is induced (Archambault et al., 2005; Green and Li, 2005). Both groups proposed that one likely source for damaged DNA was fork collapse after two replication forks followed one another too closely. This idea is supported by data mapping replication intermediates from the chorion amplicon in Drosophila melanogaster, which suggested that multiple initiation events impeded fork movement (Claycomb et al., 2002). Consistent with the latter model, our density transfer experiments show that multiple rounds of reinitiation occur at a subset of origins (Figure 5).
What Determines Origin Sensitivity to Reinitiation?
Our studies clearly show that the sequences within a few hundred base pairs of an origin are sufficient to direct reinitiation. This is in contrast to the sequence determinants that control replication timing (Friedman et al., 1996), which include large regions of DNA (>10 kb) surrounding the origin. Consistent with this difference in sequence determinants, we did not observe a correlation between an origin's ability to re-replicate and its time of replication in S-phase.
Although origin-proximal sequences are sufficient to direct ectopic re-replication, the site of insertion may influence the extent of the resulting re-replication. For example, we see that the efficiency of re-replication directed by ARS418 is reduced when ARS418 is inserted at the ectopic locus (Figure 3b). This difference suggests that the surrounding chromatin structure influences the efficiency of re-replication. We noted that a nearby site showed increased re-replication after ARS418 insertion (Figure 3B). We do not know if this increase is due to passive re-replication by replication forks derived from the ectopic origin or if the ectopic origin stimulated reinitiation at this neighboring site.
One case in which there may be a more global influence on the sensitivity to re-replication is at the telomeres. We found that proximity to telomeres was associated with an increased likelihood of re-replication. One possible reason for this particular sensitivity to re-replication is the high density of pre-RC formation at telomeres (Supplementary Figure 2; Wyrick et al., 2001).
Formation of a pre-RC Is Not Sufficient to Induce Re-replication during G2
Only a subset of the sites that assemble pre-RCs during the induction of re-replication go on to initiate. It is possible that the origins that load pre-RCs but do not reinitiate are simply S-phase inactive origins. Unlike many inactive origins in S-phase, however, in numerous instances these sites of pre-RC formation are never re-replicated and therefore are not inactivated by passive replication. Additionally, several of the sites that assemble pre-RCs but do not reinitiate overlap with active S-phase origins (e.g., Chromosome V at 406,996 base pairs and Chromosome XI at 257,488 base pairs; MacAlpine and Bell, 2005). Thus far, the described mechanisms in every organism that prevent re-replication target pre-RC formation (Blow and Dutta, 2005; Machida et al., 2005). As discussed below, our results indicate the existence of unidentified mechanisms in S. cerevisiae that prevent inappropriate pre-RC formation and activation of licensed origins outside of S-phase.
The presence of many ORC binding sites during re-replication that are not associated with Mcm2-7 suggests that even in the re-replication-sensitive strain there are still intact controls preventing pre-RC formation. There are several possible targets for this residual regulation. For example, only two of the three ORC subunits that have CDK phosphorylation sites are mutated to be nonphosphorylatable in the re-replication-sensitive strain. It is possible that CDK-dependent phosphorylation of the third phosphorylated ORC subunit, Orc1, can prevent pre-RC formation at some potential origins. Pre-RC formation also requires the presence of Cdt1 (Maiorano et al., 2000). In S. cerevisiae a role for Cdt1 in preventing re-replication has not been identified, although it has been shown to be important in other organisms (Blow and Dutta, 2005). If Cdt1 is limiting during re-replication, this could prevent efficient Mcm2-7 loading onto origins.
Although residual mechanisms preventing pre-RCs from forming likely exist, they do not explain why Mcm2-7 can load more efficiently at some origins rather than others. One possibility is that pre-RC components, other than ORC, are excluded from associating with certain origins due to a change in chromatin structure. As cells proceed toward mitosis, the chromatin undergoes structural changes due to cohesion, condensation and changes in the transcriptional program. The effect of these changes may alter the local chromatin structure surrounding certain origins, making them inaccessible to pre-RC formation.
The numerous sites of pre-RCs formation that do not reinitiate indicate that there are levels of re-replication control that prevent pre-RC activation rather than pre-RC formation. One such control could be the alterations in chromatin structure as discussed above. Although local changes may not hinder pre-RC association, they may exclude association of downstream replication factors (Mendez and Stillman, 2003). Alternatively, factors required for pre-RC activation may be limiting during G2/M. Finally, recent reports (Archambault et al., 2005; Green and Li, 2005) have shown that the DNA damage response is activated, including the Rad53 kinase, in S. cerevisiae during re-replication. Because activated Rad53 has been shown to suppress origin activation in some circumstances, it is possible that the activation of Rad53 during the DNA damage response suppresses reinitiation from some origins. Future experiments will be necessary to address whether these or other, as of yet unknown mechanisms, provide further safeguards against activation of initiation to prevent re-replication during the same cell cycle.
Supplementary Material
[Supplemental Material]
Acknowledgments
We thank Rick Young for providing access to the design of the 44K Agilent DNA microarrays and Tony Lee for technical help using the DNA microarrays; Milan de Vries, Erik Andersen, Joachim Li, Terry Orr-Weaver, and Angelika Amon for helpful discussion and comments on the manuscript; and Brian Green and Joachim Li for sharing unpublished data. The work described here was supported by a grant from the National Institutes of Health to S.P.B. (GM52339). S.P.B. and D.M.M. are employees of the Howard Hughes Medical Institute. H.G.B. was supported by a predoctoral fellowship from the Howard Hughes Medical Institute.
Abbreviations used: ORC, origin recognition complex; pre-RC, pre-replicative complex; CDK, cyclin-dependent kinase; ACS, ARS consensus sequence; HU, hydroxyurea.
References
- Aparicio, J. G., Viggiani, C. J., Gibson, D. G., and Aparicio, O. M. (2004). The Rpd3-Sin3 histone deacetylase regulates replication timing and enables intra-S origin control in Saccharomyces cerevisiae. Mol. Cell Biol. 24, 4769-4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio, O. M., Weinstein, D. M., and Bell, S. P. (1997). Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell 91, 59-69. [DOI] [PubMed] [Google Scholar]
- Archambault, V., Ikui, A. E., Drapkin, B. J., and Cross, F. R. (2005). Disruption of mechanisms that prevent rereplication triggers a DNA damage response. Mol. Cell Biol. 25, 6707-6721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, S. P., and Dutta, A. (2002). DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71, 333-374. [DOI] [PubMed] [Google Scholar]
- Blow, J. J., and Dutta, A. (2005). Preventing re-replication of chromosomal DNA. Nat. Rev. Mol. Cell Biol. 6, 476-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breier, A. M., Chatterji, S., and Cozzarelli, N. R. (2004). Prediction of Saccharomyces cerevisiae replication origins. Genome Biol. 5, R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claycomb, J. M., MacAlpine, D. M., Evans, J. G., Bell, S. P., and Orr-Weaver, T. L. (2002). Visualization of replication initiation and elongation in Drosophila. J. Cell Biol. 159, 225-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dershowitz, A., and Newlon, C. S. (1993). The effect on chromosome stability of deleting replication origins. Mol. Cell Biol. 13, 391-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley, J. F. (2004). Regulation of early events in chromosome replication. Curr. Biol. 14, R778-R786. [DOI] [PubMed] [Google Scholar]
- Donaldson, A. D. (2005). Shaping time: chromatin structure and the DNA replication programme. Trends Genet. 21, 444-449. [DOI] [PubMed] [Google Scholar]
- Donaldson, A. D., Raghuraman, M. K., Friedman, K. L., Cross, F. R., Brewer, B. J., and Fangman, W. L. (1998). CLB5-dependent activation of late replication origins in S. cerevisiae. Mol. Cell 2, 173-182. [DOI] [PubMed] [Google Scholar]
- Drury, L. S., Perkins, G., and Diffley, J. F. (2000). The cyclin-dependent kinase Cdc28p regulates distinct modes of Cdc6p proteolysis during the budding yeast cell cycle. Curr. Biol. 10, 231-240. [DOI] [PubMed] [Google Scholar]
- Elsasser, S., Chi, Y., Yang, P., and Campbell, J. L. (1999). Phosphorylation controls timing of Cdc6p destruction: a biochemical analysis. Mol. Biol. Cell 10, 3263-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman, K. L., Diller, J. D., Ferguson, B. M., Nyland, S. V., Brewer, B. J., and Fangman, W. L. (1996). Multiple determinants controlling activation of yeast replication origins late in S phase. Genes Dev. 10, 1595-1607. [DOI] [PubMed] [Google Scholar]
- Gopalakrishnan, V., Simancek, P., Houchens, C., Snaith, H. A., Frattini, M. G., Sazer, S., and Kelly, T. J. (2001). Redundant control of rereplication in fission yeast. Proc. Natl. Acad. Sci. USA 98, 13114-13119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, B. M., and Li, J. J. (2005). Loss of rereplication control in Saccharomyces cerevisiae results in extensive DNA damage. Mol. Biol. Cell 16, 421-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, B. M., Morreale, R. J., Özaydin, B., DeRisi, J. L., and Li, J. J. (2006). Genome-wide mapping of DNA synthesis in Saccharomyces cerevisiae reveals that mechanisms preventing reinitiation of DNA replication are not redundant. Mol. Biol. Cell 17, 2401-2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, F., and Aparicio, O. M. (2005). Swe1 regulation and transcriptional control restrict the activity of mitotic cyclins toward replication proteins in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 102, 8910-8915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib, K., Diffley, J. F., and Kearsey, S. E. (1999). G1-phase and B-type cyclins exclude the DNA-replication factor Mcm4 from the nucleus. Nat. Cell Biol. 1, 415-422. [DOI] [PubMed] [Google Scholar]
- MacAlpine, D. M., and Bell, S. P. (2005). A genomic view of eukaryotic DNA replication. Chromosome Res. 13, 309-326. [DOI] [PubMed] [Google Scholar]
- Machida, Y. J., Hamlin, J. L., and Dutta, A. (2005). Right place, right time, and only once: replication initiation in metazoans. Cell 123, 13-24. [DOI] [PubMed] [Google Scholar]
- Maiorano, D., Moreau, J., and Mechali, M. (2000). XCDT1 is required for the assembly of pre-replicative complexes in Xenopus laevis. Nature 404, 622-625. [DOI] [PubMed] [Google Scholar]
- Melixetian, M., Ballabeni, A., Masiero, L., Gasparini, P., Zamponi, R., Bartek, J., Lukas, J., and Helin, K. (2004). Loss of Geminin induces rereplication in the presence of functional p53. J. Cell Biol. 165, 473-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez, J., and Stillman, B. (2003). Perpetuating the double helix: molecular machines at eukaryotic DNA replication origins. Bioessays 25, 1158-1167. [DOI] [PubMed] [Google Scholar]
- Mihaylov, I. S., Kondo, T., Jones, L., Ryzhikov, S., Tanaka, J., Zheng, J., Higa, L. A., Minamino, N., Cooley, L., and Zhang, H. (2002). Control of DNA replication and chromosome ploidy by geminin and cyclin A. Mol. Cell Biol. 22, 1868-1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, V. Q., Co, C., Irie, K., and Li, J. J. (2000). Clb/Cdc28 kinases promote nuclear export of the replication initiator proteins Mcm2-7. Curr. Biol. 10, 195-205. [DOI] [PubMed] [Google Scholar]
- Nguyen, V. Q., Co, C., and Li, J. J. (2001). Cyclin-dependent kinases prevent DNA re-replication through multiple mechanisms. Nature 411, 1068-1073. [DOI] [PubMed] [Google Scholar]
- Pokholok, D. K. et al. (2005). Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122, 517-527. [DOI] [PubMed] [Google Scholar]
- Raghuraman, M. K., Winzeler, E. A., Collingwood, D., Hunt, S., Wodicka, L., Conway, A., Lockhart, D. J., Davis, R. W., Brewer, B. J., and Fangman, W. L. (2001). Replication dynamics of the yeast genome. Science 294, 115-121. [DOI] [PubMed] [Google Scholar]
- Santocanale, C., and Diffley, J. F. (1998). A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature 395, 615-618. [DOI] [PubMed] [Google Scholar]
- Shirahige, K., Hori, Y., Shiraishi, K., Yamashita, M., Takahashi, K., Obuse, C., Tsurimoto, T., and Yoshikawa, H. (1998). Regulation of DNA-replication origins during cell-cycle progression. Nature 395, 618-621. [DOI] [PubMed] [Google Scholar]
- Stotz, A., and Linder, P. (1990). The ADE2 gene from Saccharomyces cerevisiae: sequence and new vectors. Gene 95, 91-98. [DOI] [PubMed] [Google Scholar]
- Takahashi, T. S., Wigley, D. B., and Walter, J. C. (2005). Pumps, paradoxes and ploughshares: mechanism of the MCM2-7 DNA helicase. Trends Biochem. Sci. 30, 437-444. [DOI] [PubMed] [Google Scholar]
- Vogelauer, M., Rubbi, L., Lucas, I., Brewer, B. J., and Grunstein, M. (2002). Histone acetylation regulates the time of replication origin firing. Mol. Cell 10, 1223-1233. [DOI] [PubMed] [Google Scholar]
- Wilmes, G. M., Archambault, V., Austin, R. J., Jacobson, M. D., Bell, S. P., and Cross, F. R. (2004). Interaction of the S-phase cyclin Clb5 with an “RXL” docking sequence in the initiator protein Orc6 provides an origin-localized replication control switch. Genes Dev. 18, 981-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlschlegel, J. A., Dwyer, B. T., Dhar, S. K., Cvetic, C., Walter, J. C., and Dutta, A. (2000). Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science 290, 2309-2312. [DOI] [PubMed] [Google Scholar]
- Wuarin, J., Buck, V., Nurse, P., and Millar, J. B. (2002). Stable association of mitotic cyclin B/Cdc2 to replication origins prevents endoreduplication. Cell 111, 419-431. [DOI] [PubMed] [Google Scholar]
- Wyrick, J. J., Aparicio, J. G., Chen, T., Barnett, J. D., Jennings, E. G., Young, R.A., Bell, S. P., and Aparicio, O. M. (2001). Genome-wide distribution of ORC and MCM proteins in S. cerevisiae: high-resolution mapping of replication origins. Science 294, 2357-2360. [DOI] [PubMed] [Google Scholar]
- Yabuki, N., Terashima, H., and Kitada, K. (2002). Mapping of early firing origins on a replication profile of budding yeast. Genes Cells 7, 781-789. [DOI] [PubMed] [Google Scholar]
- Yanow, S. K., Lygerou, Z., and Nurse, P. (2001). Expression of Cdc18/Cdc6 and Cdt1 during G2 phase induces initiation of DNA replication. EMBO J. 20, 4648-4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, W., Chen, Y., and Dutta, A. (2004). Re-replication by depletion of geminin is seen regardless of p53 status and activates a G2/M checkpoint. Mol. Cell Biol. 24, 7140-7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental Material]