The Kinesin KIF1C and Microtubule Plus Ends Regulate Podosome Dynamics in Macrophages (original) (raw)
Abstract
Microtubules are important for the turnover of podosomes, dynamic, actin-rich adhesions implicated in migration and invasion of monocytic cells. The molecular basis for this functional dependency, however, remained unclear. Here, we show that contact by microtubule plus ends critically influences the cellular fate of podosomes in primary human macrophages. In particular, we identify the kinesin KIF1C, a member of the Kinesin-3 family, as a plus-end–enriched motor that targets regions of podosome turnover. Expression of mutation constructs or small interfering RNA-/short hairpin RNA-based depletion of KIF1C resulted in decreased podosome dynamics and ultimately in podosome deficiency. Importantly, protein interaction studies showed that KIF1C binds to nonmuscle myosin IIA via its PTPD-binding domain, thus providing an interface between the actin and tubulin cytoskeletons, which may facilitate the subcellular targeting of podosomes by microtubules. This is the first report to implicate a kinesin in podosome regulation and also the first to describe a function for KIF1C in human cells.
INTRODUCTION
Cross-talk between cytoskeletal systems is essential for processes such as cell motility, wound healing, or cell division. In particular, functional coupling of the actin and microtubule cytoskeletons has been the focus of much interest in recent years (reviewed in Goode et al., 2000; Rodriguez et al., 2003; Schliwa and Woehlke, 2003). Here, we investigate the regulatory relationship between actin-based podosomes and microtubule plus ends as well as of motor proteins that may allow the functional linkage of both structures.
Podosomes are actin-rich adhesions typical for cells of the monocytic lineage, but they are also formed in other cell types (reviewed in Linder and Aepfelbacher, 2003; Buccione et al., 2004). Prominent features that distinguish podosomes (and the related invadopodia) from other adhesions are their two-part architecture—a core of F-actin and associated proteins embedded in a ring structure containing plaque proteins such as paxillin or vinculin—and their ability to concentrate matrix metalloproteases and actively engage in matrix degradation (reviewed in Linder and Kopp, 2005). Podosomes are thought to play important roles in migration and invasion of cells. Podosome-mediated adhesion provides local anchorage and may thus stabilize cellular protrusions and support directional migration. Importantly, podosome-localized release of matrix metalloproteases probably contributes to the invasive abilities of podosome-forming cells in physiological and pathological conditions.
Podosomes are highly dynamic organelles with a lifetime of 2–12 min (Kanehisa et al., 1990; Destaing et al., 2003), and their regulation comprises multiple signal transduction pathways, including Src kinase activity (Marchisio et al., 1988) as well as RhoGTPase signaling and actin-regulatory mechanisms (Linder et al., 1999; Burns et al., 2001). Importantly, microtubules have been shown to influence the dynamics (Evans et al., 2003), positioning (Babb et al., 1997; Destaing et al., 2005), and formation (Linder et al., 2000) of podosomes. However, despite the observed close association of podosomes and microtubules, it remained unclear whether both structures indeed interact in living cells. Moreover, the molecular basis for this functional dependency remained unresolved.
Here, we show that the kinesin kinesin-like-family (KIF)1C, a member of Kinesin-3 family (Linder et al., 2000), is critically involved in the microtubule-dependent regulation of podosome dynamics in macrophages. KIF1C has been implicated in Golgi–endoplasmic reticulum (ER) transport (Dorner et al., 1998) and in mouse macrophage resistance to anthrax toxin (Watters et al., 2001). Knockout experiments in mice, however, suggested that KIF1C activity is dispensable for Golgi–ER transport (Nakajima et al., 2002). So far, no function for KIF1C has been described in human cells.
We now show that KIF1C is enriched at a subset of microtubule plus ends that contact podosomes. Contact by KIF1C is followed by enhanced podosome dynamics, i.e., fission or dissolution. Conversely, expression of mutation or truncation constructs of KIF1C resulted in decreased podosome dynamics and ultimately in podosome deficiency. We further identify nonmuscle myosin IIA as an interaction partner of KIF1C. Myosin IIA is enriched in regions of high podosome turnover, and its binding to KIF1C may contribute to the efficient targeting of podosomes by microtubule plus ends.
MATERIALS AND METHODS
Cell Isolation and Cell Culture
Human peripheral blood monocytes were isolated and differentiated into macrophages as described previously (Linder et al., 1999).
Microinjection of Proteins
Cells for microinjection experiments were cultured for 5–8 d. Proteins were expressed in Escherichia coli, as described previously (Linder et al., 2000). For microinjection, proteins were dialyzed against microinjection buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, and 5 mM MgCl2), concentrated in Centricon filters (Millipore, Bedford, MA), shock-frozen, and stored at −80°C. Microinjection was performed using transjector 5246 (Eppendorf, Hamburg, Germany) and a Compic Inject micromanipulator (Cell Biology Trading, Hamburg, Germany). Glutathione _S_-transferase (GST)-KIF1C-protein tyrosine phosphatase-binding domain (PBD) was injected into the cytoplasm at 1, 2, and 3 μg/μl. (Inhibitory) antibodies m74-2, SUK4 (Cytoskeleton, Denver, CO) and K2.4 (HiSS Diagnostics, Freiburg, Germany) against dynein heavy chain, KIF5B or KIF3B, respectively, were injected at 4 or 5 μg/μl (K2.4). Control experiments were performed with GST or microinjection buffer. Injected cells were identified by labeling coinjected rat IgG (5 mg/ml; Dianova, Hamburg, Germany) with fluorescein isothiocyanate-labeled goat anti-rat IgG antibody (Dianova).
Transfection of Cells
Cells were transiently transfected using a Nucleofector I (amaxa, Köln, Germany) according to the manufacturer's instructions and seeded on coverslips at a density of 5 × 105.
Immunofluorescence and Microscopy
Cells were fixed for 10 min in 3.7% formaldehyde solution and permeabilized for 1 min in ice-cold acetone. F-actin was stained with Alexa 568-labeled phalloidin (Molecular Probes, Leiden, The Netherlands), KIF1C was stained with specific primary antibody (Ab) (Cytoskeleton), mitochondria were stained with OxPhos-Ab against cytochrome-c oxidase subunit I (Molecular Probes), myosin IIA was stained with specific primary polyclonal Ab (DakoCytomation Denmark, Glostrup, Denmark), and tubulin was stained with specific primary polyclonal Ab (Cytoskeleton; Note: This antibody also works sufficiently with formaldehyde fixation, which was used when costaining of other proteins in tubulin-specific fixations such as glutaraldehyde/PHEM was not feasible.) Secondary antibodies were Alexa 488- or Alexa 568-labeled goat anti-mouse, goat anti-rabbit, or goat anti-sheep (Molecular Probes). Coverslips were mounted in Mowiol (Calbiochem, Darmstadt, Germany) containing _p_-phenylendiamine (Sigma-Aldrich, St. Louis, MO) as antifading reagent and sealed with nail polish.
Microscopy was performed as described previously (Linder et al., 2000). Immunofluorescence-stained preparations were observed with a Leica DM RBE microscope using a 100× plan fluotar objective. Images were captured with a Spot-camera (Leica, Wetzlar, Germany). Confocal microscopy was performed with a Leica DM IRB confocal scanning microscope and a 100× plan apo objective.
Live Cell Imaging
Confocal time-lapse microscopy of transiently transfected cells was performed using an UltraView LCI live cell imaging system with UltraView, version 5.5.0.2 (PerkinElmer Life and Analytical Sciences, Boston, MA), images were captured using a scan interline digital charge-coupled device camera, model Orca ER (Hamamatsu, Herrsching, Germany) with a resolution of 1344 × 1024 pixels and converted to 12-bit-images. Filter settings were 488-nm excitation, 525/50-nm emission, 568-nm excitation, and 697/45-nm emission. Cells were seeded on glass-bottomed dishes (MatTek, Ashland, MA) at a density of 4–8 × 105 and incubated 20 h before the start of the experiment. During the experiment, cells were incubated in a chamber connected to an incubator controller (EMBL, Heidelberg, Germany) at 37°C and 5% CO2.
To be scored as a contact between podosomes (labeled with monomeric red fluorescent protein [mRFP]-actin) and microtubule plus ends (labeled with green fluorescent protein [GFP]-cytoplasmic linker protein [CLIP]170 or KIF1C-GFP), the respective mRFP and GFP signals had to be directly adjacent or overlapping, without intermediate black pixels. Contact in most cases included 1) apparently directed, not random, movement of plus ends toward podosomes, 2) a momentary stop of movement at podosomes, and 3) a subsequent change or reversal of direction. A correlation between microtubule contact and podosome fate was scored if an alteration of podosome behavior (fission or dissolution) was discernible within 1–2 min after contact by a microtubule plus end (most alterations were observed within shorter periods).
Plasmid Construction and Mutagenesis
For cloning of GST-KIF1C-P, part of the coding sequence of human KIF1C was amplified and cloned into pGEX-1λT, resulting in a construct coding for aa 713-811. For cloning and expression of GST-KIF1C-U, the coding sequence of KIF1C was amplified and cloned into pGEX5X-3, resulting in a construct coding for aa 441-623. For cloning and expression of wild-type GFP-KIF1C, the 5′ half of the coding sequence up to nucleotide 1847 was amplified from pRKS-KIF1C generating a 5′XhoI and a 3′EcoRI restriction site. The 3′ half of the KIF1C coding sequence was amplified generating a 5′EcoRI and a 3′BamHI restriction site. The two regions were cloned successively into vector pEGFP-N1 (Clontech, Mountain View, CA). GFP-KIF1C-K103A was generated by removing the newly generated EcoRI restriction site and by introducing a K103A mutation using QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA).
Vector encoding GFP-KIF3BΔNT was a kind gift from V. Gelfand (University of Illinois, Urbana, IL); MBP-kinesin-associated protein (KAP)3 was a kind gift from Y. Takai (Osaka University, Osaka, Japan); GFP-KAP3-wild type (wt) and GFP-KAP3ΔArm5 were kind gifts from T. Akiyama (University of Tokyo, Tokyo, Japan); KIF4 was a kind gift from Y. Lee (Ajou University, Suwon, South Korea); Myc-KHC wt, Myc-KHC H582, HA-kinesin light chain (KLC) wt, HA-KLC-TPR6, and HA-KLC-L176 were kind gifts from B. Schnapp (Harvard Medical School, Boston, MA); FLAG-KLC2 was a kind gift from T. Ichimura (Tokyo Metropolitan University, Tokyo, Japan); DsRed-EB1 was a kind gift from A. Barth (Stanford University, Stanford, CA); and GFP-CLIP170 was a kind gift from N. Galjart (Erasmus Medical Center, Rotterdam, The Netherlands).
GST-Pull Down Assay
GST-pull downs were prepared as described previously (Linder et al., 1999). Briefly, 12 × 106 cells cultured for 6–12 d were lysed by addition of 1.2 ml of lysis buffer (10 mM Tris-HCl, pH 8, 1% Triton X-100, 75 mM NaCl, 5 mM EDTA, and 1 mM dithiothreitol), containing Complete mini protease inhibitors (Roche Diagnostics, Penzberg, Germany) and incubated for 30 min at 4°C. After centrifugation at 3500 × g for 10 min at 4°C, the lysate was added to glutathione-Sepharose beads (GE Healthcare, München, Germany), previously incubated with 150 μg of GST-fusion proteins or GST at comparable molar ratios. Beads were incubated with lysate for 1 h at 4°C, washed three times with lysis buffer, pelleted, and mixed with 100 μl of SDS-sample buffer.
Immunoblotting
Immunolabeling was performed by standard procedure, using the above-mentioned primary antibodies. Secondary antibodies were horseradish peroxidase-coupled anti-mouse or anti-rabbit IgG (Dianova). Protein bands were visualized by using Super Signal kit (Pierce Chemical, Rockford, IL) and X-Omat AR film (Kodak, Stuttgart, Germany).
Validation of Small Interfering RNA by Quantitative Real-Time PCR
Small interfering RNAs (siRNAs) were validated by quantitative real-time PCR (q-PCR) using lysates of transfected HeLa cells, as described previously (Machuy et al., 2005). Briefly, 0.1–0.25 μg of siRNA (final concentration, 80–200 nM) directed against KIF1C, KIF3A, KAP3A, KIF5B, or Luciferase as control and 2 μl Transmessenger reagent (QIAGEN, Hilden, Germany) were added to 10 × 104 cells seeded in 96-well plates. RNA was isolated 48 h later using the RNeasy 96 BioRobot 8000 system (QIAGEN). The relative amount of target mRNA was determined by q-PCR using Quantitect SYBR Green reverse transcriptase-PCR kit following manufacturer's instructions (QIAGEN).
siRNA/Short Hairpin RNA Knockdown Experiments
Duplex siRNA (650 ng) was transfected transiently in primary human macrophages. For visualization of transfected cells, 1.2 μg of pEGFP-N1-vector was cotransfected, and cells were cultured for 24–72 h. For vector-encoded short hairpin RNA (shRNA), different target regions in the KIF1C coding sequence were chosen and cloned into psiSTRIKE U6 (Promega, Madison, WI). Cells were transfected and cultured for additional 24–72 h. Target sequence corresponds to the nucleotides 426–442 in KIF1C. As a control, a scrambled sequence was used.
Reverse Transcriptase Reaction
Cells (6 × 106) were cultured for 7 d, and mRNA was isolated using QuickPrep Micro-mRNA purification kit (GE Healthcare). DNA was removed by DNAse digestion (Novagen, Madison, WI). For cDNA-synthesis, 1 μg of random primer (Promega) was annealed to 2 μg of RNA for 5 min at 70°C, and second strand synthesis was performed using Moloney murine leukemia virus reverse transcriptase (Promega) using an oligonucleotide primer pair corresponding to nucleotides 3030–3059 and 3369–3397 of the KIF1C coding sequence, respectively. As a control for quantitative removal of residual DNA, oligonucleotide primers specific for an exon in the human β-actin gene were used, corresponding to nucleotides 1161–1142 and 716–735, respectively.
Podosome Reformation
Podosomes were disrupted by addition of tyrosine kinase inhibitor PP2 (Sigma-Aldrich) at 25 μM for 1 h with subsequent washout. Podosome disruption/reformation does not interfere with microtubule integrity or distribution (Linder et al., 2000). Macrophages normally display 50–150 podosomes per cell. The threshold value for a macrophage containing podosomes was set to 10. This allowed an immediate and clear distinction between cells “containing” or “not containing” podosomes.
Immunoprecipitation
Immunoprecipitations were performed with the μMACS-protein isolation kit (Miltenyi Biotec, Bergisch-Gladbach, Germany). Seven- to 14-d-old macrophages were lysed in lysis buffer A (50 mM Tris-HCl, pH 7.5, 100 mM KCl, and 0.5% Triton X-100) or B (20 mM HEPES, pH 7.5, 150 mM NaCl, 250 mM sucrose, 1 mM Na-molybdate, and 1% Igepal), both with Complete protease inhibitors (Roche Diagnostics). Two micrograms of antibody was added to 50 μl of protein A-coupled magnetic beads, incubated with the cleared lysate for 1 h, and applied to the μMACS separation column. After washing four times with washing buffer (20 mM HEPES, 150 mM NaCl, 2 mM peroxovanadate, 0,1% Triton X-100, and 10% glycerol), the immunoprecipitate was eluted with 1× Laemmli buffer. Immunoprecipitations of GFP fusion proteins were performed using the GFP-μMACS epitope tag Protein isolation kit (Miltenyi Biotec) according to the manufacturer's protocol. The cells were lysed in (50 mM Tris-HCl, pH 8.0, 50 mM NaCl, and 1% Igepal), with Complete Mini protease inhibitors.
Mass Spectrometry
Silver-stained sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel bands were washed twice with water and then covered with a 1:1 mixture of 30 mM potassium ferricyanide and 100 mM sodium thiosulfate and washed twice with water. Clear gel pieces were then treated twice with 50% acetonitrile for 5 min. Trypsin (sequencing grade modified; Promega, Mannheim, Germany) was added, and proteins were digested overnight in 40 mM NH4HCO3 buffer, pH 8.0, at 37°C and 650 rpm. For protein identification, probes were used for matrix-assisted laser desorption ionization/time of flight (MALDI-TOF) experiments. Ten microliters of each sample was first purified and concentrated on a C18 reverse phase pipette tip (ZipTip; Millipore, Schwalbach, Germany). Peptides were eluted with 1 μl of α-cyano-4-hydroxycinnamic acid (Sigma-Aldrich) and directly spotted on a MALDI sample plate (Applied Biosystems, Foster City, CA). MALDI-TOF measurements were performed on a Voyager-DE STR TOF mass spectrometer (Applied Biosystems). The resulting spectra were then analyzed via Mascot software (Matrix Science, London, United Kingdom) using the NCBInr Protein Databank.
Microtubule Cosedimentation Assay
Cells (1.2 × 107) were lysed in (80 mM PIPES, pH 6.8, 1 mM MgCl2, and 1 mM EGTA, with protease inhibitors) and centrifuged at 100 000 × g for 1 h. Tubulin in the cytosol was polymerized by addition of (2 mM GTP, 2 mM MgCl2, and 20 μM taxol, end concentrations), and motor proteins were allowed to bind for 30 min at room temperature in the presence of 1.5 mM adenyl-5′-yl imidodiphosphate (AMP-PNP). Motor–tubulin complexes were pelleted by centrifugation at 165 000 × g for 1 h, and aliquots of resuspended pellets were analyzed with SDS-PAGE and Western blots.
RESULTS
Podosomes Are Contacted by Microtubule Plus Ends
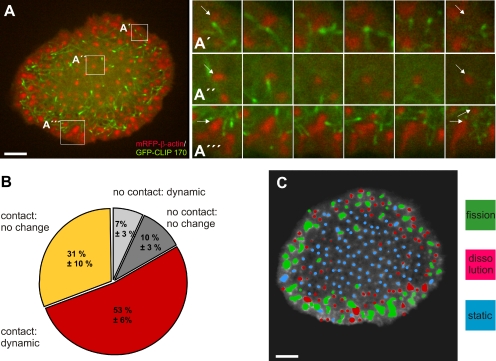
Podosomes are cell–matrix contacts that localize exclusively to the ventral cell side of substrate-attached macrophages. In contrast, microtubules originate mostly from the microtubule-organizing center (MTOC), which is localized in the dome-shaped central part of the cell containing most of the cytoplasm and also the nucleus. To investigate the spatiotemporal relationship between podosomes and microtubules in living cells, primary human macrophages were transfected with constructs encoding mRFP–β-actin, to label podosomes, and GFP-fused CLIP170, a microtubule plus end–binding protein (reviewed in Galjart, 2005). Cells were subsequently analyzed using time-lapse confocal microscopy (Figure 1A). Confirming previous results gained with fixed specimens (Linder et al., 2000), microtubules often extended to the podosome-containing ventral cell side. Moreover, podosomes were found to be targeted, often repeatedly, by microtubule plus ends (Figure 1A; for full appreciation of repeated contacts between microtubules and podosomes, see Supplemental Videos 1–4). Further analysis revealed three basic modes of podosome behavior: dissolution, fission, or static behavior (Figure 1A and Supplemental Videos 2–4).
Figure 1.
Podosome fate is influenced by contact with microtubule plus ends. (A) Confocal laser scanning micrographs of substrate-attached part of human primary macrophage expressing mRFP-β-actin (red) and GFP-CLIP170 (green). White boxes in overview image indicate corresponding areas enlarged at the right, showing dissolution (A′), static behavior (A″), or fission of podosomes (A‴. Note repeated contact of CLIP170-labeled microtubule plus ends with β-actin–labeled podosomes. Images shown were taken every 60 s from start of the experiment (Supplemental Videos 1–4). White arrows indicate single podosomes. Bar, 10 μm. Podosome behavior and concurrent contact by CLIP170-labeled microtubule plus ends was evaluated in a total of 731 podosomes from three independent experiments. Values are given as mean percentage ± SD of total counts. Podosomes contacted by CLIP170-labeled microtubule plus ends: 31 ± 10% for no change, 53 ± 6% for dynamic behavior; podosomes not contacted by CLIP170-labeled microtubule plus ends: 10 ± 3% for no change, 7 ± 3% for dynamic behavior. (C) Analysis of podosome dynamics in a human macrophage. Confocal laser scanning micrograph of primary macrophage expressing mRFP–β-actin (image of time lapse series). Podosomes are color coded according to their behavior during the course of the experiment: static (blue), fission (green), or dissolution (red). Note that small, static podosomes cluster in the cell center, whereas in the cell periphery, larger precursor clusters mostly undergo fission, and smaller podosomes mostly dissolve. Bar, 10 μm.
In a typical experiment (20 min), more than 80% of podosomes were contacted at least once by microtubule plus ends, and this was preferentially followed by dynamic behavior (dissolution or fission; ∼60%). By contrast, <20% of podosomes were not contacted by microtubules, and the majority of these podosomes (∼60%) remained static (Figure 1B; Note: Dynamic podosomes showed the typical life time of 2–8 min, whereas static podosomes persisted for at least 20 min.) Interestingly, subcellular analysis revealed areas in which one type of podosome behavior prevailed, because dynamic behavior occurred mostly in the outer regions of the cell, whereas static podosomes localized preferentially in the center of the substrate-attached cell side (Figure 1C).
Podosomes Are Influenced by ATP Hydrolysis but Not by KIF5B or Dynein
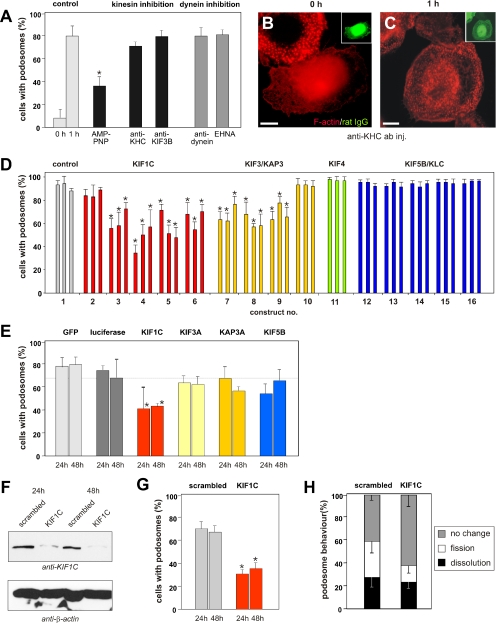
Contact by microtubules raised the possibility of motor proteins being involved in podosome regulation. To investigate this question, we evaluated podosome reformation under conditions of motor protein inhibition. This assay takes advantage of a microinjection artifact, because injection of macrophages with high pressure leads to disruption of podosomes, which are subsequently reformed within 1 h (with microtubule integrity not being compromised; Linder et al., 2000). Microinjection with the nonhydrolysable ATP analogue AMP-PNP led to a reduction of ∼60% in podosome reformation (Figure 2A and Table 1), indicating the general influence of ATP hydrolysis-dependent processes. An antibody (Ingold et al., 1988) against conventional kinesin (KIF5B in the unified nomenclature; Miki et al., 2001), which has been shown to inhibit focal adhesion regulation by microtubules in Xenopus fibroblasts (Krylyshkina et al., 2002), did not influence podosome reformation (Figure 2, A–C, and Table 1). However, in an established control assay testing its functionality (Krylyshkina et al., 2002), the antibody effectively inhibited the plus end–directed transport of mitochondria in macrophages (Supplemental Figure S1). Also, microinjection of a potentially inhibiting antibody against heterotrimeric kinesin (Morris and Scholey, 1997) did not interfere with podosome reformation (our unpublished data). Moreover, inhibition of dynein, either by an inhibitory antibody (Steffen et al., 1997) or by the inhibitor erythro-9-[3-(2-hydroxynonyl)]adenine (EHNA; Schliwa et al., 1984) did not influence podosome reformation (Figure 2A and Table 1), although, in an established functionality assay (Krylyshkina et al., 2002), both substances inhibited minus end–directed lysosome transport in macrophages (Supplemental Figure S2; our unpublished data). These results indicate that neither conventional kinesin nor dynein are important for the reestablishment of regular podosome dynamics in macrophages.
Figure 2.
Podosome formation is influenced by KIF1C but not by conventional kinesin or dynein. (A) Evaluation of podosome formation after microinjection (0 or 1 h). Values are given as mean percentage ± SD of total counts in Table 1. For differences between control values and values gained with motor protein inhibitors, a p value < 0.01 was considered significant (indicated by asterisk). (B and C) Fluorescence micrographs of macrophages injected with anti-KIF5B antibody, subsequently fixed at 0 or 1 h and stained for F-actin (red) or rat IgG as an
Table 1.
Values for podosome formation after various treatments
| Cells injected with | Cells with podosomes (%) | |
|---|---|---|
| 0 h | 1 h | |
| Mock | 8.9 ± 7.2 | 79.2 ± 11.0 |
| AMP-PNP (50 mM) | 36.7 ± 9.0 | |
| KIF5B antibody (4 μg/μl) | 72.2 ± 3.0 | |
| KIF3B antibody (4 μg/μl) | 78.9 ± 5.1 | |
| dynein antibody (4 μg/μl) | 80.0 ± 6.8 | |
| EHNA (1 mM) | 81.1 ± 3.9 |
| Cells transformed with | 4 h | 6 h | 8 h |
|---|---|---|---|
| Kinesin construct (no.) | |||
| 1 (Control) | 93.3 ± 3.3 | 94.4 ± 6.9 | 87.8 ± 1.9 |
| 2 | 84.4 ± 5.1 | 83.3 ± 12.0 | 88.9 ± 1.9 |
| 3 | 55.6 ± 8.4 | 57.8 ± 11.7 | 72.2 ± 5.1 |
| 4 | 34.4 ± 6.9 | 50.0 ± 8.8 | 56.7 ± 15.3 |
| 5 | 71.1 ± 5.1 | 51.1 ± 7.7 | 47.8 ± 8.4 |
| 6 | 67.8 ± 10.2 | 54.4 ± 6.9 | 70.0 ± 6.7 |
| 7 | 63.3 ± 6.7 | 62.2 ± 6.9 | 76.7 ± 6.7 |
| 8 | 67.8 ± 10.2 | 56.7 ± 3.3 | 57.8 ± 10.7 |
| 9 | 63.3 ± 6.7 | 77.8 ± 5.1 | 65.6 ± 8.4 |
| 10 | 93.3 ± 5.8 | 93.3 ± 3.3 | 92.2 ± 5.1 |
| 11 | 97.8 ± 1.9 | 96.7 ± 3.3 | 96.7 ± 3.3 |
| 12 | 95.6 ± 1.9 | 95.6 ± 1.9 | 92.2 ± 1.9 |
| 13 | 92.2 ± 1.9 | 95.6 ± 1.9 | 91.1 ± 5.1 |
| 14 | 94.4 ± 1.9 | 91.1 ± 5.1 | 94.4 ± 1.9 |
| 15 | 95.6 ± 1.9 | 95.6 ± 1.9 | 94.4 ± 3.8 |
| 16 | 94.4 ± 3.8 | 96.7 ± 0.0 | 96.7 ± 0.0 |
| siRNA | 24 h | 48 h |
|---|---|---|
| Vector control (pEGFP-N1) | 78.0 ± 7.0 | 80.0 ± 7.0 |
| Luciferase | 74.0 ± 4.0 | 68.0 ± 17.0 |
| KIF1C | 40.0 ± 18.0 | 42.0 ± 2.0 |
| KIF3A | 63.3 ± 6.7 | 62.2 ± 6.9 |
| KAP3A | 67.8 ± 10.2 | 56.7 ± 3.3 |
| KIF5B | 54.4 ± 8.0 | 65.6 ± 11.0 |
| shRNA | ||
|---|---|---|
| Scrambled | 68.9 ± 5.9 | 65.6 ± 5.3 |
| KIF1C | 32.2 ± 3.9 | 36.7 ± 5.1 |
| Dissolution (%) | Fission (%) | No change (%) | |
|---|---|---|---|
| Scrambled | 29.6 ± 9.0 | 30.6 ± 10.7 | 42.5 ± 3.9 |
| KIF1C | 23.1 ± 5.0 | 13.6 ± 7.4 | 63.0 ± 5.9 |
Inhibition of KIF1C Leads to Podosome Loss
To further investigate the potential role of kinesins in podosome regulation, macrophages were transfected with various wild-type or mutation constructs (for complete list, see Supplemental Table S1). Podosome reformation was induced by podosome disruption with tyrosine kinase inhibitor PP2 and subsequent washout. This assay has been shown earlier to specifically target podosomes and to not interfere with microtubule integrity or distribution or with cortical actin filaments (Linder et al., 2000). Podosome reformation was tested 4, 6, and 8 h after transfection. Importantly, expression of KIF1C or KIF3A/KAP3A constructs interfered with podosome reformation, whereas KIF4 or KIF5B/KLC (kinesin light chain) constructs showed no influence (Figure 2D and Table 1).
In a next step, we synthesized siRNAs against KIF1C, KIF3A, KAP3A, and KIF5B, which were validated in HeLa cells and found to be highly effective (>84.0% knockdown; see Materials and Methods). Primary macrophages were transfected with the respective siRNA and evaluated for podosome content 24 and 48 h after transfection. Cotransfected pEGFP-N1 (at substoichiometric concentrations; see Materials and Methods) served as a transfection marker. Compared with controls, KIF3A-, KAP3A- and KIF5B-specific siRNA showed a slight effect on podosome numbers. By contrast, KIF1C siRNA-transfected macrophages showed a marked and significant reduction in podosome content to ∼60% of controls (Figure 2E and Table 1).
This was further confirmed by transfection of a construct which allows bicistronic expression of KIF1C-specific shRNA and enhanced green fluorescent protein (EGFP) for unequivocal identification of transfected cells (STRIKE-KIF1C). Test transfection of human umbilical vein endothelial cells (HUVECs) showed efficient reduction of endogenous KIF1C protein expression levels, compared with a construct encoding a scrambled sequence (STRIKE-scrambled; Figure 2F). Primary macrophages transfected with the KIF1C-specific construct showed a (gradual) reduction of podosomes (Supplemental Figure S3A), to levels similar to those of cells directly transfected with siRNA (Figure 2G). In addition, those cells transfected with the KIF1C-specific construct but still containing podosomes showed decreased podosome fission (13.8 ± 7.4% for STRIKE-KIF1C versus 30.6 ± 10.7% for STRIKE-scrambled; Figure 2H). In sum, interference with KIF1C function during podosome reformation either by expression of mutant constructs or through transfection/expression of specific siRNA or shRNA seemed to disrupt regular podosome dynamics and led to increasing numbers of cells devoid of podosomes.
KIF1C Is Enriched at Podosome-contacting Microtubule Plus Ends
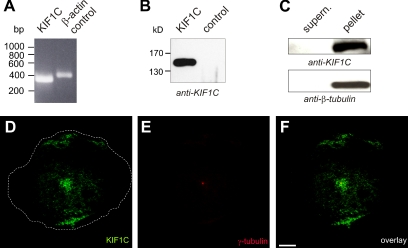
To confirm endogenous expression of KIF1C in primary human macrophages, several assays were performed. The presence of KIF1C mRNA in macrophages was confirmed by reverse transcriptase-PCR using specific primers and subsequent sequencing of the PCR product (Figure 3A), whereas Western blots of KIF1C immunoprecipitations, developed with a specific antibody, confirmed expression of the corresponding polypeptide in macrophage lysates (Figure 3B). Moreover, in a microtubule cosedimentation assay, KIF1C was present in the microtubule-containing pellet (Figure 3C), indicating a microtubule-binding ability of the protein. Immunofluorescence analysis of endogenous KIF1C showed an enrichment at the periphery of the ventral cell side (Figure 3, D and F) and also at a juxtanuclear position, which, by staining for γ-tubulin, was shown to overlap with the centrosome/MTOC (Figure 3, E and F).
Figure 3.
KIF1C is expressed in primary human macrophages. (A) Reverse transcriptase-PCR using primers specific for KIF1C (left), control reaction using primers for an exon in the β-actin gene (right), agarose gel stained with ethidium bromide; size in base pairs on left. (B) Immunoprecipitation of macrophage lysates using anti-KIF1C antibody (left), rabbit IgG was used as control (right); Western Blot probed with anti-KIF1C antibody. Molecular mass in kilodaltons is indicated on the left. (C) Microtubule cosedimentation assay using macrophage lysates. KIF1C is pelleted together with polymerized microtubules stabilized by taxol treatment. Western blot of cytosolic supernatant and pellet, probed with anti-KIF1C or anti-β-tubulin antibody, respectively. (D and E) Primary macrophage stained with specific primary antibodies for KIF1C (D) and γ-tubulin (E), overlay shown in (F). Superimposition of each time two confocal laser scanning micrographs with a z-distance of 4 μm. Cell circumference depicted by dashed white line. White bar, 10 μm.
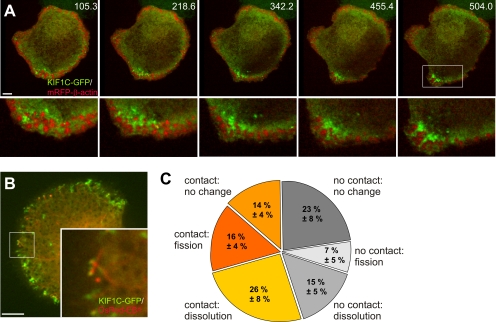
Localization of KIF1C was further analyzed by confocal time-lapse microscopy of macrophages cotransfected with a construct encoding GFP-labeled KIF1C and a construct coding for mRFP-labeled β-actin. Importantly, expression of KIF1C-GFP did not significantly influence podosome content of cells, compared with controls (Supplemental Figure S3B). KIF1C-GFP was found to accumulate in punctate structures at the podosome-containing ventral cell side and especially at regions of high podosome turnover (Figure 4A; Supplemental Videos 5 and 6). Similar to microtubule plus ends, dot-like KIF1C-GFP accumulations repeatedly contacted podosomes (Figure 4A). Indeed, coexpression of DsRed-labeled EB1, highlighting microtubule plus ends (Louie et al., 2004), confirmed that KIF1C-GFP was enriched at microtubule plus ends (Figure 4B and Supplemental Videos 7 and 8), especially in the cell periphery (Supplemental Figure S4A). Enrichment of KIF1C-GFP at microtubule ends was further confirmed by staining fixed specimens for β-tubulin (Supplemental Figure S4B–D).
Figure 4.
KIF1C at microtubule plus ends contacts podosomes. (A) KIF1C-GFP is enriched at sites of high podosome turnover. Confocal micrograph series of substrate-attached part of primary macrophage expressing KIF1C-GFP (green) and mRFP-β-actin (red; Supplemental Videos 5 and 6). Time since start of experiment is indicated in top right corners. White box indicates enlarged area. Bar, 10 μm. (B) KIF1C-GFP (green) localizes to DsRed-EB1–labeled (red) microtubule plus ends. Confocal laser scanning micrograph of substrate-attached part of cell (Supplemental Videos 7 and 8). White box indicates enlarged area. Bar, 10 μm. (C) Statistical analysis of podosome development in primary macrophage. Podosomes were analyzed for behavior (static, fission, or dissolution) and concurrent contact by KIF1C-GFP–containing punctate structures. For each value, a total of 601 podosomes from three different experiments were evaluated. Values are given as mean percentage ± SD of total counts. Contact by KIF1C-GFP accumulations: 14 ± 4% for no change, 16 ± 4% for fission, 26 ± 8% for dissolution; no contact by KIF1C-GFP accumulations: 23 ± 8% for no change, 7 ± 5% for fission, 15 ± 5% for dissolution.
During a typical experiment, ∼60% of all podosomes were contacted by KIF1C-GFP accumulations (Figure 4C). Compared with GFP-CLIP170–labeled microtubule plus ends (∼84% contact), these data indicate that only a subset of microtubule plus ends show KIF1C enrichment. Interestingly, KIF1C-decorated microtubule plus ends mostly contact podosomes in the cell periphery (Supplemental Figure S5). Contact of podosomes by this microtubule subset is apparently highly effective in inducing changes in podosome behavior, because ∼75% of KIF1C-contacted podosomes subsequently dissolved (26 ± 8% of all podosomes) or split (16 ± 4% of all podosomes). Conversely, 25% of KIF1C-GFP–contacted podosomes showed no subsequent change, compared with 50% of the podosomes that were not contacted by KIF1C-GFP (Figure 4C).
Expression of a P-Loop Mutant of KIF1C Leads to Podosome Loss
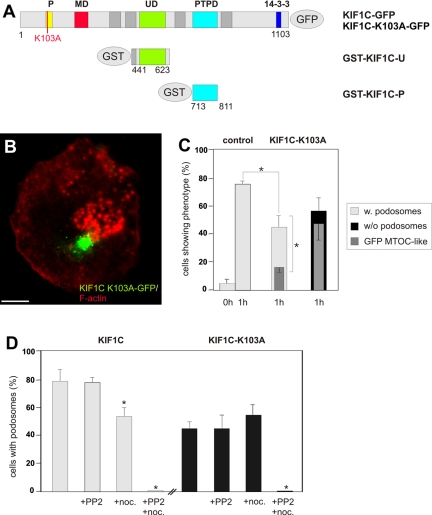
Typical for kinesins, KIF1C features a P-loop sequence (GQTGAGKS; aa 97-104; Dorner et al., 1998), which is supposedly involved in nucleotide binding (Sack et al., 1999). Mutations in the corresponding motif of KIF5B have been shown to abrogate ATP hydrolysis and induce a rigor state (Nakata and Hirokawa, 1995). Therefore, attempting to generate a dominant negative construct of KIF1C, we introduced a K103A mutation (Figure 5A). Mutant KIF1C-K103A-GFP was mostly dislocalized from microtubule plus ends and instead concentrated at a dot-like juxtanuclear position (Figure 5B), which, by costaining for γ-tubulin, was identified as the centrosome/MTOC (our unpublished data). The expression of KIF1C-K103A-GFP led to a clear reduction in cellular podosome content (Figure 5B), without apparently compromising microtubule number, length or dynamics (our unpublished data). Quantifying this phenomenon in the podosome reformation assay, we found that only ∼45% of KIF1C-K103A–expressing cells formed podosomes, compared with ∼75% of control cells (Figure 5C). The inability of KIF1C-K103A–expressing cells to reform podosomes was clearly coupled to an aberrant localization of the mutant protein, because ∼80% of cells without podosomes showed an accumulation of KIF1C-K103A at the MTOC, in contrast to ∼35% of the cells that were still able to form podosomes (Figure 5C).
Figure 5.
A KIF1C P-loop mutant is dislocalized and leads to podosome loss. (A) Domain organization of KIF1C and expression constructs used in this study: P-loop sequence (P; aa 97-104), motor domain signature (MD; aa 242-253), Unc104 domain (UD; aa 483-593), PTPD-1–binding domain (PTPD; aa 714-809), unspecified 14-3-3 binding region (14-3-3). Numbers indicate first and last amino acid residues of constructs. (B) Primary macrophage expressing GFP-labeled KIF1C K103A mutant (green), labeled for F-actin, confocal laser scanning micrograph of substrate-attached part of cell (superimposition of 4 images with z-distances of 1 μm). Note MTOC-like localization of GFP signal and scarcity of podosomes. (C) Evaluation of podosome reformation in macrophages expressing the KIF1C K103A mutant construct. Cells were treated with PP2 (0 h), followed by washout (1 h). Values for podosome formation are given as mean percentage ± SD of total counts. For each bar, 3 × 30 cells were evaluated. Control (pEGFP-C1): 0 h, 2.2 ± 1.5%; 1 h, 74.4 ± 1.5% for cells with podosomes; KIF1C-K103A: 1 h, 44.4 ± 7.8% for cells with podosomes, concurrent with 16.7 ± 2.6% of total cells showing a MTOC-like localization of the GFP signal; 55.6 ± 7.8% for cells without podosomes, concurrent with 45.6 ± 14.1% of total cells showing a MTOC-like localization of the GFP signal. For differences between control values and values gained with KIF1C-K103A expression (light gray bars), a p value < 0.01 was considered significant (indicated by asterisk); for a correlation between absence of podosomes (black bar) with an MTOC-like accumulation of GFP-KIF1C-K103A (dark gray bar), a p value < 0.02 was considered significant (indicated by asterisk). (D) Effect of microtubule disruption in cells expressing either KIF1C-GFP wt or KIF1C-K103A-GFP. Podosome formation was evaluated in untreated cells (unlabeled bars), after disruption with 25 μM PP2 and subsequent washout (+PP2), after addition of nocodazole (1 μM; +noc.), or after disruption with PP2 and addition of nocodazole to washout (+PP2+noc.). For each bar, 3 × 30 cells were evaluated. Values are given as mean percentage ± SD of total counts in Table 2, a p value < 0.04 was considered significant (indicated by asterisk).
We further assessed the influence of microtubule disruption under conditions of KIF1C inhibition. Similar to previous results (Linder et al., 2000), we found that reformation of podosomes (after addition of podosome-disrupting PP2) can be blocked by addition of nocodazole to the washout medium. This was observed both in cells expressing KIF1C-K103A or KIF1C wt (Figure 5D and Table 2). Direct addition of nocodazole to cells without prior disruption of podosomes did not change podosome numbers of KIF1C-K103A expressing cells. However, it clearly reduced the level of podosome-containing cells in KIF1C wt-expressing cells, to levels comparable with those of KIF1C-K103A cells (Figure 5D and Table 2). These results indicate that intact microtubules are absolutely necessary for reformation of podosomes in human macrophages, whereas KIF1C is not. However, in cells already containing podosomes, both microtubules and KIF1C have an impact of 30–40% on the upkeep of these structures.
Table 2.
Effect of KIF1C inhibition after microtubule disruption
| Cells transfected with | Cells incubated with | Cells with podosomes (%) | |
|---|---|---|---|
| PP2 (25 μM) | Nocodazole (1 μM) | ||
| KIF1C-GFP | − | − | 78.8 ± 11.7 |
| KIF1C-GFP | + | − | 77.8 ± 5.1 |
| KIF1C-GFP | − | + | 53.3 ± 8.8 |
| KIF1C-GFP | + | + | 0.0 ± 0.0 |
| KIF1C-K103A-GFP | − | − | 43.3 ± 13.3 |
| KIF1C-K103A-GFP | + | − | 43.3 ± 6.7 |
| KIF1C-K103A-GFP | − | + | 54.4 ± 10.7 |
| KIF1C-K103A-GFP | + | + | 0.0 ± 0.0 |
KIF1C Interacts with Nonmuscle Myosin IIA
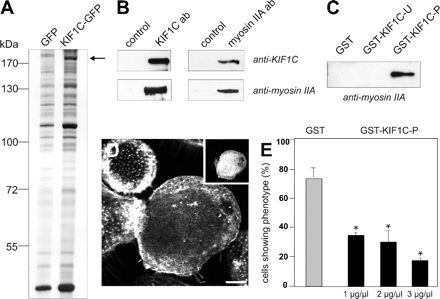
Together, our data suggest that a subset of microtubule plus ends, which are decorated by KIF1C, facilitate podosome dynamics. To investigate the role of KIF1C in this process in more detail, we tried to identify interaction partners of KIF1C by immunoprecipitation of GFP-KIF1C with subsequent MALDI analysis. Because the transfection efficiency of primary macrophages does not result in sufficient yield for MALDI, we used HUVECs, which show high transfection rates and are also able to form podosomes (Osiak et al., 2005). GFP-immunoprecipitations under conditions of low salt (50 mM NaCl) revealed a prominent band of ∼200 kDa (Figure 6A), which was identified as nonmuscle myosin IIA. This was confirmed by cross-immunoprecipitation of cell-internal KIF1C and myosin IIA from macrophage lysates under physiological salt conditions (150 mM NaCl; Figure 6B), indicating an interaction between both motors.
Figure 6.
KIF1C interacts with nonmuscle myosin IIA. (A) HUVEC lysates immunoprecipitated with anti-GFP antibody coupled to magnetic beads. Silver-stained PAA gel, left lane: cells transfected with pEGFP-N1 as control; right lane: cells transfected with KIF1C-GFP construct. Arrow indicates band subsequently identified by MALDI as nonmuscle myosin IIA. Molecular mass in kilodaltons is indicated on the left. (B) KIF1C and myosin IIA coprecipitate from macrophage lysates. Immunoprecipitation of macrophage lysates, using rabbit IgG as control, KIF1C-specific antibody or myosin IIA-specific antibody. Western blots developed with antibody indicated at the right. (C) GST-pull down of macrophage lysates. Western blot probed with anti-myosin IIA antibody. Denominations of GST-fused polypeptides used for pull down are given above each lane. (D) Microinjection of macrophages with GST-KIF1C-P disrupts podosomes. Laser scanning confocal micrograph of substrate-attached part of cell, specimen stained for F-actin and rat IgG (inset) as an injection marker. Bar, 10 μm. (E) Evaluation of podosome formation in macrophages microinjected with GST-KIF1C-P polypeptide. Values are given as mean percentage ± SD of total counts. For each value, 3 × 30 cells were evaluated. Cells with podosomes: GST control (0.5 μg/μl), 73.3 ± 7.7%; GST-KIF1C-P (1 μg/μl), 34.4 ± 1.5%; GST-KIF1C-P (2 μg/μl), 30.0 ± 7.3%; GST-KIF1C-P (3 μg/μl), 17.8 ± 1.5%. For differences between control values and values gained with KIF1C-P injections, a p value < 0.03 was considered significant (indicated by asterisk).
To investigate the KIF1C–myosin IIA interaction in more detail, we performed GST-pull down assays of macrophage lysates using GST fusions of potential interaction sites: GST-KIF1C-U, containing the Unc104 domain (aa 441-623) and GST-KIF1C-P, containing the protein tyrosine phosphatase (PTPD)-1–binding domain (aa 713-811; Figure 5A). Indeed, the GST-KIF1C-P polypeptide was able to bind myosin IIA from macrophage lysates (Figure 6C), indicating an interaction between the two motor proteins through the corresponding domain. Furthermore, microinjection of macrophages with GST-KIF1C-P, thought to compete with the interaction of cellular KIF1C and myosin IIA, induced disruption of podosomes (Figure 6D) in a dose-dependent manner (Figure 6E). The interaction between KIF1C and nonmuscle myosin IIA is therefore clearly of functional significance for an equilibrium in podosome turnover.
Inhibition of Nonmuscle Myosin IIA Leads to Podosome Loss
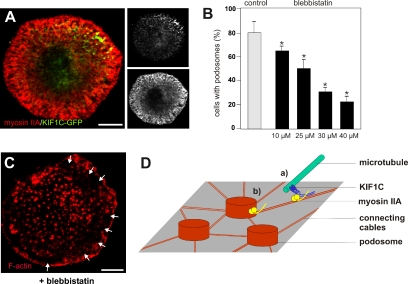
This is further underscored by the fact that both nonmuscle myosin IIA and KIF1C-GFP localize preferentially to the periphery of the ventral cell side in macrophages (Figure 7A). Moreover, inhibition of nonmuscle myosin II (A and B) via the specific inhibitor blebbistatin (Limouze et al., 2004) resulted in a dose-dependent decrease of podosome-containing cells (Figure 7B). Interestingly, even in cells that still contain podosomes, podosome disruption could be observed in the cell periphery (Figure 7C), where myosin IIA is mostly concentrated (Figure 7A). In this context, it is noteworthy that 1) dynamic podosomes also localize to this region (Figure 1C) and 2) KIF1C-GFP–decorated plus ends preferentially contact podosomes in the cell periphery, whereas GFP-CLIP170–decorated microtubule plus ends contact podosomes irrespective of their subcellular localization (see Supplemental Figure S4). In sum, both KIF1C-decorated microtubule plus ends and myosin IIA preferentially localize to the region of dynamic podosomes, and inhibition of either motor protein disrupts podosome turnover.
Figure 7.
Inhibition of nonmuscle myosin II leads to podosome loss. (A) Both myosin IIA and KIF1C-GFP localize preferentially to the cell periphery. Laser scanning confocal micrograph of substrate-attached part of a cell expressing KIF1C-GFP (green), specimen stained for myosin IIA (red), small black-and-white images show localization of KIF1C-GFP (top) or of myosin IIA (bottom). Bar, 10 μm. (B and C) Myosin II inhibitor blebbistatin impairs podosome formation. (B) Statistical evaluation of cells containing podosomes 30 min after addition of respective reagent. Values for podosome formation are given as mean percentage ± SD of total counts. For each bar, 3 × 30 cells were evaluated. Control (medium + 1% dimethyl sulfoxide), 80.0 ± 9.2%; blebbistatin, 10 μM, 65.6 ± 3.9%; 25 μM, 51.1 ± 7.8%, 30 μM, 31.1 ± 3.9%; 40 μM, 22.2 ± 5.7%. For differences between control values and values gained with blebbistatin, a p value < 0.03 was considered significant (indicated by asterisk). (C) Primary macrophage 30 min after addition of 30 μM blebbistatin. Confocal laser scanning micrograph of substrate-attached cell side, specimen stained for F-actin. Note loss of podosomes in the cell periphery. Bar, 10 μm. (D) Model of KIF1C-dependent podosome regulation. Individual podosomes are connected by actin cables. KIF1C at microtubule plus ends binds (directly or indirectly) to myosin IIA, thereby coupling the actin and tubulin cytoskeletons. This may enable an actomyosin/kinesin-based “homing mechanism” for microtubules to podosomes (a). Alternatively, KIF1C could bind podosome-localized myosin IIA, which may temporarily stabilize the contact between both structures (b). Both possibilities are not mutually exclusive.
DISCUSSION
Here, we investigate the regulatory relationship between actin-rich podosomal adhesions and microtubules in primary human macrophages. In quiescent cells, two populations of podosomes exist at distinct subcellular locations: dynamic podosomes at the cell periphery, and more static podosomes, which are concentrated in the center of the substrate-attached cell side. These findings correlate well with previous data of mouse macrophages demonstrating the localization of dynamic precursor clusters of podosomes to the leading lamella (Evans et al., 2003). Microtubules have been implicated in the regulation of podosomes previously (Babb et al., 1997; Linder et al., 2000; Evans et al., 2003). Interestingly, distinct arrangements of podosomes such as podosome precursor clusters in macrophages or podosome rings or belts in osteoclasts exist at different subcelluar locations and display a different dependency on microtubule integrity (Evans et al., 2003; Evans et al., 2003). Together, this seems to argue for the existence of a microtubule-based, subcellularly fine-tuned regulation that targets only a subset of podosomes in a cell.
Our data now show that macrophage podosomes are targeted, sometimes repeatedly, by microtubule plus ends. Interestingly, contacted podosomes, and especially those located in the cell periphery, show a certain tendency toward dynamic behavior, i.e., fission or dissolution. This correlation is not very stringent, but together with previous data showing the overall importance of microtubules for human macrophage podosomes (Linder et al., 2000), it seems to imply that contact by microtubules may be necessary for podosome regulation, whereas not all contacts are equally productive. Indeed, experiments using nocodazole showed the absolute necessity of microtubules for podosome reformation after disruption, whereas in cells already containing podosomes, microtubules have only an impact of 30–40% on the upkeep of these structures. The latter observation may indeed reflect microtubule-dependent fission of podosome precursors leading to the formation of new podosomes.
Overall, these results are reminiscent of work on focal adhesions, which shows that microtubules repeatedly target these structures, thereby inducing their dissolution (Kaverina et al., 1999; Krylyshkina et al., 2003). In human macrophages, however, microtubule plus ends influence both dissolution and fission of podosomes, i.e., not only the breakdown but also the generation of podosomes. Ultimately, both events might be based on the same phenomenon, namely, the localized or overall dissolution of the podosome structure. For fission, for example, it could be argued that a dissolution process would have to affect only the connecting structure between future daughter podosomes. However, the factors inducing or even fine-tuning such disruption processes are unknown at the moment.
Surprisingly, and in contrast to previous results showing roughly equal rates of podosome fission and fusion in IC-21 mouse macrophages (Evans et al., 2003), we could only rarely detect fusion events (<1% of all observed podosomes) in our system. It is not clear why this difference occurs; possible explanations are the use of different species (mouse versus human), the use of a cell line versus primary cells, or the observation of migrating versus quiescent cells. Regardless, this demonstrates that podosome behavior is complex, and specifics may differ in various cell systems.
Contact with microtubules suggested the involvement of microtubule-based motor proteins in podosome regulation—a possibility unexplored so far. Initial experiments using microinjection of inhibitory antibodies or inhibitors against conventional kinesin or dynein showed no effect on podosome reformation. This is in clear contrast to the regulation of focal adhesions, where microtubule-dependent disassembly has been shown to involve conventional kinesin (Krylyshkina et al., 2002). However, we cannot rule out a possible long-term role for dynein in podosome dynamics.
A broader approach, combining overexpression of diverse kinesin constructs and siRNA transfection, led to the identification of KIF1C, a member of the Kinesin-3 family, as a motor protein potentially involved in podosome regulation. The involvement of other kinesins such as heterotrimeric kinesin in this process is possible and even likely. However, in all assays, interference with KIF1C expression or function gave the most consistent and significant results. This evaluation was further confirmed by time-lapse confocal microscopy, which revealed KIF1C accumulations being localized at microtubule plus ends and contacting regions of high podosome turnover. A comparison with CLIP170-labeled plus ends showed that KIF1C was only present at a subset of microtubules, and contact of podosomes with these plus ends was coupled with high percentages of podosomes showing fission or dissolution. Conversely, transfection of KIF1C-specific shRNA or expression of a mutant carrying a point mutation in the P-loop, which has been shown to inactive ATPase activity (Honegger et al., 1987), led to decreased podosome dynamics and podosome deficiency. Together, these data argue for KIF1C being a microtubule plus end–localized regulator of podosome dynamics.
In addition to microtubule plus ends, KIF1C also localizes to a region surrounding the MTOC. Although endogenous KIF1C can be found at both locations, the KIF1C P-loop mutant localized preferentially around the MTOC, and this was accompanied by a drastic decrease in podosomes. Interestingly, a rigor mutant of the related KIF1Bβ shows a similar localization (Honegger et al., 1987). Rigor mutations may therefore lead to nonprocessive motors that are still able to bind microtubules/minus ends but can no longer move along the filaments to reach the plus ends. In this context, it is also noteworthy that overexpressed KIF1C has been shown to form heterodimers with the endogenous motor (Dorner et al., 1999). In consequence, functional defects in overexpressed forms of KIF1C, such as the P-loop mutant we used here, probably lead to functional impairment of the endogenous form, resulting in defective regulation of podosomes.
In theory, also KIF1C unassociated to microtubules may have cellular effects. However, at least in the confocal plane of podosomes, KIF1C was almost exclusively associated with microtubules or their plus ends. Moreover, expression of the P-loop mutant, which is only defective in ATP hydrolysis, also results in podosome disruption. Therefore, the movement of KIF1C along microtubules (toward their plus ends) seems to be essential for KIF1C to exert its effect on podosomes.
The search for a potential interaction partner of KIF1C in macrophages led to the identification of nonmuscle myosin IIA. Functional coupling of kinesin and myosin is needed for the coordination of cellular processes such as vesicle transport, for example through binding of both motors to the same cargo vesicle (Schliwa and Woehlke, 2003). However, kinesins and myosins can also interact directly, as shown for KhcU and MyoVa (Huang et al., 1999). Here, we show that KIF1C can interact with nonmuscle myosin IIA through its PTPD-binding domain, thus providing an interface between the actin and tubulin cytoskeletons. The possible connection between both motors is underscored by the fact that microinjection of the myosin IIA binding domain of KIF1C, thought to compete with the interaction of both proteins, led to a dose-dependent decrease of podosomes.
Interestingly, myosin IIA is enriched in the cell periphery, where also dynamic podosomes localize, and inhibition of myosin II by blebbistatin leads to loss of podosomes preferentially in this region. Actomyosin-generated force has been shown to regulate the formation and size of focal adhesions, and it has been suggested that microtubules might trigger the dissolution of focal adhesions through the local inhibition of myosin II-dependent contractility (Kaverina et al., 2002; Bershadsky et al., 2003). A similar mechanism could be envisaged for podosomes. Alternatively, myosin IIA could also act as a bridging molecule between podosomes and microtubules. Indeed, myosins have been shown to be involved in binding or capturing microtubule plus ends and targeting them to the cell cortex (reviewed in Gundersen et al., 2004).
Considering previous findings of 1) podosomes being interconnected by a fine meshwork of actin filaments (Gavazzi et al., 1989), 2) microtubule growth sometimes occurring along actin bundles (Salmon et al., 2002), and 3) microtubules targeting focal adhesions along established “tracks” (Krylyshkina et al., 2003), one can speculate that KIF1C binding to myosin IIA could couple microtubule plus ends to podosome-connecting actin cables, thus establishing a “homing mechanism” for microtubules to efficiently target podosomes (Figure 7D). A similar model has been proposed for microtubule targeting of focal adhesions (reviewed in Small et al., 2002; Rodriguez et al., 2003). Alternatively, podosome-localized myosin IIA may enable a stabilized, productive contact by microtubules without being directly involved in microtubule targeting per se (Figure 7D). Of course, both alternatives would not be mutually exclusive.
In sum, our data demonstrate that the kinesin KIF1C is a central player in the microtubule-dependent regulation of podosomes and that the KIF1C–myosin IIA interface may play a role in facilitating podosome dynamics in a subcellularly fine-tuned manner. KIF1C is the first kinesin protein to be implicated in podosome regulation, and conversely, regulation of podosome dynamics is the first function of KIF1C described in human cells. It will be interesting to determine whether KIF1C simply acts as a “tether” for microtubule plus ends or whether it also transports regulatory factors to podosomes. Indeed, it is very likely that further molecules other than KIF1C or myosin IIA are involved in (functionally or physically) linking podosomes and microtubule plus ends. The identification of these factors and of the regulatory signals supposedly being delivered by microtubules are interesting challenges to be solved in future experiments.
Supplementary Material
[Supplemental Material]
ACKNOWLEDGMENTS
We thank Tetsu Akiyama, Angela Barth, Niels Galjart, Vladimir Gelfand, Tohru Ichimura, Young Mi Lee, Bruce Schnapp, Kazuya Shimizu, and Yoshimi Takai for various constructs; the EURIT team for help with siRNA validation; Lars Israel and Tilman Schlunck for mass spectrometry analysis; Andreas Schröder for help with live cell imaging; Raphael Kland for help with video processing; Manfred Schliwa for critical reading of the manuscript; Jürgen Heesemann and Peter C. Weber for continuous support; and Barbara Böhlig for expert technical assistance. This work is part of the doctoral thesis of P. K. Work of our laboratories has been supported by grants to S. L. and M. A. from the Deutsche Forschungsgemeinschaft (SFB 413, GRK 438), by Friedrich Baur Stiftung, and by August Lenz Stiftung.
Abbreviations used:
AMP-PNP
adenyl-5′-yl imidodiphosphate
CLIP
cytoplasmic linker protein
EHNA
erythro-9-[3-(2-hydroxynonyl)]adenine
KAP
kinesin-associated protein
KIF
kinesin-like-family
KLC
kinesin light chain
MALDI-TOF
matrix-assisted laser desorption ionization/time of flight
mRFP
monomeric red fluorescent protein
MTOC
microtubule-organizing center
PBD
protein tyrosine phosphatase-binding domain
PTPD
protein tyrosine phosphatase
siRNA
small interfering RNA
shRNA
short hairpin RNA
Footnotes
REFERENCES
- Babb S. G., Matsudaira P., Sato M., Correia I., Lim S. S. Fimbrin in podosomes of monocyte-derived osteoclasts. Cell Motil. Cytoskeleton. 1997;37:308–325. doi: 10.1002/(SICI)1097-0169(1997)37:4<308::AID-CM3>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Bershadsky A., Balaban N. Q., Geiger B. Adhesion-dependent cell mechanosensitivity. Annu. Rev. Cell Dev. Biol. 2003;19:677–695. doi: 10.1146/annurev.cellbio.19.111301.153011. [DOI] [PubMed] [Google Scholar]
- Buccione R., Orth J., McNiven M. Foot and mouth: podosomes, invadopodia and circular dorsal ruffles. Mol. Cell. Biol. 2004;5:647–657. doi: 10.1038/nrm1436. [DOI] [PubMed] [Google Scholar]
- Burns S., Thrasher A. J., Blundell M. P., Machesky L., Jones G. E. Configuration of human dendritic cell cytoskeleton by Rho GTPases, the WAS protein, and differentiation. Blood. 2001;98:1142–1149. doi: 10.1182/blood.v98.4.1142. [DOI] [PubMed] [Google Scholar]
- Destaing O., Saltel F., Geminard J. C., Jurdic P., Bard F. Podosomes display actin turnover and dynamic self-organization in osteoclasts expressing actin-green fluorescent protein. Mol. Biol. Cell. 2003;14:407–416. doi: 10.1091/mbc.E02-07-0389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Destaing O., Saltel F., Gillquin B., Chabadel A., Khochbin S., Ory S., Jurdic P. A novel Rho-mDia2-HDAC6 pathway controls podosome patterning through microtubule acetylation in osteoclasts. J. Cell Sci. 2005;118:2901–2911. doi: 10.1242/jcs.02425. [DOI] [PubMed] [Google Scholar]
- Dorner C., Ciossek T., Müller S., Møller N., Ullrich A., Lammers R. Characterization of KIF1C, a new kinesin-like protein involved in vesicle transport from the Golgi apparatus to the endoplasmic reticulum. J. Biol. Chem. 1998;273:20267–20275. doi: 10.1074/jbc.273.32.20267. [DOI] [PubMed] [Google Scholar]
- Dorner C., Ullrich A., Häring H., Lammers R. The kinesin-like motor protein KIF1C occurs in intact cells as a dimer and associates with proteins of the 14-3-3 family. J. Biol. Chem. 1999;274:33654–33660. doi: 10.1074/jbc.274.47.33654. [DOI] [PubMed] [Google Scholar]
- Evans J., Correia I., Krasavina O., Watson N., Matsudaira P. Macrophage podosomes assemble at the leading lamella by growth and fragmentation. J. Cell Biol. 2003;161:697–705. doi: 10.1083/jcb.200212037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galjart N. CLIPs and CLASPs and cellular dynamics. Nat. Rev. Mol. Cell. Biol. 2005;6:487–498. doi: 10.1038/nrm1664. [DOI] [PubMed] [Google Scholar]
- Gavazzi I., Nermut M. V., Marchisio P. C. Ultrastructure and gold-immunolabelling of cell-substratum adhesions (podosomes) in RSV-transformed BHK cells. J. Cell Sci. 1989;94:85–99. doi: 10.1242/jcs.94.1.85. [DOI] [PubMed] [Google Scholar]
- Goode B., Drubin D., Barnes G. Functional cooperation between the microtubule and actin cytoskeletons. Curr. Opin. Cell Biol. 2000;12:63–71. doi: 10.1016/s0955-0674(99)00058-7. [DOI] [PubMed] [Google Scholar]
- Gundersen G. G., Gomes E. R., Wen Y. Cortical control of microtubule stability and polarization. Curr. Opin. Cell Biol. 2004;16:106–112. doi: 10.1016/j.ceb.2003.11.010. [DOI] [PubMed] [Google Scholar]
- Honegger A. M., Dull T. J., Felder S., Van Obberghen E., Bellot F., Szapary D., Schmidt A., Ullrich A., Schlessinger J. Point mutation in the ATP binding site of EGF receptor abolishes protein-tyrosine kinase activity and alters cellular routing. Cell. 1987;51:199–209. doi: 10.1016/0092-8674(87)90147-4. [DOI] [PubMed] [Google Scholar]
- Huang J., Brady S., Richards B., Stenoien D., Resau J., Copeland N., Jenkins N. Direct interaction of microtubule- and actin-based transport motors. Nature. 1999;397:267–270. doi: 10.1038/16722. [DOI] [PubMed] [Google Scholar]
- Ingold A. L., Cohn S., Scholey J. Inhibition of kinesin-driven microtubule motility by monoclonal antibodies to kinesin heavy chains. J. Cell Biol. 1988;107:2657–2667. doi: 10.1083/jcb.107.6.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa J., Yamanaka T., Dio S., Turksen K., Heersche J., Aubin F., Takeuchi H. A band of F-actin containing podosomes is involved in bone resorption by osteoclasts. Bone. 1990;11:287–293. doi: 10.1016/8756-3282(90)90082-a. [DOI] [PubMed] [Google Scholar]
- Kaverina I., Krylyshkina O., Small J. Microtubule targeting of substrate contacts promotes their relaxation and dissociation. J. Cell Biol. 1999;146:1033–1043. doi: 10.1083/jcb.146.5.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaverina I., Krylyshkina O., Small J. Regulation of substrate adhesion dynamics during cell motility. Int. J. Biochem. Cell Biol. 2002;34:746–761. doi: 10.1016/s1357-2725(01)00171-6. [DOI] [PubMed] [Google Scholar]
- Krylyshkina O., Anderson K., Kaverina I., Upmann I., Manstein D., Small J., Toomre D. Nanometer targeting of microtubules to focal adhesions. J. Cell Biol. 2003;161:853–859. doi: 10.1083/jcb.200301102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krylyshkina O., Kaverina I., Kranewitter W., Steffen W., Alonso M., Cross R., Small J. Modulation of substrate adhesion dynamics via microtubule targeting requires kinesin-1. J. Cell Biol. 2002;156:349–359. doi: 10.1083/jcb.200105051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence C. J., et al. A standardized kinesin nomenclature. J. Cell Biol. 2004;167:19–22. doi: 10.1083/jcb.200408113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limouze J., Straight A. F., Mitchison T., Sellers J. F. Specificity of blebbistatin, an inhibitor of myosin II. J. Muscle Res. Cell Motil. 2004;25:337–341. doi: 10.1007/s10974-004-6060-7. [DOI] [PubMed] [Google Scholar]
- Linder S., Aepfelbacher M. Podosomes: adhesion hot-spots of invasive cells. Trends Cell Biol. 2003;13:376–385. doi: 10.1016/s0962-8924(03)00128-4. [DOI] [PubMed] [Google Scholar]
- Linder S., Hufner K., Wintergerst U., Aepfelbacher M. Microtubule-dependent formation of podosomal adhesion structures in primary human macrophages. J. Cell Sci. 2000;113:4165–4176. doi: 10.1242/jcs.113.23.4165. [DOI] [PubMed] [Google Scholar]
- Linder S., Kopp P. Podosomes at a glance. J. Cell Sci. 2005;118:2079–2082. doi: 10.1242/jcs.02390. [DOI] [PubMed] [Google Scholar]
- Linder S., Nelson D., Weiss M., Aepfelbacher M. Wiskott-Aldrich syndrome protein regulates podosomes in primary human macrophages. Proc. Natl. Acad. Sci. USA. 1999;96:9648–9653. doi: 10.1073/pnas.96.17.9648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louie R. K., Bahmanyar S., Siemers K. A., Votin V., Chang P., Stearns T., Nelson W. J., Barth A. I. Adenomatous polyposis coli and EB1 localize in close proximity of the mother centriole and EB1 is a functional component of centrosomes. J. Cell Sci. 2004;117:1117–1128. doi: 10.1242/jcs.00939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machuy N., Thiede B., Rajalingam K., Dimmler C., Thieck O., Meyer T. F., Rudel T. A global approach combining proteome analysis and phenotypic screening with RNA interference yields novel apoptosis regulators. Mol. Cell Proteomics. 2005;4:44–55. doi: 10.1074/mcp.M400089-MCP200. [DOI] [PubMed] [Google Scholar]
- Marchisio P. C., D'Urso N., Comoglio P. M., Giancotti F. G., Tarone G. Vanadate-treated baby hamster kidney fibroblasts show cytoskeleton and adhesion patterns similar to their Rous sarcoma virus-transformed counterparts. J. Cell. Biochem. 1988;37:151–159. doi: 10.1002/jcb.240370203. [DOI] [PubMed] [Google Scholar]
- Miki H., Setou M., Kaneshiro K., Hirokawa N. All kinesin superfamily protein, KIF, genes in mouse and human. Proc. Natl. Acad. Sci. USA. 2001;98:7004–7011. doi: 10.1073/pnas.111145398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris R., Scholey J. Heterotrimeric Kinesin-II is required for the assembly of motile 9+2 ciliary axonemes on sea urchin embryos. J. Cell Biol. 1997;138:1009–1022. doi: 10.1083/jcb.138.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K., Takei Y., Tanaka Y., Nakagawa T., Nakata T., Noda Y., Setou M., Hirokawa N. Molecular motor KIF1C is not essential for mouse survival and motor-dependent retrograde Golgi apparatus-to-endoplasmic reticulum transport. Mol. Cell. Biol. 2002;22:866–873. doi: 10.1128/MCB.22.3.866-873.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata T., Hirokawa N. Point mutation of adenosine triphosphate-binding motif generated rigor kinesin that selectively blocks anterograde lysosome membrane transport. J. Cell Biol. 1995;131:1039–1053. doi: 10.1083/jcb.131.4.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osiak A.-E., Zenner G., Linder S. Subconfluent endothelial cells form podosomes downstream of cytokine and RhoGTPase signaling. Exp. Cell Res. 2005;307:342–353. doi: 10.1016/j.yexcr.2005.03.035. [DOI] [PubMed] [Google Scholar]
- Rodriguez O. C., Schaefer A., Mandato C., Forscher P., Bement W., Waterman-Storer C. Conserved microtubule-actin interactions in cell movement and morphogenesis. Nat. Cell Biol. 2003;5:599–609. doi: 10.1038/ncb0703-599. [DOI] [PubMed] [Google Scholar]
- Sack S., Kull F. J., Mandelkow E. Motor proteins of the kinesin family. Structures, variations, and nucleotide binding sites. Eur. J. Biochem. 1999;262:1–11. doi: 10.1046/j.1432-1327.1999.00341.x. [DOI] [PubMed] [Google Scholar]
- Salmon W. C., Adams M. C., Waterman-Storer C. M. Dual-wavelength fluorescent speckle microscopy reveals coupling of microtubule and actin movements in migrating cells. J. Cell Biol. 2002;158:31–37. doi: 10.1083/jcb.200203022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliwa M., Ezzell R. M., Euteneuer U. erythro-9-[3-(2-Hydroxynonyl)]adenine is an effective inhibitor of cell motility and actin assembly. Proc. Natl. Acad. Sci. USA. 1984;81:6044–6048. doi: 10.1073/pnas.81.19.6044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliwa M., Woehlke G. Molecular motors. Nature. 2003;422:759–765. doi: 10.1038/nature01601. [DOI] [PubMed] [Google Scholar]
- Small J. V., Geiger B., Kaverina I., Bershadsky A. How do microtubules guide migrating cells? Nat. Rev. Mol. Cell. Biol. 2002;3:957–964. doi: 10.1038/nrm971. [DOI] [PubMed] [Google Scholar]
- Steffen W., Karki S., Vaughan K. T., Vallee R. B., Holzbaur E.L.F., Weiss D. G., Kuznetsov S. A. The involvement of the intermediate chain of cytoplasmic dynein in binding the motor complex to membranous organelles of Xenopus oocytes. Mol. Biol. Cell. 1997;8:2077–2088. doi: 10.1091/mbc.8.10.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watters J. W., Dewar K., Lehoczky J., Boyartchuk V., Dietrich W. F. Kif1C, a kinesin-like motor protein, mediates mouse macrophage resistance to anthrax lethal factor. Curr. Biol. Bull. 2001;11:1503–1511. doi: 10.1016/s0960-9822(01)00476-6. [DOI] [PubMed] [Google Scholar]
- Zhao C., et al. Charcot-Marie-Tooth disease type 2A caused by mutation in a microtubule motor KIF1Bβ. Cell. 2001;105:587–597. doi: 10.1016/s0092-8674(01)00363-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental Material]