Specific Effects of Ribosome-Tethered Molecular Chaperones on Programmed −1 Ribosomal Frameshifting (original) (raw)
Abstract
The ribosome-associated molecular chaperone complexes RAC (Ssz1p/Zuo1p) and Ssb1p/Ssb2p expose a link between protein folding and translation. Disruption of the conserved nascent peptide-associated complex results in cell growth and translation fidelity defects. To better understand the consequences of deletion of either RAC or Ssb1p/2p, experiments relating to cell growth and programmed ribosomal frameshifting (PRF) were assayed. Genetic analyses revealed that deletion of Ssb1p/Ssb2p or of Ssz1p/Zuo1p resulted in specific inhibition of −1 PRF and defects in Killer virus maintenance, while no effects were observed on +1 PRF. These factors may provide a new set of targets to exploit against viruses that use −1 PRF. Quantitative measurements of growth profiles of isogenic wild-type and mutant cells showed that translational inhibitors exacerbate underlying growth defects in these mutants. Previous studies have identified −1 PRF signals in yeast chromosomal genes and have demonstrated an inverse relationship between −1 PRF efficiency and mRNA stability. Analysis of published DNA microarray experiments reveals conditions under which Ssb1, Ssb2, Ssz1, and Zuo1 transcript levels are regulated independently of those of genes encoding ribosomal proteins. Thus, the findings presented here suggest that these _trans_-acting factors could be used by cells to posttranscriptionally regulate gene expression through −1 PRF.
Programmed ribosomal frameshifting (PRF) is a posttranscriptional regulatory mechanism in which elongating ribosomes are directed to shift the reading frame in response to specific _cis_-acting signals in mRNAs. Many viruses, including human immunodeficiency virus type 1, use PRF to optimize the stoichiometric ratios between their structural and enzymatic proteins (5, 11). Changes in PRF efficiency alter those ratios, inhibiting virion morphogenesis (9). In particular, retroviruses appear to be very susceptible to such changes (4, 21, 33, 37). Thus, elucidation of the molecular mechanisms underlying PRF can aid in the rational design of antiviral therapeutics (reviewed in reference 8). Though first discovered in viruses, it is also becoming apparent that programmed −1 ribosomal frameshifting (−1 PRF) is also used to regulate expression of cellular genes (reviewed in reference 34). Given the widespread use of PRF, it is important to understand the molecular mechanisms underlying PRF and to identify cellular factors that might be used to regulate these processes.
Ribosomes can be directed to shift by one base in either the 5′ or 3′ direction depending on the _cis_-acting signal. Programmed −1 ribosomal frameshifting is the result of a net shift of the translational reading frame by 1 base in the 5′ direction. The shift is typically directed by a tripartite _cis_-acting mRNA element composed of a (from 5′ to 3′) heptameric slippery site, a spacer, and a thermodynamically stable structure, typically an mRNA pseudoknot (reviewed in reference 5). It is generally accepted that −1 PRF requires a change in the forward kinetics of elongation, e.g., that the stable secondary structure forces elongating ribosomes to pause with their A- and P-site tRNAs paired with bases 2 through 7 of the slippery site. The nature of the slippery sites tends to be such that slippage of tRNAs by 1 base in the 5′ direction results in codon-anticodon pairing in the nonwobble positions.
Yeast cells commonly harbor a symbiotic double-stranded RNA virus called Killer (reviewed in reference 54). Killer is composed of the L-A helper virus, which uses −1 PRF to produce the correct ratio of structural (Gag) to enzymatic (Gag-Pol) proteins and a satellite called M1, which encodes a secreted toxin and relies on L-A as a source of viral particles and replicase function. The Killer+ phenotype of cells harboring L-A and M1 is due to the action of the secreted toxin, which kills nearby uninfected cells (a proteolytic intermediate of the protoxin confers immunity to infected cells). While changes in −1 PRF tend to have profound effects on the propagation of M1, L-A is much more resistant to these effects. Though the reasons for this remain unclear, one explanation could be that the L-A plus strand, which is the substrate for packaging into nascent viral particles, is also the mRNA from which the viral proteins are translated. The potentially high local concentrations of viral proteins may allow the L-A mRNA plus strand to package itself in cis, perhaps buffering the effects of changes in the ratios of Gag to Gag-Pol. Nonetheless, the extreme sensitivity of M1 (which must be packaged in trans) to changes in −1 PRF efficiency makes the yeast Killer virus an excellent model system for studies of −1 PRF.
Programmed +1 ribosomal frameshifting (+1 PRF) is the result of a net shift of the translational reading frame by 1 base in the 3′ direction. In Saccharomyces cerevisiae, the Ty1 retrotransposable element heptameric sequence CUU AGG C directs a +1 PRF event that results in synthesis of a TYA-TYB fusion protein (reviewed in reference 11). This mechanism is driven by a ribosomal pause at the “hungry” AGG-Arg codon which corresponds to a very low abundance Arg-tRNACCU. The low abundance of the cognate A-site tRNA is thought to cause a translational pause during which the P-site Leu-tRNAUAG, which typically decodes the CUU codon in the 0-frame P-site, slips into the UUA codon in the overlapping +1 frame. This creates a new A-site codon, GGC, a commonly used Gly codon that is decoded by an abundant cognate tRNA. The establishment of this codon-anticodon pair establishes the +1 frameshift. Changes in Ty1 +1 efficiency also have profound negative effects on Ty1 propagation.
A recurring motif in the history of modern molecular biology is that basic molecular mechanisms are first identified in viral systems. It is becoming apparent that PRF is widely used to regulate the expression of cellular as well as viral genes. Programmed +1 frameshifting is widely used to regulate expression of ornithine decarboxylase antizyme (reviewed in reference 23), and in yeast, two other genes are known to use +1 PRF (2, 26). In mice, −1 PRF is required for translation of the 3′ portions of the edr mRNA (27, 46), and in humans the paraneoplastic Ma3 gene also uses a −1 frameshift (55).
Computational approaches have also been used to identify −1 PRF signals in genome databases. A recent search of the yeast genome for fusion genes produced by a −1 PRF event employing a two-step approach identified 189 candidate genes (3). Of 58 mRNAs tested, almost half expressed full-length mRNAs encompassing both open reading frames, and sequences derived from 11 of these promoted highly efficient −1 PRF. An earlier study designed to identify sequence and structure motifs resembling viral −1 PRF signals identified significant numbers of putative −1 PRF signals in many genomes, found evolutionarily conserved −1 PRF signals in homologous genes, identified known human disease alleles that colocalized with putative −1 PRF signals, and demonstrated efficient −1 PRF promoted by sequences in two yeast genes in vivo (16).
Notably, nearly all of the −1 PRF signals were predicted to direct elongating ribosomes into premature termination events, generating the hypothesis that −1 PRF could be used to target mRNAs for rapid degradation via the nonsense-mediated mRNA decay pathway. Proof-of-principle experiments using a viral −1 PRF signal inserted in the “genomic organization” into a cellular reporter mRNA confirmed this notion (41). Additional experiments demonstrating an inverse relationship between −1 PRF efficiency and mRNA half-lives suggested that regulation of −1 PRF could be employed to posttranscriptionally regulate gene expression. More recent investigations along these lines have identified a significant number of functional −1 PRF signals that can act as mRNA destabilizing elements (J. D. Dinman, unpublished). Thus, _trans_-acting factors that affect −1 PRF efficiency could potentially regulate gene expression.
Genetic, biochemical, pharmacological, and molecular modeling studies suggest that −1 PRF occurs either during or after accommodation of aminoacyl-tRNA (aa-tRNA) into the ribosomal A-site and prior to peptidyltransfer (reviewed in reference 19). Alterations in the kinetics of aa- or peptidyl-tRNA binding or in peptidyltransfer can affect the frequency of −1 PRF (29-32, 39). In contrast, the Ty1 +1 PRF occurs while the ribosomal A-site is unoccupied, i.e., after translocation but prior to delivery of aa-tRNA to the A-site. Since the −1 and +1 PRF mechanisms occur at different stages of the elongation cycle, they can be used as foils in mechanistic studies of translation elongation (19).
A search of the literature suggested the ribosome-associated chaperone complex as candidate _trans_-acting factors that might be used to regulate PRF. During the process of protein synthesis, unfolded peptides are inevitably produced. Cells have evolved a ribosome-associated chaperone complex to ensure that nascent peptides are correctly folded and cleared from the ribosome peptide exit tunnel (reviewed in reference 44). In yeast, the functional chaperone is a triad composed of a functional interaction between the Zuo1p/Hsp40 J-protein and Ssb1p/Ssb2p, coupled with the Ssz1p Hsp70 homolog (14), an arrangement that is universally conserved from bacteria to humans (20). The Ssz1p-Zuo1p-Ssb1/2p chaperone system is not present in bacteria. Bacteria have trigger factor instead. Ribosomes isolated from mammalian cells possess a zuotin homolog called MPP11 (20). MPP11 forms a complex with a specific Hsp70 homolog (Hsp70L1), and the combination of MPP11 and Hsp70L1 (mammalian RAC) can complement the lack of RAC in yeast (36). However, the issue of whether or not Ssb1/2p is conserved from yeast to humans remains unclear.
The current study was motivated by the observation that deletion of this complex has been shown to have functional consequences. Specifically, ssb1Δ ssb2Δ and ssz1Δ zuo1Δ cells grow slowly, are cold sensitive, are hypersensitive to high osmolarity and to aminoglycoside antibiotics (13, 35, 56), and promote increased rates of nonsense and missense suppression under certain conditions in yeast (42). Here, data are presented showing that −1 PRF is specifically repressed in ssb1Δ ssb2Δ and ssz1Δ zuo1Δ cells and that these cells are hypersensitive to two translational inhibitors that were previously shown to affect this process. The implications of these findings with respect to their relevance to posttranscriptional regulation of gene expression are discussed.
MATERIALS AND METHODS
Yeast strains, media, and genetic methods.
The S. cerevisiae strains used in this study are presented in Table 1. All experiments were carried out at 30°C unless otherwise indicated. The ssz1Δ zuo1Δ (Rac−) and ssb1Δ ssb2Δ strains were provided by Sabine Rospert. Escherichia coli DH5α was used to amplify plasmids, and transformations were performed by using standard calcium chloride methods (45). Yeast strains were transformed by using the alkali cation method (22), and yeast media were prepared as previously described (10). The L-A and M1 viruses were transferred to [_rho_0] cells by cytoplasmic mixing (cytoduction) using a kar1-1 donor strain, and killer assays were performed as described previously (10). Total nucleic acids were isolated from the cytoductants as previously described (12) and treated with RNase A under high-salt conditions (500 mM NaCl) to degrade single-stranded RNA. Nucleic acids were extracted, separated through a 1% agarose-Tris-acetate-EDTA gel, and visualized with ethidium bromide.
TABLE 1.
Yeast strains used in this study
| Strain | Description | Source |
|---|---|---|
| 5×47 | Standard diploid killer tester | J. D. Dinman |
| JD758 | MATakar1-1 arg1 [L-AHN M1] | J. D. Dinman |
| MH272 3fα | _MAT_α leu2 ura3 trp1 ade2 Δhis3 GAL+HMLa rme1 | S. Rospert |
| IDA12 | _MAT_α leu2 ura3 rme1 trp1 Δhis3 GAL+HMLa ade2 ssz1::LEU2 zuo1::TRP1 | S. Rospert |
| IDA56A | _MAT_α leu2 ura3 trp1 ade2 Δhis3 GAL+HMLa rme1 ssb1::KAN ssb2::HIS3 | S. Rospert |
Plasmids and programmed ribosomal frameshifting assays.
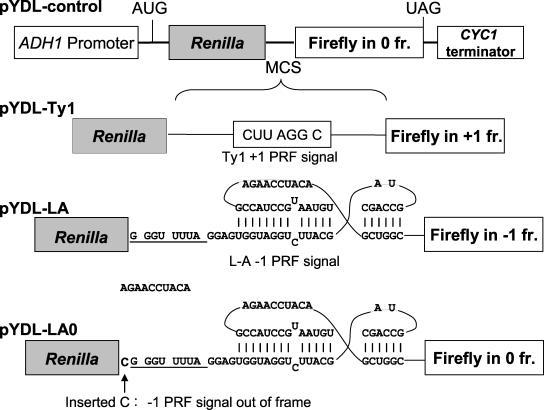
Reporter plasmids used to quantitatively monitor PRF in yeast cells are shown in Fig. 1. pYDL-control, pYDL-LA, and pYDL-Ty1 have been described previously (18). These are dual-luciferase reporter plasmids based on the pRS series of yeast shuttle vectors (6, 47). From 5′ to 3′, each reporter contains the ADH1 promoter, a translational start site, the Renilla luciferase gene, a multiple cloning site, the firefly luciferase gene, and the CYC1 transcription termination signal. In the 0-frame control pYDL-control, the firefly luciferase gene is in the 0 frame with respect to the Renilla gene. In pYDL-LA, the firefly luciferase gene is in the −1 frame respective to the Renilla gene and is 3′ of the L-A viral −1 PRF signal. In pYDL-Ty1 the firefly luciferase gene is in the +1 frame relative to the Renilla luciferase gene and is 3′ of the Ty1 +1 PRF signal. Thus, in pYDL-LA and pYDL-Ty1, firefly luciferase is expressed only consequent to a −1 or +1 PRF event, respectively.
FIG. 1.
Dual-luciferase reporters for in vivo determination of PRF efficiencies in yeast. In all reporters, transcription is initiated from the yeast ADH1 promoter and terminated at sequence derived from the CYC1 3′ untranslated region. The Renilla and firefly luciferase genes are cloned in tandem and are separated by a multiple cloning site (MCS). In pYDL-control, the Renilla and firefly luciferase genes are in frame with one another and produce a fusion of the two proteins. In pYDL-Ty1 and pYDL-LA, the Ty1 +1 and L-A −1 PRF signals are inserted in the MCS and the firefly luciferase gene is in the +1 or −1 reading frame, respectively, relative to the Renilla luciferase gene. pYDL-LA0 is an alternative 0-frame control reporter in which a cytosine residue was inserted upstream of the L-A −1 PRF signal to inactivate frameshifting while also bringing the firefly and Renilla luciferase genes into frame with one another. PRF efficiencies were calculated by dividing the ratio of firefly to Renilla luciferase generated from cells harboring the −1 or +1 test vector by the ratio of firefly to Renilla luciferase generated from cells harboring either of the 0-frame control plasmids.
Another 0-frame control reporter was made to control for potential effects of the L-A mRNA pseudoknot on ribosome processivity. Oligonucleotide site-directed mutagenesis was performed to insert a cytosine 5′ of the L-A −1 PRF signal in pYDL-LA. The resulting construct, pYDL-LA0, maintains the sequence and mRNA pseudoknot structure of pYDL-LA, but the −1 PRF signal is inactivated by placing the slippery site out of frame with respect to the incoming ribosomes. The single base insertion moves the firefly luciferase gene into the same reading frame as the Renilla luciferase gene, making the resulting construct a 0-frame control for pYDL-LA.
Luciferase assays were performed as described previously (18). Frameshifting data were analyzed using the statistical method described previously (24) to ensure that all data sets were normally distributed and passed the 95% confidence levels.
In vivo and in vitro DMS modification and RNA sequencing.
rRNAs were modified in vivo using dimethyl sulfate (DMS) as previously described (28). Briefly, logarithmically growing wild-type and mutant cultures were collected and treated with either 80 mM or 160 mM dimethyl sulfate (molecular biology grade; Sigma, St. Louis, MO). Both untreated and stop controls were included. Treatment with DMS proceeded for 3 min, β-mercaptoethanol was added to a final concentration of 0.7 M, and samples were then treated with ice-cold water-saturated isoamyl alcohol. Cells were washed with water and collected by centrifugation, and nucleic acids were extracted with acid phenol and treated with DNase, and RNAs were precipitated with ethanol.
For in vitro DMS protection studies, ribosomes were purified from logarithmically growing cells by glass bead lysis and purified by ultracentrifugation as previously described (32). Purified ribosomes were treated with 1 mM puromycin and 1 mM GTP for 30 minutes at 37°C to remove aa-tRNAs and elongation factors. Treated ribosomes were then incubated with DMS as previously described (25). RNAs isolated from DMS-treated and untreated cells and ribosomes were annealed with 32P-end-labeled primers (Table 2) by incubation at 70°C followed by slow cooling and holding for 5 min to 40°C followed by immediate transfer to ice. These oligonucleotides were designed to provide full coverage of the A-site finger (helix 38), the peptidyltransferase center, the A- and P-loops, and the sarcin/ricin loop. Primer extension and RNA sequencing were performed using avian myeloblastosis virus reverse transcriptase and nucleoside triphosphates (Roche, Mannheim, Germany) at 42°C for 30 min. Products were separated through 10% urea-acrylamide gels followed by visualization on a Kodak phosphorimager.
TABLE 2.
Oligonucleotides used for primer extension
| Primer | Sequence (5′→3′) | 25S rRNA coverage |
|---|---|---|
| 25-1 | CTTACCAAAAATGGCCCGTC | Expansion segment 12, helix 38 |
| 25-4 | TAGGCCACACTTTCATGGT | Helix 86-80 |
| 25-6 | AACCTGTCTCACGACGG | Helix 93-89 |
| 25-7 | CTTGATCAGACAGCCGC | Helix 95-93 |
| 25-8 | GACGCCTTATTCGTATCCA | Helix 98-95 |
| 25-9 | GGGACAGTGGAAATCTC | Helix 61-75 |
Growth curves and drug sensitivity assays.
Growth and drug sensitivity assays were performed in duplicate with constant high-intensity shaking at 30°C. Optical density (OD) measurements were recorded automatically using a Synergy HT microplate reader (Bio-Tek Instruments, Inc., Winooski, VT) at 595 nm and sampled every 17 min for 28 h. Cultures were inoculated from saturated overnight cultures to a starting OD595 of 0.05 into prewarmed 500-μl wells with or without drugs. Anisomycin and sparsomycin were obtained from Sigma Aldrich (St. Louis, MO).
Data analysis was done as previously described (51, 52). Optical density data were smoothed and log transformed. Estimation of lag phase was determined by calculating the slope over eight time points, approximately 2.5 h. The intercept of the highest calculated slope was designated the lag phase. Doubling times were determined by calculating the slope between every third data point on the growth curve. The lowest five of the seven highest slopes were averaged, giving the mean doubling time (DT). Doubling time was calculated as (log10 2)/mean DT. Stationary-phase OD was calculated by first determining the standard deviation of the final six OD measurements. A standard deviation of <2% of the final OD was used to designate the culture as being in stationary phase. The OD change is determined by subtracting the initial OD from the final OD.
RESULTS
Specific repression of −1 but not +1 PRF.
A previous study demonstrated that mutants of the RAC-Ssb1p/2p chaperone complex exhibited increased rates of both nonsense and missense suppression in the presence of paromomycin (42). Based on these observations, we assayed the effects of these mutants on a different aspect of translational fidelity, PRF. Isogenic wild-type, ssz1Δ zuo1Δ (Rac−), and ssb1Δ ssb2Δ cells were transformed with the reporter plasmids shown in Fig. 1 and PRF efficiencies were determined as previously described (18, 24). The mutants had little effect on +1 PRF (1.1- and 0.9-fold difference with respect to the wild type, respectively). In contrast, translational fidelity as monitored by −1 PRF was actually enhanced, i.e., deletion of either portion of the Rac-Ssb1/2 chaperone complex inhibited −1 PRF to approximately half of the wild-type level (Table 3). Given the previous findings of decreased translational fidelity by these mutants, the results of these analyses were not anticipated.
TABLE 3.
Inactivation of the RAC-Ssb1/2 chaperone complex specifically inhibits −1 PRFa
| Strain | +1 PRF (% of wt) | −1 PRF (% of wt) | −1 PRFb (% of wt) |
|---|---|---|---|
| MH272 3fα (wild type) | 7.6 ± 0.1 | 9.0 ± 0.3 | 7.3 ± 0.3 |
| IDA12 (_ssz1_Δ _zuo1_Δ) | 8.1 ± 0.3 (1.1) | 4.5 ± 0.2 (0.5) | 4.3 ± 0.1 (0.6) |
| IDA56A (_ssb1_Δ _ssb2_Δ) | 6.8 ± 0.3 (0.9) | 4.4 ± 0.2 (0.5) | 4.4 ± 0.1 (0.6) |
It is possible that a defect in ribosome processivity, e.g., decreased ability to resolve and transit through the mRNA pseudoknot in the −1 PRF signal, could result in an apparent but false inhibition of −1 PRF. pYDL-LA0 was designed and utilized to address this possibility. This 0-frame control reporter is identical to pYDL-LA except for the addition of one cytosine residue 5′ of the L-A −1 PRF signal. This base insertion inactivates the −1 PRF signal by moving the slippery site out of frame with regard to incoming ribosomes while retaining the mRNA pseudoknot structure. Isogenic wild-type and mutant cells were transformed with pYDL-LA0 and pYDL-LA, and −1 PRF efficiencies were determined. These results confirmed that the absence of the ribosome-tethered chaperone complex resulted in repression of −1 PRF (Table 3).
Killer virus maintenance defects.
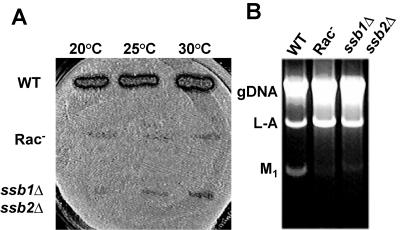
The efficiency of −1 PRF determines the ratio of structural to enzymatic proteins available for virus particle assembly, and altering −1 PRF efficiency has a severe negative impact on the ability of yeast to propagate the L-A and M1 Killer viruses (9). Thus, if inactivation of the ribosome-tethered chaperone truly inhibited −1 PRF, then such cells would not be able to propagate the Killer virus. The L-A helper and M1 satellite viruses were introduced into isogenic [_rho_0] wild-type and mutant cells by cytoduction, and cytoductants were then assayed for the killer phenotype. While wild-type cells were Killer+, neither the ssz1Δ zuo1Δ nor the ssb1Δ ssb2Δ strain was able to maintain the Killer phenotype (Fig. 2A).
FIG. 2.
Deletion of the Rac-Ssb1/2 chaperone complex results in killer virus maintenance defects. The endogenous yeast L-A and M1 killer viruses were introduced into isogenic [_rho_0] wild-type and mutant strains by cytoduction at the indicated temperatures. A. Killer virus maintenance phenotypes. Cytoductants were replica plated onto a lawn of diploid, Killer− 5X47 indicator cells and incubated at 20°C for 3 days. Killer activity is scored by the zone of growth inhibition around colonies. B. Analysis to determine the presence of viral double-stranded RNAs. Total nucleic acids were extracted from isogenic cells, subjected to RNase A treatment in 500 mM NaCl, and separated through a 1% Tris-acetate-EDTA-agarose gel. Genomic DNA (gDNA) and the L-A and M1 double-stranded RNAs are indicated.
To determine whether the Killer− phenotype was due to virus maintenance defects, as opposed to, e.g., defects in the processing or secretion of the killer toxin (reviewed in reference 53), total nucleic acids were extracted from isogenic wild-type and mutant cells, separated through an agarose-Tris-acetate-EDTA gel, and visualized by ethidium bromide staining. Compared to those in wild-type cells, M1 double-stranded RNA viral copy numbers were significantly decreased in the mutant cells; in fact, trace amounts of M1 double-stranded RNA could be visualized in the mutants only by loading approximately five times more sample than extracts of wild-type cells (Fig. 2B). The inability of the RAC mutants to maintain M1 provides independent confirmation that they confer a defect in −1 PRF.
Quantitative growth defect analyses.
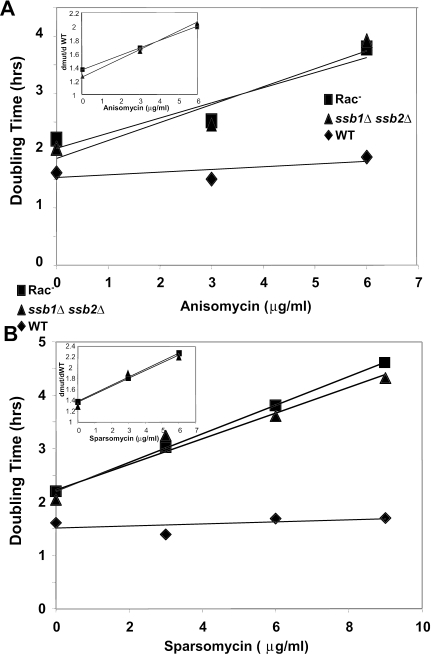
The ssb1Δ ssb2Δ and Rac− mutants are characterized by slow growth, cold sensitivity, hypersensitivity to high osmolarity, and hypersensitivity to aminoglycosides (13, 35, 56). To better understand the functional significance of the ribosome-associated chaperone complex, growth profiles of isogenic wild-type and mutant cells were quantitatively determined and compared. As summarized in Fig. 3 and Table 4, almost twice as much time was required for mutant cells to exit lag phase, and doubling times were similarly delayed in log phase. Mutant cells also reached stationary phase at significantly lower densities than did their wild-type counterparts. These findings suggest that inefficient utilization of energy resources consequent to defective protein translation resulted in decreased rates of cell growth and premature medium depletion by the mutants.
FIG. 3.
Growth curves of isogenic cells in the presence and absence of translational inhibitors. Cultures were inoculated from saturated overnight cultures into prewarmed 500-μl wells with or without the indicated drugs to an OD595 of 0.05. OD595 measurements were taken every 17 min for 28 h and recorded automatically using a Synergy HT microplate reader. Doubling times in the presence of anisomycin (A) or sparsomycin (B) during log-phase growth were calculated for increasing drug concentrations. Inset graphs indicate the fold changes in doubling times of the mutants in the presence of increasing drug concentrations normalized to that of wild-type cells under the same conditions.
TABLE 4.
Quantitative analyses of growth characteristics of cells lacking the RAC− Ssb1/2 chaperone complexa
| Parameter | Wild type (MH272 3fα) | _ssz1_Δ _zuo1_Δ mutant (IDA12) | _ssb1_Δ _ssb2_Δ mutant (IDA56A) |
|---|---|---|---|
| Lag phase (h) | 8.3 | 15.2 | 14.3 |
| Doubling time (log phase) (h) | 1.5 | 2.2 | 2.0 |
| Increase in OD (units) | 5.0 | 3.8 | 2.9 |
| Anisomycin lethal dose (μg/ml) | 25 | 9 | 9 |
| Sparsomycin lethal dose (μg/ml) | 25 | 12 | 9 |
Anisomycin and sparsomycin are small molecules that bind to the peptidyltransferase center of ribosomes, interfering with normal peptidyl transfer and tRNA binding (reviewed in reference 38). Sensitivity to these drugs is indicative of changes within the functional center of the ribosomes that affect either peptidyl transfer or tRNA binding. Quantitative analyses of cell growth in the presence of these drugs were used to monitor the effects of the RAC mutants on this region of the ribosome. While the rate of increase in the doubling times in the mutants increased, the wild-type doubling time was essentially unaffected at the same drug concentrations. This indicates that the mutants are sensitive to both anisomycin and sparsomycin, consistent with a defect in translation. (Fig. 3A and B).
DMS protection assays.
Empirical studies and modeling of the mechanism of −1 PRF suggested that defects in the ability to accommodate aa-tRNAs into the ribosomal A-site should promote specific inhibition of −1 PRF (reviewed in reference 19). Potentially, if nascent peptides are not efficiently chaperoned upon exit from the ribosome, the ribosome exit tunnel could become congested, backing up into the peptidyltransferase center. This could result in displacement of the 3′ ends of peptidyl-tRNAs toward the A-site, which in turn might inhibit the ability of new aa-tRNA to properly accommodate the A-site. Alternatively, the absence of the ribosome-associated chaperone complex could in some way affect ribosome assembly and hence its final structure. In either event, changes in tRNA positioning or overall ribosome structure could be reflected by changes in the accessibility of rRNA bases in this region to small chemical probes.
In vivo and in vitro DMS probings of rRNA were employed to test this hypothesis. While in vitro probing can identify structural changes in the nontranslating ribosome population, in vivo analysis provides a means to sample all of the ribosomes in all states of translation (nontranslating ones as well), enabling a global view of ribosome status. This can potentially allow detection of innate structural differences as well as identifying elusive differences that may occur during specific stages of translation elongation.
Logarithmically growing cells were incubated with two different concentrations of DMS (or no-DMS controls), total RNAs were extracted, and primer extension reactions using 32P-labeled oligonucleotides sufficient to probe the entire peptidyltransferase center, the sarcin/ricin loop, and helix 38 were employed as described in Materials and Methods. Puromycin-treated purified ribosomes were also probed with DMS in vitro.
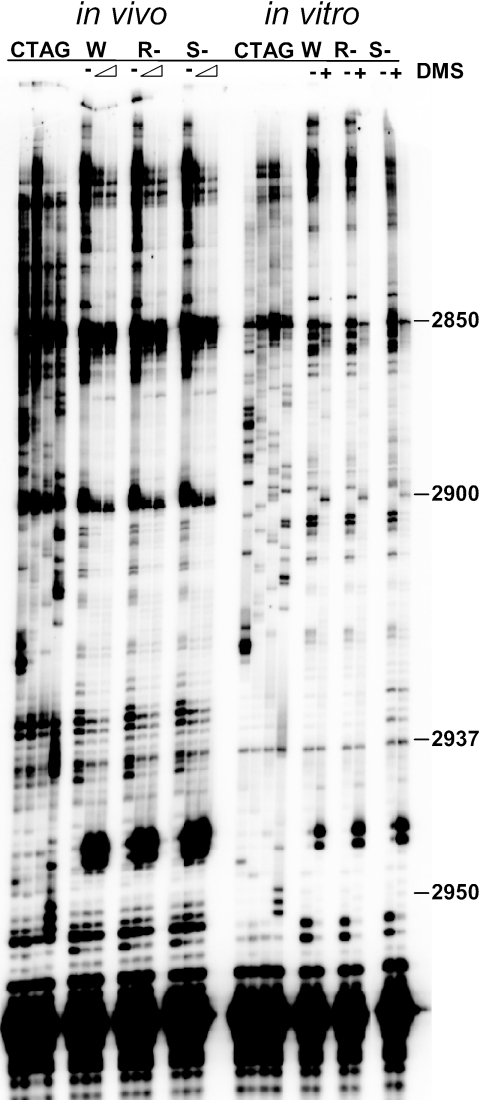
Figure 4 shows a representative autoradiogram of in vivo and in vitro DMS protection assays using primer 25-6. Although these autoradiograms show DMS-specific changes in base protection patterns, there were no detectable changes between wild-type and mutant ribosomes probed either in vivo or in vitro. The apparent ssb1Δ ssb2Δ mutant-specific changes at positions 2932 and 2935 in the in vitro assay are artifacts of this particular autoradiogram. The in vitro probing experiments demonstrate that lack of the ribosome-associated chaperone complex does not affect overall ribosome structure. As discussed below, however, the in vivo DMS protection assay may not be sensitive enough to detect changes in protection patterns if a given defect affects only a small fraction of elongating ribosomes.
FIG. 4.
DMS protection experiments: representative autoradiograms probing the vicinity of the peptidyltransferase center using primer 25-6. In vivo probing of intact cells and in vitro probing using puromycin-treated purified ribosomes are indicated. Sequencing reactions indicated by CTAG are located to the left of each set. W, wild type; R−, ssz1Δ zuo1Δ mutant; and S−, ssb1Δ ssb2Δ mutant. Untreated samples are denoted by dashes, and DMS treatment in the in vivo assays is denoted by a wedge (80 mM or 160 mM DMS) and in the in vitro assays by + symbols. Representative bases of yeast 25S rRNA are numbered.
DISCUSSION
The ribosome-associated molecular chaperones interact with nascent polypeptide chains, and it is hypothesized that they help them to fold properly and prevent their aggregation. The data presented here indicate that these chaperones are not only involved in nascent polypeptide binding and protein folding, but also affect innate ribosomal processes involved in translational fidelity. It has been suggested that nascent polypeptides can signal backwards to the peptidyltransferase center (44). The lack of effects of the mutants on +1 PRF, coupled with the inhibition of −1 PRF, and stimulation of missense and nonsense suppression can be used to infer the mechanistic consequences of such signaling on ribosome function.
As discussed in the integrated model, programmed ribosomal frameshifting must be considered within the context of the translation elongation cycle (19). In turn, the hybrid-states model of the translation elongation cycle has been described in terms of a series of nine discrete steps (reviewed in reference 15). −1 PRF occurs while the A- and P-sites of the large ribosomal subunit are occupied by tRNAs. Viewed within the context of the hybrid-states model and taking other genetic and biochemical data into account, this event was narrowed down to a window occurring after delivery of aa-tRNA to the A-site by elongation factor 1A (eEF1A) and prior to peptidyl transfer (19).
The 9-Å model, which examines the mechanism of −1 PRF, further narrows this window, predicting that inhibition of the accommodation step of the elongation cycle would decrease the steady-state abundance of the substrate for −1 PRF, decreasing the frequency of this event (40). This prediction is supported by the observation that anisomycin, which inhibits accommodation by competing with the 3′ end of the aa-tRNA for the A-site of the peptidyltransferase center (17), specifically inhibits −1 PRF (7, 24). Similarly, as accommodation is the rate-limiting step of the elongation cycle (reviewed in reference 43), such defects would also be expected to result in decreased fidelity of other events that rely on proper A-site occupancy states, whether it be the ability to discriminate between cognate and near-cognate aa-tRNAs (missense suppression) or between release factors and near- or noncognate aa-tRNAs (termination suppression). In contrast, since +1 PRF occurs when the A-site of the small subunit is unoccupied (19), it should not be affected by these mutants.
Figure 5 shows a set of cartoons describing how accommodation may be inhibited by this class of mutants. In normal circumstances (Fig. 5A, lower panel), the 3′ end of the acceptor tRNA moves from the A- to the P-site of the large subunit upon peptidyl transfer, occupying the A/P hybrid state, which is then followed by translocation, moving the peptidyl-tRNA to the P/P hybrid state. This in turn pushes the nascent peptide up into the exit tunnel. The presumed role of the ribosome-associated chaperone complex is to act at the other end of the exit tunnel, helping to ensure smooth passage of the nascent peptide out of the ribosome (Fig. 5A, upper panel).
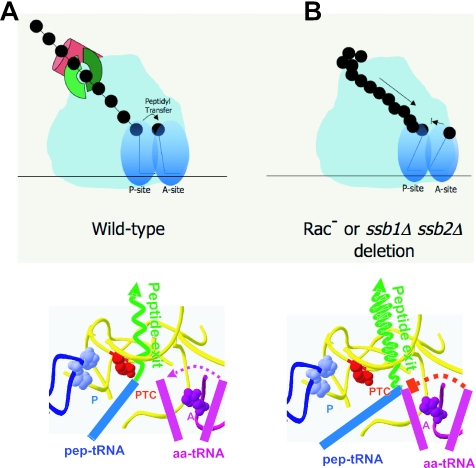
FIG. 5.
Modeling the effects of the Rac− and ssb1Δ ssb2Δ mutants on ribosome structure and function. The upper illustrations depict cartoons of translating ribosomes in wild-type and mutant cells. The lower illustrations focus on events in the peptidyltransferase center at the molecular scale. Color-coded features include the nascent peptide (green), the 3′ ends of the aa- and peptidyl-tRNAs, the A- and P-loops (purple and blue, respectively), and the peptidyltransferase center (PTC) (red). A. Wild-type ribosomes. Incoming aa-tRNAs are accommodated in the A-site of the peptidyltransferase center along the path indicated by the dotted purple line. Peptidyl transfer between peptidyl- and aa-tRNAs then occurs, and nascent peptide continues to be extruded through the exit tunnel. B. Rac− and ssb1Δ ssb2Δ mutants. Impaired chaperoning causes nascent peptides to back up into the exit tunnel, mispositioning the peptidyl-tRNA 3′ end and thus inhibiting accommodation of the aa-tRNA in the A-site (blocked dotted red line).
Akin to the proposed activity of erythromycin, which binds to the inside of the polypeptide exit tunnel, thus blocking peptide synthesis (49), the absence of ribosome-tethered molecular chaperones could result in peptide aggregation, blocking the polypeptide exit tunnel and creating problems inside the ribosome (Fig. 5B, upper panel). If movement of the nascent peptide into the tunnel were inhibited, the 3′ end of the acceptor tRNA would also be prevented from fully occupying the P-site (Fig. 5B, lower panel). Such misplacement of the peptidyl-tRNA 3′ end would in turn inhibit the accommodation of the 3′ end of the incoming aa-tRNAs into the A-site.
Although we had hoped that changes in the tRNA occupancy status at the peptidyltransferase center might have been detectable as alterations in the accessibility of rRNA residues in this area, the in vivo DMS modification data were inconclusive. This is not surprising given the fact that only approximately 5% to 10% of translating ribosomes participate in the highly transient −1 PRF event. However, the absence of gross changes in rRNA modification patterns in purified ribosomes supports the model discussed above by arguing against the notion that the observed effects were indirectly due to gross changes in ribosome structure.
The quantitative analyses of the effects of the mutants on cell growth point to the biological importance of the ribosome-associated chaperone complex not only as cellular chaperones, but also as direct effectors of translation elongation. In a logarithmically growing yeast cell, an overwhelming fraction of resources is devoted to protein synthesis (reviewed in reference 50). In the growth conditions normally used in the laboratory, i.e., unlimited nutritional resources, ideal temperatures, osmolarity, and aeration, the absence of this complex inhibits rates of cell growth and division. However, conditions in the real world are less than ideal, and the ability to posttranscriptionally regulate the translational machinery can significantly extend valuable resources and/or activate new biosynthetic pathways. Evidence that the RAC complex may be involved in such regulation comes from DNA microarray experiments in which mRNAs encoding components of the RAC complex are specifically downregulated independently of those encoding components of the translational apparatus under conditions that affect chromatin remodeling and peroxisome assembly (1, 48). Thus, in addition to the obvious function of ensuring proper folding of nascent peptides, another intriguing possibility is that this complex may be used to regulate protein translation by, e.g., regulating −1 PRF efficiency or rates of nonsense and missense suppression. As such, it may also present an attractive target for drugs directed against viruses that utilize these mechanisms.
Acknowledgments
We are indebted to Sabine Rospert for generous sharing of yeast strains. We also thank Ewan Plant and Jonathan Jacobs for critical review of the manuscript and Arturas Meskauskas for valuable conceptual discussions.
This work was supported by a grant to J.D.D. from the National Institutes of Health (GM58859).
REFERENCES
- 1.Angus-Hill, M. L., A. Schlichter, D. Roberts, H. Erdjument-Bromage, P. Tempst, and B. R. Cairns. 2001. A Rsc3/Rsc30 zinc cluster dimer reveals novel roles for the chromatin remodeler RSC in gene expression and cell cycle control. Mol. Cell 7**:**741-751. [DOI] [PubMed] [Google Scholar]
- 2.Asakura, T., T. Sasaki, F. Nagano, A. Satoh, H. Obaishi, H. Nishioka, H. Imamura, K. Hotta, K. Tanaka, H. Nakanishi, and Y. Takai. 1998. Isolation and characterization of a novel actin filament-binding protein from Saccharomyces cerevisiae. Oncogene 16**:**121-130. [DOI] [PubMed] [Google Scholar]
- 3.Bekaert, M., H. Richard, B. Prum, and J. P. Rousset. 2005. Identification of programmed translational −1 frameshifting sites in the genome of Saccharomyces cerevisiae. Genome Res. 15**:**1411-1420. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4.Biswas, P., X. Jiang, A. L. Pacchia, J. P. Dougherty, and S. W. Peltz. 2004. The human immunodeficiency virus type 1 ribosomal frameshifting site is an invariant sequence determinant and an important target for antiviral therapy. J. Virol. 78**:**2082-2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brierley, I. 1995. Ribosomal frameshifting on viral RNAs. J. Gen. Virol. 76**:**1885-1892. [DOI] [PubMed] [Google Scholar]
- 6.Christianson, T. W., R. S. Sikorski, M. Dante, J. H. Shero, and P. Hieter. 1992. Multifunctional yeast high-copy-number shuttle vectors. Yeast 110**:**119-122. [DOI] [PubMed] [Google Scholar]
- 7.Dinman, J. D., M. J. Ruiz-Echevarria, K. Czaplinski, and S. W. Peltz. 1997. Peptidyl transferase inhibitors have antiviral properties by altering programmed −1 ribosomal frameshifting efficiencies: development of model systems. Proc. Natl. Acad. Sci. USA 94**:**6606-6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dinman, J. D., M. J. Ruiz-Echevarria, and S. W. Peltz. 1998. Translating old drugs into new treatments: identifying compounds that modulate programmed −1 ribosomal frameshifting and function as potential antiviral agents. Trends Biotechnol. 16**:**190-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dinman, J. D., and R. B. Wickner. 1992. Ribosomal frameshifting efficiency and Gag/Gag-Pol ratio are critical for yeast M1 double-stranded RNA virus propagation. J. Virol. 66**:**3669-3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dinman, J. D., and R. B. Wickner. 1994. Translational maintenance of frame: mutants of Saccharomyces cerevisiae with altered −1 ribosomal frameshifting efficiencies. Genetics 136**:**75-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farabaugh, P. J. 1996. Programmed translational frameshifting. Microbiol. Rev. 60**:**103-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fried, H. M., and G. R. Fink. 1978. Electron microscopic heteroduplex analysis of “killer” double-stranded RNA species from yeast. Proc. Natl. Acad. Sci. USA 75**:**4224-4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gautschi, M., H. Lilie, U. Funfschilling, A. Mun, S. Ross, T. Lithgow, P. Rucknagel, and S. Rospert. 2001. RAC, a stable ribosome-associated complex in yeast formed by the DnaK-DnaJ homologs Ssz1p and zuotin. Proc. Natl. Acad. Sci. USA 98**:**3762-3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gautschi, M., A. Mun, S. Ross, and S. Rospert. 2002. A functional chaperone triad on the yeast ribosome. Proc. Natl. Acad. Sci. USA 99**:**4209-4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green, R., and H. F. Noller. 1997. Ribosomes and translation. Annu. Rev. Biochem. 66**:**679-716. [DOI] [PubMed] [Google Scholar]
- 16.Hammell, A. B., R. L. Taylor, S. W. Peltz, and J. D. Dinman. 1999. Identification of putative programmed −1 ribosomal frameshift signals in large DNA databases. Genome Res. 9**:**417-427. [PMC free article] [PubMed] [Google Scholar]
- 17.Hansen, J. L., P. B. Moore, and T. A. Steitz. 2003. Structures of five antibiotics bound at the peptidyl transferase center of the large ribosomal subunit. J. Mol. Biol. 330**:**1061-1075. [DOI] [PubMed] [Google Scholar]
- 18.Harger, J. W., and J. D. Dinman. 2003. An in vivo dual-luciferase assay system for studying translational recoding in the yeast Saccharomyces cerevisiae. RNA 9**:**1019-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harger, J. W., A. Meskauskas, and J. D. Dinman. 2002. An ‘integrated model’ of programmed ribosomal frameshifting and posttranscriptional surveillance. Trends Biochem. Sci. 27**:**448-454. [DOI] [PubMed] [Google Scholar]
- 20.Hundley, H. A., W. Walter, S. Bairstow, and E. A. Craig. 2005. Human Mpp11 J protein: ribosome-tethered molecular chaperones are ubiquitous. Science 308**:**1032-1034. [DOI] [PubMed] [Google Scholar]
- 21.Hung, M., P. Patel, S. Davis, and S. R. Green. 1998. Importance of ribosomal frameshifting for human immunodeficiency virus type 1 assembly and replication. J. Virol. 72**:**4819-4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ito, H., Y. Fukuda, K. Murata, and A. Kimura. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153**:**163-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ivanov, I. P., C. B. Anderson, R. F. Gesteland, and J. F. Atkins. 2004. Identification of a new antizyme mRNA +1 frameshifting stimulatory pseudoknot in a subset of diverse invertebrates and its apparent absence in intermediate species. J. Mol. Biol. 339**:**495-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacobs, J. L., and J. D. Dinman. 2004. Systematic analysis of bicistronic reporter assay data. Nucleic Acids Res. 32**:**e160-e170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kiparisov, S., A. Petrov, A. Meskauskas, P. V. Sergiev, O. A. Dontsova, and J. D. Dinman. 2005. Structural and functional analysis of 5S rRNA. Mol. Genet. Genomics 27**:**235-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lundblad, V., and D. K. Morris. 1997. Programmed translational frameshifting in a gene required for yeast telomere replication. Curr. Biol. 7**:**969-976. [DOI] [PubMed] [Google Scholar]
- 27.Manktelow, E., K. Shigemoto, and I. Brierley. 2005. Characterization of the frameshift signal of Edr, a mammalian example of programmed −1 ribosomal frameshifting. Nucleic Acids Res. 33**:**1553-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mereau, A., R. Fournier, A. Gregoire, A. Mougin, P. Fabrizio, R. Luhrmann, and C. Branlant. 1997. An in vivo and in vitro structure-function analysis of the Saccharomyces cerevisiae U3A snoRNP: protein-RNA contacts and base-pair interaction with the pre-ribosomal RNA. J. Mol. Biol. 273**:**552-571. [DOI] [PubMed] [Google Scholar]
- 29.Meskauskas, A., J. L. Baxter, E. A. Carr, J. Yasenchak, J. E. G. Gallagher, S. J. Baserga, and J. D. Dinman. 2003. Delayed rRNA processing results in significant ribosome biogenesis and functional defects. Mol. Cell. Biol. 23**:**1602-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meskauskas, A., and J. D. Dinman. 2001. Ribosomal protein L5 helps anchor peptidyl-tRNA to the P-site in Saccharomyces cerevisiae. RNA 7**:**1084-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meskauskas, A., J. W. Harger, K. L. M. Jacobs, and J. D. Dinman. 2003. Decreased peptidyltransferase activity correlates with increased programmed −1 ribosomal frameshifting and viral maintenance defects in the yeast Saccharomyces cerevisiae. RNA 9**:**982-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meskauskas, A., A. N. Petrov, and J. D. Dinman. 2005. Identification of functionally important amino acids of ribosomal protein L3 by saturation mutagenesis. Mol. Cell. Biol. 25**:**10863-10874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moosmayer, D., H. Reil, M. Ausmeier, J. G. Scharf, H. Hauser, K. D. Jentsch, and G. Hunsmann. 1991. Expression and frameshifting but extremely inefficient proteolytic processing of the HIV-1 gag and pol gene products in stably transfected rodent cell lines. Virology 183**:**215-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Namy, O., J. P. Rousset, S. Napthine, and I. Brierley. 2004. Reprogrammed genetic decoding in cellular gene expression. Mol. Cell 13**:**157-168. [DOI] [PubMed] [Google Scholar]
- 35.Nelson, R. J., T. Ziegelhoffer, C. Nicolet, M. Werner-Washburne, and E. A. Craig. 1992. The translation machinery and 70 kd heat shock protein cooperate in protein synthesis. Cell 71**:**97-105. [DOI] [PubMed] [Google Scholar]
- 36.Otto, H., C. Conz, P. Maier, T. Wolfle, C. K. Suzuki, P. Jeno, P. Rucknagel, J. Stahl, and S. Rospert. 2005. The chaperones MPP11 and Hsp70L1 form the mammalian ribosome-associated complex. Proc. Natl. Acad. Sci. USA 102**:**10064-10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park, J., and C. D. Morrow. 1991. Overexpression of the gag-pol precursor from human immunodeficiency virus type 1 proviral genomes results in efficient proteolytic processing in the absence of virion production. J. Virol. 65**:**5111-5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pestka, S. 1977. Inhibitors of protein synthesis, p. 467-553. In H. Weissbach and S. Pestka (ed.), Molecular mechanisms of protein biosynthesis. Academic Press, New York, N.Y.
- 39.Petrov, A., A. Meskauskas, and J. D. Dinman. 2004. Ribosomal protein L3: influence on ribosome structure and function. RNA Biol. 1**:**59-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Plant, E. P., K. L. M. Jacobs, J. W. Harger, A. Meskauskas, J. L. Jacobs, J. L. Baxter, A. N. Petrov, and J. D. Dinman. 2003. The 9-angstrom solution: how mRNA pseudoknots promote efficient programmed −1 ribosomal frameshifting. RNA 9**:**168-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Plant, E. P., P. Wang, J. L. Jacobs, and J. D. Dinman. 2004. A programmed −1 ribosomal frameshift signal can function as a cis-acting mRNA destabilizing element. Nucleic Acids Res. 32**:**784-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rakwalska, M., and S. Rospert. 2004. The ribosome-bound chaperones RAC and Ssb1/2p are required for accurate translation in Saccharomyces cerevisiae. Mol. Cell. Biol. 24**:**9186-9197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodnina, M. V., K. B. Gromadski, U. Kothe, and H. J. Wieden. 2005. Recognition and selection of tRNA in translation. FEBS Lett. 579**:**938-942. [DOI] [PubMed] [Google Scholar]
- 44.Rospert, S. 2004. Ribosome function: governing the fate of a nascent polypeptide. Curr. Biol. 14**:**R386-R388. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 46.Shigemoto, K., J. Brennan, E. Walls, C. J. Watson, D. Stott, P. W. J. Rigby, and A. D. Reith. 2001. Identification and characterisation of a developmentally regulated mammalian gene that utilises −1 programmed ribosomal frameshifting. Nucleic Acids Res. 29**:**4079-4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122**:**19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith, J. J., M. Marelli, R. H. Christmas, F. J. Vizeacoumar, D. J. Dilworth, T. Ideker, T. Galitski, K. Dimitrov, R. A. Rachubinski, and J. D. Aitchison. 2002. Transcriptome profiling to identify genes involved in peroxisome assembly and function. J. Cell Biol. 158**:**259-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tu, D., G. Blaha, P. B. Moore, and T. A. Steitz. 2005. Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell 121**:**257-270. [DOI] [PubMed] [Google Scholar]
- 50.Warner, J. R. 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24**:**437-440. [DOI] [PubMed] [Google Scholar]
- 51.Warringer, J., and A. Blomberg. 2003. Automated screening in environmental arrays allows analysis of quantitative phenotypic profiles in Saccharomyces cerevisiae. Yeast 20**:**53-67. [DOI] [PubMed] [Google Scholar]
- 52.Warringer, J., E. Ericson, L. Fernandez, O. Nerman, and A. Blomberg. 2003. High-resolution yeast phenomics resolves different physiological features in the saline response. Proc. Natl. Acad. Sci. USA 100**:**15724-15729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wickner, R. B. 1991. Yeast RNA virology: the killer systems, p. 263-296. In J. R. Broach, E. W. Jones, and J. R. Pringle (ed.), The molecular and cellular biology of the yeast Saccharomyces: genome dynamics, protein synthesis, and energetics, vol. 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [Google Scholar]
- 54.Wickner, R. B. 1996. Prions and RNA viruses of Saccharomyces cerevisiae. Annu. Rev. Genet. 30**:**109-139. [DOI] [PubMed] [Google Scholar]
- 55.Wills, N. M., B. Moore, A. Hammer, R. F. Gesteland, and J. F. Atkins. 5January2006. A functional −1 ribosomal frameshift signal in the human paraneoplastic Ma3 gene. J. Biol. Chem. [Epub ahead of print.] [DOI] [PubMed]
- 56.Yan, W., B. Schilke, C. Pfund, W. Walter, S. Kim, and E. A. Craig. 1998. Zuotin, a ribosome-associated DnaJ molecular chaperone. EMBO J. 17**:**4809-4817. [DOI] [PMC free article] [PubMed] [Google Scholar]