Analysis of Molecular Alterations in Left- and Right-Sided Colorectal Carcinomas Reveals Distinct Pathways of Carcinogenesis: Proposal for New Molecular Profile of Colorectal Carcinomas (original) (raw)
Abstract
To clarify distinct genetic profiles of colorectal cancers based on tumor location (left- and right-sided), we evaluated the status of loss of heterozygosity (LOH), CpG islands methylation phenotype (CIMP), microsatellite instability (MSI), and mutations of p53, Ki-ras, and APC genes in 119 colorectal cancers. Statuses of LOH (at 5q, 8p, 17p, 18q, and 22q), MSI, and CIMP (MINT1, MINT2, MINT31, MLH-1, MGMT, p14, p16, and RASSF1A) were determined using microsatellite polymerase chain reaction and methylation-specific polymerase chain reaction coupled with a crypt isolation method, respectively. In addition, mutations of p53, Ki-ras, and APC genes were also examined. LOH, MSI, and CIMP status allowed us to classify samples into two groups: low or negative and high or positive. Whereas the frequency of p53 mutations in the LOH-high status was significantly higher in left-sided cancers than in right-sided cancers, CIMP-high in the LOH-high status and MSI-positive status were more frequently found in right-sided cancers compared with left-sided cancers. Finally, location-specific methylated loci were seen in colorectal cancers: type I (dominant in right-sided cancer) and type II (common in both segments of cancer). Our data confirm that distinct molecular pathways to colorectal cancer dominate in the left and right sides of the bowel.
Recent studies have described two forms of genetic instability that are associated with colorectal cancer: chromosomal instability (CIN) and microsatellite instability (MSI).1,2 CIN is represented in most cancers and is characterized by frequent Ki-ras and p53 gene mutations and loss of heterozygosity (LOH) in a number of cancer-related genes (LOH type).1,2,3,4 CIN represents chromosomal level, but not genetic level, instability. Therefore, CIN is not identical to LOH type. MSI has a distinct genetic profile that differs from CIN in that mutations of the Ki-ras and p53 genes, as well as LOH, occur only infrequently.1,2,3,4 In a previous study, MSI was found in 10% of sporadic colorectal cancers.5 These findings suggest that the two genetic types are independent of each other and that most colorectal cancers can be classified as either MSI or CIN.6 On the other hand, microsatellite stable (MSS) is used to classify colorectal tumors that do not show MSI.7 MSI and MSS are also independent classifications that can be used to separate colorectal cancers into two largely nonoverlapping groups.7 However, MSS is not the same as CIN. MSS includes two types of genetic alterations: one with a low frequency of LOH (LOH-low) and one with a high frequency of LOH (LOH-high). CIN indicates a high frequency of LOH (LOH-high type).8 Therefore, MSS should be further broken down to indicate the two genetic types of status, LOH-high and LOH-low, when genetic profiles are analyzed in human tumors.
Another recently established molecular alteration that is commonly present in colorectal cancers is the CpG island methylator phenotype (CIMP).9 This phenotype has been identified at certain sites in the genome that are preferentially methylated in tumors.9 CIMP is recognized as an important mechanism for gene inactivation, as an alternative to gene mutation or allelic deletion, in tumorigenesis.9 Previous studies have clearly shown that CIMP is frequently found in MSI cancers, although it is not restricted to this group.7,9 However, the relationship of CIMP to high-LOH-type and low-LOH-type tumors is still not fully understood.
It has been suggested that left- and right-sided colorectal cancers differ in their associated genetic alterations in neoplastic transformation based on studies in Western countries, especially Australia and Europe.6,10 For example, MSI-positive cancers are preferentially found in right-sided colon cancer and in older women.5,6 Hawkins et al7 indicated a higher incidence of CIMP-positive tumors in right-sided colon cancer compared with left-sided colon cancers. On the other hand, CIN is said to characterize left-sided colorectal cancers.10 These findings have provided further evidence for the existence of at least two mechanisms of colorectal cancer.6 One group (CIMP+/MSI+) occurs predominantly in the right-sided colon and the other (CIN) in the left-sided colon.6,10 However, the underlying molecular features are likely to show considerable overlap between the two cancers.6 Therefore, it is important to study genetic colorectal carcinogenesis while taking into account whether the tumor is located in the left- or right-sided colon.
In this study, we examined sporadic colorectal cancers in an attempt to determine their genetic profiles in terms of LOH status, MSI, CIMP, and tumor location. Mutations of p53, Ki-ras, and APC genes that are associated with LOH-high status (which occurs with almost the same frequency as CIN) were evaluated to clarify their relationships to the genetic profiles at the two sites.
Materials and Methods
A total of 119 primary colorectal cancers and corresponding normal tissue specimens were obtained from patients at the Iwate Medical University School of Medicine. Pathological diagnosis and staging were performed according to a combination of a Japanese classification and the modified Dukes’ classification.11,12 Tumor locations were noted as left- or right-sided.
DNA Extraction
Crypt isolation from the tumor and normal mucosa was performed in accordance with a previously reported method to obtain pure glands.13,14 The isolated gland was processed routinely to confirm its nature using paraffin-embedded histological sections. Contamination by other materials such as interstitial cells was not evident in the samples that were examined, as described in previous reports.15,16 DNA from the tumor and from corresponding normal crypts was extracted by standard sodium dodecyl sulfate proteinase K treatment.
Microsatellite Analysis
LOH studies were performed by polymerase chain reaction (PCR) amplification of 13 highly polymorphic microsatellite markers (D5S107, D5S346, D5S299, D5S82, D8S201, D8S513, D8S532, TP53, D18S487, D18S34, D22S274, D22S1140, and D22S1168) located at five chromosomal loci (5q, 17p, 18q, 8p, and 22q).17 Microsatellite sequences were obtained from specific primers reported in the GDB Human Genome Database (http://gdbwww.gdb.org/gdb/). PCR amplification was performed in a 25-μl reaction volume, containing approximately 10 ng of genomic DNA, 1 μmol/L of each primer, 0.2 mmol/L deoxynucleotide triphosphate, 1× reaction buffer containing 1.5 mmol/L MgCl2, and 1.5 U of Taq polymerase (Boehringer Mannheim Co., Mannheim, Germany). Samples were processed for 25 to 30 cycles, with each cycle consisting of 30 seconds at 94°C, 1 minute at 55 to 58°C, and 2 minutes at 72°C, followed by a final 10-minute extension at 72°C. PCR products were loaded onto 6% polyacrylamide gels and run on an ABI PRISM 377 DNA Sequencer (Applied Biosystems, Foster City, CA). The data were collected automatically and analyzed by GeneScan 3.1 software (Applied Biosystems). LOH was determined by calculating the ratio of the peak areas of the constitutional alleles, as described previously.16 In this study, we defined LOH as more than a 50% difference in this ratio.
Scoring of LOH Status
LOH status was scored according to following criteria. A tumor sample was considered to be LOH-high if two or more of the markers showed LOH. When one or none of the markers showed LOH, the tumor was designated as LOH-low.
Analysis of MSI
The primers proposed by the National Cancer Institute Workshop on Microsatellite Instability (BAT25, BAT26, D5S346, D2S123, and D17S250) were used in this study.18 Products were run on an ABI PRISM 377 fluorescent DNA sequencer. PCR conditions are described elsewhere.16 MSI was defined as the presence of an additional peak. A tumor sample was considered to be MSI-high (MSI-H) when two or more of the markers demonstrated instability and MSI-low when only one marker was unstable. However, tumors showing one alteration using the above criteria and categorized as MSI-low were considered MSI-negative or MSS in this analysis.
Mutation Analysis of the p53, APC, and Ki-ras Genes
Sequencing of PCR-amplified products was used to detect mutations of exons 5 to 8 of the p53 gene, exon 1 of the Ki-ras gene, and the mutation cluster region of the APC gene in patients’ normal mucosa and tumor DNA samples. PCR conditions and sequencing of mutations were performed as described previously.15,17 Direct sequencing was performed using fluorescently labeled dideoxynucleotide triphosphates by automated DNA sequence analysis (373A sequencer; Applied Biosystems).
CIMP of Carcinomas
CIMP status was determined for carcinomas, which were evaluated at eight loci (MINT-1, MINT-2, MINT-31, p14, p16, MGMT, MLH-1, and RASSF-1A) after bisufite treatment. These loci are frequently methylated in colorectal carcinomas.7,9 Methylation-specific PCR was performed using specific primers for either the methylated or the unmethylated examined primers. Primers and conditions have been described previously.19,20 In vitro methylated DNA was used as a positive control for methylation, and water was used as a negative control. The results of methylation-specific PCR were scored when there was a clearly visible band on the gel after PCR with methylated and unmethylated primers. Electrophoresis results were interpreted by two independent investigators. When a discrepancy between the two was found, a third opinion was sought. Carcinomas were classified as CIMP-negative/low if less than three loci were methylated and CIMP-high if more than two loci were methylated. Alternatively, CIMP-negative/low and -high were classified into CIMP-low and -high, respectively.
Statistical Analysis
The data were analyzed using the χ2 test with the aid of StatView-IV software (Abacus Concepts, Berkeley, CA). Samples were determined to be significantly different at P ≥ 0.05.
Results
In the present study, differences in clinicopathological findings and genetic alterations in left-sided and right-sided colorectal carcinomas were analyzed. Molecular profiles were categorized into four types: LOH-high, low-LOH, MSI, and CIMP. Clinicopathological findings for left- and right-sided colorectal cancers are listed in Table 1.
Table 1.
Clinicopathological Findings between Left- and Right-Sided Colorectal Cancers
| Left-sided (%) | Right-sided (%) | |
|---|---|---|
| Total | 84 (70.6) | 35 (29.4) |
| Sex (male/female) | 56/28 | 20/15 |
| Age (mean) | 46–93 (64.6) | 22–94 (62.8) |
| Histological type | ||
| WDA | 18 (21.4) | 6 (17.1) |
| MDA | 65 (77.4) | 22 (62.9) |
| PDA | 1 (1.2) | 2 (5.7) |
| MC | 0 | 5 (14.3) |
| Stage | ||
| A | 14 (16.7) | 4 (11.4) |
| B | 33 (39.3) | 16 (45.7) |
| C | 22 (26.2) | 10 (28.6) |
| D | 15 (17.9) | 5 (14.3) |
Molecular Alterations in Left- and Right-Sided Cancers
Although the frequency of LOH-high in left-sided colorectal cancer (72 of 84 cases, 85.7%) was higher than that of right-sided cancer (24 of 35 cases, 68.6%), this difference was not statistically significant. There was a significant difference in the frequencies of p53 mutations found in left-sided (49 of 84, 57.6%) and right-sided (9 of 35, 25.7%) cancers (P < 0.05). However, no differences were found in the frequencies of Ki-ras and APC mutations. The CIMP-high status was found in 38 of 119 colorectal cancers that were examined (31.9%). The CIMP-high status was more common in right-sided cancers (22 of 35, 62.9%) compared with left-sided cancers (21 of 84, 25%) (P < 0.01).
Mutations of p53, Ki-ras, and APC Genes and CpG Islands Methylation Phenotype in LOH-High Status
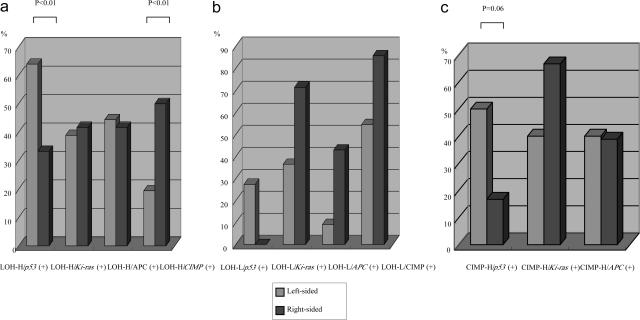
The frequency of p53 mutations in LOH-high status cancers was significantly higher for left-side cancers than for right-side cancers (Figure 1a, P < 0.01). In contrast, no significant differences in the frequencies of Ki-ras and APC mutations in LOH-high status cancers were observed for left- versus right-sided cancers (Table 2). On the other hand, the CIMP-high status in LOH-high cancers was statistically higher in right-sided (12 of 24, 50%) than in left-sided colon cancers (14 of 72, 19.4%) (Figure 1a, P < 0.01). In addition, there were no differences in the frequency of LOH at each chromosomal locus between left-sided and right-sided cancers, as shown in Table 2.
Figure 1.
a: Relationship of mutations in p53, Ki-ras, and APC in LOH-high status tumors based on tumor location. b: Relationship of mutations in p53, Ki-ras, and APC in LOH-low status tumors based on tumor location. c: Relationship of mutations in p53, Ki-ras, and APC in CIMP-high status tumors based on tumor location.
Table 2.
Frequencies of Allelic Imbalances at Cancer-Related Chromosomal Loci in Left- and Right-Sided Colorectal Cancers
| Total (left/right) | Left-sided (%) | Right-sided (%) | |
|---|---|---|---|
| 17p | 84/35 | 54/77 (70.1) | 15/25 (60) |
| 5q | 84/35 | 51/83 (61.4) | 16/30 (53.3) |
| 18q | 84/35 | 69/79 (87.3) | 24/28 (85.7) |
| 8p | 84/35 | 51/81 (60.7) | 16/30 (53.3) |
| 22q | 84/35 | 42/81 (51.9) | 15/29 (51.7) |
Mutations of p53, Ki-ras, and APC Genes and CpG Islands Methylation Phenotype in LOH-Low Status
The frequency of LOH-low status was 18 of 119 (15.1%) in our study. Although no p53 mutations were detected in right-sided colorectal cancers with LOH-low status (0 of 7), no significant difference was found compared with the frequency of p53 mutations in the left-sided cancers (3 of 11, 27.3%). Ki-ras and APC gene mutations in the LOH-low status were found more frequently in right-sided colorectal cancers (5 of 7, 71.4%; and 3 of 7, 42.9%, respectively) compared with left-sided cancers (4 of 11, 36.4%; and 1 of 11, 9.1%, respectively), but again these differences did not reach statistical significance. The frequency of CIMP status in the LOH-low cancers was high for left-sided cancers (6 of 11, 54.5%) as well as for right-sided cancers (6 of 7, 85.7%). These findings are shown in Figure 1b.
Mutations of p53, Ki-ras, and APC Genes and LOH-High Status in CpG Islands Methylation Phenotype Excluding MSI Tumors
The frequencies of CIMP-high status were significantly higher in right-sided cancers (20 of 84, 24.7%) than in left-sided cancers (18 of 35, 51.4%) (P < 0.01). Although p53 gene mutations in the CIMP-high status cancers were found more frequently in left-sided (10 of 20, 50%) compared with right-sided colorectal cancers (3 of 18, 16.7%), the difference did not reach a statistically significant level, as shown in Figure 1c (P = 0.068). On the other hand, although the frequency of Ki-ras mutations was relatively higher in right-sided cancers (12 of 18, 66.7%) than in left-sided cancers (8 of 20, 40%), no significant difference between the two sites was found. There was also no significant difference in the frequency of APC gene mutations in the CIMP-high status cancers between the two locations (left-sided, 8 of 20, 40%; right-sided, 7 of 18, 38.9%).
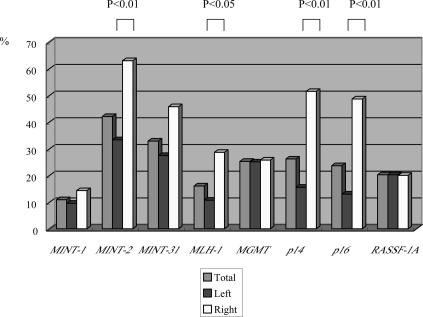
The overall frequencies of promoter hypermethylation in right-side cancers were higher, with the exception of MINT1, MGMT, and RASSF-1A. In particular, the frequencies of MINT-2, MLH-1, p14, and p16 hypermethylations in right-sided cancers were significantly higher than for left-sided cancers (P < 0.01, P < 0.05, P < 0.01, and P < 0.01, respectively). The differences for MINT31 did not reach a significant level (P = 0.052). The data are shown in Figure 2. In the present study, among the eight CpG islands occurring within known promoter regions in colorectal cancers, the hypermethylation patterns fell into two types. One type was found to be frequently methylated in right-sided colon cancers (type I). The other type was methylated in both types of colorectal cancers (type II). Whereas MINT-2, MLH-1, p14, and p16 methylations were classified as type I, MINT-1, MGMT, and RASSF-1A methylations were grouped into type II.
Figure 2.
Frequencies of methylation of MINT1, MINT2, MINT31, MLH-1, MGMT,p14, p16, and RASSF1A promoters for left- and right-sided cancers.
p53, Ki-ras, and APC Gene Mutations, CIMP, and LOH Status in MSI-Positive Status
A significant difference in the frequency of MSI between left- and right-sided colorectal cancers was found (1 of 84 cases vs. 4 of 35 cases, P < 0.05). In the present study, all MSI-positive cases that were observed had a CIMP-high status. Mutations of Ki-ras and APC genes were found in one left-sided cancer. In addition, a p53 mutation was detected in one right-sided cancer. The other three MSI-positive tumors showed no mutations. LOH-status could not be confirmed because of the presence of MSI-positive at individual chromosomal loci. Finally, representative examples of molecular analysis of left- and right-sided cancers are shown in Figures 3and 4.
Figure 3.
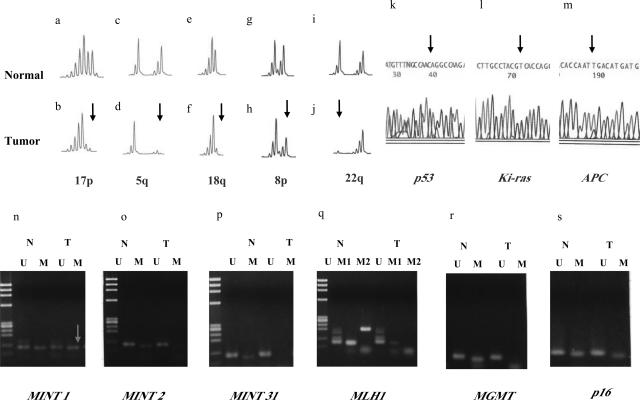
A representative example of molecular alterations in left-sided cancer. b, d, f, h, and i: Multiple LOHs were seen (arrowhead). Although methylation at MINT 1 was found, MINT2, MINT31, p16, p14 (not shown), and RASSF1A promoters were not methylated. Furthermore, mutations of p53, Ki-ras, and APC genes were found (arrowhead). k: A GGC to GAC transition in Ki-ras codon 12 was found, resulting in a Gly to Asp substitution (sequence by reverse primer). l: A CTG to CAG transversion in exon 5 of p53 gene was observed. m: Finally, a CGA to TGA transition in codon 805 of APC gene was found, resulting in a stop codon. The three detectable mutated nucleotides in tumor DNA are indicated by arrows in the photograph (k, l, and m).
Figure 4.
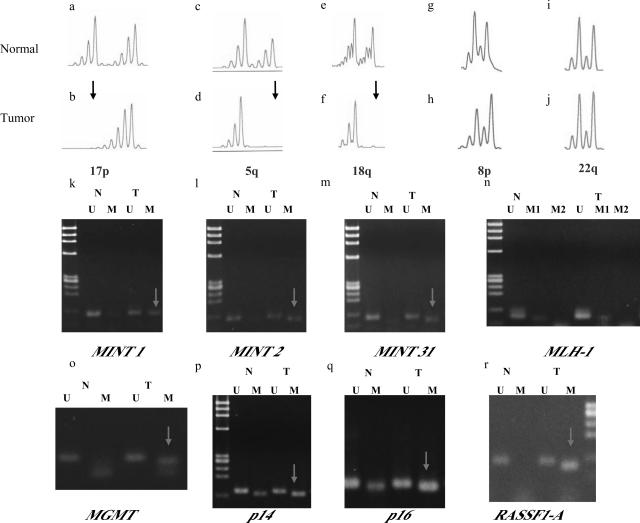
A representative example of molecular alterations in right-sided cancer. b, d, and f: Multiple LOHs were seen (arrowhead). Arrowheads indicate LOH. k, l, m, o, p, q, and r: In addition, multiple promoter regions were methylated (arrowhead). However, the p53 gene was not mutated (not shown).
Tumor Mutation Spectra of p53, Ki-ras, and APC Mutations in Left- and Right-Sided Cancers
Although the proposed sequence of genetic alterations leading to the development of colorectal cancer involves mutations in all three genes (p53, Ki-ras, and APC), a significant percentage of the tumors contained mutations in only one of these genes: p53 (22 of 119, 18.5%), Ki-ras (18 of 119, 15.1%), and APC (11 of 119, 9.3%). In our study, we characterized tumor mutation spectra in left- and right-sided cancers. The detailed results are shown in Table 3. Only 8 (9.5%) and 2 (5.7%) tumors of left- and right-sided cancers, respectively, contained mutations in all 3 genes, whereas 12 tumors (14.3%) of left-sided and 6 (8.6%) of right-sided cancers contained no mutations. The most common combination of mutations in left-sided tumors was p53 and APC (14 of 84, 16.7%), whereas that of right-sided tumors was Ki-ras and APC mutations (4 of 35, 11.4%).
Table 3.
Mutation Frequencies of p53, Ki-ras, and APC Genes in Left- and Right-Sided Cancers
| Total (%) | Left-sided (%) | Right-sided (%) | |
|---|---|---|---|
| Total | 119 | 84 | 35 |
| p53 (+)/Ki-ras (+)/APC (+) | 10 (8.4) | 8 (9.5) | 2 (5.7) |
| p53 (+)/Ki-ras (+) | 6 (5.0) | 5 (6.0) | 1 (2.9) |
| p53 (+)/APC (+) | 14 (11.8) | 14 (16.7) | 0 |
| Ki-ras (+)/APC (+) | 11 (9.2) | 7 (8.3) | 4 (11.4) |
| p53 (−)/Ki-ras (−)/APC (−) | 18 (15.1) | 12 (14.3) | 6 (8.6) |
Discussion
In this study, analysis of LOH and MSI status was performed for 119 colorectal cancers to assess the possible differences between left- and right-sided colorectal cancers. Although we did not observe significant differences in LOH status between the two locations, LOH-high status was found in 84.5% of left-sided colorectal cancers and 68.6% of right-sided colorectal cancers. In addition, we observed a tendency toward a similar frequency of LOH at each individual locus in the two segments of colorectal cancers. On the other hand, the frequency of MSI was significantly higher in right-sided colorectal cancers than in left-sided colorectal cancers.7,21 However, MSI-positive cancer is not generally representative of right-sided colorectal cancers, given that it was found in only 11.4% of right-sided colorectal cancers in our study. We note that the remainder, approximately 90%, of right-sided colorectal cancers consisted of MSS-type tumors, especially LOH-high-type tumors (approximately 70% of right-sided cancers). These findings suggest that LOH-high status is a common genetic event in both segments of colorectal cancers and that the MSI type is a minor genetic type in right-sided cancers.
In the present study, mutations in the p53 gene were predominantly present in left-sided carcinomas. The overall percentage of p53 mutated tumors and the higher incidence of p53 mutations in left-sided carcinomas is consistent with reports by other investigations,22,23,24 although a contrasting result has also been reported.25 In addition, the frequencies of p53 mutations were correlated with LOH-high status in left-sided colorectal cancers. This finding suggests that LOH-high status in left-sided colorectal cancers is characterized by p53 gene mutations. Judging from the fact that this alteration is found more frequently in the left-sided colorectal cancers, a p53 mutation may confer a selective advantage to carcinoma cells in the left-sided colon. On the other hand, interestingly, the incidence of mutations in Ki-ras and APC genes did not show regional differences, suggesting that these mutations may be relevant to the progression of colorectal cancer in both segments of the colon, although contrasting findings have been reported.25,26,27 These findings suggest that a p53 mutation is a key molecular event leading to development of left-sided colorectal cancer, and mutations of Ki-ras and APC genes are common molecular events that may be found through the entire colon.
Previous studies have shown that CIMP is more likely to occur in right-sided colon cancers compared with left-sided colorectal cancers.28 This tendency is consistent with our result that CIMP-high was more frequently observed in right-sided colon cancers than in left-sided colorectal cancers. In the present study, CIMP-high in LOH-high status occurred at a high rate in right-sided cancers compared with left-sided cancers. This finding suggests that a tumor showing LOH-high status in right-sided cancers acquires a further growth advantage by gaining CIMP-high status. This suggestion is supported by the finding that p16 and p14 tumor suppressor genes were heavily methylated in right-sided cancers.7,9 Through its ability to silence multiple genes simultaneously, CIMP in LOH-high status would functionally lead to genetic instability, resulting in the rapid accumulation of molecular alterations with a potential to accelerate the neoplastic process.
The process of molecular carcinogenesis in LOH-low status colorectal cancers is still unclear. Bell et al29 indicated that the accumulation of genetic alterations determines the nature of the neoplasm. On the other hand, another study has shown that approximately 90% of colorectal cancers contain two or more genetic alterations, and no single genetic alteration has been detected by existing techniques in all cancers.30 This finding suggests that tumors having subtle molecular alterations or even no alterations actually exist in human colorectal cancers. In the present study, the association of mutations in p53, Ki-ras, and APC genes and CIMP-high with LOH-low status between the two locations was assessed for 18 patients with LOH-low status. Although these associations showed no statistical differences, we speculate that whereas p53 mutations may be implicated in the development of some left-sided colonic cancers, Ki-ras and APC may play an important role in tumorigenesis of a subset of right-sided colorectal cancers. In addition, we suggest that CIMP-high status may be associated with development of both segments of colorectal cancer having LOH-low status. These findings suggest the existence of molecular pathways besides accumulations of multiple LOHs and MSI in at least some human colorectal cancer cases. Description of other molecular mechanisms may be required for understanding the development of such tumors.31
In the present study, whereas p53 mutations in CIMP-high status were frequently detected in left-sided cancers, the frequency of Ki-ras mutations in CIMP-high status cancers was relatively high in right-sided cancers, although these associations did not reach a significant level. In contrast, APC mutations in CIMP-high status cancers were commonly found in both segments of the colorectum. Interestingly, these findings suggest the possibility that a specific gene alteration in CIMP-high status cancers is associated with development of left- and right-sided cancers. Toyota et al28 and Hawkins et al7 indicated that Ki-ras mutations are more frequent in cancers showing CIMP-high status. Although in the present study, CIMP-high excluding MSI had higher rates of Ki-ras mutations than those seen in the overall samples (20 of 38, 52.6% versus 48 of 119, 40.3%), no significant difference between the two samples was found. It is said that CIMP is responsible for mutations of this gene.7 A recent study has shown that the O6-MGMT gene is associated with increased Ki-ras mutations.32 However, in the present study, this kind of relationship was not apparent (data not shown). Finally, Kambara et al33 have shown that BRAF mutation is frequently seen in sporadic MSI-H colorectal cancer and in sessile serrated adenomas. Their results indicated that sporadic MSI-H cancers may originate in sessile serrated adenomas and not conventional adenomas. Unfortunately, we did not assay for BRAF mutations in the series of colorectal cancers that we tested. However, MSI-H cancers that originate from serrated adenomas may be included among the MSI-H cancers that we examined, because one of the five MSI-H cancers showed histological serrated components within the cancer tissue (profile of the case: age, 71 years; sex, female; tumor location, cecum; MLH-1, methylated; CIMP status, high; Ki-ras mutation, negative; and p53 mutation, positive). We would like to test for an association between CIMP-H cancers and BRAF mutation in the near future.
We also examined the methylation status at eight individual loci in 119 colorectal carcinomas. Analysis of the 119 colorectal cancers revealed two types of hypermethylation in the cancers (type I for right-sided colon cancer specific and type II for both types of colorectal cancer specific), which may have distinct causes as well as different roles in cancer development. Type I methylation was seen more frequently in right-sided colon cancers than in left-sided colorectal cancers. In particular, the frequency of methylations at MLH1, p14, and p16 was significantly higher in right-sided colon cancers than in left-sided colorectal cancers.7,28 This result suggests that a specific type of CpG island in the human genome is incrementally methylated during tumor development in the right-sided colon. Although we do not know the mechanism of type I methylation, this type of methylation may play a major role in the progression of right-sided colon cancers.28 In contrast to type I methylation, type II methylation is commonly found through the entire colon. We suggest that these alterations commonly occur in both types of colonic cancers.
A genetic model for colorectal cancer has been proposed in which sequential accumulation of mutations in specific genes, namely the p53, Ki-ras, and APC genes, plays a major role in the development of colorectal cancer.30,34 According to the theory, mutations in the three genes are expected to accumulate in normal colonic epithelia throughout the progression of colorectal adenoma to colorectal cancer. If this theory is correct, progressive acquisition of alterations in all three genes should occur in the same tumor tissue. However, in the present study, the occurrence of mutations in all three genes was very rare among tumors of both segments, and the expected pattern of combinations of the three genes was found in a very limited subset of the cases we examined. These findings suggest that the established model of colorectal genetic carcinogenesis will only explain tumor formation in an extremely small number of colorectal cancers.35 Furthermore, our data suggest that the occurrence of multiple mutations is a causal event and does not represent a synergistic accumulation of multiple mutations occurring in the same tumor. This finding was common to both left-sided and right-sided cancers. Although the present study is not prospective, the progressive accumulation of multiple mutations in these genes may not be a prerequisite for the development of colorectal tumors in both segments.35 Therefore, the present results suggest an alterative genetic pathway to colorectal cancer.
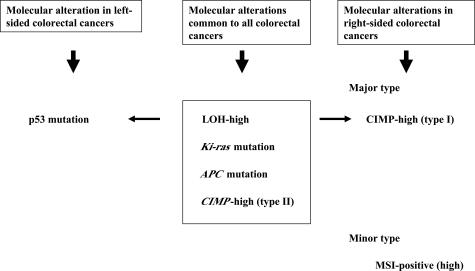
In conclusion, we propose two major categories of carcinogenesis in the case of colorectal carcinomas. One major type is LOH-high status characterized by multiple LOHs, Ki-ras, and APC mutations.1,2,3,4 The alternative type is MSI closely associated with CIMP-high status.7 The former (LOH-high status) may be subclassified into two types. The first type is defined by p53 gene mutations that are frequently found in left-sided colorectal cancers. The second type is characterized by CIMP-high status and this occurs frequently in right-sided colorectal cancers. MSI was a rather minor subcategory of right-sided cancers. Therefore, there appear to be two types of molecular alterations, designated LOH-high/CIMP-high and MSI/CIMP-high, in right-sided cancers. This molecular profile is depicted in Figure 5. CIMP was also subdivided into two groups in the present study, namely type I and type II. Whereas type I is specific to right-sided colonic cancers, type II is a common event to both segments of colorectal cancers. We suggest that there are distinct molecular carcinogenesis pathways that occur in left-sided and right-sided colorectal cancers, respectively. However, the cause of the observed differences between the two types of tumors remains unknown. Finally, accumulation of mutations in all three major genes p53, Ki-ras, and APC is a rare event in the development of colorectal carcinoma, suggesting multiple alterative genetic pathways to colorectal cancer in both segments. Further studies will be required to clarify the origins of human colorectal carcinogenesis.
Figure 5.
The molecular profile of left- and right-sided colorectal cancers.
Acknowledgments
We thank the technical assistance of Miss E. Sugawara and Mr. T. Kasai. We also thank members of the Division of Pathology, Central Clinical Laboratory, and the Department of First Internal Medicine, Iwate Medical University, for their support.
Footnotes
Supported in part by a grant-in-aid from the “Open Research” project for private universities and a matching fund subsidy from the Ministry of Education, Culture, Sport, Science, and Technology, 2002–2006.
References
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- Sugai T, Habano W, Nakamura S, Sato H, Uesugi N, Takahashi H, Jiao Y-F, Yoshida T, Itoh C. Genetic alterations in DNA diploid, aneuploid and multiploid colorectal carcinomas identified by the crypt isolation technique. Int J Cancer. 2000;88:614–619. doi: 10.1002/1097-0215(20001115)88:4<614::aid-ijc15>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- Sugai T, Habano W, Uesugi N, Jiao Y-F, Nakamura S, Yoshida T, Higuchi T. Frequent allelic imbalance at the ATM locus in DNA multiploid colorectal carcinomas. Oncogene. 2001;20:6095–6101. doi: 10.1038/sj.onc.1204731. [DOI] [PubMed] [Google Scholar]
- Jass JR, Whitehall VL, Young J, Leggett BA. Emerging concepts in colorectal neoplasia. Gastroenterology. 2002;123:862–876. doi: 10.1053/gast.2002.35392. [DOI] [PubMed] [Google Scholar]
- Iacopetta B. Are there two sides to colorectal cancer? Int J Cancer. 2002;101:403–408. doi: 10.1002/ijc.10635. [DOI] [PubMed] [Google Scholar]
- Hawkins N, Norrie M, Cheong K, Mokany E, Ku SL, Meagher A, O’Connor T, Ward R. CpG island methylation in sporadic colorectal cancers and its relationship to microsatellite instability. Gastroenterology. 2002;122:1376–1387. doi: 10.1053/gast.2002.32997. [DOI] [PubMed] [Google Scholar]
- Sugai T, Takahashi H, Habano W, Nakamura S, Sato K, Orii S, Suzuki K. Analysis of genetic alterations, classified according to their DNA ploidy pattern, in the progression of colorectal adenomas and early colorectal carcinomas. J Pathol. 2003;200:168–176. doi: 10.1002/path.1340. [DOI] [PubMed] [Google Scholar]
- Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci USA. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblom A. Different mechanisms in the tumorigenesis of proximal and distal colon cancers. Curr Opin Oncol. 2001;13:63–69. doi: 10.1097/00001622-200101000-00013. [DOI] [PubMed] [Google Scholar]
- Turnbull RB, Kyle K, Watson FR, Spratt J. Cancer of the colon; the influence of the no-touch isolation technique on survival rates. Ann Surg. 1967;166:420–427. doi: 10.1097/00000658-196709000-00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Japanese Society for Cancer of the Colon and Rectum Tokyo: Kanehara Co.; Japanese Classification of Colorectal Carcinoma, (First English Edition.) 1997:30–63. [Google Scholar]
- Arai T, Kino I. Morphometrical and cell kinetic studies of normal human colorectal mucosa: comparison between the proximal and the distal large intestine. Acta Pathol Jpn. 1989;39:725–730. doi: 10.1111/j.1440-1827.1989.tb02421.x. [DOI] [PubMed] [Google Scholar]
- Nakamura S, Goto J, Kitayama M, Kino I. Application of the crypt-isolation technique to flow-cytometric analysis of DNA content in colorectal neoplasms. Gastroenterology. 1994;106:100–107. doi: 10.1016/s0016-5085(94)94651-5. [DOI] [PubMed] [Google Scholar]
- Sugai T, Habano W, Nakamura S, Uesugi N, Sasou S, Itoh C. A unique method for mutation analysis of tumor suppressor genes in colorectal carcinomas using a crypt isolation technique. Arch Pathol Lab Med. 2000;124:382–386. doi: 10.5858/2000-124-0382-AUMFMA. [DOI] [PubMed] [Google Scholar]
- Sugai T, Habano W, Nakamura S, Yoshida T, Uesugi N, Sasou S, Itoh C, Katoh R. Use of crypt isolation to determine loss of heterozygosity of multiple tumor suppressor genes in colorectal carcinoma. Pathol Res Pract. 2000;196:145–150. doi: 10.1016/s0344-0338(00)80094-1. [DOI] [PubMed] [Google Scholar]
- Sugai T, Habano W, Uesugi N, Jiao YF, Nakamura S, Sato K, Chiba T, Ishii M. Molecular validation of the modified Vienna classification of colorectal tumors. J Mol Diagn. 2002;4:191–200. doi: 10.1016/s1525-1578(10)60703-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN, Srivastava S. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–5257. [PubMed] [Google Scholar]
- Chan AO, Broaddus RR, Houlihan PS, Issa JP, Hamilton SR, Rashid A. CpG island methylation in aberrant crypt foci of the colorectum. Am J Pathol. 2002;160:1823–1830. doi: 10.1016/S0002-9440(10)61128-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid A, Shen L, Morris JS, Issa JP, Hamilton SR. CpG island methylation in colorectal adenomas. Am J Pathol. 2001;159:1129–1135. doi: 10.1016/S0002-9440(10)61789-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyakura Y, Sugano K, Konishi F, Ichikawa A, Maekawa M, Shitoh K, Igarashi S, Kotake K, Koyama Y, Nagai H. Extensive methylation of hMLH1 promoter region predominates in proximal colon cancer with microsatellite instability. Gastroenterology. 2001;121:1300–1309. doi: 10.1053/gast.2001.29616. [DOI] [PubMed] [Google Scholar]
- Samowitz WS, Curtin K, Ma KN, Edwards S, Schaffer D, Leppert MF, Slattery ML. Prognostic significance of p53 mutations in colon cancer at the population level. Int J Cancer. 2002;99:597–602. doi: 10.1002/ijc.10405. [DOI] [PubMed] [Google Scholar]
- Soong R, Grieu F, Robbins P, Dix B, Chen D, Parsons R, House A, Iacopetta B. p53 alterations are associated with improved prognosis in distal colonic carcinomas. Clin Cancer Res. 1997;3:1405–1411. [PubMed] [Google Scholar]
- Breivik J, Lothe RA, Meling GI, Rognum TO, Borresen-Dale AL, Gaudernack G. Different genetic pathways to proximal and distal colorectal cancer influenced by sex-related factors. Int J Cancer. 1997;74:664–669. doi: 10.1002/(sici)1097-0215(19971219)74:6<664::aid-ijc18>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- Bleeker WA, Hayes VM, Karrenbeld A, Hofstra RM, Hermans J, Buys CC, Plukker JT. Impact of KRAS and TP53 mutations on survival in patients with left- and right-sided Dukes’ C colon cancer. Am J Gastroenterol. 2000;95:2953–2957. doi: 10.1111/j.1572-0241.2000.02327.x. [DOI] [PubMed] [Google Scholar]
- Bleeker WA, Hayes VM, Karrenbeld A, Hofstra RM, Verlind E, Hermans J, Poppema S, Buys CH, Plukker JT. Prognostic significance of K-ras and TP53 mutations in the role of adjuvant chemotherapy on survival in patients with Dukes C colon cancer. Dis Colon Rectum. 2001;44:358–363. doi: 10.1007/BF02234733. [DOI] [PubMed] [Google Scholar]
- Luchtenborg M, Weijenberg MP, Roemen GM, de Bruine AP, van den Brandt PA, Lentjes MH, Brink M, van Engeland M, Goldbohm RA, de Goeij AF. APC mutations in sporadic colorectal carcinomas from The Netherlands Cohort Study. Carcinogenesis. 2004;25:1219–1226. doi: 10.1093/carcin/bgh117. [DOI] [PubMed] [Google Scholar]
- Toyota M, Ohe-Toyota M, Ahuja N, Issa JP. Distinct genetic profiles in colorectal tumors with or without the CpG island methylator phenotype. Proc Natl Acad Sci USA. 2000;97:710–715. doi: 10.1073/pnas.97.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SM, Scott N, Cross D, Sagar P, Lewis FA, Blair GE, Taylor GR, Dixon MF, Quirke P. Prognostic value of p53 overexpression and c-Ki-ras gene mutations in colorectal cancer. Gastroenterology. 1993;104:57–64. doi: 10.1016/0016-5085(93)90835-z. [DOI] [PubMed] [Google Scholar]
- Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- Sugai T, Habano W, Jiao Y-F, Suzuki M, Takagi R, Otsuka K, Higuchi T, Nakamura S. Analysis of allelic imbalances at multiple cancer-related chromosomal loci and microsatellite instability within the same tumor using a single tumor gland from colorectal carcinomas. Int J Cancer. 2005;114:337–345. doi: 10.1002/ijc.20689. [DOI] [PubMed] [Google Scholar]
- Whitehall VL, Walsh MD, Young J, Leggett BA, Jass JR. Methylation of O-6-methylguanine DNA methyltransferase characterizes a subset of colorectal cancer with low-level DNA microsatellite instability. Cancer Res. 2001;61:827–830. [PubMed] [Google Scholar]
- Kambara T, Simms LA, Whitehall VL, Spring KJ, Wynter CV, Walsh MD, Barker MA, Arnold S, McGivern A, Matsubara N, Tanaka N, Higuchi T, Young J, Jass JR, Leggett BA. BRAF mutation is associated with DNA methylation in serrated polyps and cancers of the colorectum. Gut. 2004;53:1137–1144. doi: 10.1136/gut.2003.037671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AMM, Bos JL. Genetic alterations during colorectal tumor development. N Eng J Med. 1988;319:525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- Smith G, Carey FA, Beattie J, Wilkie MJ, Lightfoot TJ, Coxhead J, Garner RC, Steele RJ, Wolf CR. Mutations in APC, Kirsten-ras, and p53-alternative genetic pathways to colorectal cancer. Proc Natl Acad Sci USA. 2002;99:9433–9438. doi: 10.1073/pnas.122612899. [DOI] [PMC free article] [PubMed] [Google Scholar]