Tethering KSRP, a Decay-Promoting AU-Rich Element-Binding Protein, to mRNAs Elicits mRNA Decay (original) (raw)
Abstract
Inherently unstable mRNAs contain AU-rich elements (AREs) in their 3′ untranslated regions that act as mRNA stability determinants by interacting with ARE-binding proteins (ARE-BPs). We have destabilized two mRNAs by fusing sequence-specific RNA-binding proteins to KSRP, a decay-promoting ARE-BP, in a tethering assay. These results support a model that KSRP recruits mRNA decay machinery/factors to elicit decay. The ability of tethered KSRP to elicit mRNA decay depends on functions of known mRNA decay enzymes. By targeting the Rev response element of human immunodeficiency virus type 1 by using Rev-KSRP fusion protein, we degraded viral mRNA, resulting in a dramatic reduction of viral replication. These results provide a foundation for the development of novel therapeutic strategies to inhibit specific gene expression in patients with acquired or hereditary diseases.
mRNA stability varies considerably from one mRNA species to another and plays an important role in determining levels of gene expression (19, 46, 47). Differential mRNA decay rates are determined by specific _cis_-acting elements within the mRNA molecule. The most common cis element responsible for rapid mRNA decay in mammalian cells is the AU-rich element (ARE) present within the 3′ untranslated regions (UTRs) of a variety of short-lived mRNAs (2, 10). Recent computational analysis of the 3′ UTRs revealed that as many as 8% of human mRNAs contain AREs (2). This finding suggests that AREs may account for degradation of most unstable mRNAs in human cells.
A number of proteins bind AREs and are important for regulating ARE-directed mRNA decay (4). These proteins can be divided into two groups by function: decay-promoting/destabilizing and stabilizing ARE-binding proteins (ARE-BPs). Tristetraprolin (TTP) binds to the AREs of tumor necrosis factor alpha (TNF-α) and other cytokine mRNAs and promotes their deadenylation and decay (5). Mice lacking TTP exhibit decreased TNF-α and granulocyte-macrophage colony-stimulating factor mRNA turnover (5). BRF1, a relative of TTP, also promotes ARE-directed mRNA decay (38). AUF1 modulates mRNA decay and either stabilizes or destabilizes ARE-containing mRNAs depending on the experimental system used (26, 35, 36, 49). KSRP (K homology splicing regulatory protein), originally identified as a component of a protein complex that assembles on an intronic c-src neuronal-specific splicing enhancer (30), is required for decay of ARE-containing mRNAs (9, 18). In contrast, HuR stabilizes ARE-containing mRNAs (6).
Understanding the mechanisms that control mRNA decay requires identification of the relevant enzymatic machinery. Two mRNA decay pathways exist in Saccharomyces cerevisiae (39). Both pathways initially involve poly(A) shortening which is mediated predominantly by an enzyme complex containing Ccr4p and Caf1p (40). Subsequently, the mRNA is degraded in either a 5′-to-3′ or a 3′-to-5′ direction. In the 5′-to-3′ decay pathway, 7mGDP is removed from the 7mGpppN cap by the Dcp1p/Dcp2p decapping complex, followed by rapid degradation of the transcript by the 5′-to-3′ exonuclease Xrn1p (39). In the 3′-to-5′ decay pathway, mRNA is rapidly degraded by the cytoplasmic exosome (21), a complex containing at least nine exoribonucleases (31). Human homologs of the yeast mRNA decay enzymes have been identified and include the decapping complex DCP1/DCP2 (27, 43, 45), the 5′-to-3′ exonuclease XRN1 (3), the exosome (1), and the deadenylases CCR4 and PAN2 (12, 41). Human cells also contain an additional deadenylase, the poly(A) RNase PARN (22).
The first step in mammalian mRNA decay, including ARE-directed decay, appears to be poly(A) shortening (10, 37). However, the mechanistic steps in mammalian mRNA decay after deadenylation are not well defined. The AREs were shown to stimulate deadenylation, 3′-to-5′ decay, and 5′ decapping (9, 11, 16, 17, 32). Whether the human mRNA decay enzymes described above are involved in decay of ARE-containing mRNAs in vivo remains elusive. Despite intensive investigation, the mechanism underlining the regulation of these decay enzymes by the interactions between the AREs and decay-promoting ARE-BPs remains nebulous. We have suggested that the mRNA decay machinery is recruited to ARE-containing mRNAs through its interaction with decay-promoting ARE-BPs. This recruitment provides the basic mechanism responsible for rapid decay of the mRNAs (9, 18).
In the present study, we investigate whether KSRP triggers mRNA decay in a tethering assay. We demonstrate that tethering of KSRP elicits mRNA decay. We also tether different domains of KSRP and find that two separate regions of KSRP are able to elicit mRNA decay. KSRP is found to form a complex with mRNA decay enzymes, including the poly(A) RNase PARN, the exosome components, and the decapping enzyme DCP2. Silencing expression of PARN, DCP2, and the exosome components by RNA interference (RNAi) inhibits decay of KSRP-tethered mRNA. Overexpression of a non-RNA-binding mutant of KSRP impairs ARE-directed mRNA decay likely by preventing the access of decay enzymes (or factors) to the mRNA. Finally, we demonstrate that human immunodeficiency virus type 1 (HIV-1) mRNAs are targeted for degradation by tethering of KSRP, which results in a dramatic decrease in viral replication. These results strongly suggest that mRNAs are targeted for degradation by tethered KSRP through the recruitment of mRNA decay enzymes. This tethering system can be used as a therapeutic approach to shut off specific gene expression.
MATERIALS AND METHODS
Plasmids.
Constructs expressing human β-globin reporter mRNAs were described previously (28) and were kindly provided by J. Lykke-Andersen and J. Steitz. To express reporter mRNAs containing AREs, a polylinker (5′-AGATCTATCGATCTGCAGGATATCGCGGCCGCGTCGACAAGCTTGCATGC-3′) was inserted into a BglII site immediately downstream of the stop codon of the rabbit β-globin gene, which was previously subcloned into a tetracycline (Tet)-regulatory vector, pTRE (BD Biosciences), and was kindly provided by G. Brewer. The TNF-α ARE (nucleotides 1221 to 1310; GenBank accession no. M10988) or a fragment of the c-fos 3′ UTR containing the ARE (nucleotides 1876 to 2084; GenBank accession no. NM005252) was then inserted between EcoRV and HindIII sites. To express MS2 fusion proteins, a cDNA encoding an oligomerization-defective mutant of the MS2 coat protein (provided by J. Lykke-Andersen and J. Steitz) was amplified by PCR using a 5′ primer containing a sequence encoding the FLAG epitope and subcloned between HindIII and EcoRV sites of pcDNA3 to obtain pFLAG-MS2. The open reading frames of KSRP, BRF1, and hnRNP A1 were inserted between EcoRV and XhoI sites of pFLAG-MS2. Xpress-tagged and FLAG-tagged KSRP were constructed by subcloning KSRP cDNA between EcoRI and XhoI sites of pcDNA3-HisB (Invitrogen) and pCMV-Tag 2B (Stratagene), respectively. Constructs expressing different regions of KSRP were made by PCR and inserted into pFLAG-MS2, pcDNA3-HisB, or pCMV-Tag 2B. The expressed polypeptides and the amino acids of KSRP that they contain are as follows: KSRPN (amino acids 47 to 132), KSRPC (amino acids 501 to 711), KSRPC1 (amino acids 501 to 568), KSRPC2 (amino acids 570 to 711), KH1-4 (amino acids 133 to 500), KH1-3 (amino acids 133 to 412), KH2-4 (amino acids 219 to 500), KH12 (amino acids 133 to 316), KH23 (amino acids 219 to 412), KH34 (amino acids 317 to 500), KH3 (amino acids 317 to 412), KSRPN-KH1-3-KSRPC (amino acids 47 to 412 and 501 to 711), KSRPN-KH1-3 (amino acids 47 to 412), and KH1-3-KSRPC (amino acids 133 to 412 and 501 to 711). To express Rev fusion proteins, a cDNA encoding HIV-1 Rev was amplified by PCR and subcloned between KpnI and EcoRV sites of pcDNA3/His vector. The open reading frame of KSRP and fragments encoding KH1-4 of KSRP or enhanced green fluorescent protein (GFP) (Invitrogen) were inserted between EcoRV and XhoI sites of the resultant vector to obtain Rev-KSRP, Rev-KH1-4, and Rev-GFP, respectively.
Transfection, RNA isolation, and Northern blot analysis.
HeLa-Tet Off (HeLa-TO) cells were plated onto six-well plates and transfected using Lipofectamine. For analysis of steady-state mRNA levels, constructs expressing reporter mRNAs under the control of the cytomegalovirus promoter were used. Constructs expressing reporter mRNAs under the control of a Tet-regulatory promoter were used for analysis of mRNA half-life (_t_1/2). Thirty-six hours after transfection, cytoplasmic RNA was isolated for analysis of steady-state levels. To examine mRNA decay, 16 h after transfection, cells were collected and replated onto 35-mm plates. After another 3 h, the cells were treated with medium containing doxycycline (2 μg/ml), and cytoplasmic RNA was isolated at different times. For HIV-1 mRNA analysis, 293T cells were transfected with an HIV-1 proviral DNA (HIV-1NL4-3) and constructs expressing various Rev fusion proteins. Cytoplasmic or nuclear RNA was isolated 48 h after transfection. Northern blot analysis using a 32P-labeled RNA probe was done as previously described (18).
siRNAs.
Small interfering RNAs (siRNAs) against mRNA decay enzymes were purchased from QIAGEN. Sequences of siRNAs used to silence gene expression are GGAGCUCUGUCCUAUGUAU (for PARN), CAACCUGUUACGAGUUCUA (for CCR4), CACGGAAACUUCAGGAUAA (for DCP2), GUAGGAGACAUCGUAGUGG (for RRP4), CAAGGCCACACUCGAAGUG (for RRP46), GCAGAGUAAUGCAGUACCA (for PM-Scl100), GGAGACCAUGUGAUUGGCA (for RRP40), UCAUGCCUGUGCUGAAUCA (for MTR3), and UGAAGACUCACCAAUUAUA (for XRN1).
Immunoprecipitation analysis.
HeLa-TO cells were transfected with constructs expressing different FLAG-tagged proteins. Cytoplasmic extracts were prepared from transfected cells (9) and subjected to RNase A treatment (0.2 mg/ml at room temperature for 10 min). The treated extracts were incubated with 10 μl (bed volume) of anti-FLAG agarose (Sigma) for 4 h at 4°C. The beads were washed eight times with buffer (50 mM Tris, 150 mM NaCl, and 0.05% NP-40), and immunoprecipitated materials were eluted with sodium dodecyl sulfate sample buffer. The eluted fractions were subjected to immunoblot analysis.
RNA-binding analysis and immunoprecipitation of RNA-protein complexes.
RNA-binding assays were performed as described previously (9). After UV cross-linking, the reactions were diluted with buffer (500 μl) containing 50 mM Tris-Cl (pH 7.6), 150 mM NaCl, and 0.5% NP-40 and incubated with anti-FLAG agarose (10 μl) at 4°C for 4 h. The immunoprecipitates were washed six times and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and autoradiography.
Analysis of virus infectivity.
Virus infectivity was analyzed with the JC53-BL cell line as described previously (33). This cell line enables quantitative measurement of HIV-1 infection in a single cycle of infection based on activation of an integrated long terminal repeat-β-galactosidase expression cassette. Supernatants from transfected cultures were also analyzed for HIV-1 p24 antigen concentration by enzyme-linked immunosorbent assay as described previously (33).
RESULTS
Tethering KSRP to a non-ARE-containing mRNA elicits mRNA decay.
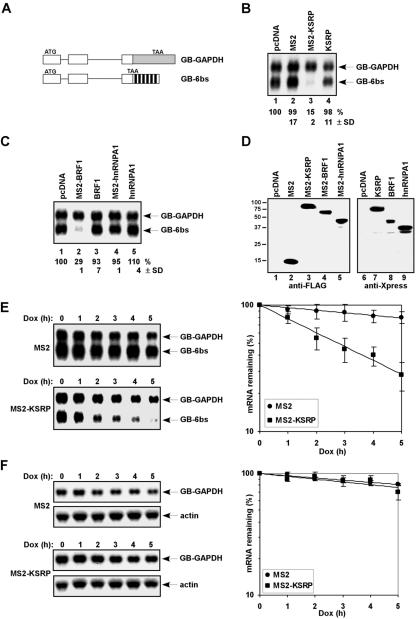
To determine whether KSRP promotes mRNA decay in a tethering assay, we fused it with the bacteriophage MS2 coat protein. Two additional ARE-BPs, including BRF1, which promotes decay of ARE-containing mRNAs (38), and hnRNP A1, whose role in ARE-directed mRNA decay is unknown (20), were also fused with the MS2 coat protein. We coexpressed these MS2-ARE-BP fusion proteins in HeLa-TO cells with β-globin reporter mRNAs containing either six MS2 coat protein binding sites (GB-6bs) or a fragment of GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (GB-GAPDH) in the 3′ UTR (Fig. 1A). As the MS2 coat protein binds to its cognate site as a dimer, at most six MS2-KSRP dimers could bind to the MS2 sites on the reporter mRNA, as opposed to a KSRP dimer which may likely bind to an ARE (7). Expression levels of reporter mRNAs were examined by Northern blot analysis. Coexpression of MS2-KSRP dramatically decreased the steady-state levels of GB-6bs mRNA compared to that in cells coexpressing the MS2 coat protein (Fig. 1B, compare lane 3 to lane 2). In contrast, coexpression of KSRP lacking the MS2 coat protein did not significantly decrease GB-6bs mRNA levels (Fig. 1B, lane 4). A decrease in the expression levels of GB-6bs mRNA was also detected when BRF1, but not hnRNP A1, was tethered to the mRNA (Fig. 1C, lanes 2 and 4). The reduction was specific to the tethering, as no significant decrease in the GB-6bs mRNA levels was detected in cells coexpressing BRF1 lacking the MS2 coat protein (Fig. 1C, lane 3). Thus, we conclude that tethering decay-promoting ARE-BPs, including KSRP and BRF1, to a heterologous non-ARE-containing mRNA causes a marked reduction in the expression levels.
FIG. 1.
Tethering KSRP to a non-ARE-containing mRNA triggers mRNA decay. (A) Schematic diagram of constructs expressing human β-globin reporter mRNAs. Exons and introns are shown as boxes and lines, respectively. The fragment containing part of the coding region and 3′ UTR of GAPDH inserted is shown as a gray box. The six copies of the MS2 binding site are depicted as black boxes. (B and C) Northern blot analysis of the steady-state levels of reporter mRNAs in HeLa-TO cells expressing various effectors. Numbers below lanes represent the levels of GB-6bs normalized to that of GB-GAPDH. GB-6bs levels in cells transfected with pcDNA were set at 100%. Mean values with standard deviations (SDs) are shown. (D) Immunoblot analysis shows expression of transfected proteins by use of anti-FLAG (lanes 1 to 5) or anti-Xpress (lanes 6 to 9) antibody. (E) HeLa-TO cells were transfected with a construct expressing GB-6bs mRNA, under the control of a Tet-regulatory promoter, a construct constitutively expressing GB-GAPDH mRNA, and constructs expressing either MS2 or MS2-KSRP. The decay of GB-6bs mRNA was analyzed (left panel). Varied levels of GB-GAPDH mRNA are due to loading variations but not degradation of the mRNA. Signals of GB-6bs were quantitated by a phosphorimager, normalized to that of GB-GAPDH, and plotted as mean values ± SDs against time (right panel). (F) HeLa-TO cells were transfected with a reporter expressing GB-GAPDH mRNA, under the control of a Tet-regulatory promoter, and constructs expressing either MS2 or MS2-KSRP. The decay of GB-GAPDH mRNA and levels of endogenous β-actin mRNA were analyzed (left panel). Signals of GB-GAPDH were quantitated, normalized to β-actin mRNA levels, and plotted as mean values ± SDs against time (right panel). Dox, doxycycline.
To determine whether the reduction in mRNA levels caused by tethered KSRP is indeed due to a consequence of increased mRNA decay, we expressed GB-6bs and GB-GAPDH mRNAs under the control of a Tet-regulatory promoter in HeLa-TO cells and analyzed decay of the mRNAs in the presence of coexpressed MS2 or MS2-KSRP after addition of doxycycline. GB-6bs mRNA was degraded slowly in control cells expressing the MS2 coat protein (_t_1/2 = 15 h) but was degraded rapidly in cells expressing MS2-KSRP (_t_1/2 = 2.5 h) (Fig. 1E). In contrast, the half-life of GB-GAPDH mRNA was not affected by coexpressing MS2-KSRP (_t_1/2 = 15 h) (Fig. 1F). Thus, we conclude that tethering of KSRP reduces mRNA levels by increasing decay rate.
The KH domain and the C-terminal domain of KSRP trigger decay when tethered to an mRNA.
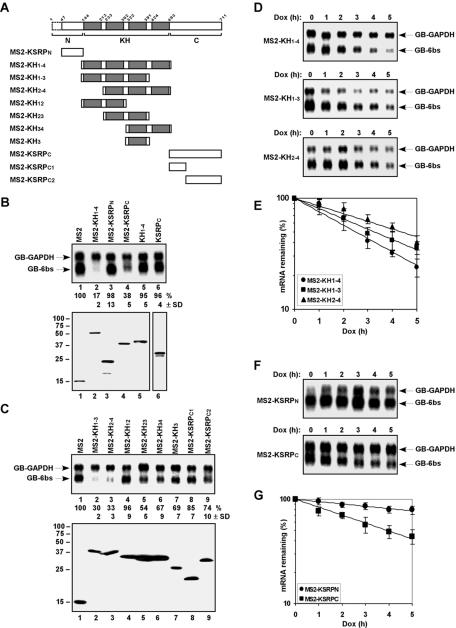
We previously showed that the central KH domain of KSRP was sufficient to promote ARE-directed mRNA decay. This domain is composed of activities for RNA binding and interaction with mRNA decay enzymes (18). We investigated which domain(s) of KSRP is able to elicit decay when tethered to an mRNA. KSRP comprises a 143-amino-acid N terminus, a central 350-amino-acid RNA-binding KH domain composed of four KH motifs (each motif comprises ∼70 amino acids), and a 218-amino-acid C terminus (Fig. 2A). Whereas the KH motifs recognize specific RNA sequences, functions of the N and C termini are still unclear. We first constructed three MS2 fusion proteins comprising the N terminus (KSRPN; amino acids 47 to 132), the central four KH motifs (KH1-4; amino acids 133 to 500), or the C terminus (KSRPC; amino acids 501 to 711) of KSRP. We coexpressed these MS2 fusion proteins with reporter mRNAs and examined their ability to reduce mRNA levels. Coexpression of MS2-KH1-4, but not MS2-KSRPN, dramatically decreased the steady-state levels of GB-6bs mRNA (Fig. 2B, compare lane 2 with lane 3). The C-terminal domain of KSRP also efficiently reduced the levels of tethered mRNA (Fig. 2B, lane 4). The reduction in GB-6bs mRNA levels by MS2-KH1-4 and MS2-KSRPC was specific to the tethering, as no significant decrease in the mRNA levels was detected in cells coexpressing their counterparts lacking the MS2 coat protein (Fig. 2B, lanes 5 and 6).
FIG. 2.
The central KH motifs or the C-terminal domain of KSRP is capable of triggering mRNA decay. (A) Schematic diagram of KSRP and different fragments used to prepare expression vectors. The N and C termini are shown as open boxes, and the four KH motifs are depicted as gray boxes. Our KSRP clone lacks the first 46 amino acids (indicated as dashed lines). (B and C) Northern blot analysis of the steady-state levels of reporter mRNAs in HeLa-TO cells expressing various effectors (top panels). The levels of GB-6bs mRNA were quantified as described in the legend for Fig. 1. Mean values with standard deviations (SDs) are shown. Expression of transfected proteins analyzed by anti-FLAG or anti-Xpress (B, lane 6) immunoblotting is shown (bottom panels). (D and F) The decay of GB-6bs mRNA was analyzed in cells coexpressing (D) different KH motifs or (F) either the N or the C terminus of KSRP fused with the MS2 coat protein. Varied levels of GB-GAPDH mRNA are due to loading variations but not degradation of the mRNA. (E and G) Signals of GB-6bs shown in panels D and F (for panels E and G, respectively) were quantitated, normalized to that of GB-GAPDH, and plotted as mean values ± SDs against time. Dox, doxycycline.
To further define the central KH and C-terminal domains responsible for decrease in mRNA levels, we fused fragments consisting of different KH motifs or different regions of the C terminus with the MS2 coat protein (Fig. 2A). MS2 fusion polypeptides containing KH1-3 or KH2-4 efficiently decreased GB-6bs mRNA levels (Fig. 2C, lanes 2 and 3). MS2 fusion polypeptides comprising KH23, KH34, or KH3 also caused moderate reduction in GB-6bs mRNA levels, whereas KH12 was inactive when tethered to the mRNA (Fig. 2C, lanes 4 to 7). Removal of either the distal two-third (KSRPC1) or the proximal one-third (KSRPC2) of the KSRP C terminus significantly impaired the ability to decrease tethered mRNA levels (Fig. 2C, lanes 8 and 9). These data suggest that multiple sequences within KH1-3 and KH2-4 motifs and both sequences residing in KSRP C1 and C2 fragments are required for efficiently triggering decay of tethered mRNA.
Half-life studies indicated that the reduction in GB-6bs mRNA levels by MS2 fusion polypeptides results from increased mRNA decay rate. Coexpression of MS2-KH1-4, -KH1-3, or -KH2-4 accelerated decay of GB-6bs mRNA (Fig. 2D and E). The C terminus (KSRPC), but not the N terminus (KSRPN), of KSRP increased turnover rate of the tethered mRNA (Fig. 2F and G). Thus, consistent with our previous results on the role of the central KH domain of KSRP in ARE-directed mRNA decay (18), KH1-4 constitutes an mRNA decay activation domain in tethering assays. Additionally, the C terminus of KSRP constitutes a second mRNA decay activation domain in these tethering assays.
The KH and the C-terminal domains of KSRP associate with mRNA decay enzymes.
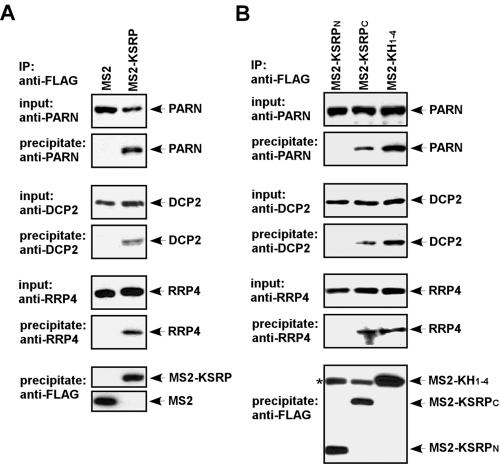
We previously demonstrated that KSRP associates with mRNA decay enzymes, including PARN and the exosome components (18). We examined whether MS2-KSRP exists in a complex containing mRNA decay enzymes. We transiently expressed FLAG-tagged MS2 or MS2-KSRP and examined their association with mRNA decay enzymes in RNase-treated extracts by coimmunoprecipitation assays (Fig. 3). MS2 or MS2-KSRP was immunopurified with anti-FLAG antibody, and the presence of decay enzymes was analyzed by immunoblotting. PARN, DCP2, and an exosome component, RRP4, were coimmunopurified with MS2-KSRP but not with MS2 (Fig. 3A). To determine which regions of KSRP are responsible for the association, we expressed FLAG-tagged MS2-KSRP polypeptides containing the N or C terminus or KH1-4 and examined their association with mRNA decay enzymes. The tested decay enzymes were coimmunopurified with MS2-KH1-4 and MS2-KSRPC but not with MS2-KSRPN (Fig. 3B). Based on these results, we suggest that MS2-KSRP recruits mRNA decay enzymes onto the mRNA through both the KH motifs and the C terminus.
FIG. 3.
MS2-KSRP associates with human mRNA decay enzymes. (A) HeLa-TO cells were transfected with constructs expressing FLAG-tagged MS2 or MS2-KSRP. RNase-treated cell extracts were subjected to anti-FLAG immunoprecipitation. (B) HeLa-TO cells were cotransfected with constructs expressing FLAG-tagged MS2-KSRPN, MS2-KSRPC, or MS2-KH1-4. Cell extracts were subjected to anti-FLAG immunoprecipitation. For both panels, the precipitates were analyzed by anti-PARN, anti-DCP2, anti-RRP4, or anti-FLAG immunoblotting. A 2.5% input for immunoprecipitation reactions was also analyzed. The asterisk indicates the heavy chain of anti-FLAG immunoglobulin G, which comigrates with MS2-KH1-4 and cross-reacts with the secondary antibody. IP, immunoprecipitate.
Decay of KSRP-tethered transcripts relies on functions of known mRNA decay enzymes.
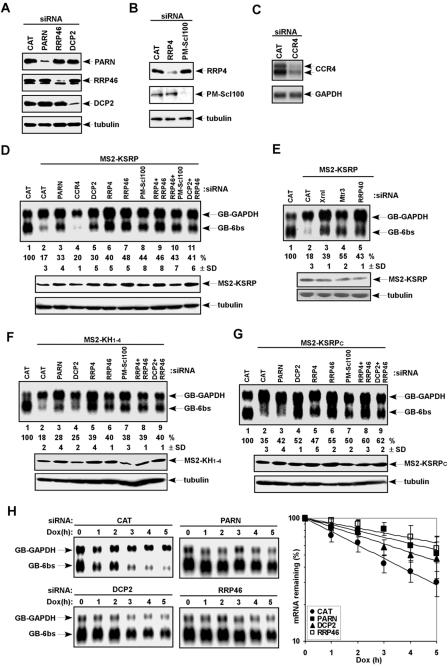
We investigated whether mRNA decay elicited by tethered KSRP requires mRNA decay enzymes that associate with KSRP. We downregulated the expression of PARN, CCR4, DCP2, and the exosome components RRP4, RRP46, and PM-Scl100 by RNAi and examined the levels of KSRP-tethered mRNA. When expression of RRP4, RRP46, PM-Scl100, PARN, or DCP2 was decreased to 10 to 20% of the control levels (Fig. 4A and B), KSRP-tethered mRNA levels were restored (Fig. 4D). Similarly, downregulation of the 5′-to-3′ mRNA decay enzyme XRN1 and two more exosome components, RRP40 and MTR3, led to restoration of KSRP-tethered mRNA levels (Fig. 4E). The varied levels of restoration in cells treated with exosome siRNAs may reflect the silencing efficiency of the siRNAs. In contrast, downregulation of CCR4 (Fig. 4C), a major deadenylase involved in mRNA decay in yeast (40), did not increase the mRNA levels (Fig. 4D). The overall levels of GB-GAPDH and GB-6bs mRNAs were reduced in cells treated with CCR4 siRNA, perhaps reflecting a role of CCR4 in transcription, as suggested with yeast (15). In similar experiments, silencing expression of PARN, DCP2, and exosome components restored the levels of the mRNA tethered by KH1-4 or KSRPC. While downregulation of PARN or DCP2 moderately increased the levels of the mRNA tethered by KH1-4, downregulation of exosome components significantly restored tethered mRNA levels (Fig. 4F). The levels of KSRPC-tethered mRNA were equally restored in cells depleted of DCP2 or exosome components (Fig. 4G).
FIG. 4.
mRNA decay triggered by tethered KSRP requires human mRNA decay enzymes. (A and B) HeLa-TO cells were transfected with siRNAs targeting chloramphenicol acetyltransferase (CAT), PARN, RRP46, or DCP2 (A) and siRNAs targeting CAT, RRP4, or PM-Scl100 (B). Total extracts were subjected to immunoblot analysis with anti-PARN, anti-RRP46, anti-DCP2, or anti-α-tubulin antibody (A) and with anti-RRP4, anti-PM-Scl100, or anti-α-tubulin antibody (B). (C) HeLa-TO cells were transfected with siRNAs targeting CAT or CCR4. poly(A)+ mRNAs were subjected to Northern blot analysis with probes against CCR4 or GAPDH. (D to G) The levels of β-globin mRNAs were analyzed in cells coexpressing MS2 (lanes 1), MS2-KSRP (D, lanes 2 to 11, and E, lanes 2 to 5), MS2-KH1-4 (F, lanes 2 to 9), or MS2-KSRPC (G, lanes 2 to 9) along with siRNAs targeting different mRNA decay enzymes as indicated (top panels). The levels of GB-6bs mRNA were quantified as described in the legend for Fig. 1. Mean values with standard deviations (SDs) are shown. The expression of MS2-KSRP (D and E), MS2-KH1-4 (F), and MS2-KSRPC (G) in siRNA-treated cells was analyzed by anti-FLAG immunoblotting (bottom panels). (H) The decay of GB-6bs mRNA was analyzed in cells coexpressing MS2-KSRP along with siRNAs targeting CAT, PARN, DCP2, or RRP46. Varied levels of GB-GAPDH mRNA are due to loading variations. Signals of GB-6bs were quantitated, normalized to that of GB-GAPDH, and plotted as mean values ± SDs against time. Dox, doxycycline.
We also examined whether the restoration of KSRP-tethered mRNA levels results from decreased mRNA decay. The half-life of KSRP-tethered mRNA was prolonged from 2.5 h to 5.5 h (2.2-fold), to 4.5 h (1.8-fold), and to 7 h (2.8-fold) in cells depleted of PARN, DCP2, and RRP46, respectively (Fig. 4H). These results confirm a mechanistic role for PARN, DCP2, and exosome components in KSRP-mediated mRNA decay. We did not observe complete restoration of KSRP-tethered mRNA levels in cells treated with siRNAs against two exosome components simultaneously (RRP4 and RRP46 or RRP46 and PM-Scl100) or against DCP2 and RRP46 together (Fig. 4D). It is likely that residual activities of DCP2 and the exosome are still sufficient for decay of the tethered mRNA.
Overexpression of non-RNA-binding KSRP polypeptide impairs decay of ARE-containing mRNAs.
If functions of the KH motifs and C terminus of KSRP are to recruit mRNA decay _trans_-acting factors (including decay enzymes), expression of non-RNA-binding polypeptides comprising these regions might impair ARE-directed mRNA decay. We previously suggested that the KH4 motif of KSRP is required for binding to AREs (18). We expressed FLAG-tagged polypeptides containing different regions of KSRP in HeLa-TO cells and examined their ability to interact with an ARE by UV cross-linking assays. We immunoprecipitated specific ARE-KSRP polypeptide complexes by anti-FLAG agarose (Fig. 5A). We detected strong ARE-binding activity of KSRP or a polypeptide composed of the four KH motifs (KH1-4) (Fig. 5A, lanes 2 and 6). In contrast, no ARE-binding activity was detected with KSRP polypeptides lacking the KH4 motif, such as those consisting of KH1-3 (Fig. 5A, lane 4), the N terminus and KH1-3 (N-KH1-3) (Fig. 5A, lane 8), KH1-3 and the C terminus (KH1-3-C) (Fig. 5A, lane 10), or the N terminus, KH1-3, and the C terminus (N-KH1-3-C) (Fig. 5A, lane 12). Upon longer exposure, we could detect ARE binding of KSRP polypeptides deleted of KH4 (data not shown). Their binding activities were estimated to be 30-fold lower than that of KH1-4. We immunopurified FLAG-tagged KSRP polypeptides and examined the presence of exosome component by immunoblotting. We detected RRP4 in the precipitates of KSRP polypeptides composed of KH1-3 or the C terminus, including KH1-3, KSRPC, N-KH1-3, KH1-3-C, and N-KH1-3-C (Fig. 5B). Together with the above data, we conclude that deletion of the KH4 motif dramatically reduces ARE-binding activity of KSRP and that polypeptides containing KH1-3 or the C terminus associate with mRNA decay enzymes.
FIG. 5.
Overexpression of non-RNA-binding KSRP polypeptide impairs ARE-directed mRNA decay. (A) HeLa-TO cells were transfected with vectors expressing different FLAG-tagged KSRP polypeptides. Cytoplasmic extracts were prepared and incubated with 32P-labeled AREtnf RNA, and UV cross-linking assays were performed. The UV cross-linking reactions were immunoprecipitated with anti-FLAG agarose (lanes 2, 4, 6, 8, 10, and 12), and precipitates were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and autoradiography (top panel). A portion (25%) of each UV cross-linking reaction was loaded onto lanes 1, 3, 5, 7, 9, and 11. The immunoprecipitates were also analyzed by anti-FLAG immunoblotting (bottom panel). (B) HeLa-TO cells were transfected with constructs expressing FLAG-tagged KSRP polypeptides. Cell extracts were subjected to anti-FLAG immunoprecipitation. The precipitates were analyzed by anti-RRP4 or anti-FLAG immunoblotting. A 5% extract used for immunoprecipitation reactions was also analyzed. The asterisk indicates the heavy chain of anti-FLAG immunoglobulin G. (C and D) The decay of GB-AREtnf (C) and GB-AREfos (D) mRNAs in HeLa-TO cells expressing various KSRP polypeptides was analyzed. Signals of GB-AREtnf and GB-AREfos mRNAs were quantitated and normalized with that of GB-GAPDH. The calculated half-lives (n = 3) are indicated as mean values ± standard deviations. IP, immunoprecipitate; IgG, immunoglobulin G; Dox, doxycycline.
Finally, we determined whether expression of non-ARE-binding KSRP polypeptides could impair ARE-directed mRNA decay. We coexpressed KSRP polypeptides with a β-globin reporter mRNA containing the TNF-α ARE (GB-AREtnf) and examined decay of the mRNA. We did not detect significant inhibition in the decay of GB-AREtnf mRNA in cells individually or simultaneously expressing polypeptides composed of KH1-3 or KSRPC (data not shown) or in cells expressing polypeptides containing the N terminus and KH1-3 or KH1-3 and the C terminus (Fig. 5C). Interestingly, coexpression of a KSRP polypeptide when the N and C termini and KH1-3 are fused together (N-KH1-3-C) significantly inhibited decay of GB-AREtnf mRNA (Fig. 5C). Similarly, coexpression of the N-KH1-3-C polypeptide inhibited the decay of a β-globin mRNA containing the c-fos ARE (GB-AREfos) (Fig. 5D). However, the inhibition effect on GB-AREfos mRNA was smaller than that on GB-AREtnf mRNA, perhaps because an additional factor specifically required for decay of GB-AREfos mRNA could not be titrated by the KSRP mutant. These results suggest that expression of a non-ARE-binding KSRP mutant deleted of the KH4 motif inhibits ARE-directed mRNA decay by titrating out of limiting factors important for the decay.
Tethering of KSRP targets HIV-1 mRNAs for degradation and dramatically inhibits viral replication.
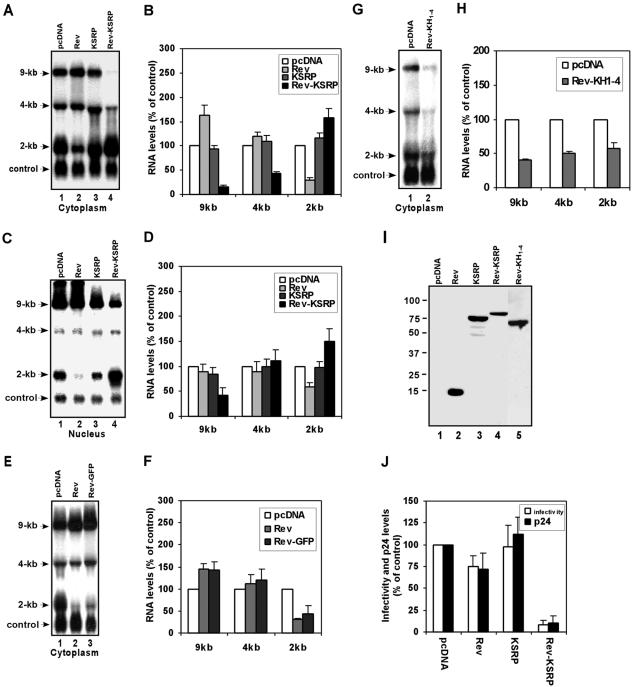
Finally, we asked whether tethering of KSRP could target the transcripts of HIV-1 for degradation. Three classes of HIV-1 mRNAs, an unspliced 9-kb mRNA, singly spliced 4-kb mRNAs, and fully spliced 2-kb mRNAs, are expressed in infected cells (14, 34). Nuclear export of the unspliced and singly spliced mRNAs is dependent on an RNA-binding protein, Rev (13, 14). Rev interacts with a specific RNA sequence, the Rev response element (RRE), present in the 9-kb and 4-kb mRNAs (34). We expressed Rev-KSRP fusion protein in cells cotransfected with an HIV-1 proviral DNA and analyzed cytoplasmic levels of HIV-1 mRNAs (Fig. 6A). The levels of the 9-kb and 4-kb mRNAs were dramatically decreased in cells coexpressing Rev-KSRP (Fig. 6A and B). The reduction in the 9- and 4-kb mRNA levels by Rev-KSRP was specific to the tethering; KSRP lacking Rev had no significant effect on the mRNA levels (Fig. 6A and B). Furthermore, mRNAs were targeted for decay in an RRE-dependent manner. The levels of 2-kb mRNA as well as a cotransfected, truncated HIV-1 mRNA containing a large internal deletion, both of which lack the RRE, were not affected by Rev-KSRP (Fig. 6A). Cytoplasmic levels of the 9-kb and 4-kb mRNAs were slightly increased in cells coexpressing Rev. This effect is likely due to increased nuclear export of the mRNAs by overexpressing Rev, resulting in depletion of the 2-kb mRNA (14).
FIG. 6.
Tethering of KSRP targets HIV-1 mRNAs for degradation and dramatically inhibits viral replication. (A) 293T cells were transfected with an HIV-1 proviral DNA, a construct expressing a truncated HIV-1 mRNA (in which most of the sequences [nucleotides 715 to 8887], including the RRE, are deleted), and constructs expressing Rev, KSRP, or Rev-KSRP. Cytoplasmic levels of HIV-1 mRNAs (labeled 9-kb, 4-kb, and 2-kb) and control mRNA (labeled control) were analyzed by Northern blotting with a probe against the U3 sequences. (B) Levels of cytoplasmic 9-, 4-, and 2-kb mRNAs were quantitated and normalized to levels of control mRNA. (C) Nuclear RNA was isolated and analyzed by Northern blotting as described for panel A. (D) Levels of nuclear 9-, 4-, and 2-kb mRNAs were quantitated. (E) Cytoplasmic levels of HIV-1 mRNAs and control mRNA were analyzed by Northern blotting with cells cotransfected with pcDNA or constructs expressing Rev or Rev-GFP. (F) Levels of 9-, 4-, and 2-kb mRNAs were quantitated and normalized to levels of control mRNA. (G) Cytoplasmic levels of HIV-1 mRNAs and control mRNA were analyzed by Northern blotting with cells cotransfected with pcDNA or a construct expressing Rev-KH1-4. (H) Levels of 9-, 4-, and 2-kb mRNAs were quantitated and normalized to levels of control mRNA. (I) Expression of transfected proteins was analyzed by anti-Xpress immunoblotting. (J) Supernatants withdrawn from transfected cells were analyzed for viral infectivity and p24 antigen levels. For all bar graphs, the levels of each class of HIV-1 mRNA in cells transfected with pcDNA are set at 100 and mean values with standard deviations (error bars) are shown.
The reduction of the cytoplasmic levels of 9- and 4-kb mRNAs, but not 2-kb mRNAs, by Rev-KSRP (Fig. 6A) could also be explained as inhibition of mRNA nuclear export by fusion of KSRP with Rev. We performed additional experiments to provide support that the reduction of cytoplasmic 9-kb and 4-kb mRNA levels is not completely due to inhibition of viral mRNA nuclear export by coexpression of Rev-KSRP. We first examined nuclear mRNA levels. The levels of the 9-kb mRNA in the nucleus were reduced by coexpression of Rev-KSRP (Fig. 6C and D), not increased, which would be expected if the Rev functions were inhibited by fusion with KSRP. We next examined cytoplasmic levels of HIV-1 mRNAs in cells coexpressing Rev or Rev-GFP. We did not detect a significant difference in the levels of all three classes of HIV-1 mRNAs between cells coexpressing Rev and Rev-GFP (Fig. 6E and F), indicating that fusion of GFP with Rev does not compromise its functions in mRNA nuclear export. We also coexpressed Rev-KH1-4 and examined cytoplasmic HIV-1 mRNA levels. The levels of 9- and 4-kb mRNAs were significantly decreased by coexpression of Rev-KH1-4. Most importantly, the cytoplasmic levels of 2-kb mRNAs were dramatically reduced as well (Fig. 6G, lane 2, and H). These data suggest that the 9- and 4-kb mRNAs are degraded by Rev-KH1-4, which also reduces the levels of 2-kb mRNA by depleting the pre-mRNAs for the production of the fully spliced 2-kb mRNAs. Altogether, our data suggest that tethering of KSRP or KH1-4 to HIV-1 mRNAs decreases the HIV-1 mRNAs by eliciting mRNA decay. Finally, we examined virus particle production and infectivity. The viral infectivity as well as the HIV-1 p24 antigen levels was dramatically decreased in cells expressing Rev-KSRP (Fig. 6J). These data indicate that targeting of KSRP to HIV-1 mRNAs via Rev results in inhibition of viral replication.
DISCUSSION
The ARE is one of the most prevalent _cis_-acting elements responsible for mRNA decay in human cells (2). Although identified more than 15 years ago, the mechanism of ARE-directed mRNA decay regulated by decay-promoting ARE-BPs is still not completely understood. How does a decay-promoting ARE-BP target bound mRNA for decay? Which mRNA decay enzymes are involved in degradation of the bound mRNA? Here, using a tethering assay, we provide strong evidence for a previously suggested model that mRNA decay enzymes/factors are recruited to KSRP-bound transcripts to elicit mRNA decay. Four lines of data support the recruitment model: (i) tethering KSRP to non-ARE-containing mRNAs elicits decay (Fig. 1 and 6), (ii) KSRP associates with mRNA decay enzymes (Fig. 3), (iii) silencing expression of some mRNA decay enzymes impairs decay of KSRP-tethered mRNA, and (iv) interfering with the access of mRNA decay factors/enzymes to ARE-containing mRNAs by overexpression of a non-RNA-binding KSRP mutant inhibits their decay (Fig. 5). Recent studies of TTP and BRF1 also support the recruitment model (29).
Using MS2 tethering assays, we found that two individual domains of KSRP are able to trigger decay when tethered to a reporter mRNA. The central KH domain, which constitutes four KH motifs, is one of the decay activation domains and associates with mRNA decay enzymes. Further dissecting the KH motifs indicates that the third KH motif (KH3) moderately elicits decay of tethered transcripts. However, efficient mRNA decay requires additional KH motifs, including the first and second or the second and fourth KH motifs, to be present together with KH3. These results suggest that multiple sequences within the KH motifs or a proper conformation formed by the KH motifs is necessary for efficiently triggering mRNA decay. Since KH1-4 elicits decay of tethered mRNA more efficiently than KH1-3 or KH2-4 (Fig. 2D and E), our results also suggest that no individual KH motifs can be fully excluded for promoting mRNA decay. These sequences may be required for stabilizing the interactions between the KH motifs and mRNA decay factors (or enzymes).
The C-terminal domain, which is dispensable for ARE-directed decay in our previous depletion and reconstitution experiments using ARE-containing mRNA as a substrate (18), is another decay activation domain in the tethering assays. The C-terminal domain may promote decay of tethered mRNA by recruiting decay machinery, as it also associates with mRNA decay enzymes (Fig. 3B and 5B). Our findings on KSRP domains required for mRNA decay are somewhat consistent with the recent conclusion from the studies of TTP and BRF1 by use of similar assays. TTP and BRF1 contain two domains, the N- and C-terminal domains, that can promote mRNA decay when tethered to a reporter mRNA. While the N terminus of TTP promotes decay by recruiting mRNA decay enzymes, the C terminus does so likely by recruitment of factors other than known mRNA decay enzymes (29). Whether the KH motifs or the C terminus of KSRP interacts with mRNA decay enzymes directly or indirectly awaits further investigation. Our results suggest that mRNA decay enzymes are recruited onto KSRP-bound mRNA via both domains.
We assessed functions of known mRNA decay enzymes in KSRP-mediated decay by RNAi-mediated gene silencing. The decay of tethered transcripts relies on functions of PARN, DCP2, XRN1, and the exosome components. In contrast, CCR4 does not appear to be involved in decay of KSRP-tethered mRNA in our system. Although PARN is required for decay of KSRP-tethered mRNA, it does not appear to play a major role in the decay, as we detected only moderate restoration in the levels of tethered mRNA in cells depleted of its activity. These data suggest that there are additional enzymes involved in deadenylation of the tethered mRNA. This conclusion is consistent with the findings that at least seven ribonucleases are predicted or known to possess deadenylation activity in mammalian cells, including PARN, PAN2, CCR4-related families (CCR4, nocturnin, angel, and 3635), and the CCR4-associated factor CAF1 (12, 15, 22, 41, 44). Some of these deadenylases may be redundant or specific to certain transcripts. Furthermore, tethered KSRP appears to rely primarily on function of the exosome components and depend moderately on functions of DCP2 and XRN1 after deadenylation, as downregulation of exosome components restores tethered mRNA levels better than those restored by downregulation of DCP2 or XRN1. This suggests that the tethered mRNA may be preferentially degraded through a 3′-to-5′ direction. Nevertheless, we did not observe complete restoration of KSRP-tethered mRNA levels in cells treated with siRNAs against two exosome components simultaneously or against DCP2 and RRP46 together (Fig. 4D), suggesting that residual DCP2 and exosome activities in siRNA-treated cells are still sufficient for the decay. It is also possible that downregulation of both DCP2 and the exosome component or two exosome components together is lethal, as suggested with yeast (21, 31); therefore, no further restoration in KSRP-tethered mRNA levels could be detected. Furthermore, additional unidentified or untested mRNA decay enzymes may be involved in decay of KSRP-tethered mRNA, but this awaits further investigation.
Recent studies of regulation of mRNA decay in mammalian cells by use of different experimental systems have implicated human homologs of yeast mRNA decay enzymes in different mRNA decay pathways. Overexpression of a catalytic mutant of CCR4, but not mutants of PARN or PAN2, alters mRNA decay rate mediated by the c-fos major protein-coding determinant of instability (8). PAN2 and CCR4 are the major poly(A) nucleases that act in concert in the decay of normal and nonsense-containing (nonsense-mediated decay [NMD]) mRNAs (50). Overexpression of DCP2 facilitates ARE-directed mRNA decay (29). NMD requires PARN, components of the human exosome, and DCP2 (24). mRNA decay triggered by tethered SMG7, which mimics NMD, requires functions of DCP2 and XRN1 (42). Regardless of systems used, all of these studies suggest that mRNA decay in mammalian cells is complex and may involve several specific as well as redundant decay pathways which are activated simultaneously or distinctly by _trans_-acting factors involved in different mRNA decay mechanisms. Some of the decay pathways require the same enzymes; others require distinct enzymes. In addition, there may be more enzymes and pathways involved in mammalian mRNA decay. A complete understanding of mammalian mRNA decay requires the identification and characterization of these additional factors (or enzymes).
We demonstrated that ARE-directed mRNA decay is impaired by expression of non-RNA-binding KSRP polypeptides. Despite their association with tested mRNA decay enzymes, non-ARE-binding polypeptides comprising only KH1-3 or the C terminus of KSRP or both domains expressed in the same context did not significantly inhibit ARE-directed mRNA decay. Efficient inhibition of mRNA decay was observed to occur only when the N and C termini and the KH1-3 motifs were expressed in the same context. The mechanism of the inhibition by this polypeptide is not known, but possible scenarios can be envisaged. When the N and C termini and KH1-3 are fused together in one polypeptide, a proper structure is formed to sequester factors in addition to the tested, known mRNA decay enzymes from the ARE-containing mRNA. These factors, which are important for eliciting mRNA decay, can still access the mRNA when KSRP domains are expressed separately. Our data also imply that the recruitment of these factors by ARE-bound KSRP or other decay-promoting ARE-BPs can elicit mRNA decay. Although these additional sequestered factors await further characterization, they may be other decay-promoting ARE-BPs. A support for this is that TTP was shown to interact with KSRP (25). Alternatively, the association of mRNA decay enzymes with the KSRP polypeptide lacking KH4 only, but not with separately expressed KSRPN, KH1-3, or KSRPC, may prevent the access of mRNA decay enzymes to other decay-promoting ARE-BPs that bind to the same ARE as KSRP but recruit decay machinery through their association with distinct components of the machinery. We propose that the association of the KSRP polypeptide lacking only KH4 with the mRNA decay machinery may form a larger block to prevent the recruitment of the machinery by other decay-promoting ARE-BPs.
Finally, we provide proof of principle that tethering of KSRP reduces the abundance of unspliced and singly spliced HIV-1 mRNAs. Although we cannot provide definite evidence at this stage that the reduction of HIV-1 mRNAs by Rev-KSRP or Rev-KH1-4 is due to mRNA degradation and likely that other effects on the Rev-KSRP-tethered mRNAs, such as mRNA nuclear export or splicing, may cause the reduction of cytoplasmic 9- and 4-kb mRNA levels, our results support, at least in part, that the tethered mRNAs are targeted for decay by Rev-KSRP and Rev-KH1-4. Three lines of data suggest that the reduction of viral transcripts is due to mRNA degradation. We did not detect a large increase in nuclear 9- and 4-kb mRNA levels in cells coexpressing Rev-KSRP compared with that in control cells (transfected with a control vector) (Fig. 6C and D), which would be expected if fusion of KSRP with Rev inhibited its functions. Fusion of Rev with GFP does not impair Rev functions in mRNA nuclear export (Fig. 6E and F), which provides indirect support that fusion of KSRP unlikely inhibits Rev's activity. Coexpression of Rev-KH1-4 significantly decreases the cytoplasmic levels of 9- and 4-kb mRNAs as well as those of the 2-kb mRNAs. These data (Fig. 6G and H) suggest that Rev-KH1-4 is functionally active in mRNA nuclear export as Rev and Rev-KH1-4-bound 9- and 4-kb mRNAs are degraded in the cytoplasm. As the nuclear 2-kb mRNA levels are increased in cells coexpressing Rev-KSRP, we cannot completely exclude that KSRP fusion may somewhat inhibit Rev's nuclear export activity, which provides a favored condition for the unspliced 9-kb and 4-kb mRNAs to be spliced. It is also possible that Rev-KSRP alters the splicing activity of the 9-kb and 4-kb mRNAs in the nucleus.
Targeting HIV-1 mRNAs for degradation by various approaches, such as antisense RNAs, ribozymes, and RNA interference, successfully inhibits HIV-1 replication (23, 48). Several gene therapies against HIV-1 have been developed and have been shown to inhibit viral replication (23, 48). However, additional therapeutic strategies must be developed continuously to inhibit newly evolved strains. Our tethering approach complements existing therapeutic strategies for AIDS treatment and may have advantages over other approaches. Any HIV-1 strains that escape recognition by Rev-KSRP by altering the RRE will not replicate in infected cells because the interaction between the RRE and Rev is essential for viral replication (34). Strains bearing mutations in the RRE would require simultaneous, compensatory mutations in Rev in order to replicate, which would be extremely unlikely. The tethering method described here to degrade specific HIV-1 mRNAs can be adapted to target other mRNAs that are overexpressed in acquired or hereditary diseases for degradation.
Acknowledgments
We thank P. Detloff and T. Townes for their valuable comments on the manuscript; J. Lykke-Andersen, J. Steitz, and G. Brewer for kindly providing previous reagents; and E. Wahle for anti-PARN, G. Pruijn for anti-RRP46 and -PM-Scl100, P. Mitchell for anti-RRP4, and M. Kiledjian for anti-DCP2 antibodies.
Work was supported by NIH grant no. GM68758 (C.-Y.C.) and AI47714 (J.K.) and a grant from Associazione Italiana Ricerca sul Cancro (R.G.).
REFERENCES
- 1.Allmang, C., E. Petfalski, A. Podtelejnikov, M. Mann, D. Tollervey, and P. Mitchell. 1999. The yeast exosome and human PM-Scl are related complexes of 3′→5′ exonucleases. Genes Dev. 13**:**2148-2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bakheet, T., B. R. Williams, and K. S. Khabar. 2003. ARED 2.0: an update of AU-rich element mRNA database. Nucleic Acids Res. 31**:**421-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bashkirov, V. I., H. Scherthan, J. A. Solinger, J. M. Buerstedde, and W. D. Heyer. 1997. A mouse cytoplasmic exoribonuclease (mXRN1p) with preference for G4 tetraplex substrates. J. Cell Biol. 136**:**761-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bevilacqua, A., M. C. Ceriani, S. Capaccioli, and A. Nicolin. 2003. Post-transcriptional regulation of gene expression by degradation of messenger RNAs. J. Cell. Physiol. 195**:**356-372. [DOI] [PubMed] [Google Scholar]
- 5.Blackshear, P. J. 2002. Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Biochem. Soc. Trans. 30**:**945-952. [DOI] [PubMed] [Google Scholar]
- 6.Brennan, C. M., and J. A. Steitz. 2001. HuR and mRNA stability. Cell Mol. Life Sci. 58**:**266-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Briata, P., S. V. Forcales, M. Ponassi, G. Corte, C. Y. Chen, M. Karin, P. L. Puri, and R. Gherzi. 2005. p38-dependent phosphorylation of the mRNA decay-promoting factor KSRP controls the stability of select myogenic transcripts. Mol. Cell 20**:**891-903. [DOI] [PubMed] [Google Scholar]
- 8.Chang, T. C., A. Yamashita, C. Y. Chen, Y. Yamashita, W. Zhu, S. Durdan, A. Kahvejian, N. Sonenberg, and A. B. Shyu. 2004. UNR, a new partner of poly(A)-binding protein, plays a key role in translationally coupled mRNA turnover mediated by the c-fos major coding-region determinant. Genes Dev. 18**:**2010-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, C. Y., R. Gherzi, S. E. Ong, E. L. Chan, R. Raijmakers, G. J. Pruijn, G. Stoecklin, C. Moroni, M. Mann, and M. Karin. 2001. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 107**:**451-464. [DOI] [PubMed] [Google Scholar]
- 10.Chen, C. Y., and A. B. Shyu. 1995. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 20**:**465-470. [DOI] [PubMed] [Google Scholar]
- 11.Chen, C.-Y. A., N. Xu, and A.-B. Shyu. 1995. mRNA decay mediated by two distinct AU-rich elements from c-fos and granulocyte-macrophage colony-stimulating factor transcripts: different deadenylation kinetics and uncoupling from translation. Mol. Cell. Biol. 15**:**5777-5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, J., Y. C. Chiang, and C. L. Denis. 2002. CCR4, a 3′-5′ poly(A) RNA and ssDNA exonuclease, is the catalytic component of the cytoplasmic deadenylase. EMBO J. 21**:**1414-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cullen, B. R. 2003. Nuclear mRNA export: insights from virology. Trends Biochem. Sci. 28**:**419-424. [DOI] [PubMed] [Google Scholar]
- 14.Cullen, B. R. 1998. Retroviruses as model systems for the study of nuclear RNA export pathways. Virology 249**:**203-210. [DOI] [PubMed] [Google Scholar]
- 15.Dupressoir, A., A. P. Morel, W. Barbot, M. P. Loireau, L. Corbo, and T. Heidmann. 2001. Identification of four families of yCCR4- and Mg2+-dependent endonuclease-related proteins in higher eukaryotes, and characterization of orthologs of yCCR4 with a conserved leucine-rich repeat essential for hCAF1/hPOP2 binding. BMC Genomics 2**:**9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ford, L. P., J. Watson, J. D. Keene, and J. Wilusz. 1999. ELAV proteins stabilize deadenylated intermediates in a novel in vitro mRNA deadenylation/degradation system. Genes Dev. 13**:**188-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao, M., C. J. Wilusz, S. W. Peltz, and J. Wilusz. 2001. A novel mRNA-decapping activity in HeLa cytoplasmic extracts is regulated by AU-rich elements. EMBO J. 20**:**1134-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gherzi, R., K. Y. Lee, P. Briata, D. Wegmuller, C. Moroni, M. Karin, and C. Y. Chen. 2004. A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol. Cell 14**:**571-583. [DOI] [PubMed] [Google Scholar]
- 19.Guhaniyogi, J., and G. Brewer. 2001. Regulation of mRNA stability in mammalian cells. Gene 265**:**11-23. [DOI] [PubMed] [Google Scholar]
- 20.Hamilton, B. J., C. M. Burns, R. C. Nichols, and W. F. Rigby. 1997. Modulation of AUUUA response element binding by heterogeneous nuclear ribonucleoprotein A1 in human T lymphocytes. The roles of cytoplasmic location, transcription, and phosphorylation. J. Biol. Chem. 272**:**28732-28741. [DOI] [PubMed] [Google Scholar]
- 21.Jacobs, J. S., A. R. Anderson, and R. P. Parker. 1998. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 17**:**1497-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Körner, C. G., M. Wormington, M. Muckenthaler, S. Schneider, E. Dehlin, and E. Wahle. 1998. The deadenylating nuclease (DAN) is involved in poly(A) tail removal during the meiotic maturation of Xenopus oocytes. EMBO J. 17**:**5427-5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee, N. S., and J. J. Rossi. 2004. Control of HIV-1 replication by RNA interference. Virus Res. 102**:**53-58. [DOI] [PubMed] [Google Scholar]
- 24.Lejeune, F., X. Li, and L. E. Maquat. 2003. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol. Cell 12**:**675-687. [DOI] [PubMed] [Google Scholar]
- 25.Linker, K., A. Pautz, M. Fechir, T. Hubrich, J. Greeve, and H. Kleinert. 2005. Involvement of KSRP in the post-transcriptional regulation of human iNOS expression-complex interplay of KSRP with TTP and HuR. Nucleic Acids Res. 33**:**4813-4827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loflin, P., C. Y. Chen, and A. B. Shyu. 1999. Unraveling a cytoplasmic role for hnRNP D in the in vivo mRNA destabilization directed by the AU-rich element. Genes Dev. 13**:**1884-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lykke-Andersen, J. 2002. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol. Cell. Biol. 22**:**8114-8121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lykke-Andersen, J., M. D. Shu, and J. A. Steitz. 2000. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell 103**:**1121-1131. [DOI] [PubMed] [Google Scholar]
- 29.Lykke-Andersen, J., and E. Wagner. 2005. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev. 19**:**351-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Min, H., C. W. Turck, J. M. Nikolic, and D. L. Black. 1997. A new regulatory protein, KSRP, mediates exon inclusion through an intronic splicing enhancer. Genes Dev. 11**:**1023-1036. [DOI] [PubMed] [Google Scholar]
- 31.Mitchell, P., E. Petfalski, A. Shevchenko, M. Mann, and D. Tollervey. 1997. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell 91**:**457-466. [DOI] [PubMed] [Google Scholar]
- 32.Mukherjee, D., M. Gao, J. P. O'Connor, R. Raijmakers, G. Pruijn, C. S. Lutz, and J. Wilusz. 2002. The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. EMBO J. 21**:**165-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mulky, A., S. G. Sarafianos, E. Arnold, X. Wu, and J. C. Kappes. 2004. Subunit-specific analysis of the human immunodeficiency virus type 1 reverse transcriptase in vivo. J. Virol. 78**:**7089-7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pollard, V. W., and M. H. Malim. 1998. The HIV-1 Rev protein. Annu. Rev. Microbiol. 52**:**491-532. [DOI] [PubMed] [Google Scholar]
- 35.Raineri, I., D. Wegmueller, B. Gross, U. Certa, and C. Moroni. 2004. Roles of AUF1 isoforms, HuR and BRF1 in ARE-dependent mRNA turnover studied by RNA interference. Nucleic Acids Res. 32**:**1279-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sarkar, B., Q. Xi, C. He, and R. J. Schneider. 2003. Selective degradation of AU-rich mRNAs promoted by the p37 AUF1 protein isoform. Mol. Cell. Biol. 23**:**6685-6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shyu, A. B., J. G. Belasco, and M. E. Greenberg. 1991. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev. 5**:**221-231. [DOI] [PubMed] [Google Scholar]
- 38.Stoecklin, G., M. Colombi, I. Raineri, S. Leuenberger, M. Mallaun, M. Schmidlin, B. Gross, M. Lu, T. Kitamura, and C. Moroni. 2002. Functional cloning of BRF1, a regulator of ARE-dependent mRNA turnover. EMBO J. 21**:**4709-4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tucker, M., and R. Parker. 2000. Mechanisms and control of mRNA decapping in Saccharomyces cerevisiae. Annu. Rev. Biochem. 69**:**571-595. [DOI] [PubMed] [Google Scholar]
- 40.Tucker, M., M. A. Valencia-Sanchez, R. R. Staples, J. Chen, C. L. Denis, and R. Parker. 2001. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell 104**:**377-386. [DOI] [PubMed] [Google Scholar]
- 41.Uchida, N., S. Hoshino, and T. Katada. 2004. Identification of a human cytoplasmic poly(A) nuclease complex stimulated by poly(A)-binding protein. J. Biol. Chem. 279**:**1383-1391. [DOI] [PubMed] [Google Scholar]
- 42.Unterholzner, L., and E. Izaurralde. 2004. SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol. Cell 16**:**587-596. [DOI] [PubMed] [Google Scholar]
- 43.van Dijk, E., N. Cougot, S. Meyer, S. Babajko, E. Wahle, and B. Seraphin. 2002. Human Dcp2: a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J. 21**:**6915-6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Viswanathan, P., T. Ohn, Y. C. Chiang, J. Chen, and C. L. Denis. 2004. Mouse CAF1 can function as a processive deadenylase/3′-5′-exonuclease in vitro but in yeast the deadenylase function of CAF1 is not required for mRNA poly(A) removal. J. Biol. Chem. 279**:**23988-23995. [DOI] [PubMed] [Google Scholar]
- 45.Wang, Z., X. Jiao, A. Carr-Schmid, and M. Kiledjian. 2002. The hDcp2 protein is a mammalian mRNA decapping enzyme. Proc. Natl. Acad. Sci. USA 99**:**12663-12668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilusz, C. J., and J. Wilusz. 2004. Bringing the role of mRNA decay in the control of gene expression into focus. Trends Genet. 20**:**491-497. [DOI] [PubMed] [Google Scholar]
- 47.Wilusz, C. J., M. Wormington, and S. W. Peltz. 2001. The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell Biol. 2**:**237-246. [DOI] [PubMed] [Google Scholar]
- 48.Wolkowicz, R., and G. P. Nolan. 2005. Gene therapy progress and prospects: novel gene therapy approaches for AIDS. Gene Ther. 12**:**467-476. [DOI] [PubMed] [Google Scholar]
- 49.Xu, N., C.-Y. A. Chen, and A.-B. Shyu. 2001. Versatile role for hnRNP D isoforms in the differential regulation of cytoplasmic mRNA turnover. Mol. Cell. Biol. 21**:**6960-6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamashita, A., T. C. Chang, Y. Yamashita, W. Zhu, Z. Zhong, C. Y. Chen, and A. B. Shyu. 2005. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat. Struct. Mol. Biol. 12**:**1054-1063. [DOI] [PubMed] [Google Scholar]