The two Escherichia coli chromosome arms locate to separate cell halves (original) (raw)
Abstract
DNA replication divides the circular Escherichia coli chromosome into equal arms (replichores). Visualization of pairwise combinations of multiple genetic loci reveals that the two replichores occupy separate nucleoid halves, with the replication origin between; positions of loci on each replichore recapitulate the genetic map. Sequential replication–segregation regenerates the <left–right> structure by sequentially layering newly replicated replichore DNA to specific inner and outer edges of the developing sister nucleoids. Replication fork-dependent locus positions are imprinted, so that in most generations the <left–right> chromosome orientation in a mother cell is recreated as a <left–right–left–right> arrangement of sister chromosomes in daughter cells.
Keywords: E. coli, chromosome, replichore, segregation
Although it has been known for many years that bacterial circular chromosomes are replicated bidirectionally from a defined origin, precisely how this DNA is organized into the bacterial nucleoid, and how and when chromosomal DNA is segregated, has remained elusive (Sherratt 2003; Wu 2004; Gitai et al. 2005). Replication divides the chromosome into two equal arms (replichores), each being transcribed predominantly in the same direction as replication, and each containing distinctive asymmetric base-skewed DNA sequences that are important for chromosome processing (Blattner et al. 1997; Rocha 2004; Lesterlin et al. 2005). In recent years a combination of improved cytological methods, in which specific genetic loci were visualized using either fluorescent in situ hybridization (FISH), or the binding of fluorescent DNA-binding proteins to their cognate DNA-binding sites (FROS), have begun to reveal that bacterial chromosomes are highly organized yet dynamic structures (e.g., see Gordon et al. 1997; Niki et al. 2000; Li et al. 2002; Bates and Kleckner 2005; Wang et al. 2005), with DNA replication apparently being confined to specific cellular “replication factories” (Lemon and Grossman 2000; Adachi et al. 2005; Bates and Kleckner 2005).
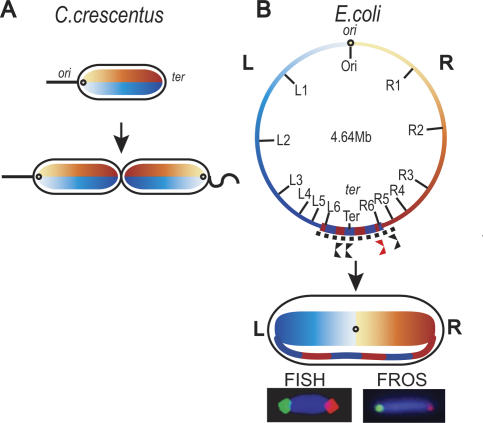
Models of chromosome organization based on these analyses for Escherichia coli, Bacillus subtilis, and Caulobacter crescentus have favored an arrangement in which the ori–ter chromosome axis runs longitudinally along the cell, with left and right chromosome arms (replichores) placed above and below this axis, or interwound (Teleman et al. 1998; Niki et al. 2000; Viollier et al. 2004; Gitai et al. 2005; Thanbichler et al. 2005). Such an organization is clear in C. crescentus, where in a resting cell, the replication origin region, ori, localizes to one cell pole, and ter, the replication termination region, to the other pole, and other loci lie between the two. Sequential replication–segregation generates two daughter cells that are mirror symmetrical with respect to chromosome orientation (Fig. 1; Viollier et al. 2004; Gitai et al. 2005). The E. coli data have provided conflicting views of how DNA is organized in vivo and how/when it segregates. Nevertheless, the idea of chromosome organization about a longitudinal axis has been implicit (Niki and Hiraga 1998; Niki et al. 2000; Bates and Kleckner 2005), with controversy about whether newly replicated sisters remain cohesed for much of the cell cycle (Sunako et al. 2001; Li et al. 2002; Bates and Kleckner 2005; Wang et al. 2005).
Figure 1.
E. coli chromosome organization recapitulates the genetic map. The left panel shows a schematic of chromosome organization in C. crescentus; left and right replichores are organized about a longitudinal axis (Viollier et al. 2004; Gitai et al. 2005). The right panel shows the E. coli chromosome with the markers used, and its cellular organization below. The left and right replichores flank a transverse axis. The left replichore (L) of each chromosome is shown in blue and the right replichore (R) is shown in orange. We imagine that the bulk of DNA in left and right replichores is organized into independent domains. Normal replication termination sites within the ter region (dotted line; alternating blue–orange) are shown in black below the map and the ectopic site is shown in red. The left replichore is positioned left of ori and the right replichore to the right of ori. The ter region spans the two outer nucleoid edges; much of it can be replicated by either fork. The dif site-specific recombination site (Blakely et al. 1991) is located 18 kb counterclockwise of the Ter locus. Representative FISH (left) and DNA-bound fluorescent repressor (FROS) (right) images are shown for L3 (green) and R3 (red).
Here we demonstrate that the left (L) and right (R) replichores lie on separate nucleoid halves (<L–R> organization), with a transverse ori–ter axis at mid-cell rather than a longitudinal axis. Furthermore, we show that most sister nucleoids are not related by mirror symmetry, as previously believed, but rather by a <L–R–L–R> translational symmetry. This is imprinted during the sequential replication–segregation layering process, so that a <L–R> mother chromosome organization is recreated as a <L–R–L–R> organization of sister chromosomes in the two daughter cells. This validates our proposal from a recent study, which focused on the E. coli ter region (Wang et al. 2005).
Results and Discussion
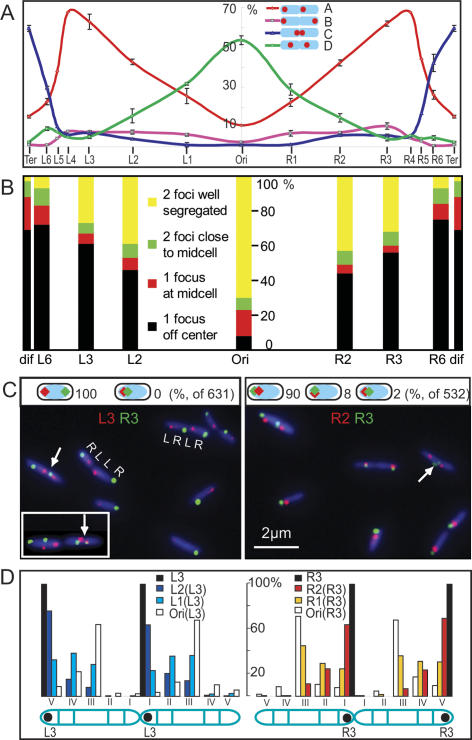
Initially, the relative positioning of sister foci for 14 genetic loci was examined by FISH (Fig. 2A). After measuring the relative positions of sister foci for each locus, cells were grouped into one of four informative patterns on the basis of relative focus position and the frequency of each pattern was compared with chromosome position.
Figure 2.
Snapshot analysis. (A) Single-locus FISH survey of the whole genome. Focus pairs (14 loci) were binned into groups depending on pattern (see D). The four patterns shown, which account for >65% of all cells, were most informative with regard to asymmetric–symmetric positioning of sister foci. The frequency of each pattern with respect to genetic map was plotted. For each locus, ∼300 two-focus cells were analyzed. Nucleoid pairs were maximized by treating an exponential culture (τ 100 min) for 60 min with cephalexin to block cell division. This does not significantly perturb segregation pattern. (B) Sequential replication–segregation of eight loci, using FROS snapshot analysis of an exponential culture (τ 100 min). The position of a locus on the _X_-axis with respect to genetic map is shown. Cells were binned into four groups in terms of number and position of the foci: one focus off-center; one focus at mid-cell; two foci close to the mid-cell; and two foci well segregated. Time-lapse analysis (Fig. 3; Supplementary Fig. S3) confirms that loci move to the middle third of the cell prior to duplication, where the duplicated foci appear close together before generally segregating away from the mid-cell region. At least 2000 cells were analyzed for each locus. (C, left panel) Representative L3 (red)–R3 (green) FROS snapshots of τ 100-min and τ 26-min cells (inset). (Right panel) Representative R2 (red)–R3 (green) snapshots of τ 100-min cells. White arrows show closely separated sister foci that appear to have duplicated close to mid-cell. Schematics showing frequencies of different patterns in cells with only one focus for each locus (631 of 1112 cells in L3–R3, and 532 of 1243 in R2–R3) are above the micrographs. (D) Pairwise FISH analysis. Cells were divided into five arbitrary regions (I–V; III was one-third of cell length; others were one-sixth of cell length). Six locus pairs, using L3 or R3 as a reference, were examined (Ori–L3, L1–L3, L2–L3; Ori–R3, R1–R3, R2–R3). For each locus pair, ∼200 four-focus cells (two sisters for the target locus and two for the reference, L3 or R3) in which the L3 or R3 sisters have the pattern (V, I) were extracted and then oriented with respect to the reference pattern (V, I). The relative positions of the target sister loci on the oriented nucleoids were plotted. The pairwise L replichore (plus Ori) analysis is shown on the left and the R replichore (plus Ori) analysis is shown on the right. Further snapshot analysis is shown in Supplementary Figure S2.
The frequency of the most asymmetric pattern (pattern A), increased linearly from a low level at oriC to a maximum at L4–R4 at the boundaries of ter, and then declined in the 300 kb preceding the center of ter (Ter). This decrease in ter was mirrored by an increase of sister foci located close to mid-cell (pattern C). A symmetrical pattern with sister foci at cell quarters (pattern D), was maximal at oriC, consistent with previous results (Lau et al. 2003; Bates and Kleckner 2005; Fekete and Chattoraj 2005; Wang et al. 2005), although a significant level (∼10%) of asymmetry was also observed for oriC (Bates and Kleckner 2005; Wang et al. 2005). The symmetry of the curves with a transverse axis about oriC is striking, and suggests that the two replichores are organized and segregated by the same mechanism, despite differences in DNA sequence and content (Blattner et al. 1997; Rocha 2004).
In a parallel initial survey, eight loci were tracked by fluorescent repressor DNA binding (FROS) (Fig. 2B). We infer from the patterns of focus positioning, and their frequencies, that there is a sequential recruitment of loci to the mid-cell region, where they duplicate and segregate, with time of locus duplication proportional to position on genetic map. Any sister cohesion must be local rather than extending over much of the chromosome. This conclusion is in accord with the C. crescentus analysis (Viollier et al. 2004), but is inconsistent with conclusions from more limited E. coli studies that proposed extensive sister cohesion (Sunako et al. 2001; Bates and Kleckner 2005).
The initial surveys allowed us to choose loci for a more detailed analysis in which pairwise combinations of genetic loci were simultaneously visualized in exponentially growing cells (generation time [τ] 100 min [Fig. 2C; Supplementary Fig. S2] or 26 min [Fig. 2C, inset, left]) and in exponential cultures (τ 100 min) that had been treated for 60 min with cephalexin, which inhibits cell division and thereby maintains sister nucleoids together so as to maximize the number of analyzable sister nucleoids (Fig. 2D; Supplementary Fig. S2; Wang et al. 2005). The results from all methods of visualization were in agreement, thereby mitigating any concerns about artifacts and vindicating the methods of analysis. Previous work has shown that under the conditions used here to visualize specific repressor-bound DNA, there is little or no perturbation of cell cycle parameters (Wang et al. 2005). Our analysis of pairwise combinations of loci in sister nucleoids has been crucial in revealing the chromosome organization identified here. Additional conclusions from the data summarized in Figure 2 (detailed in Supplementary Fig. S2) are as follows:
(1) In cells with τ 100 min or 26 min, loci on opposite replichores locate to opposite nucleoid halves, and loci on the same replichore locate to the same nucleoid half (Fig. 2C). Such <L–R> localization of replichores is also evident for each sister nucleoid in a dividing cell (Fig. 2C,D; Supplementary Fig. S2).
(2) Whenever sister foci are close together for any locus, consistent with recent duplication, they are positioned in the middle third of the cell for τ 100-min cells and at both quarter positions in τ 26-min cells (Fig. 2B,C). Segregation of most newly replicated loci is asymmetric with respect to mid-cell. The data indicate that one sister locus migrates to the nucleoid edge adjacent to an old cell pole while the other remains close to a newly developing sister nucleoid edge at mid-cell. The asymmetrical segregation of sisters on different replichores directs them in opposite directions, thus generating a <L–R–L–R> pattern. The asymmetry is particularly strong for markers in the L2–L5 and R2–R5 segments of the chromosome (Fig. 2A,C,D; Supplementary Fig. S2). A minority of sister nucleoids (for L3–R3, 19%–24% in exponentially growing cells [FROS]; 12%–15% in cephalexin [FROS and FISH]) have a <L–R–R–L> or <R–L–L–R> pattern, as compared with the majority with the <L–R–L–R> pattern (Supplementary Fig. S2).
(3) Loci on the same replichore have positions within the nucleoid that reflect their time of replication–segregation, with earlier replicating loci internal to later replicating loci (Fig. 2C [right panel], D; Supplementary Fig. S2). Thus, the genetic map is recapitulated within the cell. The sequential segregation of the loci on each replichore (Fig. 2B), and the loci position gradient from ori to both nucleoid edges (Fig. 2D), indicate a layering process during replication segregation, with a sequential and bilateral deposition of newly replicated loci.
The extensive data set presented here, which required analysis of relative focus positions in >5 × 104 cells, provides strong experimental support for the organization model outlined in Figure 1. We now provide evidence that strengthens the notion that the segregation fate of a locus is determined by which fork replicates it rather than by the DNA sequence context of the locus and its surrounding DNA. Loci L6–R6 in ter, 200 kb either side of Ter (Fig. 1), segregate asymmetrically to opposite poles 67% of the time and asymmetrically to the same pole 23% of the time (Supplementary Fig. S1; Wang et al. 2005). L6 is expected to be replicated by the counterclockwise fork, since progress of the clockwise fork is blocked by two Tus-dependent replication termination sites (Neylon et al. 2005). R6 is accessible to either fork, although proximity should lead to it being replicated by the clockwise fork most frequently (Fig. 1). If replication fork determines segregation fate, then inserting an ectopic termination site between R6 and Ter (Fig. 1) will increase the frequency of replication of the locus by the clockwise fork, and therefore increase segregation to opposite poles and decrease cosegregation to the same poles. This was observed; the frequency of asymmetric segregation to the same poles decreased from 23% to 6%, while that to opposite poles increased from 77% to 94% (Supplementary Fig. S1). We conclude that replication rather than DNA sequence directs segregation fate.
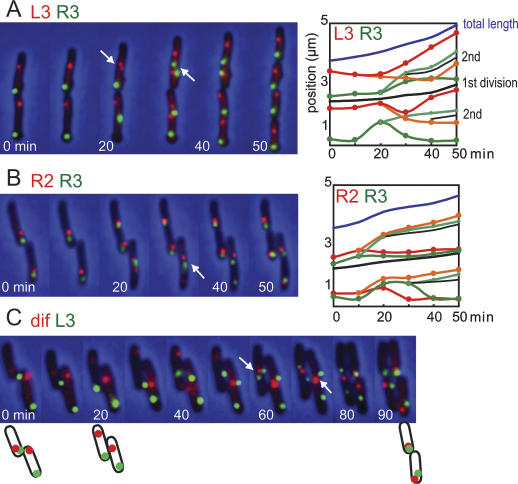
In order to test the inferences from the snapshot analyses, extensive time-lapse analysis was performed on selected combinations of loci in media in which τ is 100 min in liquid culture (Fig. 3; Supplementary Fig. S3). This analysis confirmed that the all regions appear to duplicate in the middle third of the cell at τ 100 min, consistent with reports that replisome markers start as a single mid-cell focus, which subsequently duplicates and moves to the quarter positions (Adachi et al. 2005; Bates and Kleckner 2005). Loci on opposite replichores segregate to opposing positions (Fig. 3A), while loci on the same replichore cosegregate (Fig. 3B), as inferred from the snapshot analysis. The relative positioning of R2–R3 foci confirms sequential deposition during replication–segregation; R3 sisters are located closer than R2 to the old nucleoid edge and the newly developing inner nucleoid edge in ∼90% of cells in the time lapses (Fig. 3B; data not shown).
Figure 3.
Time-lapse analysis. (A) Representative FROS time-lapse progressions (10-min intervals) are shown for L3 (red)–R3 (green). The right panel shows the position of L3 (red) and R3 (green) as a function of time in the left time lapse. The black line positions the division site when it is visible during invagination and the new poles after division. (B,C) The position of sister L3s are tracked by red and orange lines, and R3s are tracked by green and light-green lines. (B) R2 (red)–R3 (green). (C) L3 (green)–dif (red). Schematics of cells at 0, 20, and 90 min are shown. Further analysis is in Supplementary Figure S3. The dif locus array is located 8 kb clockwise of dif and 11 kb from Ter.
In addition, time-lapse analysis shows directly that in the majority of cell generations, an imprinting mechanism directs polar migrations of a newly replicated locus to the same pole that the mother locus occupied in the previous generation (Fig. 3A,B; Supplementary Fig. S3). The predominant imprinted <L–R–L–R> pattern for L3–R3 is maintained in 12 of 17 cell divisions (Fig. 3A; Supplementary Fig. S3a). In one of 17 divisions, the pattern switched from <L–R–L–R> to <R–L–R–L> (Supplementary Fig. S3d), while in four of 17 cell divisions, it switched to <L–R–R–L> or <R–L–L–R> patterns (Supplementary Fig. S2B,C). These data are in excellent agreement with the snapshots of live sister nucleoid L3–R3 pairs and with FISH-visualized L3–R3 and L4–R4 pairs, where ∼85% of sister nucleoids had the <L–R–L–R> pattern, and ∼15% had a <L–R–R–L> or <R–L–L–R> pattern (Supplementary Fig. S2). Irrespective of the pattern, sister nucleoids always retain a <L–R>, or a <R–L> arrangement necessary to successfully complete chromosome segregation.
It follows from our model for chromosome organization that ter DNA must cross from one nucleoid edge to the other (Fig. 1). The 28-base-pair (bp) dif DNA recombination site, which lies close to the center of ter (18 kb counterclockwise of the Ter locus in Fig. 1; Blakely et al. 1991; Kuempel et al. 1991), is essential for chromosome dimer resolution by XerCD-FtsK site-specific recombination, and may be used in the final stages of decatenation, by TopoIV (Hojgaard et al. 1999) and by XerCD (Ip et al. 2003). Consistent with this, replication may frequently terminate in the region of dif since base composition skew, resulting at least in part from different mutation rates on the leading and lagging strand templates, reverses close to dif (Lesterlin et al. 2005). It seems likely that the locus that is replicated last will give sister loci that remain at mother mid-cell, and become located at the nucleoid edges adjacent to the new poles after division. Unless there is a very precise site of replication termination, for example at dif, this locus will differ in different cell generations. A locus in ter that is not replicated last in any given replication cycle may have more of an opportunity to segregate asymmetrically, the direction of the polar segregation being determined by which replication fork replicates it. Time-lapse analysis (Fig. 3C) shows that over successive generations, a locus 8 kb from dif is initially present as two new pole-associated sisters (with L3 asymmetrically segregated), followed in the subsequent generations by cosegregation with the L replichore and then the R replichore. Furthermore, detailed analysis of dif segregation relative to L3/R3, by extensive snapshot analysis (Supplementary Fig. S4A) and over 23 cell generations by time lapse (Supplementary Fig. S4B), leads us to conclude that during asymmetric dif segregation, it cosegregates with the L and R replichore markers in equal proportion. This behavior of dif is consistent with it lying in the ter region that crosses from one nucleoid edge to the other; any marker in ter has the potential to be colocalized with either the left or right replichore or in between.
In the snapshot analysis, sister dif loci at mid-cell predominate (Fig. 2A,B; Supplementary Fig. S4A), and asymmetrical segregation accounts for 12% in exponential cells and 22% in cephalexin-treated cells (Supplementary Fig. S4A). In time-lapse analysis of 23 cell generations, asymmetric segregations account for 14 of 23 dif sister segregations, while in seven of 23 segregations, the newly replicated dif sisters remain at mid-cell–new pole until they migrate to the mid-cell position of the daughter cells prior to rereplication (Supplementary Fig. S4B). The ratio of dif asymmetrical segregation is higher in cephalexin-treated cells (22%) than in exponential cells (12%), and is higher in time-lapse analysis (61%) than in snapshot analysis (12%–22%). These differences indicate that the asymmetrical segregation of the dif locus may occur after cell division.
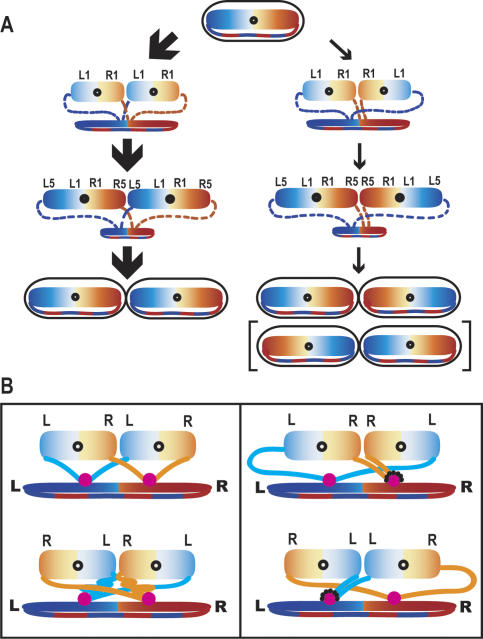
Our extensive analysis has revealed the organization of the E. coli circular chromosome at the replichore level and provides insight into how replication–segregation–deposition creates such an organization (Fig. 4A). We demonstrate that opposite replichores map to opposite nucleoid halves, with no evidence for replichore interwrapping. This organization around a transverse ori_–_ter axis, differs from that in C. crescentus, which places the replichores about a longitudinal axis. We suspect that, in C. crescentus, the two replichores are also not interwound. It seems likely that in all bacteria, the replichore is the unit of chromosome organization, directed by replication–segregation.
Figure 4.
(A) Schematic of E. coli chromosome organization and segregation. Slow-growing E. coli in which a single pair of sister replication forks replicate the whole chromosome. The major <L–R–L–R> pathway (∼85%) and the minor <L–R–R–L> pathways (∼15%) are indicated by thick and thin arrows. The L replichore is in blue and the R replichore is in orange. The two sister nucleoids, expanding through replication, are shown on top and the contracting mother nucleoid is shown on the bottom. The deposition of nascent strands to both edges of the expanding sister nucleoids is indicated by dashed lines. The relative positions of L1, L5, R1, and R5 on the two sister nucleoids are also indicated. The product of the <R–L–L–R> pathway, which occurs at equal probability to <L–R–R–L>, is shown in parentheses. (B) The “split replisome” model for E. coli sister nucleoid segregation. Organization is directed as a consequence of sister replisomes moving apart along the longitudinal axis. Left panels show the two possible situations for generating <L–R–L–R> (frequent), or <R–L–R–L> (rare) sister nucleoids from a <L–R> mother. Right panels show pathways for generating the minority <L–R–R–L> or <R–L–L–R> sister nucleoids. These may be favored when one of the replication forks stalls or collapses (dotted lines around the replisome, shown in pink).
Recent studies have proposed that foci representing replisome markers or newly replicated DNA, initially positioned at mid-cell in slow-growing newborn cells, duplicate and move toward the quarter positions after replication initiation (Adachi et al. 2005; Bates and Kleckner 2005). A “splitting” of sister replisomes along the longitudinal cell axis, and segregation of newly replicated DNA from the separated replisomes may help explain the relative proportions of the four sister nucleoid arrangements observed (Fig. 4B). In the dominant <L–R–L–R> arrangement (Fig. 4B, left panel, top), the nascent strands from L and R replisome travel similar distances to their respective L and R edge of each sister nucleoid, which is compatible with a normal parallel progression of replication in the two replisomes (Breier et al. 2005). However, in the rarely observed <R–L–R–L> arrangement (Fig. 4B, left panel, bottom), such a balanced situation is broken. We propose that the <L–R–R–L> and <R–L–L–R> arrangements (Fig. 4B, right panel) become permitted when replication at one replisome is delayed during the critical period that determines nucleoid orientation. In these situations, the nascent strands from the undisturbed replisome are proposed to go to the outer edges of both sister nucleoids, while the strands from the delayed replisome travel less distance and go to the inner edges of both sister nucleoids. Consistent with this view, one fork has been estimated to arrest in ∼18% of E. coli replication cycles, with arrest occurring equally frequently on both replichores (Maisnier-Patin et al. 2001).
If the sister replisomes do not “split” and remain at mid-cell, it is necessary to invoke a true asymmetric segregation process; for example, one in which leading and lagging strand sisters are segregated differentially (Wang et al. 2005).
The model proposed here suggests an intimate connection between replisome dynamics and chromosome organization, and allows the coexistence of two pathways, while at the same time strongly favoring the imprinted <L–R–L–R> pathway. Such replication–segregation organization is not compatible with models in which replication is followed by extensive sister cohesion and then subsequent segregation, as in eukaryotes (Sunako et al. 2001; Bates and Kleckner 2005). The characterization of chromosome organization and behavior presented here will facilitate studies that reveal the underlying mechanisms that position replication and segregate DNA.
Material and methods
E. coli AB1157 strains containing lacO and tetO arrays were constructed as in Lau et al. (2003). Conditions for bacterial growth and for visualizing fluorescent protein foci in living cells have been described in Wang et al. (2005). During exponential growth in M9 minimal glycerol medium, cells had a τ of 100 min at 37°C, with nonoverlapping sequential G1 (B), S (C), and G2/M (D) phases, with S being ∼55 min and G2/M being ∼35 min (Wang et al. 2005). FISH was performed as in Bates and Kleckner (2005).
Acknowledgments
This work was funded by the Wellcome Trust. C.P. was supported by an EMBO long-term fellowship, and X.L. was supported by a Clarendon post-graduate award.
Footnotes
References
- Adachi S., Kohiyama M., Onogi T., Hiraga S. Localization of replication forks in wild-type and mukB mutant cells of. Escherichia coli. Mol. Genet. Genomics. 2005;274:264–271. doi: 10.1007/s00438-005-0023-6. [DOI] [PubMed] [Google Scholar]
- Bates D., Kleckner N. Chromosome and replisome dynamics in E. coli: Loss of sister cohesion triggers global chromosome movement and mediates chromosome segregation. Cell. 2005;121:899–911. doi: 10.1016/j.cell.2005.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakely G., Colloms S., May G., Burke M., Sherratt D. Escherichia coli XerC recombinase is required for chromosomal segregation at cell division. New Biol. 1991;3:789–798. [PubMed] [Google Scholar]
- Blattner F.R., Plunkett G., III, Bloch C.A., Perna N.T., Burland V., Riley M., Collado-Vides J., Glasner J.D., Rode C.K., Mayhew G.F., et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Breier A.M., Weier H.U., Cozzarelli N.R. Independence of replisomes in Escherichia coli chromosomal replication. Proc. Natl. Acad. Sci. 2005;102:3942–3947. doi: 10.1073/pnas.0500812102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekete R.A., Chattoraj D.K. A cis-acting sequence involved in chromosome segregation in. Escherichia coli. Mol. Microbiol. 2005;55:175–183. doi: 10.1111/j.1365-2958.2004.04392.x. [DOI] [PubMed] [Google Scholar]
- Gitai Z., Thanbichler M., Shapiro L. The choreographed dynamics of bacterial chromosomes. Trends Microbiol. 2005;13:221–228. doi: 10.1016/j.tim.2005.03.006. [DOI] [PubMed] [Google Scholar]
- Gordon G.S., Sitnikov D., Webb C.D., Teleman A., Straight A., Losick R., Murray A.W., Wright A. Chromosome and low copy plasmid segregation in E. coli: Visual evidence for distinct mechanisms. Cell. 1997;90:1113–1121. doi: 10.1016/s0092-8674(00)80377-3. [DOI] [PubMed] [Google Scholar]
- Hojgaard A., Szerlong H., Tabor C., Kuempel P.L. Norfloxacin-induced DNA cleavage occurs at the dif resolvase locus in E. coli and is the result of interaction with topoisomerase IV. Mol. Microbiol. 1999;33:1027–1036. doi: 10.1046/j.1365-2958.1999.01545.x. [DOI] [PubMed] [Google Scholar]
- Ip S.Y.P., Bregu M., Barre F.-X., Sherratt D.J. Decatenation of DNA circles by FtsK-dpendent Xer site-specific recombination. EMBO J. 2003;22:6399–6407. doi: 10.1093/emboj/cdg589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuempel P.L., Henson J.M., Dircks L., Tecklenburg M., Lim D.F. dif, a recA-independent recombination site in the terminus region of the chromosome of. Escherichia coli. New Biol. 1991;3:799–811. [PubMed] [Google Scholar]
- Lau I.F., Filipe S.R., Soballe B., Okstad O.A., Barre F.X., Sherratt D.J. Spatial and temporal organization of replicating Escherichia coli chromosomes. Mol. Microbiol. 2003;49:731–743. doi: 10.1046/j.1365-2958.2003.03640.x. [DOI] [PubMed] [Google Scholar]
- Lemon K.P., Grossman A.D. Movement of replicating DNA through a stationary replisome. Mol. Cell. 2000;6:1321–1330. doi: 10.1016/s1097-2765(00)00130-1. [DOI] [PubMed] [Google Scholar]
- Lesterlin C., Mercier R., Boccard F., Barre F.X., Cornet F. Roles for replichores and macrodomains in segregation of the Escherichia coli chromosome. EMBO Rep. 2005;6:557–562. doi: 10.1038/sj.embor.7400428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Sergueev K., Austin S. The segregation of the Escherichia coli origin and terminus of replication. Mol. Microbiol. 2002;46:985–996. doi: 10.1046/j.1365-2958.2002.03234.x. [DOI] [PubMed] [Google Scholar]
- Maisnier-Patin S., Nordstrom K., Dasgupta S. Replication arrests during a single round of replication of the Escherichia coli chromosome in the absence of DnaC activity. Mol. Microbiol. 2001;42:1371–1382. doi: 10.1046/j.1365-2958.2001.02718.x. [DOI] [PubMed] [Google Scholar]
- Neylon C., Kralicek A.V., Hill T.M., Dixon N.E. Replication termination in Escherichia coli: Structure and antihelicase activity of the Tus–Ter complex. Microbiol. Mol. Biol. Rev. 2005;69:501–526. doi: 10.1128/MMBR.69.3.501-526.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niki H., Hiraga S. Polar localization of the replication origin and terminus in Escherichia coli nucleoids during chromosome partitioning. Genes & Dev. 1998;12:1036–1045. doi: 10.1101/gad.12.7.1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niki H., Yamaichi Y., Hiraga S. Dynamic organization of chromosomal DNA in. Escherichia coli. Genes & Dev. 2000;14:212–223. [PMC free article] [PubMed] [Google Scholar]
- Rocha E.P. Order and disorder in bacterial genomes. Curr. Opin. Microbiol. 2004;7:519–527. doi: 10.1016/j.mib.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Sherratt D.J. Bacterial chromosome dynamics. Science. 2003;301:780–785. doi: 10.1126/science.1084780. [DOI] [PubMed] [Google Scholar]
- Sunako Y., Onogi T., Hiraga S. Sister chromosome cohesion of. Escherichia coli. Mol. Microbiol. 2001;42:1233–1241. doi: 10.1046/j.1365-2958.2001.02680.x. [DOI] [PubMed] [Google Scholar]
- Teleman A.A., Graumann P.L., Lin D.C., Grossman A.D., Losick R. Chromosome arrangement within a bacterium. Curr. Biol. 1998;8:1102–1109. doi: 10.1016/s0960-9822(98)70464-6. [DOI] [PubMed] [Google Scholar]
- Thanbichler M., Viollier P.H., Shapiro L. The structure and function of the bacterial chromosome. Curr. Opin. Genet. Dev. 2005;15:153–162. doi: 10.1016/j.gde.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Viollier P.H., Thanbichler M., McGrath P.T., West L., Meewan M., McAdams H.H., Shapiro L. Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. Proc. Natl. Acad. Sci. 2004;101:9257–9262. doi: 10.1073/pnas.0402606101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Possoz C., Sherratt D.J. Dancing around the divisome: Asymmetric chromosome segregation in. Escherichia coli. Genes & Dev. 2005;19:2367–2377. doi: 10.1101/gad.345305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L.J. Structure and segregation of the bacterial nucleoid. Curr. Opin. Genet. Dev. 2004;14:126–132. doi: 10.1016/j.gde.2004.01.006. [DOI] [PubMed] [Google Scholar]