Hemizygous subtelomeres of an African trypanosome chromosome may account for over 75% of chromosome length (original) (raw)
Abstract
African trypanosomes are parasitic protozoa that infect a wide range of mammals, including humans. These parasites remain extracellular in the mammalian bloodstream, where antigenic variation allows them to survive the immune response. The Trypanosoma brucei nuclear genome sequence has been published recently. However, the significant chromosome size polymorphism observed among strains and subspecies of T. brucei, where total DNA content may vary up to 30%, necessitates a comparative study to determine the underlying basis and significance of such variation between parasites. In addition, the sequenced strain (Tb927) presents one of the smallest genomes analyzed among T. brucei isolates; therefore, establishing polymorphic regions will provide essential complementary information to the sequencing project. We have developed a Tb927 high-resolution DNA microarray to study DNA content variation along chromosome I, one of the most size-variable chromosomes, in different strains and subspecies of T. brucei. Results show considerable copy number polymorphism, especially at subtelomeres, but are insufficient to explain the observed size difference. Additional sequencing reveals that >50% of a larger chromosome I consists of arrays of variant surface glycoprotein genes (_VSG_s), involved in avoidance of acquired immunity. In total, the subtelomeres appear to be three times larger than the diploid core. These results reveal that trypanosomes can utilize subtelomeres for amplification and divergence of gene families to such a remarkable extent that they may constitute most of a chromosome, and that the VSG repertoire may be even larger than reported to date. Further experimentation is required to determine if these results are applicable to all size-variable chromosomes.
African trypanosomes include several species of parasitic protozoa such as Trypanosoma brucei, which is responsible for African sleeping sickness (trypanosomiasis) and kills many thousands of people each year in sub-Saharan Africa (WHO 2002). There are no vaccines against this parasite, and novel drugs and diagnostics are required (Burri et al. 2004). Unlike related parasites such as T. cruzi and Leishmania, African trypanosomes remain extracellular. In the mammalian bloodstream the parasites are encased in a dense coat of surface glycoprotein and they have a sophisticated mechanism of antigenic variation, involving switching expression from one surface protein variant to another, which allows the infecting population to avoid complete destruction by the mammalian immune system (for review, see Barry and McCulloch 2001). The genome contains a large repertoire of non-expressed variant surface glycoprotein genes (_VSG_s) that may be copied into one of several VSG expression sites containing VSG promoters, only one of which is active at a time.
Recent publication of the T. brucei genome sequence (Berriman et al. 2005) has revealed that the 11 pairs of megabase chromosomes of T. brucei share a common architecture: a diploid central chromosome core containing closely spaced genes showing a high level of synteny with other trypanosomatids (Ghedin et al. 2004; El-Sayed et al. 2005), and subtelomeres containing _T. brucei_-specific genes and other sequences. Among these are rapidly evolving gene families such as RHS, highly variable genes of unknown function that contain a hot spot of insertion for retroelements (Bringaud et al. 2002), LRRP (Leucine Rich Repeat Protein), and _VSG_s.
However, comparison of different T. brucei isolates reveals considerable genetic variability in the form of chromosomal size variation (Gottesdiener et al. 1990; Gibson and Garside 1991; Turner et al. 1997; Melville et al. 1998, 1999, 2000). Chromosome size polymorphism is not unique to African trypanosomes: It has been observed in Plasmodium and Leishmania, where size variation of 10%–25% has been reported (Janse 1993; Wincker et al. 1996), and in T. cruzi, where twofold variation has been observed (Henriksson et al. 1995). In the diploid kinetoplastid parasites, variation is also observed between homologs within a single parasite (references as above). But the range of size variation we observe in African trypanosomes far exceeds anything reported in other organisms to date. Apparently homologous chromosomes (i.e., containing the same coding sequences and, where investigated, in the same order) vary up to fourfold in size between isolates (Melville et al. 1998, 2000; V. Leech and S. Melville, unpubl.), and we have observed variation of up to twofold between diploid homologs within a parasite (Melville et al. 2000). Yet, mapping projects have failed to identify major DNA translocations or loss of synteny (Melville et al. 1998, 1999, 2000), and only one instance is reported in the literature (Gibson and Garside 1991).
The sequenced T. brucei isolate originated from a tsetse fly in Kenya, and the cloned laboratory stock derived from this (Tb927) is fully competent in differentiation and genetic exchange (Hope et al. 1999; van Deursen et al. 2001). There is no reason to suppose that essential genes may be missing. However, this genome may not be fully representative of genetic variation in the field as it contains chromosomes that are mostly smaller in size than their homologs in other isolates studied in our laboratory (Melville et al. 1998, 2000; V. Leech and S. Melville, unpubl.). In all pairwise comparisons of karyotypes, Tb927 contains the least megabase chromosomal DNA, and only three cases of smaller homologs in other independent field isolates have been identified: _chr_Va in Tb386 (Fig. 2 in Melville et al. 1998) and _chrs_IIa and IXa in Tb427 (Table 1 in Melville et al. 2000). To understand the significance of this chromosome size polymorphism, and to place the T. brucei genome sequence into the context of field variation, it is necessary to know which chromosomal regions and which genes are affected by these polymorphisms.
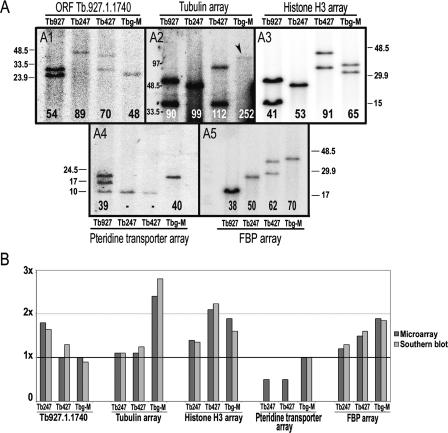
Figure 2.
Validation of data derived from hybridization to the Tb927-_chr_Ia tiling path genomic microarray. (A) Hybridization of five gene-specific probes to Southern-blotted genomic fragments containing intact tandem arrays. Numbers at the bottom of each lane indicate (in kb) total DNA content of each array in each genome, estimated from these gels. Two bands in the same lane indicate that the array varies in length in the _chr_I homologs. The 10-kb band in panel 4 probably is due to cross-hybridization with a similar sequence in a different chromosome, and therefore has not been included in the final length for the _chr_I region containing pteridine transporter genes. DNA size markers are in kb. (B) Comparison between microarray results and Southern blot results. _y_-axis represents the amount of DNA compared with Tb927. Value of 1× in the _y_-axis indicates equal DNA content compared with Tb927.
To analyze chromosome size polymorphism within T. brucei strains and subspecies we chose to compare homologs of chromosome I (_chr_I), which are among the most size-variant, using resources available from the sequencing project. The smaller homolog (_chr_Ia) of Tb927 was sequenced by chromosome shotgun (Hall et al. 2003), involving end-sequencing of cloned 1-to 2-kb sheared DNA fragments of gel-eluted Tb927-_chr_Ia DNA, and is one of only two chromosomes sequenced from telomere to telomere (Berriman et al. 2005). We selected 1055 sequenced clones to create a minimally overlapping tiling path microarray of _chr_Ia. We then co-hybridized fluorescently labeled genomic DNA from stock 927 with differentially labeled DNA from other cloned stocks, to identify Tb927-_chr_Ia sequences that are present in higher copy number in the genomes containing larger _chr_I homologs. This technique allows us, for the first time, to detect DNA content variation continuously along the whole length of a T. brucei chromosome in different cloned isolates with a resolution high enough to accurately determine which genes or regions are involved in these polymorphisms. Results show that regions of _chr_I containing tandemly repeated genes are very polymorphic in size. However, the most significant copy number polymorphism is observed in the 200-kb subtelomeric region on the left end of the chromosome containing gene families and retrotransposons. Nevertheless, the observed variation does not explain the final length of _chr_I homologs in some isolates. Additional sequencing reveals that ∼55% of a greater than threefold larger _chr_I homolog consists of arrays of VSG genes, typically located in subtelomeric regions. Additional sequencing of clones derived from two other size-variable chromosomes also revealed no evidence of DNA translocation from elsewhere in the genome and provided indirect evidence of VSG array expansion. Nevertheless, further experimentation is required to determine whether amplification of subtelomeric families is the primary basis of all chromosomal size variation in the T. brucei genome.
Results
Comparison between strains
We have analyzed variation in DNA content in different African trypanosome strains and subspecies (Tb247, Tb427, and Tbg-M; we will refer to these genomes as “test genomes”) compared with the sequenced strain Tb927 (“reference genome”) using a Tb927-_chr_Ia genomic microarray. A total of 1055 PCR products amplified from a Tb927-_chr_Ia plasmid library were spotted onto the slides. The tiling path microarray generated covers most of the 1.1 Mb length of Tb927-_chr_Ia (see Methods).
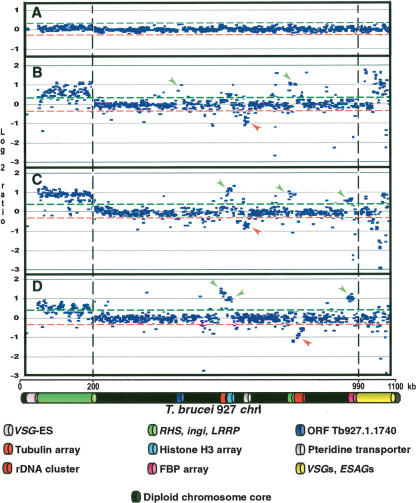
Figure 1 shows the log2 ratio (test genome/reference genome) of the intensity of hybridization to each spot on the array presented in the linear context of Tb927-_chr_Ia. The top panel shows co-hybridization of Tb927 DNA populations labeled separately with Cy-3 and Cy-5, to determine the level of noise in the experiment. Red and green lines represent 2× standard deviation (SD) from the zero line in this control experiment, and these lines have been transposed to panels B–D. Panels B, C, and D show the results of hybridization of total DNA from Tb247, Tb427, and Tbg-M, respectively, onto the Tb927-_chr_Ia array (Tb247 _chr_I sizes: 1.3 and 1.5 Mb; Tb427: 1.85 and 3.6 Mb;
Figure 1.
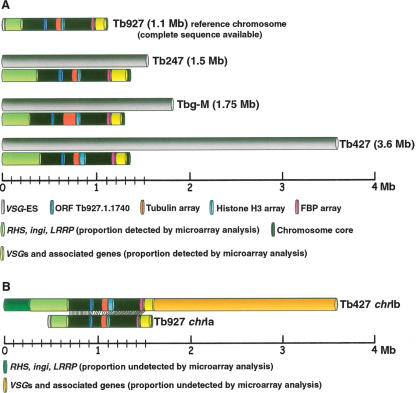
Copy number polymorphism in chromosome I homologs of T. brucei, assayed by comparative genomic hybridization to a microarray of T. brucei 927 _chr_Ia DNA fragments. (A) Co-hybridization of Tb927 genomic DNA differentially labeled with Cy5 and Cy3. (B) Cohybridization of differentially labeled Tb247 and Tb927 genomic DNA; (C) Tb427 and Tb927; (D) _T.b. gambiense_-M (Tbg-M) and Tb927. (Green and red lines) Two SD from the zero line in experiment A. Location of a fragment above the green line indicates copy number amplification in the test genome relative to Tb927; below the red line indicates DNA loss or sequence divergence in the test genome. (Vertical discontinuous lines) Subtelomere limits. Left subtelomere contains primarily retrotransposons (ingi) and gene families RHS and LRRP. Right subtelomere contains primarily genes involved in antigenic variation (_VSG_s) and associated genes (_ESAG_s). (_VSG_-ES) A VSG expression site; (FBP) a gene containing an F-box motif. (Green arrows) Gains of DNA; (red arrows) losses of DNA. Tandemly repeated gene coordinates: Tubulin array Tb927.1.2330–2410; Histone H3 array Tb927.1.2430–2550; Pteridine transporter array Tb927.1.2820, 2850, 2880; FBP array Tb927.1.4540–4650.
Tbg-M: 1.65 and 1.75 Mb). In all cases we co-hybridized total DNA from Tb927 as the reference DNA (Tb927 _chr_I sizes: 1.1 and 1.2 Mb).
Around 90% of spots consisting of DNA derived from the _chr_I diploid core show a log2 ratio within two SD of controls, suggesting little copy number variation in the central core of these homologous chromosomes. Significant variation in DNA content in this region is mainly restricted to tandemly repeated genes such as tubulin, histone H3, pteridine transporter, and an array of genes containing an F-box motif (genes 4540–4650 in _chr_I, here called FBP) (Fig. 1). Other significant variations observed within the diploid chromosome region correspond to the ORF Tb927.1.1740, which contains a 123-bp sequence repeated 169 times in Tb927, and the rDNA cluster. Although variation is restricted to the same regions in all the strains, its extent varies between them. For example, there is little variation in the histone H3 array in Tb247, but a significant gain of DNA in that region in Tb427 and Tbg-M. All gains of DNA observed in the chromosome core fall between log2 ratios of 0.4 and 1.5. Except for the variation in DNA content corresponding to the pteridine transporter in Tb247 and Tb427 and the rDNA cluster in Tbg-M, we did not detect losses of DNA in this region spanning more than 1.5–2 kb compared with the reference genome. Gains and losses of DNA in short, isolated regions are observed throughout, involving single spots on the array and corresponding to intergenic regions and hypothetical genes. For more details about the exact position represented by each spot, see Supplemental Figure S1.
The ∼200-kb subtelomeric region at the left end of Tb927-_chr_I contains RHS, LRRP genes, and retrotransposon elements such as ingi (Hall et al. 2003). Figure 1 shows that DNA spots in the micro-array representing this region exhibit a significant positive log2 ratio in all cases, ranging between 0.4 and 1 within and between subtelomeres. Overall, these data suggest that the sequences found in this chromosomal region are amplified in the test genomes by about 1.5-to twofold compared with the reference genome.
The ∼70 kb subtelomeric region at the right end of Tb927-_chr_I contains VSG genes and VSG expression site-associated genes (_ESAG_s 1, 2, 9, and 11) with one ingi (not indicated in Fig. 1). Array spots on the microarray representing _VSG_s and _ESAG_s show very high log2 ratios in some cases, such as Tb247 (up to 1.8), while in others, such as Tb427, we observe some spots with negative log2 ratios (up to −3). In Tbg-M we observe no significant variation. _VSG_s and some _ESAG_s may vary greatly in DNA sequence both within and between genomes. _VSG_s occur as gene families that may share very little primary sequence conservation, and different families may be found in different strains; therefore, a high level of variation in this region most likely reflects this.
Validation of the Tb927-_chr_Ia genomic array
In order to verify the accuracy of these data, digested genomic DNA containing intact arrays of the Tb.927.1.1740 ORF repeats, and the tubulin, histone H3, pteridine transporter, and FBP genes was Southern blotted and probed with clones derived from each of these genes (Fig. 2A). Some of the strains present two different bands for an array, indicating they have different lengths in each homologous chromosome (_chr_Ia and _chr_Ib). Where one band is visible, homologs carry arrays of the same length. However, in the case of the pteridine transporter gene array (panel 4 in Fig. 2A), a third band of 10 kb appears in Tb927, presumably due to hybridization of a putative pteridine transporter gene located in chromosome X. We also observe this band in Tb247 and Tb427 where the pteridine transporter array in _chr_I appears to be absent, indicating that the signal picked up from the spots representing the pteridine transporter in the microarray experiments must be due to hybridization of the DNA corresponding to this third band. At the bottom of each lane we show the estimated total DNA content (in kb) for each array in the entire genome (i.e., including copies from both homologs). Differences in tandem repeat array lengths between isolates observed in these Southern blots are unlikely to be due to restriction site polymorphism, since most have been verified using combinations of different restriction enzymes and also an enzyme cutting once inside the repeat unit to check that expansion is not due to unique sequence insertions (Hall et al. 2003).
We illustrate the comparison between these values and the microarray results in Figure 2B. The _y_-axis represents the amount of DNA compared with Tb927 (i.e., 1× indicates an equal amount compared with Tb927). We found all estimations of DNA content for tandemly repeated sequences consistently similar in both microarray and Southern blot experiments, confirming the high accuracy of the microarray experiment.
Search for novel sequences in Tb427-_chr_Ib
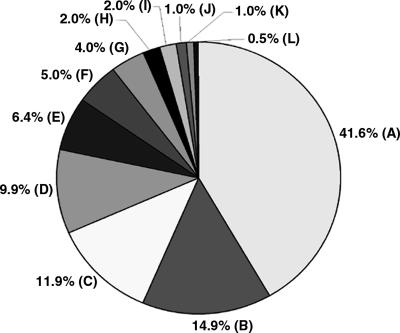
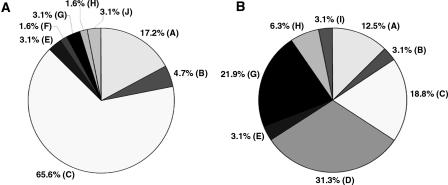
To investigate whether Tb427-_chr_Ib (3.6 Mb) contains sequences absent in Tb927-_chr_Ia (1.1 Mb), we created a Tb427-_chr_Ib-enriched plasmid library using DNA excised from a pulsed field gel (PFG) and amplified by DOP-PCR (Telenius et al. 1992), and produced an array of 3072 clones. We hybridized four replicates of this array separately with Tb927 and Tb427 genomic DNA and with gel-eluted Tb927-_chr_Ia and Tb427-_chr_Ib DNA (comparison of chromosomal and genomic probes would reveal straightforward DNA translocation to Tb427-_chr_Ib from elsewhere in the genome, if that were the case). We selected and end-sequenced 192 non-redundant clones that hybridized to 427-_chr_Ib but not to 927 (Supplemental Table S1). Then we used the sequence tags (EMBL Nucleotide Sequence Database, accession nos. AJ969959–AJ970139) to search the T. brucei sequence databases, and almost all identified matches in Tb927 (Fig. 3; Supplemental Table S2), albeit at a lower level of similarity than would be detected by our initial hybridization screen. Ten percent identified RHS and retrotransposons, but most (>70%) showed the greatest similarity with VSG gene sequences, or sequences known to be associated with silent VSG arrays in Tb927, such as _ESAG_s, UDP-dependent glycosyl transferase genes, or intergenic sequences in VSG arrays (Berriman et al. 2005). The remaining 20% correspond to genes and intergenic regions from other chromosomes (those adjacent to Tb427-_chr_Ib in the PFG), failed sequences, genes from the _chr_Ia diploid core, and sequences with no significant matches in the databases.
Figure 3.
Identity of highest-scoring pairs for 192 sequence tags from clones selected from the Tb427-_chr_Ib-enriched plasmid library, derived by BLASTN comparison of sequence tags with Tb927 sequence in GeneDB and trypanosome sequence in the EMBL database. (A) _VSG_s, (B) VSG expression site-associated genes, (88% of these sequences are variants of ESAG3, most copies of which are associated with non-expressed VSG gene arrays in the Tb927 genome sequence [Berriman et al. 2005]). (C) intergenic sequences in VSG arrays in Tb927, (D) RHS genes and retrotransposons, (E) genes from other chromosomes, (F) UDP-dependent glycosyl transferase, (G) failed sequences, (H) _chr_I genes (chromosome core), (I) intergenic sequences (chromosome core), (J) telomeric and telomere-associated sequences, (K) no significant match, (L) UDP-gal-phosphoglycan transferase. (The genes in F and L are associated with VSG gene arrays in Tb927 [Berriman et al. 2005]. See Supplemental Table S2 for additional details.)
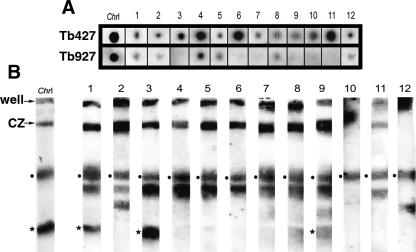
To determine whether these sequences or very similar sequences (sufficient for cross-hybridization) are present in the Tb927 genome, and to investigate whether they are found on both Tb427-_chr_I homologs, we hybridized 12 of the sequenced fragments to dot blots of genomic DNA from Tb927 and Tb427 and to chromosome blots from Tb427. Figure 4A shows that all hybridize to Tb427 DNA, and most either do not hybridize to Tb927 or the hybridization is much weaker. Figure 4B shows that all sequences hybridize to _chr_Ib. Of the 12 probes, nine identified _VSG_s in the databases, one identified an intergenic sequence from a Tb927 VSG array, one identified an ESAG, and one showed no significant match (see Supplemental Table S2). Two of the _VSG_s and the ESAG probe hybridize to both _chr_I homologs, but the remaining clones do not hybridize to the smaller _chr_Ia. The observed hybridization to other chromosome bands, including the large amount of DNA remaining in the compression zone, presumably indicates cross-hybridization to other members of the gene family. We also hybridized these clones to a gel in which Tb427-_chr_Ia was resolved (Melville et al. 2000) to confirm that only three hybridized to _chr_Ia (data not shown). All of them hybridized to several different additional chromosomes in this lower size range, although not necessarily to both homologs.
Figure 4.
Hybridization of 12 Tb427_-chr_Ib-derived probes to dot-blots of Tb927 and Tb427 total DNA (A) and Southern blots of Tb427 chromosomes (B). In dot-blot experiments most probes hybridize only to Tb427, although in some cases the lower stringency of this experiment (compared with the initial screening) allows cross-hybridization to similar VSG families in Tb927. (Black dots) Hybridization to _chr_Ib (fully resolved in this gel), (black stars) hybridization to the _chr_Ia homolog (here compressed with other chromosomes), (_Chr_I) _chr_I-specific gene probe, (CZ) compression zone containing _chr_s VIIIb, IXb, Xab, and XIab (∼40% of megabase genome). See Supplemental Table S2 for additional information on each of the probes.
Selecting homolog-specific sequences from other size-variable chromosomes
In order to determine whether the amplification of subtelomeric sequences, especially of VSG gene families, also underlies the increased size of other chromosomes, we prepared additional chromosome-enriched clone libraries. The number of chromosomes for which we can perform this experiment is somewhat restricted, since in all strains many chromosomes co-migrate with others in PFGs. In Tb927, the sequenced reference genome, only _chrs_I, II, and IX homologs can be resolved sufficiently for excision. _Chr_II varies little in size between strains, but _chr_IX is very variable, and we decided to compare Tb927-_chr_IXa (3.4 Mb) and Tb427-_chr_IXb (4.5 Mb). In addition, we identified a well resolved homolog of _chr_VI in an F1 hybrid clone (mcl4) from a cross between Tb927 and Tb247. The smaller mcl4-_chr_VIa homolog (1.75 Mb) derives from Tb927, and we compared this with Tb427-_chr_VIa (2.8 Mb), as the larger Tb427 homolog of 3.4 Mb cannot be resolved. Although we anticipated that the large VSG arrays on these chromosomes in Tb927 (Berriman et al. 2005) would reduce our ability to select chromosome-specific _VSG_s, due to cross-hybridization between Tb927 and Tb427 families, we reasoned that this experiment should allow us to detect alternative bases for chromosome expansion, such as translocation of DNA from the diploid core of other chromosomes.
We created and arrayed Tb427-_chr_VIa- and Tb427-_chr_IXb-enriched plasmid libraries and selected Tb427 homolog-specific clones by hybridizing DNA derived from the Tb427 homologs and from the Tb927 homologs, exactly as described in the previous section for _chr_I. In the _chr_IX experiment we were able to identify only 19 Tb427-specific clones (accession nos. AM263038–AM263056), of which nine identified subtelomeric sequences in the Tb927 database. This raised our fears that the large number of _VSG_s in both subtelomeres of the Tb927 chromosome prevented selection of Tb427-specific _VSG_s by cross-hybridization between family members.
When we performed the comparative hybridization experiment using the Tb427-_chr_VI array, we identified 64 that did not hybridize to mcl4-_chr_VIa and derived sequence tags from each of these (Fig. 5A; see Supplemental Table S3 for accession nos.). We observed 22% VSG and ESAG sequences and no RHS, but an astonishing 66% of clones that identified intergenic sequences from Tb927 VSG arrays as their highest-scoring pair in BLASTN analyses (see Supplemental Table S3 for more details). This result was so striking that we decided to check the clone content of the arrayed library by sequencing a set of randomly selected clones. Comparison of Figure 5, A and B, shows that the comparative hybridization of _chr_VI homologs had indeed primarily selected putative intergenic sequence from VSG arrays, but relatively few Tb427-_chr_VI-specific _VSG_s.
Figure 5.
Identity of highest-scoring pairs for sequence tags from 64 Tb427-_chr_VIa-specific (panel A) and 32 randomly selected clones from the Tb427-_chr_VIa-enriched plasmid library (panel B), derived by BLASTN comparison of sequence tags with Tb927 sequence in GeneDB and trypanosome sequence in the EMBL database. (A) _VSG_s, (B) VSG expression site-associated genes, (C) intergenic sequences in VSG arrays in Tb927, (D) RHS genes and retrotransposons, (E) genes from other chromosomes, (F) UDP-dependent glycosyl transferase, (G) _chr_VI genes (chromosome core), (H) intergenic sequences (chromosome core), (I) telomeric and telomere-associated sequences, (J) no significant match. See Supplemental Tables S3 and S4 for additional details.
Discussion
The sequencing of the Tb927 genome has provided an invaluable amount of information about genomic structure and organization, the size of the VSG repertoire, and metabolic pathways (Berriman et al. 2005). It also has made possible the development of new molecular tools such as the high-resolution Tb927-_chr_Ia genomic microarray we have used in this study to determine the underlying basis of DNA content variation in different isolates of T. brucei.
_Chr_Ia has a total length of 1.1 Mb in Tb927 (Hall et al. 2003), while in Tb247, Tb427, and Tbg-M all the _chr_I homologs are larger (the largest homologs are 1.5, 3.6, and 1.75 Mb, respectively) (Melville et al. 1998; 2000; V. Leech and S. Melville, unpubl.). Our data show that variation in DNA content in _chr_I homologs compared with Tb927-_chr_Ia is significantly concentrated in specific regions: the so-called subtelomeres, which contain large and diverse aneuploid gene families (Melville et al. 1999; Berriman et al. 2005), and tandemly repeated sequences within the diploid chromosome core (see Fig. 1).
All arrays of tandemly repeated sequences in _chr_I show variation in size. In most cases, tandemly repeated genes located in the chromosomal core consist of almost perfect repeats (e.g., α-and β-tubulin and histone H3 arrays). The almost complete absence of base substitution suggests that the process of amplification and deletion of these arrays is an ongoing process involving gene conversion (Hall et al. 2003). Since transcription is polycistronic and there is a general lack of transcriptional control, it has been speculated that expansion of gene families by tandem duplications may be a mechanism to increase expression levels of specific genes, thereby increasing the level of protein in the cell (Ivens et al. 2005). We found a wide range of array lengths in the test genomes compared with the reference genome. In the case of the FBP array there are six copies of the same gene in Tb927; the first five copies are identical, but the last ORF and its 3′ UTR are divergent (a similar case is reported in T. cruzi; Tomas and Kelly 1996). The function of this gene family is unclear, but this array is duplicated in Tbg-M and expanded 1.5 times in Tb427. However, in the three test genomes the last ORF is missing or very divergent, indicating it is more variable between isolates than the rest of the family. Variation in the size of gene arrays might be a response to different requirements for protein expression in a particular strain or environment, although we are not able at present to offer any hypothesis as to why these genes are of particular importance.
In the left subtelomeric region of _chr_I, most of the genes belong to the novel multigene family called RHS (Bringaud et al. 2002). Little is known about RHS proteins except that they are constitutively expressed and localize in or around the nucleus. Our microarray results suggest that there are about twofold more copies in the genome of each of the three isolates studied compared with Tb927. Other elements in this region such as retrotransposons and LRRP genes are amplified in a similar proportion to RHS genes in all three cases. Since related members of all these sequence families are present in other chromosomes, we cannot accurately determine the extent of amplification on _chr_I homologs; we are able only to estimate the maximum amount that this is likely to contribute. In some of the isolates, this may represent an additional 200 kb of subtelomeric sequence.
The right subtelomere of _chr_Ia in Tb927 contains mainly VSG and ESAG genes with one ingi element. In the megabase chromosomes of Tb927, the VSG arrays are always located in the subtelomeric regions, and some previously reported observations have suggested that _VSG_s may be present as haploid copies (van der Ploeg et al. 1982; Beals and Boothroyd 1992; Melville et al. 2000; Berriman et al. 2005). In the microarray experiments, we detected significant gains of DNA in this region in Tb247 but insufficient variation in copy number in Tb427 and Tbg-M to make a significant contribution to their larger _chr_I homologs. In some cases, such as Tb427, we observed losses of DNA, indicating sequence divergence or sequence absence. In total, the maximum increase in length that could be due to amplification of the sequences found in this Tb927 subtelomere, according to our microarray data, is ∼55 kb in Tb247, just 7 kb in Tb427, and almost nothing in Tbg-M.
These microarray analyses suggested that copy number variation of perfect tandem repeats and of subtelomeric gene families are important sources of chromosome size polymorphism, but their contribution to the final length of much larger _chr_I homologs appeared relatively small. In Figure 6A we compare the maximum estimated length for the largest _chr_I homologs based on the array data, (i.e., the longest of each tandem array, the maximum amplification of the left subtelomere) to that observed in PFGs, highlighting only those regions that could significantly contribute to size variation in this chromosome. Only the left subtelomeric region and, in the case of Tb247, the right subtelomeric region seem to increase appreciably the length of these _chr_I homologs. Overall, these data can only explain ∼56%, 9%, and 27% of the “extra DNA” observed by PFG in the largest Tb247, Tb427, and Tbg-M _chr_I homologs, respectively.
Figure 6.
Models of chromosome expansion based on the comparative microarray analyses and sequencing of chromosome-specific DNA clones presented here. Note that we make the assumption that all gene families are positioned at the same location in both Tb927-_chr_Ia and Tb427-_chr_Ib, and therefore that arrays expand/contract in situ. (A) Comparison of c_hr_I size as estimated by PFGE analysis of different isolates of T. brucei (gray bars) with the theoretical maximum length estimated from the microarray and Southern blot analyses comparing the test chromosomes with Tb927-_chr_Ia. Colored regions correspond to tandemly repeated genes and gene families that show a significant degree of polymorphism compared with Tb927. (B) Our proposed model of Tb427-_chr_Ib organization. The central core is highly conserved compared with the sequenced Tb927-_chr_Ia; sequences in the left subtelomeric region are amplified about twofold, but there is no _VSG_-ES. Results presented here indicate that the remainder of the additional DNA in Tb427-_chr_Ib consists primarily of VSG arrays and associated sequences. We propose that this forms a hemizygous “subtelomeric” region of ∼2 Mb, and that this is probably on the right end of the chromosome.
Therefore, in Tb427-_chr_Ib there remained 2.2 Mb of DNA consisting of sequence undetected by the microarray experiments. Yet, mapping projects failed to detect any translocations from other chromosomes (Melville et al. 2000). We hypothesized that gene families present in subtelomeric regions such as VSG and RHS, which are known to be highly divergent, may be underestimated in the microarray experiments, and that the explanation for the larger _chr_I homologs in some T. brucei strains may lie in the presence of additional divergent members of these gene families. However, it was also possible that novel sequences were present on Tb427-_chr_Ib that were absent from Tb927-_chr_Ia. If such sequences were in the subtelomeres, they may not be detected in comparative mapping projects.
We identified 192 clones carrying Tb427-_chr_Ib-specific sequences from a Tb427-_chr_Ib-enriched array (i.e., clones not detected by hybridization of Tb927 DNA). End-sequencing of these clones shows that ∼74% of these sequences share the greatest similarity with VSG genes or sequences known to be associated with VSG arrays in Tb927, and 10% show the greatest similarity with RHS genes. Therefore, 84% of these Tb427-_chr_Ib-specific sequences most likely derive from subtelomeric locations (Fig. 3). The remaining 16% are contaminating sequences from the chromosome core or other chromosomes, and failed sequences. These results strongly indicate that the 2.2 Mb of Tb427-_chr_Ib-specific DNA apparently absent in Tb927-_chr_Ia consists mostly of subtelomeric sequences (∼1.85 Mb), and in particular VSG arrays (∼1.63 Mb). This estimation, added to the expansions detected by the microarray experiments and accounting for failed and contaminating sequences, suggests that subtelomeric sequences present in Tb427-_chr_Ib may represent three-quarters of the chromosome and that VSG arrays may account for >50% of its length. Based on VSG gene density in Tb927, this could amount to an additional 400 _VSG_s in Tb427-_chr_Ib.
We also performed further rounds of selection and sequencing of homolog-specific clones to investigate whether the expansion of subtelomeric gene families, in particular the massive expansion of VSG arrays, is the primary basis for variation in other size-polymorphic T. brucei chromosomes. However, we suspected that the absence of an extensive VSG array on Tb927-_chr_I had enabled us to obtain such unequivocal data on the content of the additional DNA in Tb427-_chr_Ib. In contrast, we feared that the large arrays located in the subtelomeric regions of all the other Tb927 chromosomes (and therefore most probably other strains also) would contain members of VSG gene families also present in the larger homologs in other strains. In these cases, cross-hybridization between multiple _VSG_s would obscure any array expansion on the larger homologs. Indeed, the Tb927 arrays on many chromosomes may be larger than shown in the published sequence, as most subtelomeres are incomplete (Berriman et al. 2005). To test our hypothesis, we created clone libraries from Tb427 homologs of chromosomes VI and IX. Although Tb427-_chr_IX is 33% larger than Tb927-_chr_IX, we identified very few homolog-specific clones in the _chr_IX library, strongly suggesting that the increased size is due to amplification or translocation of common sequences, not translocation of unique DNA, and that cross-hybridization prevents identification of the additional DNA. We identified more 427-specific clones in the _chr_VI library and found that most identified intergenic sequences from VSG arrays in the Tb927 database, although we identified few additional homolog-specific VSGs. We propose the most likely reason is that the Tb427-specific _VSG_s are cross-hybridizing within the large Tb927 VSG arrays, but the lesser conservation of some intergenic sequence has led to the selection of Tb427-specific clones of these regions. We did not identify homolog-specific RHS genes in either case, suggesting that there are no additional RHS or there is a high level of cross-hybridization between family members.
None of these experiments identified any interchromosomal translocation of unique coding DNA, and the chromosome VI comparison strongly, albeit indirectly, suggests that the larger homolog contains an expanded VSG array. However, it is clear that these are not the most efficient techniques for more extensive study of subtelomere expansion in other T. brucei chromosomes. Given the apparent ease of identification of sequences from Tb427 subtelomeric gene families by comparison with the Tb927 database (we identified few sequences with no significant similarity at all to Tb927 sequence), the most rapid and cost-efficient method to compare the relative content of genomes of different size will probably be comparative shotgun sequencing. In this regard, the ongoing analysis of the genome of an isolate of T.b.gambiense (http://www.sanger.ac.uk/Projects/T_b_gambiense) may provide our first overall view of expansion of subtelomeric gene families within the genome, although this will not provide specific data on expansion of individual chromosomes.
We have not determined the structure of Tb427-_chr_Ib, but we propose that this chromosome probably carries a much larger VSG array on the right-hand end (model shown in Fig. 6B) for several reasons. Firstly, the Tb927-_chr_Ia has a short array close to this telomere, and this region is extended in Tb247-_chr_Ib (Melville et al. 1999). Also, we note that _chr_IX is the only Tb927 chromosome to have large VSG arrays at both ends (Berriman et al. 2005), and this is the only chromosome on which we have not yet detected a _VSG_-ES in any stock (Melville et al. 1998, 2000), suggesting ES and array ends may be exclusive. However, it is possible that VSG arrays may be found at both telomeres on Tb427-_chr_Ib. The schematic in Figure 6B illustrates our model, showing a striking modular organization for these chromosomes: a diploid “central core” (Melville et al. 1998, 2000; MacLeod et al. 2005) consisting of just 750–850 kb of relatively gene-dense DNA, while the remainder, almost 3 Mb in the case of the larger _chr_I homolog, consists of highly polymorphic gene families in largely aneuploid segments of the chromosome.
All 12 Tb427-_chr_Ib-specific clones that we probed onto Tb427 chromosomes in Southern blots hybridized to _chr_Ib, but only three (two _VSG_s and one ESAG) hybridized to _chr_Ia. All hybridized to other chromosomes also, although not necessarily both homologs. Little is known about the generation of diversity in VSG families, but recombination between subtelomeres may facilitate transfer of VSG family members to diverse nonhomologous chromosomes, where they continue to amplify and diverge in sequence, as reported in Plasmodium parasites (Freitas-Junior et al. 2000). Other data indicate the occurrence of ectopic recombination in subtelomeric regions of T. brucei: For example, _VSG_-expression sites are found on different chromosome ends in different isolates (Melville et al. 1998).
Most eukaryotic organisms have subtelomeric regions, forming the transition between chromosome-specific sequence and telomere repeats (Mefford and Trask 2002; Barry et al. 2003). In lower eukaryotes, their plasticity is thought to help organisms to adapt to new environmental conditions, especially in pathogenic organisms (Freitas-Junior et al. 2000; Barry et al. 2003; Fabre et al. 2005). The parasite sequencing projects have revealed a common chromosome organization where genes that are conserved and syntenic in related species are concentrated in the gene-dense chromosome core, while species-specific gene families that diverge more rapidly within populations are located at the ends of chromosomes (El-Sayed et al. 2005; Hall et al. 2005; Pain et al. 2005). However, our results indicate that African trypanosomes can utilize subtelomeres for amplification and divergence of gene families to such a remarkable extent that they may constitute most of a chromosome. If subtelomeric expansion underlies size variation in all chromosomes, then the sequenced Tb927 chromosomes may have shorter subtelomeric regions than most homologs in other isolates. Tb427 is a laboratory-maintained stock, but in terms of DNA content and chromosome size it is more representative of the range of isolates that we have karyotyped. We observe _chr_I homologs of between 2 and ≥3.6 Mb in other E. African T. brucei isolates and in T. congolense, the largest to date in a pleomorphic line of T. b. rhodesiense EATRO 2340 (provided by J.D. Barry, Univ. of Glasgow) (V. Leech and S. Melville, unpubl.). DNA content of T. brucei isolates may vary by up to 30% (Melville et al. 1998; Hope et al. 1999). Tb427 megabase chromosomes contain ∼16 Mb extra DNA compared with Tb927 (Melville et al. 2000), and, if our observations on _chr_I prove to be applicable to many or all other chromosomes, a large proportion of this could consist of VSG arrays. No other parasite that undergoes antigenic variation has been reported to devote so much of its genome content to antigen genes (Barry et al. 2003; Craig and Scherf 2003; Hall et al. 2003, 2005). The genome sequencing program revealed that most of the _VSG_s on the Tb927 megabase chromosomes are pseudogenes (Berriman et al. 2005). It is possible that these represent VSG graveyards, i.e., VSG copies left behind after recombination events during antigen-switching and undergoing sequence decay (more _VSG_s are located at telomeres of ∼100 mini-chromosomes). If that is the case, then most trypanosomes are replicating very large amounts of useless (junk) DNA during each cell cycle. Alternatively, if the large arrays of pseudogenes and gene fragments are in fact used to create novel VSG genes by recombination before or during transposition to an expression site, as occasionally reported (Roth et al. 1989), the potential size of the VSG repertoire in these huge “subtelomeres” suggests an unrivaled capacity for new antigenic types in African trypano-some populations.
In this paper we have presented data to show that in T. brucei the so-called “sub_telomeric_ regions” may account for three-quarters of the length of a chromosome.
Methods
Parasite strains, axenic culture, and DNA extraction
The origins and histories of the cloned T. brucei stocks TREU (Trypanosomiasis Research, Edinburgh University) 927/4 (GPAL/ KE/70/EATRO 1534) single VAT derivate GUTat10.1 (Tb927), STIB (Swiss Tropical Institute, Basel) 247L (WA/TZ/71/STIB 247) (Tb247), Tb427, and T. brucei gambiense Mamaissata (Tbg-M) have been described (Turner et al. 1990; Biteau et al. 2000; Melville et al. 2000; van Deursen et al. 2001). They were prepared from isolates collected in Kenya (Tb927), Tanzania (Tb247), and Guinea (T.b.gambiense Mamaissata); the collection location of Tb427 is unknown, but SNP typing suggests it may belong to the Serengeti tsetse belt (E. Bart-Delabesse and S. Melville, unpubl.). F532/72-mcl4 (mcl4) is an F1 progeny clone from a cross between TREU927/4 and STIB 247 (Turner et al. 1990; Melville et al. 1998). Genomic DNA was isolated as described in Melville et al. (1996).
Selection of a chromosome Ia (_chr_Ia) tiling clone set
A plasmid library containing Tb927-_chr_Ia DNA inserts was prepared at the Sanger Institute and end-sequenced during the _chr_I sequencing project (Hall et al. 2003). Using the Genome Assembly Program GAP4 Viewer (Bonfield et al. 1995), we selected 1055 overlapping clones to create a tiling array encompassing the entire length of the sequenced Tb927-_chr_Ia, except the telomeric VSG expression site at the left end (∼40 kb including the 50-bp repeats). Inserts from the selected clones were amplified by PCR in a standard PCR reaction. Products ranged from 1 to 2.5 kb and were purified using MultiScreen-FB plates (Millipore).
Construction of a Tb927-_chr_Ia tiling array on glass slides
Samples containing a final DNA concentration of PCR product in spotting buffer (150 mM sodium phosphate; 0.01% SDS) of ∼200 ng/μL were spotted in triplicate onto GAPS II Coated Slides (Corning). Total DNA from T. brucei 927 (100 ng/μL) was spotted as positive control, and Escherichia coli and λ phage DNA (100 ng/μL), pUC18 (100 ng/μL), pUC18 polylinker (100 ng/μL), spotting buffer, and double-distilled H2O were spotted as negative controls. Slides were baked for 2 h at 80_°_C, then exposed to UV light (450 mJ).
Preparation of fluorescently labeled DNA for hybridization
For each experiment, 300 ng of genomic DNA was labeled overnight with either Cy3-dUTP or Cy5-dUTP (Amersham Bioscience) by random priming using the Bioprime Labeling Kit (Invitrogen) following the manufacturer's instructions, except that dCTP, dGTP, and dATP were added to a final concentration of 200 μM each, dTTP to a final concentration of 50 μM, and Cy3-dUTP or Cy5-dUTP was added to a final concentration of 40 μM. The DNA was purified using micro-spin G50 columns (Amersham Bioscience). In each pairwise experiment, two hybridizations were performed with dye swap. Each pairwise experiment was performed four times.
Hybridization of labeled DNA samples to Tb927-_chr_Ia DNA on the array
Two samples of DNA, labeled with Cy-3 or Cy-5, were combined and applied to each array slide. Hybridization was carried out for 12–16 h at 60°C. The glass slides were washed to a final stringency of 0.01× SSC and dried.
Scanning and analysis
Slides were scanned on both Cy3 and Cy5 channels using a GenePix Personal scanner 4100A (Axon Instruments), and the image was analyzed using GenePix Pro 3.0 software. Spots with an irregular shape or where <95% of pixels had an intensity higher than two SD of the background in both channels were removed. The ratio of one spot was calculated as the mean intensity of all pixels in one channel divided by mean intensity of all pixels in the other channel. Further analysis of data was performed in Excel software (Microsoft). Ratios from reverse-labeled (dye-swap) experiments were reciprocated. Normalization of results from different slides was achieved by dividing the ratio of each spot by the mean of ratios of all spots on each slide. Each pairwise experiment was carried out in quadruplicate and each PCR product was spotted in triplicate so a total of 12 ratios was expected for each experiment. The final ratio for each spot with regard to each sample was calculated as the mean of the 12 ratios. To avoid error due to variation between slides, those spot readings with a coefficient of variation (CV, CV = SD/mean of ratios) >0.2 were removed. For the remaining spots, log2 of final ratios were calculated, and in Figure 1 these are displayed according to the position of the DNA sequence in Tb927-_chr_Ia.
Microarray validation
Restriction enzyme sites flanking tandem arrays, but not within the array units, were identified for digestion of genomic DNA: EcoRI for the histone H3, FBP arrays and Tb.927.1.1740 intra-ORF repeat, XmnI for the tubulin array, and HindIII for the pteridine transporter array. Fragments were separated by pulsed field electrophoresis together with markers of known size, then Southern blotted and probed with a clone of the relevant repeat unit. Southern blot and radioactive labeling of probes with 32P were carried out using standard methods (Sambrook and Russell 2001).
T. brucei 427 haploid chromosome library construction and screening
Tb427-_chr_Ib, Tb427-_chr_VIa, and Tb427-_chr_IXb were excised as single haploid chromosomes from pulsed field gels (separations as in Melville et al. 2000), and the chromosomal DNAs were each amplified directly by DOP-PCR (Telenius et al. 1992) in four independent reactions using four different random primers: 5′CC GACTCGAGNNNNNNATGTGG3′, 5′ACATCAGTGGGAC NNNNNNGCA3′, 5′AGTGACTGGTNNNNNNGGTAGT3′, 5′AAGTCGCGGCCGCNNNNNNATG3′. The reaction mixture contained 1× PCR buffer, 2 mM MgCl2, 0.2 mM dNTPs, 0.2 μM of primer, 2.5 units of AmpliTaq DNA Polymerase (Applied Biosystems), DNA template as 1 μL of agarose from the pulsed field gel, and dH2O to 50 μL. The reaction conditions were: initial denaturation for 5 min at 95°C, 5 cycles of denaturation for 1 min at 94°C, annealing for 1.5 min at 30°C, extension for 3 min at 72°C, 34 cycles of denaturation (1 min at 94°C), annealing (1 min at 56°C), and extension (3 min +1 extra second per cycle at 72°C), and final extension (10 min at 72°C). PCR products, mostly in the range 0.2–3 kb, were analyzed by standard electrophoresis in 1% agarose gels. PCR products from the four independent reactions were mixed and cloned into E. coli using the TOPO TA cloning kit (Invitrogen). White colonies were spotted onto Hybond-N+ membranes (Amersham Bioscience) using a PBA Flexys gridder to create a macroarray.
Library screening
Samples of Tb927-_chr_Ia, Tb427-_chr_Ib, Tb427-_chr_VIa, mcl4-_chr_VIa, Tb927-_chr_IXa, and Tb427-_chr_IXb were excised from PFGs (separations as in Melville et al. 1998, 2000) and amplified as above, then hybridized to two separate replicate arrays of the corresponding chromosome-enriched library; samples of total genomic DNA (Tb427 and Tb927) amplified by DOP-PCR were hybridized to two separate replicate arrays of the Tb427-_chr_Ib library. DNA samples were labeled with 32P-dCTP using the Ready To Go DNA Labeling Beads kit (Amersham Bioscience). Hybridization was carried out at 65°C using the Church and Gilbert protocol (Church and Gilbert 1984).
Sequencing
Inserts of selected clones were amplified by PCR and visualized on agarose gels in order to reduce redundancy by selecting clones of different sizes for sequencing. In total, 382 non-redundant clones were sequenced using an ABI Prism 3100 Genetic Analyzer. The DNA sequences obtained were compared using BLASTN and TBLASTX to known T. brucei 927 DNA sequences in GeneDB (www.genedb.org/genedb/tryp/) and all sequences in the EMBL database (www.ebi.ac.uk/embl/). Highest scoring BLASTN results were used to produce Supplemental Tables S2, S3, and S4. In most cases where the highest scoring pair (HSP) is a known VSG sequence, in fact multiple _VSG_s were detected at a significant level of similarity in the databases. Searches of EMBL data did not alter the HSP list derived from Tb927 databases.
Chromosomal location of probes
DNA from 12 sequenced clones was labeled using the AlkPhos Direct Labeling and Detection System (Amersham Bioscience) and hybridized to dot-blots (Fig. 4A) and to Southern blots (Fig. 4B). Low levels of hybridization to Tb927 in this experiment are due to the lower stringency of the hybridization technique (possibly also to probe concentration), again indicating there are similar VSG families in Tb927. Hybridization to Tb927 chromosomes reconfirmed that they did not hybridize to Tb927-_chr_Ia, and weakly or not at all to other chromosome bands (data not shown).
Acknowledgments
This research has been supported by a Marie Curie Fellowship of the European Community program “Improving Human Potential” under contract number MCFI-2002-00800 awarded to S. Callejas and by the UK Biotechnology and Biological Sciences Research Council (grant reference 8/G16891). We thank Paul Edwards and Maria Garcia for their collaboration on genomic microarrays, help with DOP-PCR, and comments; colleagues at the Sanger Institute for the arrayed Tb927-_chr_Ia plasmid library used for the sequencing project; and Matt Berriman for his help with GAP4.
Footnotes
References
- Barry J.D., McCulloch R., McCulloch R. Antigenic variation in trypanosomes: Enhanced phenotypic variation in a eukaryotic parasite. Adv. Parasitol. 2001;49:1–70. doi: 10.1016/s0065-308x(01)49037-3. [DOI] [PubMed] [Google Scholar]
- Barry J.D., Ginger M.L., Burton P., McCulloch R., Ginger M.L., Burton P., McCulloch R., Burton P., McCulloch R., McCulloch R. Why are parasite contingency genes often associated with telomeres? Int. J. Parasitol. 2003;33:29–45. doi: 10.1016/s0020-7519(02)00247-3. [DOI] [PubMed] [Google Scholar]
- Beals T.P., Boothroyd J.C., Boothroyd J.C. Genomic organization and context of a trypanosome variant surface glycoprotein gene family. J. Mol. Biol. 1992;225:961–971. doi: 10.1016/0022-2836(92)90097-4. [DOI] [PubMed] [Google Scholar]
- Berriman M., Ghedin E., Hertz-Fowler C., Blandin G., Renauld H., Bartholomeu D.C., Lennard N.J., Caler E., Hamlin N.E., Haas B., Ghedin E., Hertz-Fowler C., Blandin G., Renauld H., Bartholomeu D.C., Lennard N.J., Caler E., Hamlin N.E., Haas B., Hertz-Fowler C., Blandin G., Renauld H., Bartholomeu D.C., Lennard N.J., Caler E., Hamlin N.E., Haas B., Blandin G., Renauld H., Bartholomeu D.C., Lennard N.J., Caler E., Hamlin N.E., Haas B., Renauld H., Bartholomeu D.C., Lennard N.J., Caler E., Hamlin N.E., Haas B., Bartholomeu D.C., Lennard N.J., Caler E., Hamlin N.E., Haas B., Lennard N.J., Caler E., Hamlin N.E., Haas B., Caler E., Hamlin N.E., Haas B., Hamlin N.E., Haas B., Haas B., et al. The genome of the African trypanosome Trypanosoma brucei. . Science. 2005;309:416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- Biteau N., Bringaud F., Gibson W., Truc P., Baltz T., Bringaud F., Gibson W., Truc P., Baltz T., Gibson W., Truc P., Baltz T., Truc P., Baltz T., Baltz T. Characterization of Trypanozoon isolates using a repeated coding sequence and microsatellite markers. Mol. Biochem. Parasitol. 2000;105:185–201. [PubMed] [Google Scholar]
- Bonfield J.K., Smith K., Staden R., Smith K., Staden R., Staden R. A new DNA sequence assembly program. Nucleic Acids Res. 1995;23:4992–4999. doi: 10.1093/nar/23.24.4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringaud F., Biteau N., Melville S.E., Hez S., El-Sayed N.M., Leech V., Berriman M., Hall N., Donelson J.E., Baltz T., Biteau N., Melville S.E., Hez S., El-Sayed N.M., Leech V., Berriman M., Hall N., Donelson J.E., Baltz T., Melville S.E., Hez S., El-Sayed N.M., Leech V., Berriman M., Hall N., Donelson J.E., Baltz T., Hez S., El-Sayed N.M., Leech V., Berriman M., Hall N., Donelson J.E., Baltz T., El-Sayed N.M., Leech V., Berriman M., Hall N., Donelson J.E., Baltz T., Leech V., Berriman M., Hall N., Donelson J.E., Baltz T., Berriman M., Hall N., Donelson J.E., Baltz T., Hall N., Donelson J.E., Baltz T., Donelson J.E., Baltz T., Baltz T. A new, expressed multigene family containing a hot spot for insertion of retroelements is associated with polymorphic subtelomeric regions of Trypanosoma brucei . Eukaryot. Cell. 2002;1:137–151. doi: 10.1128/EC.1.1.137-151.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burri C., Stich A., Brun R., Stich A., Brun R., Brun R. Current chemotherapy of African trypanosomiasis. In: Maudlin I., et al., editors. The trypanosomiases. CABI Publishing; Wallingford, Oxfordshire UK: 2004. pp. 403–419. [Google Scholar]
- Church G.M., Gilbert W., Gilbert W. Genomic sequencing. Proc. Natl. AcadSci. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig A., Scherf A., Scherf A. Antigenic variation. Academic Press; Amsterdam: 2003. [Google Scholar]
- El-Sayed N.M., Myler P.J., Blandin G., Berriman M., Crabtree J., Aggarwal G., Caler E., Renauld H., Worthey E.A., Hertz-Fowler C., Myler P.J., Blandin G., Berriman M., Crabtree J., Aggarwal G., Caler E., Renauld H., Worthey E.A., Hertz-Fowler C., Blandin G., Berriman M., Crabtree J., Aggarwal G., Caler E., Renauld H., Worthey E.A., Hertz-Fowler C., Berriman M., Crabtree J., Aggarwal G., Caler E., Renauld H., Worthey E.A., Hertz-Fowler C., Crabtree J., Aggarwal G., Caler E., Renauld H., Worthey E.A., Hertz-Fowler C., Aggarwal G., Caler E., Renauld H., Worthey E.A., Hertz-Fowler C., Caler E., Renauld H., Worthey E.A., Hertz-Fowler C., Renauld H., Worthey E.A., Hertz-Fowler C., Worthey E.A., Hertz-Fowler C., Hertz-Fowler C., et al. Comparative genomics of trypanosomatid parasitic protozoa. Science. 2005;309:404–409. doi: 10.1126/science.1112181. [DOI] [PubMed] [Google Scholar]
- Fabre E., Muller H., Therizols P., Lafontaine I., Dujon B., Fairhead C., Muller H., Therizols P., Lafontaine I., Dujon B., Fairhead C., Therizols P., Lafontaine I., Dujon B., Fairhead C., Lafontaine I., Dujon B., Fairhead C., Dujon B., Fairhead C., Fairhead C. Comparative genomics in hemiascomycete yeasts: Evolution of sex, silencing, and subtelomeres. Mol. Biol. Evol. 2005;22:856–873. doi: 10.1093/molbev/msi070. [DOI] [PubMed] [Google Scholar]
- Freitas-Junior L.H., Bottius E., Pirrit L.A., Deitsch K.W., Scheidig C., Guinet F., Nehrbass U., Wellems T.E., Scherf A., Bottius E., Pirrit L.A., Deitsch K.W., Scheidig C., Guinet F., Nehrbass U., Wellems T.E., Scherf A., Pirrit L.A., Deitsch K.W., Scheidig C., Guinet F., Nehrbass U., Wellems T.E., Scherf A., Deitsch K.W., Scheidig C., Guinet F., Nehrbass U., Wellems T.E., Scherf A., Scheidig C., Guinet F., Nehrbass U., Wellems T.E., Scherf A., Guinet F., Nehrbass U., Wellems T.E., Scherf A., Nehrbass U., Wellems T.E., Scherf A., Wellems T.E., Scherf A., Scherf A. Frequent ectopic recombination of virulence factor genes in telomeric chromosome clusters of P. falciparum . Nature. 2000;407:1018–1022. doi: 10.1038/35039531. [DOI] [PubMed] [Google Scholar]
- Ghedin E., Bringaud F., Peterson J., Myler P., Berriman M., Ivens A., Andersson B., Bontempi E., Eisen J., Angiuoli S., Bringaud F., Peterson J., Myler P., Berriman M., Ivens A., Andersson B., Bontempi E., Eisen J., Angiuoli S., Peterson J., Myler P., Berriman M., Ivens A., Andersson B., Bontempi E., Eisen J., Angiuoli S., Myler P., Berriman M., Ivens A., Andersson B., Bontempi E., Eisen J., Angiuoli S., Berriman M., Ivens A., Andersson B., Bontempi E., Eisen J., Angiuoli S., Ivens A., Andersson B., Bontempi E., Eisen J., Angiuoli S., Andersson B., Bontempi E., Eisen J., Angiuoli S., Bontempi E., Eisen J., Angiuoli S., Eisen J., Angiuoli S., Angiuoli S., et al. Gene synteny and evolution of genome architecture in trypanosomatids. Mol. Biochem. Parasitol. 2004;134:183–191. doi: 10.1016/j.molbiopara.2003.11.012. [DOI] [PubMed] [Google Scholar]
- Gibson W., Garside L., Garside L. Genetic exchange in. Variable chromosomal location of housekeeping genes in different trypanosome stocks. Trypanosoma brucei brucei. Mol. Biochem. Parasitol. 1991;45:77–89. doi: 10.1016/0166-6851(91)90029-6. [DOI] [PubMed] [Google Scholar]
- Gottesdiener K., Garcia-Anoveros J., Lee M.G., van der Ploeg L.H., Garcia-Anoveros J., Lee M.G., van der Ploeg L.H., Lee M.G., van der Ploeg L.H., van der Ploeg L.H. Chromosome organization of the protozoan Trypanosoma brucei . Mol. Cell. Biol. 1990;10:6079–6083. doi: 10.1128/mcb.10.11.6079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall N., Berriman M., Lennard N.J., Harris B.R., Hertz-Fowler C., Bart-Delabesse E.N., Gerrard C.S., Atkin R.J., Barron A.J., Bowman S., Berriman M., Lennard N.J., Harris B.R., Hertz-Fowler C., Bart-Delabesse E.N., Gerrard C.S., Atkin R.J., Barron A.J., Bowman S., Lennard N.J., Harris B.R., Hertz-Fowler C., Bart-Delabesse E.N., Gerrard C.S., Atkin R.J., Barron A.J., Bowman S., Harris B.R., Hertz-Fowler C., Bart-Delabesse E.N., Gerrard C.S., Atkin R.J., Barron A.J., Bowman S., Hertz-Fowler C., Bart-Delabesse E.N., Gerrard C.S., Atkin R.J., Barron A.J., Bowman S., Bart-Delabesse E.N., Gerrard C.S., Atkin R.J., Barron A.J., Bowman S., Gerrard C.S., Atkin R.J., Barron A.J., Bowman S., Atkin R.J., Barron A.J., Bowman S., Barron A.J., Bowman S., Bowman S., et al. The DNA sequence of chromosome I of an African trypanosome: Gene content, chromosome organisation, recombination and polymorphism. Nucleic Acids Res. 2003;31:4864–4873. doi: 10.1093/nar/gkg674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall N., Karras M., Raine J.D., Carlton J.M., Kooij T.W., Berriman M., Florens L., Janssen C.S., Pain A., Christophides G.K., Karras M., Raine J.D., Carlton J.M., Kooij T.W., Berriman M., Florens L., Janssen C.S., Pain A., Christophides G.K., Raine J.D., Carlton J.M., Kooij T.W., Berriman M., Florens L., Janssen C.S., Pain A., Christophides G.K., Carlton J.M., Kooij T.W., Berriman M., Florens L., Janssen C.S., Pain A., Christophides G.K., Kooij T.W., Berriman M., Florens L., Janssen C.S., Pain A., Christophides G.K., Berriman M., Florens L., Janssen C.S., Pain A., Christophides G.K., Florens L., Janssen C.S., Pain A., Christophides G.K., Janssen C.S., Pain A., Christophides G.K., Pain A., Christophides G.K., Christophides G.K., et al. A comprehensive survey of the Plasmodium life cycle by genomic, transcriptomic, and proteomic analyses. Science. 2005;307:82–86. doi: 10.1126/science.1103717. [DOI] [PubMed] [Google Scholar]
- Henriksson J., Porcel B., Rydaker M., Ruiz A., Sabaj V., Galanti N., Cazzulo J.J., Frasch A.C., Pettersson U., Porcel B., Rydaker M., Ruiz A., Sabaj V., Galanti N., Cazzulo J.J., Frasch A.C., Pettersson U., Rydaker M., Ruiz A., Sabaj V., Galanti N., Cazzulo J.J., Frasch A.C., Pettersson U., Ruiz A., Sabaj V., Galanti N., Cazzulo J.J., Frasch A.C., Pettersson U., Sabaj V., Galanti N., Cazzulo J.J., Frasch A.C., Pettersson U., Galanti N., Cazzulo J.J., Frasch A.C., Pettersson U., Cazzulo J.J., Frasch A.C., Pettersson U., Frasch A.C., Pettersson U., Pettersson U. Chromosome specific markers reveal conserved linkage groups in spite of extensive chromosomal size variation in Trypanosoma cruzi . Mol. Biochem. Parasitol. 1995;73:63–74. doi: 10.1016/0166-6851(95)00096-j. [DOI] [PubMed] [Google Scholar]
- Hope M., MacLeod A., Leech V., Melville S., Sasse J., Tait A., Turner C.M., MacLeod A., Leech V., Melville S., Sasse J., Tait A., Turner C.M., Leech V., Melville S., Sasse J., Tait A., Turner C.M., Melville S., Sasse J., Tait A., Turner C.M., Sasse J., Tait A., Turner C.M., Tait A., Turner C.M., Turner C.M. Analysis of ploidy (in megabase chromosomes) in Trypanosoma brucei after genetic exchange. Mol. Biochem. Parasitol. 1999;104:1–9. doi: 10.1016/s0166-6851(99)00103-6. [DOI] [PubMed] [Google Scholar]
- Ivens A.C., Peacock C.S., Worthey E.A., Murphy L., Aggarwal G., Berriman M., Sisk E., Rajandream M.A., Adlem E., Aert R., Peacock C.S., Worthey E.A., Murphy L., Aggarwal G., Berriman M., Sisk E., Rajandream M.A., Adlem E., Aert R., Worthey E.A., Murphy L., Aggarwal G., Berriman M., Sisk E., Rajandream M.A., Adlem E., Aert R., Murphy L., Aggarwal G., Berriman M., Sisk E., Rajandream M.A., Adlem E., Aert R., Aggarwal G., Berriman M., Sisk E., Rajandream M.A., Adlem E., Aert R., Berriman M., Sisk E., Rajandream M.A., Adlem E., Aert R., Sisk E., Rajandream M.A., Adlem E., Aert R., Rajandream M.A., Adlem E., Aert R., Adlem E., Aert R., Aert R., et al. The genome of the kinetoplastid parasite. Leishmania major. Science. 2005;309:436–442. doi: 10.1126/science.1112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janse C.J. Chromosome size polymorphism and DNA rearrangements in Plasmodium . Parasitol. Today. 1993;9:19–22. doi: 10.1016/0169-4758(93)90158-c. [DOI] [PubMed] [Google Scholar]
- MacLeod A., Tweedie A., McLellan S., Taylor S., Hall N., Berriman M., El-Sayed N.M., Hope M., Turner C.M., Tait A., Tweedie A., McLellan S., Taylor S., Hall N., Berriman M., El-Sayed N.M., Hope M., Turner C.M., Tait A., McLellan S., Taylor S., Hall N., Berriman M., El-Sayed N.M., Hope M., Turner C.M., Tait A., Taylor S., Hall N., Berriman M., El-Sayed N.M., Hope M., Turner C.M., Tait A., Hall N., Berriman M., El-Sayed N.M., Hope M., Turner C.M., Tait A., Berriman M., El-Sayed N.M., Hope M., Turner C.M., Tait A., El-Sayed N.M., Hope M., Turner C.M., Tait A., Hope M., Turner C.M., Tait A., Turner C.M., Tait A., Tait A. The genetic map and comparative analysis with the physical map of Trypanosoma brucei . Nucleic Acids Res. 2005;33:6688–6693. doi: 10.1093/nar/gki980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mefford H.C., Trask B.J., Trask B.J. The complex structure and dynamic evolution of human subtelomeres. Nat. Rev. Genet. 2002;3:91–102. doi: 10.1038/nrg727. [DOI] [PubMed] [Google Scholar]
- Melville S., Shepherd N.S., Gerrard C., Le Page R.W., Shepherd N.S., Gerrard C., Le Page R.W., Gerrard C., Le Page R.W., Le Page R.W. The selection of chromosome-specific clones from African trypanosome genomic libraries. In: Birren B., Lai E., Lai E., editors. Analysis of non-mammalian genomes. Academic Press; New York: 1996. pp. 257–293. [Google Scholar]
- Melville S.E., Leech V., Gerrard C.S., Tait A., Blackwell J.M., Leech V., Gerrard C.S., Tait A., Blackwell J.M., Gerrard C.S., Tait A., Blackwell J.M., Tait A., Blackwell J.M., Blackwell J.M. The molecular karyotype of the megabase chromosomes of Trypanosoma brucei and the assignment of chromosome markers. Mol. Biochem. Parasitol. 1998;94:155–173. doi: 10.1016/s0166-6851(98)00054-1. [DOI] [PubMed] [Google Scholar]
- Melville S.E., Gerrard C.S., Blackwell J.M., Gerrard C.S., Blackwell J.M., Blackwell J.M. Multiple causes of size variation in the diploid megabase chromosomes of African tyrpanosomes. Chromosome Res. 1999;7:191–203. doi: 10.1023/a:1009247315947. [DOI] [PubMed] [Google Scholar]
- Melville S.E., Leech V., Navarro M., Cross G.A., Leech V., Navarro M., Cross G.A., Navarro M., Cross G.A., Cross G.A. The molecular karyotype of the megabase chromosomes of Trypanosoma brucei stock 427. Mol. Biochem. Parasitol. 2000;111:261–273. doi: 10.1016/s0166-6851(00)00316-9. [DOI] [PubMed] [Google Scholar]
- Pain A., Renauld H., Berriman M., Murphy L., Yeats C.A., Weir W., Kerhornou A., Aslett M., Bishop R., Bouchier C., Renauld H., Berriman M., Murphy L., Yeats C.A., Weir W., Kerhornou A., Aslett M., Bishop R., Bouchier C., Berriman M., Murphy L., Yeats C.A., Weir W., Kerhornou A., Aslett M., Bishop R., Bouchier C., Murphy L., Yeats C.A., Weir W., Kerhornou A., Aslett M., Bishop R., Bouchier C., Yeats C.A., Weir W., Kerhornou A., Aslett M., Bishop R., Bouchier C., Weir W., Kerhornou A., Aslett M., Bishop R., Bouchier C., Kerhornou A., Aslett M., Bishop R., Bouchier C., Aslett M., Bishop R., Bouchier C., Bishop R., Bouchier C., Bouchier C., et al. Genome of the host-cell transforming parasite Theileria annulata compared with T. parva . Science. 2005;309:131–133. doi: 10.1126/science.1110418. [DOI] [PubMed] [Google Scholar]
- Roth C., Bringaud F., Layden R.E., Baltz T., Eisen H., Bringaud F., Layden R.E., Baltz T., Eisen H., Layden R.E., Baltz T., Eisen H., Baltz T., Eisen H., Eisen H. Active late-appearing variable surface antigen genes in Trypanosoma equiperdum are constructed entirely from pseudogenes. Proc. Natl. Acad. Sci. 1989;86:9375–9379. doi: 10.1073/pnas.86.23.9375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Russell D.W., Russell D.W. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. Molecular cloning: A laboratory manual. [Google Scholar]
- Telenius H., Carter N.P., Bebb C.E., Nordenskjold M., Ponder B.A., Tunnacliffe A., Carter N.P., Bebb C.E., Nordenskjold M., Ponder B.A., Tunnacliffe A., Bebb C.E., Nordenskjold M., Ponder B.A., Tunnacliffe A., Nordenskjold M., Ponder B.A., Tunnacliffe A., Ponder B.A., Tunnacliffe A., Tunnacliffe A. Degenerate oligonucleotide-primed PCR: General amplification of target DNA by a single degenerate primer. Genomics. 1992;13:718–725. doi: 10.1016/0888-7543(92)90147-k. [DOI] [PubMed] [Google Scholar]
- Tomas A.M., Kelly J.M., Kelly J.M. Stage-regulated expression of cruzipain, the major cysteine protease of Trypanosoma cruzi, is independent of the level of RNA1. Mol. Biochem. Parasitol. 1996;76:91–103. doi: 10.1016/0166-6851(95)02545-6. [DOI] [PubMed] [Google Scholar]
- Turner C.M., Sternberg J., Buchanan N., Smith E., Hide G., Tait A., Sternberg J., Buchanan N., Smith E., Hide G., Tait A., Buchanan N., Smith E., Hide G., Tait A., Smith E., Hide G., Tait A., Hide G., Tait A., Tait A. Evidence that the mechanism of gene exchange in Trypanosoma brucei involves meiosis and syngamy. Parasitology. 1990;101:377–386. doi: 10.1017/s0031182000060571. [DOI] [PubMed] [Google Scholar]
- Turner C.M., Melville S.E., Tait A., Melville S.E., Tait A., Tait A. A proposal for karyotype nomenclature in Trypanosoma brucei . Parasitol. Today. 1997;13:5–6. doi: 10.1016/s0169-4758(96)20056-0. [DOI] [PubMed] [Google Scholar]
- van der Ploeg L.H., Valerio D., De Lange T., Bernards A., Borst P., Grosveld F.G., Valerio D., De Lange T., Bernards A., Borst P., Grosveld F.G., De Lange T., Bernards A., Borst P., Grosveld F.G., Bernards A., Borst P., Grosveld F.G., Borst P., Grosveld F.G., Grosveld F.G. An analysis of cosmid clones of nuclear DNA from Trypanosoma brucei shows that the genes for variant surface glycoproteins are clustered in the genome. Nucleic Acids Res. 1982;10:5905–5923. doi: 10.1093/nar/10.19.5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Deursen F.J., Shahi S.K., Turner C.M., Hartmann C., Guerra-Giraldez C., Matthews K.R., Clayton C.E., Shahi S.K., Turner C.M., Hartmann C., Guerra-Giraldez C., Matthews K.R., Clayton C.E., Turner C.M., Hartmann C., Guerra-Giraldez C., Matthews K.R., Clayton C.E., Hartmann C., Guerra-Giraldez C., Matthews K.R., Clayton C.E., Guerra-Giraldez C., Matthews K.R., Clayton C.E., Matthews K.R., Clayton C.E., Clayton C.E. Characterisation of the growth and differentiation in vivo and in vitro of bloodstream-form Trypanosoma brucei strain TREU 927. Mol. Biochem. Parasitol. 2001;112:163–171. doi: 10.1016/s0166-6851(00)00359-5. [DOI] [PubMed] [Google Scholar]
- WHO . World Health Organization. Geneva: 2002. The world health report. [Google Scholar]
- Wincker P., Ravel C., Blaineau C., Pages M., Jauffret Y., Dedet J.P., Bastien P., Ravel C., Blaineau C., Pages M., Jauffret Y., Dedet J.P., Bastien P., Blaineau C., Pages M., Jauffret Y., Dedet J.P., Bastien P., Pages M., Jauffret Y., Dedet J.P., Bastien P., Jauffret Y., Dedet J.P., Bastien P., Dedet J.P., Bastien P., Bastien P. The Leishmania genome comprises 36 chromosomes conserved across widely divergent human pathogenic species. Nucleic Acids Res. 1996;24:1688–1694. doi: 10.1093/nar/24.9.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]