CLONING AND CHARACTERIZATION OF OSTEOCLAST PRECURSORS FROM THE RAW264.7 CELL LINE (original) (raw)
. Author manuscript; available in PMC: 2010 Jun 11.
Published in final edited form as: In Vitro Cell Dev Biol Anim. 2006 Jul–Aug;42(7):182–188. doi: 10.1290/0510075.1
SUMMARY
Osteoclasts are bone-resorbing cells that differentiate from macrophage precursors in response to receptor activator of NF-κB (RANKL). In vitro models of osteoclast differentiation are principally based on primary cell culture, which are poorly suited to molecular and transgene studies due to the limitations associated with the use of primary macrophage. RAW264.7 is a transfectable macrophage cell line with the capacity to form osteoclast-like cells. In the present study we have identified osteoclast precursors among clones of RAW264.7 cells. RAW264.7 cell were cloned by limiting dilution and induced to osteoclast differentiation by treatment with recombinant RANKL. Individual RAW264.7 cell clones formed tartrate resistant acid phosphatase (TRAP) positive multinuclear cells to various degrees with RANKL treatment. All clones tested expressed the RANKL receptor RANK. Each of the clones expressed the osteoclast marker genes TRAP and cathepsin-K mRNA with RANKL treatment. However, we noted that only select clones were able to form large, well-spread, TRAP positive multinuclear cells. Clones capable of forming large TRAP positive multinuclear cells also expressed β3 integrin and calcitonin receptor mRNAs and were capable of resorbing a mineralized matrix. All clones tested activated NF-κB with RANKL treatment. cDNA expression profiling of osteoclast precursor RAW264.7 cell clones demonstrates appropriate expression of a large number of genes before and after osteoclastic differentiation. These osteoclast precursor RAW264.7 cell clones provide a valuable model for dissecting the cellular and molecular regulation of osteoclast differentiation and activation.
Keywords: RANKL, bone, resorption, differentiation
INTRODUCTION
Bone resorbing osteoclasts are terminally differentiated polykaryons of the monocyte/macrophage lineage that are specialized cells responsible for physiological bone resorption as well as pathologic bone loss. Osteoclast differentiation is primarily mediated by osteoblast lineage cells and marrow stromal cells through their expression of growth factors and cytokines, including macrophage colony stimulating factor (MCSF) and receptor activator of NFκB ligand (RANKL) (Yasuda et al., 1998). RANKL acts directly on osteoclast precursors, via the receptor RANK, to induce differentiation of precursors to multinuclear bone resorbing cells (Hsu et al., 1999; Nakagawa et al., 1998). RANK is a member of the tumor necrosis factor (TNF) receptor family and shares signaling pathways common to this family of receptors. On stimulation with RANKL, RANK signals via TRAF2 and TRAF6 to activate downstream signaling pathways, including activation of the transcription factors NF-κB and Ap1 (fos/jun) (Armstrong et al., 2002; Lee et al., 2002; Wong et al., 1998). RANKL/RANK interaction also initiates calcium signaling which leads to the activation of nuclear factor of activated T cells (NFATc1), a transcription factor crucial to osteoclast differentiation (Takayanagi et al., 2002). These transcription factors, and others, direct the expression or repression of the unique set of genes associated with osteoclast differentiation and activation. Marker genes up-regulated in early osteoclast differentiation, include tartrate resistant acid phosphatase (TRAP) and cathepsin-K (Cath-K), while the integrin β3 and the calcitonin receptor (CTR) are expressed only by mature osteoclasts (Hattersley and Chambers, 1989; Lee et al., 1995).
Biochemical studies of osteoclast differentiation and function have been hindered because a tractable cell culture model of osteoclast differentiation has not been available. To address questions relating to osteoclast differentiation, earlier in vitro studies differentiated mouse osteoclasts from primary monocyte/macrophage or splenocyte precursors by co-culture with primary osteoblasts or stromal cell lines (Quinn et al., 1997; Quinn et al., 1998b; Takahashi et al., 1988). Treatment of the co-cultures with collagenase, released the osteoblast/stromal cell component, allowing the isolation of osteoclast-like cells (Abu-Amer et al., 1997; Inoue et al., 2000; Shioi et al., 1994). Various techniques have been employed to identify the cell target in these mixed cultures, including treatment of cells prior to co-culture and utilizing mixed co-cultures from genetically modified strains of mice (Abu-Amer et al., 2000). These procedures fail to produce the pure populations of in vitro generated osteoclasts that are required for biochemical and molecular analyses. With the identification of RANKL/RANK signaling as an essential requirement for osteoclast formation, primary osteoclast precursor cells (bone marrow macrophage (BMMs), splenocytes, and peripheral blood monocytes) can now be induced to osteoclastic differentiation in vitro by culture in the presence of recombinant MCSF and RANKL (Quinn et al., 1998a). However, difficulties are associated with the use of these primary cells including limited availability and variation in response patterns between different preparations. A crucial limitation of primary cell-derived osteoclasts and their precursors is that they are virtually untransfectable and thus are poorly suited for genetic manipulation and promoter studies.
The mouse macrophage cell line RAW264.7 has been identified as a transfectable RANK expressing cell line (Hsu et al., 1999; Thompson et al., 1999). RANKL has been shown to activate NF-κB and to induce limited osteoclastic differentiation and bone resorption in RAW264.7 cells (Hsu et al., 1999; Matsuo et al., 2004; Wei et al., 2001). The RAW264.7 cell line is well characterized with regard to macrophage mediated immune, metabolic, and phagocytic functions (Li et al., 1996; Ralph and Nakoinz, 1977; Raschke et al., 1978) and is increasingly used and accepted as a cellular model of osteoclast formation and function (Battaglino et al., 2004; Collin-Osdoby et al., 2003; Liu et al., 2003; Matsumoto et al., 2004; Matsuo et al., 2004; Yu et al., 2004). This is of concern considering RAW264.7 cell derived osteoclasts have not yet been well characterized and RAW264.7 cells are polymorphic with respect to the phenotype of the individual cells comprising this cell line (Ravasi et al., 2002). In the current studies, we observed that not all RAW264.7 cells are capable of generating osteoclasts. This finding led us to clone osteoclast precursors from the RAW264.7 cell line and to characterize the capacity to form osteoclasts. The resulting osteoclast precursor RAW264.7 cells clones provide an excellent in vitro model for dissecting the cellular and molecular regulation of osteoclast differentiation and activation.
METHODS
Cell culture and osteoclastogenesis
RAW264.7 cells (cat. #TIB 71; ATCC, Manassas, VA) were cloned in 48 well plates by limiting dilution (4 viable cells /ml plated at 0.25 ml per well). After 2 weeks, cells from wells containing single colonies were resuspended by vigorous pipetting, expanded by culture in 6 well plates and followed by T-75 flasks, before freezing in DMSO cell freezing medium (Sigma, St. Louis, MO). For controls, primary mouse BMMs were prepared from 5 week old, male C3H/HeJ mice, as described (McHugh et al., 2001). All animal experiments were carried out with protocols approved by the Institutional Animal Care and Use Committee (IACUC) in accordance with all local, State, and Federal regulations. Primary macrophages and RAW264.7 cells were cultured in α-MEM (Mediatech, Herndon, VA) +10% heat inactivated fetal bovine serum (FBS) (Sigma, St. Louis, MO). Cultures of BMM and osteoclasts were supplemented with recombinant mouse MCSF (R&D Systems, Minneapolis, MN) at 20ng/ml. Osteoclast-like cells were derived from primary macrophages and RAW264.7 cells by treating with 20ng/ml recombinant mouse RANKL (R&D Systems, cat. #462-TR), and a cross-linking antibody, anti-6X-histidine at 0.5μg/ml (R&D Systems). Subsequent experiments employing non-6X-his tagged RANKL (R&D systems, cat. #462-TEC) did not require a cross-linking antibody and give equivalent results. After 3 days of culture, 50% of the medium was changed and replenished with α-MEM +10% FBS and 40ng/ml RANKL plus 0.5μg/ml anti-6X-his antibody in relevant cultures. M-CSF was replenished in the BMM cultures. After 5 days of culture, cells were fixed in 10% formalin and stained for TRAP activity using the Leukocyte Acid Phosphatase kit (Sigma).
To assess the resorption activity of osteoclasts, RAW264.7 cell clones were plated at 1X104/cm2 and cultured under differentiating conditions, as described above, on synthetic hydroxyapatite coated slides (Osteologic; BD Biosciences, San Jose, CA). After 5 days slides were washed with water and mineral was visualized with von Kossa stain (Kurihara et al., 1998).
Qualitative reverse transcription polymerase chain reaction (RT-PCR) assays
Total RNA was isolated with Trizol reagent, 0.5ml/well, using the manufacturer's protocol (Invitrogen, Carlsbad, CA). DNA contaminants were removed with RQ1 RNase free DNase (Promega, Madison, WI) followed by an additional Trizol extraction. RT-PCR was with the Advantage One-Step RT-PCR kit (BD Biosciences). For RANKL, TRAP, and Cath-K RT-PCRs, 5ng of template RNA was reverse-transcribed, and for CTR and β3 integrin RT-PCRs, 0.5μg of template RNA was transcribed. Primer sequences were as follows: TRAP, 5′(AGCAGCCAAGGAGGACTACGTT)3′ and 5′(TCGTTGATGTCGCACAGAGG)3′, giving a 220bp product; Cath-K, 5′(TTAATTTGGGAGAAAAACCT)3′ and 5′(AGCCGCCTCCACAGCCATAAT)3′, giving a 400bp product; RANK, 5′(CTCTATGCCCGTGTCCCCTGAAAA)3′ and 5′(GGCCGCGAT GTCCCGTCCTT)3′, giving a 588bp product; β3 integrin, 5′(CCTTTGCCCAGCCTTCCA)3′ and 5′(GTCCCCACAGTTACATTG)3′, giving a 305bp product; and CTR, 5′(GTCATGGTGGCTCTGGTGGTCAAC)3′ and 5′(GTTGACCACCAGAGCCACCATGAC)3′, giving a 270bp product. Reverse transcription was for 1 hour at 50°C. PCR amplification for TRAP and Cath-K was (94°C/30 sec., 56°C/30 sec., 68°C/60 sec.) X 30 cycles. PCR for RANK was (94°C/30 sec., 59°C/30 sec., 68°C/60 sec.) X 35 cycles. CTR PCR was (94°C/30 sec., 61°C/30 sec., 68°C/60 sec.) X 40 cycles. PCR for the β3 integrin was (94°C/30 sec., 57°C/30 sec., 68°C/60 sec.) X 40 cycles. PCR products were resolved by electrophoresis on a 2% agarose gel in Tris-phosphate-EDTA (TPE) buffer (Maniatis et al., 1982), stained with ethidium bromide, and photographed under UV light. PCR products were of the predicted size and sequence specificity was verified by Southern hybridization with internal probes (data not shown). For NFATc1 RT-PCR 1μg of total RNA, purified as described above, was reverse transcribed in a 20μl reaction using the SuperScript First Strand Synthesis kit (Invitrogen). For PCR amplification of cDNA, 1μl of the RT reaction was PCR amplified using Platinum Taq PCR Supermix (Invitrogen) according to manufactures suggested conditions. .
Preparation of nuclear extracts
RAW264.7 cell clones were plated in 6 well dishes at 1.5X10 5/well. After 3 days cells were treated with RANKL at 100ng/ml for 15 min, or untreated as controls. The cells were scraped in cold phosphate buffered saline suspensions collected and centrifuged at 13K rpm in an eppendorf microcentrifuge. Cell pellets were resuspended in 250μl hypotonic lysis buffer (10mM HEPES-KOH, pH 7.9; 10mM KCl; 1.5mM MgCl; 10μl/ml protease inhibitor cocktail (Sigma cat. # P-8340)) and allowed to swell on ice for 15 min. Cells were gently lysed by the addition of 16μl of 10% NP-40 and incubated on ice for 10 min.. After a 30 sec centrifugation at 13K rpm, the supernatant was removed and the nuclei were resuspended in 25μl of nuclear extraction buffer (20mM Hepes-KOH, pH7.9; 420mM NaCl; 1.2mM MgCl; 0.2mM EDTA; 20% glycerol; 10μl/ml protease inhibitor cocktail), and incubated on ice for 20 min. Nuclear debris was cleared by centrifugation at 13K rpm for 4 min at 4°C. Nuclear extract supernatants were snap-frozen and stored at –80°C.
EMSA assays
Double-stranded NF-κB consensus oligonucleotides, 5′(AGTTGAGGGGACTTTCCC AGGC)3′ (Santa Cruz), were end-labeled with γ32PATP (PerkinElmer) and polynucleotide kinase (New England Biolabs, Beverly, MA) to a specific activity of 3.5X105cpm/ng. Binding reactions were prepared with 0.2μl of nuclear extract, 1μg poly dI/dC (Amersham Biosciences, Waukesha, WI) in 10mM Tris**.**HCl (pH 7.5), 100mM NaCl, 1mM EDTA, 10% glycerol, 1mM DTT, and 3X104 cpm of oligonucleotide probe. After a 30 min incubation on ice, complexes were resolved on pre-cast 5% polyacrylamide gels (BioRad, Herculas, CA) in 1X TBE (Maniatis et al., 1982). Gels were dried and autoradiographed for 24 h before development.
cDNA array expression profiling
RAW264.7 clones C3, C6, and C11 and primary BMMs were grown and induced to osteoclastic differentiation for 5 days as described above. Total RNA was purified from cells with Trizol (Invitrogen, Carlsbad, CA) as described above. DNA was removed from RNA preparations by treatment with RQ1 RNase-free DNase (Promega) and RNA was re-purified by Trizol extraction. Total RNA was reverse-transcribed using MMLV-RT and primers specific for the 1200 genes on the mouse 1.2 Atlas cDNA array (Clonetech) using α-33PdCTP (PerkinElmer) and the manufactures specified conditions. Probes were hybridized to membranes over-night at 71°C and washed with a final wash of 0.1X SSC, 0.5% SDS, at 71°C for 30 min. Autoradiographic exposures were 3 days or 10 days. Autoradiographs were scanned and the data were analyzed using the Atlas Image software (Clonetech). Global normalization was used, which normalizes based on the signal strength of all genes expressed. For graphical analysis, results for RAW264.7 cell clones C3, C6, and C11 (with or without RANKL treatment) were averaged while duplicates of primary BMMs (with or without RANKL treatment) results were averaged. Data were plotted on x-y scatterplot, least-squares regression lines drawn, and Pearson correlation coefficient of the least-squares regression lines (R-squared values) calculated using Microsoft Excel 2000 software.
RESULTS
Osteoclast-like cell formation by RAW264.7 cell clones
When RAW264.7 cells were plated at low cell density and treated with RANKL, not all individual colonies gave rise to TRAP positive multinuclear cells (not shown). This result suggested the possible existence of clonal heterogeneity among RAW264.7 cells and prompted us to isolate clonal populations with osteoclastogenic capacity. We therefore cloned RAW264.7 cells by limited dilution cloning. Clones were expanded, and at confluence stocks were frozen in liquid nitrogen. Twelve randomly selected clones were grown out and replated at 5X105 cells per well in six well plates (C1-C12). Cultures were induced to differentiate into osteoclasts by treatment with 20ng/ml RANKL for 5 days. Cultures were then fixed and stained for TRAP activity. Untreated cultures served as controls. We observed clear differences in the capacity of individual clones to form osteoclast-like cells, as shown in figure 1 for representative clones C1, C5, and C6. Clones C1 and C7 formed few TRAP positive cells, while C5, C8, C9, and C12 formed mononuclear TRAP positive cells or TRAP positive multinuclear cells which did not spread. Clones C2, C3, C4, C6, C10 and C11 formed well-spread TRAP positive multinuclear cells. Cultures with clones C3, C6 and C11 resulted in greater than 99% TRAP positive multinuclear cell formation. Clones C3, C6, and C11, that formed TRAP positive multinuclear cells which were well-spread, were found in other studies, to be transfectable (Crotti et al., 2006) and stable in osteoclast-forming potential over several passages (tested up to 10 passages).
Figure 1.
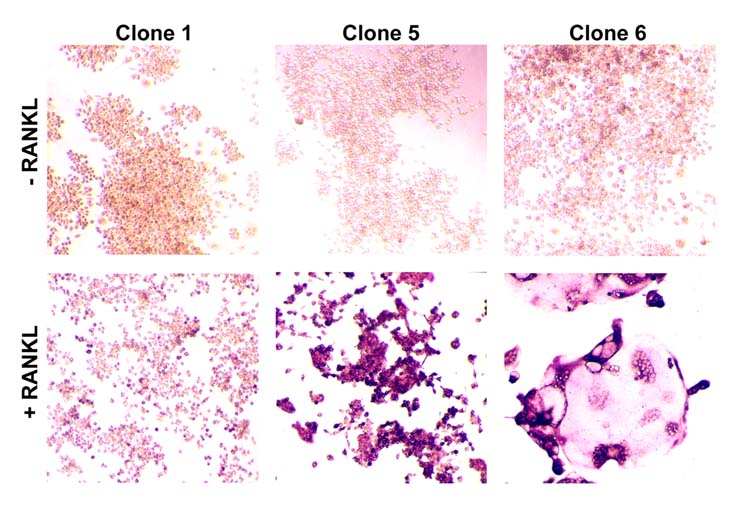
Osteoclast formation in RAW264.7 cell clones. RAW264.7 cell clones were plated in 6 well dishes at 1X105 cells / well and were grown for 5 days with RANKL (20ng/ml) or without (control). Cells were fixed and stained for TRAP activity. Clones displaying a representative range of morphologies are shown (original magnification 40X).
Osteoclast marker gene expression by RAW264.7 cell clones
To determine the pattern of osteoclast gene expression in the RAW264.7 cell clones, we used qualitative RT-PCR to assess mRNA expression of a panel of osteoclast marker genes. RT-PCR targets included: RANK, TRAP, Cath-K, β3 integrin, and CTR. RNA from primary BMMs and osteoclasts derived from BMMs were amplified as controls. Each of the clones assessed, as well as the control BMM cells, qualitatively expressed RANK (figure 2). RAW264.7 clones C1-C12 also expressed TRAP when treated with RANKL, as did the osteoclasts derived from primary macrophages. Cath-K was induced in all clones, with clones C2, C10, and C11 showing some constitutive Cath-K expression. Clones C2, C3, C4, C6, C10, C11, and C12 expressed CTR and the β3 integrin when treated with RANKL. Notably, only the clones able to express β3 integrin on RANKL-induction were capable of CTR expression. Conversely, only CTR inducible clones expressed β3 integrin with RANKL treatment, (clones: C2, C3, C4, C6, C10, C11 and C12). Coordinated expression of CTR and the β3 integrin also corresponded to clones capable of forming well-spread, TRAP positive, multinuclear cells with the exception of clone C12.
Figure 2.
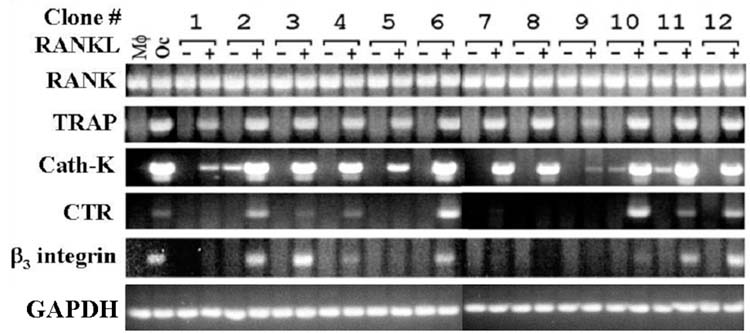
Osteoclast marker gene expression by RAW264.7 clones. Total RNA was purified from each of 12 clones, grown with (+) or without (-) 20ng/ml RANKL, as described in figure 1. Total RNA was used as template for RT-PCR reactions with primers for osteoclast gene markers, TRAP, CTR, Cath-K, β3. Controls included RT-PCR with RNA from primary bone marrow macrophage (Mφ) and osteoclasts derived from them by RANKL treatment (Oc).
Mineralized matrix resorption by osteoclasts from RAW264.7 cell clones
To determine whether the osteoclasts derived from RAW264.7 clones were functional, we tested the clones for the ability to resorb synthetic hydroxyapatite films. Cells were plated on commercially available calcium phosphate matrices and induced to differentiate by treatment with RANKL, as in the previous experiments. The clones expressing the β3 integrin and CTR (figure 2; clones: C2, C3, C4, C6, C10, and C11) were capable of resorption of mineralized matrix, as shown in figure 3. Clone C12, which expressed CTR and the β3 intergrin with RANKL treatment, did not spread and was unable to resorb calcified matrix.
Figure 3.
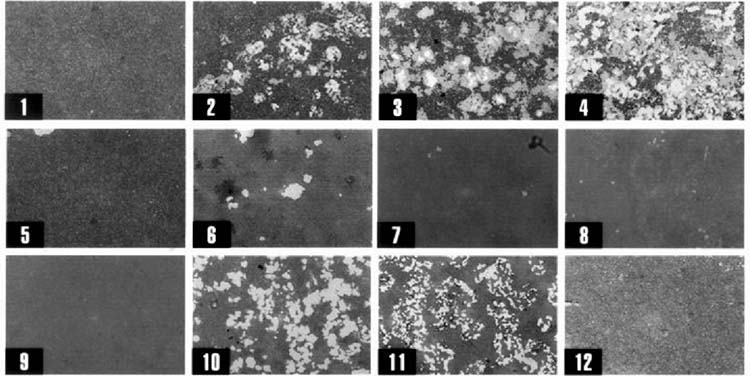
Hydroxyapatite resorption by RAW264.7 cell clones. RAW264.7 cell clones C1-C12 (numbered 1 - 12) grown on Osteologic slides (synthetic calcium phosphate thin films) with 20 ng/ml RANKL or without RANKL (not shown). Cells were removed and mineral was stained with Von Kossa's stain. Clones phenotypically similar to osteoclasts (figure 1 and figure 2) are capable of resorption (original magnification 10X).
RANK signaling in RAW264.7 cell clones
Since all RAW264.7 cell clones expressed RANK, yet not all were capable of osteoclast formation, we tested RANK signaling in the clones. To evaluate RANK signaling we assayed NF-κB activation in response to RANKL by EMSA. Nuclear extracts were prepared from RAW264.7 cell clones treated for 15 min with recombinant RANKL (100ng/ml). This treatment regime has been shown by others to activate NF-κB in primary macrophage cells and RAW264.7 cells (Wei et al., 2001; Zhang et al., 2001). Equal volumes of nuclear extracts from each clone, untreated and RANKL treated, were assayed by EMSA with a labeled NF-κB consensus oligonucleotide probe. This NF-κB consensus probe has been determined by us (data not shown), and others (Wei et al., 2001), to bind NF-κB induced by RANKL in both RAW264.7 cells and primary BMMs. Specificity of binding was demonstrated by competition with unlabeled homologous probe and by the lack of competition with the unlabeled mutated sequences (results not shown). Additionally, identity of the protein moiety was confirmed by reaction with NF-κB p50 antiserum, producing a super-shift of the retarded band (Wei et al., 2001). EMSA analysis indicated that all clones qualitatively respond to RANKL treatment by activation of NF-κB, as shown in figure 4. The clones differed in basal, unstimulated, NF-κB binding activity, however, these differences did not correlate with the ability of each clone to induce expression of osteoclast marker genes, or to form TRAP positive, mineralized matrix resorbing, multinuclear cells.
Figure 4.
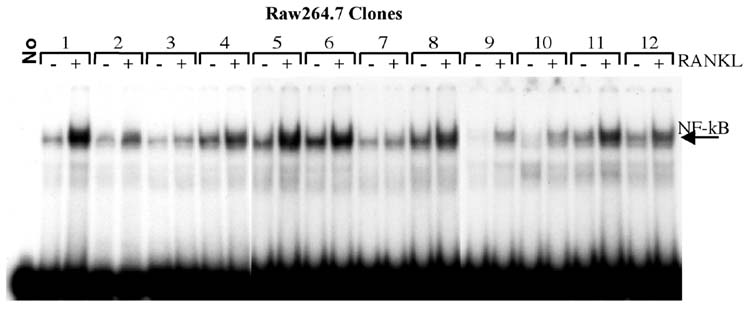
NF-κB activation by RANKL in RAW264.7 cell clones. EMSA analysis of nuclear extracts from clones C1-C12 without (-) or with (+) RANKL treatment (100ng/ml for 15 min). Equal volumes of nuclear extract from treated and control samples were incubated with radiolabeled NF-κB oligonucleotide probe for 30 min on ice. Bound complexes were separated by electorphoresis on 5% polyacrylamide in 1X TBE. The arrow indicates the position of the NF-κB containing complex. All clones are capable of RANKL-dependent NF-κB activation.
cDNA expression profiling
To compare a broader range of phenotypic markers between the RAW264.7 cell clones, the osteoclasts formed from them, and primary macrophage and osteoclasts, we profiled mRNA steady-state expression levels by cDNA array hybridization. Gene expression profiles for untreated RAW264.7 cells were compared to those of primary macrophages and expression profiles for RAW264.7 clone derived osteoclasts were compared to that of primary mouse marrow-derived osteoclasts. Dense cDNA array blots containing over 1200 genes were hybridized with cDNA probes from RNA of RAW264.7 cell clones and mouse primary BMMs. After over-night hybridization, blots were washed, and exposed to Xray film. Autoradiographs were scanned and the relative expression levels of 1187 genes were determined. The relative expression levels from 3 of the osteoclast precursor RAW264.7 clones (C3, C6, and C11) were averaged and expression levels from duplicate primary cell experiments were averaged. Primary macrophage and RAW264.7 cell genes with relative expression levels above 100 were plotted (approximately 710 genes). As shown by x-y scatterplot in figure 5A, we find that untreated osteoclastogenic RAW264.7 cell clones, compared with primary BMMs, express largely the same complement of genes at approximately the same levels over three orders of magnitude. We calculated R2 values (Pearson correlation coefficients) to quantify the similarity of the data around the least-squares regression line. The R2 value for pair-wise comparison of primary BMMs and RAW264.7 cell clones is R2 = 0.7953. This value is similar to those reported for same tissue comparisons and for comparison of the same cell line analyzed at different times using cDNA array profiling (Rhee et al., 1999; Smid-Koopman et al., 2000). The R2 value of 0.7953 is much better than concordance values reported for similar cell lines and dissimilar tissues (R2 values of 0.2 to 0.5) (Smid-Koopman et al., 2000). A single outlier identified in this experiment, the c-myc gene, is highly expressed in RAW264.7 cells but not in the primary BMMs. To compare osteoclastic gene expression in primary osteoclasts versus RAW264.7 cell derived osteoclasts, dense cDNA arrays were hybridized and analyzed as described above. cDNA probes were transcribed from total RNA purified from primary osteoclasts and RAW264.7-derived osteoclasts. The relative expression levels from 3 clones were averaged for RAW264.7-derived osteoclasts (C3, C6, and C11). Primary osteoclast and RAW264.7 osteoclast genes with relative expression levels above 100 were plotted in an x-y scatterplot (approximately 405 genes). We find that RAW264.7 cell derived osteoclasts also express largely the same complement of genes as primary osteoclasts and at similar levels (figure 5B). The correlation coefficient for the RAW264.7 clone osteoclast versus osteoclasts derived from primary BMMs is R2=0.7599. Again this is a good correlation between a cell line and primary cells using cDNA expression profiling. c-myc was induced in primary macrophage cells by RANKL treatment and is expressed at equal levels RAW264.7 cell osteoclasts as in osteoclasts from primary BMMs.
Figure 5.
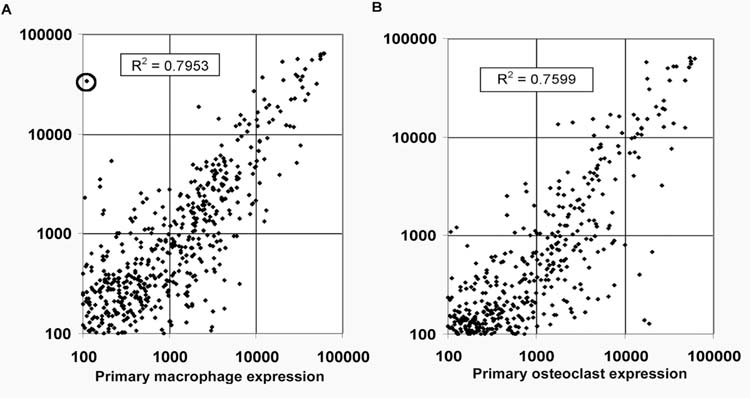
Gene expression in primary cells vs RAW267.4 cell clones. (A) Dense arrays were hybridized with cDNA probes from RNAs of RAW264.7 clones and mouse primary BMMs. Autoradiographs were scanned and relative gene expression levels were determined. The relative expression levels from 3 RAW264.7 clones (C3, C6, & C11) were averaged. The primary macrophage and RAW cell genes with relative expression levels above 100 were plotted (approximately 712 genes). The circled point corresponds to c-myc. (B) cDNA probes from primary osteoclasts and RAW264.7 cell-derived osteoclasts. Arrays were hybridized and analyzed as above. The relative expression levels from 3 clones (C3, C6, & C11) were averaged and the primary osteoclast and RAW264.7 osteoclast genes with relative expression levels above 100 were plotted (approximately 405 genes).
DISCUSSION
Using cell cloning we have demonstrated significant polymorphism within the parental RAW264.7 cell line, especially with regard to osteoclast forming potential. We found that, with RANKL treatment, all of the RAW264.7 cell clones tested were capable of expressing the osteoclast markers TRAP and the acid protease, Cath-K. A fraction of the clones go on to coordinately express both the β3 integrin and CTR mRNA, which are late markers of authentic, committed, osteoclasts (Hattersley and Chambers, 1989; Lee et al., 1995). Importantly, we demonstrate that most of the RAW264.7 cell clones expressing the β3 integrin and CTR were capable of resorption of mineralized matrix. We conclude that many of the parental RAW264.7 cells are blocked in the early stage of osteoclast differentiation. This conclusion explains the generally accepted observation that the parental RAW264.7 cell line exhibits a limited capacity to form osteoclasts.
Previous studies have gone to great lengths to isolate pure populations of osteoclasts from RAW264.7 cells. Methods have included lifting differentiated cells and sucrose gradient centrifugation (Collin-Osdoby et al., 2003). The RAW264.7 cell clones C3, C6, and C11, isolated in this study, may essentially be considered osteoclast precursors since RANKL treatment was able to induce osteoclast gene expression and osteoclast cell morphology, along with the functional ability to resorb mineralized matrix. Our osteoclast precursor RAW264.7 cell clones produced nearly 100% TRAP positive multinuclear matrix resorbing cells when treated with RANKL, making them a valuable tool for the study of osteoclast biochemistry and molecular biology. Our results also indicate that caution is required when interpreting experiments where RAW264.7 cell stable transfectants are cloned and then assayed for osteoclast formation or function. Caution is particularly warranted in the case of transgenes that are purported to inhibit osteoclast formation or function, since non-osteoclast forming cells are present at a high frequency in the parental RAW264.7 cell line and may easily be inadvertently isolated through cloning.
The interaction of RANKL with its receptor RANK is crucial to osteoclast differentiation and activation. Mice with a defect in the expression of RANKL or RANK are incapable of osteoclast differentiation and have an osteopetrotic phenotype (Kong et al., 1999; Li et al., 2000). We therefore assessed whether the RAW264.7 clones that were incapable of osteoclast differentiation lack expression of the RANKL receptor RANK. We found that this was not the case and show that each of the clones tested express RANK mRNA. In addition, all RAW264.7 clones tested must express the RANK protein since they are all capable of responding to RANKL with TRAP and Cath-K mRNA expression. Furthermore, all clones respond to RANKL treatment with NF-κB activation and therefore must express the RANK receptor. These results do not rule out differences in the absolute amount of RANK expressed by individual clones, since our RT-PCR assays are qualitative. To further characterize the osteoclast precursor RAW264.7 cell clones and to determine whether they are a valid cell line for the investigation of osteoclast differentiation and gene expression, we compared the cDNA expression profiles with that of primary BMMs and osteoclasts derived from them. Using expression profiling of osteoclast precursor RAW264.7 cell clones we demonstrate that a broad range of genes are appropriately expressed in RAW264.7 cell macrophage and osteoclasts as compared with primary BMMs and osteoclasts. These results validate the osteoclast precursor RAW264.7 cell clones as an appropriate cell model for studying osteoclast transcription. A single gene of the 1200 assayed, the c-myc gene, was found to have inappropriately high expression in RAW264.7 cell macrophages. We found however, that c-myc, which has been implicated in osteoclast differentiation and gene expression (Battaglino et al., 2002), is induced in primary macrophages by RANKL to levels similar to in unstimulated RAW264.7 cells. .
In conclusion, RAW264.7 is an immortal and transfectable cell line and the clones that exhibit phenotypic and genetic patterns of RANKL-induced osteoclast formation retain these properties. These clones are osteoclast precursors and provide a unique model for dissecting the cellular and molecular regulation of osteoclast differentiation and activation.
ACKNOWLEDGEMENTS
The authors would like to thank Deborah L Galson and Catherine Laplace for sharing PCR primer sequences and conditions for RT-PCR of osteoclast marker genes.
This work was aided by a grant from the Orthopaedic Research and Education Foundation (# 00-020) (K.P.M.) and by National Institute of Health (NIH), National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS: R01 AR47229) (K.P.M.), and a National Health and Medical Research Council (Aust.) C.J. Martin Fellowship (T.N.C.) (I.D. 200078)
REFERENCES
- Abu-Amer Y, Ross FP, Edwards J, Teitelbaum SL. Lipopolysaccharide-stimulated osteoclastogenesis is mediated by tumor necrosis factor via its P55 receptor. J Clin Invest. 1997;100:1557–1565. doi: 10.1172/JCI119679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Amer Y, Erdmann J, Alexopoulou L, Kollias G, Ross FP, Teitelbaum SL. Tumor necrosis factor receptors types 1 and 2 differentially regulate osteoclastogenesis. J Biol Chem. 2000;275:27307–27310. doi: 10.1074/jbc.M003886200. [DOI] [PubMed] [Google Scholar]
- Armstrong AP, Tometsko ME, Glaccum M, Sutherland CL, Cosman D, Dougall WC. A RANK/TRAF6-dependent signal transduction pathway is essential for osteoclast cytoskeletal organization and resorptive function. J Biol Chem. 2002;277:44347–44356. doi: 10.1074/jbc.M202009200. [DOI] [PubMed] [Google Scholar]
- Battaglino R, Kim D, Fu J, Vaage B, Fu XY, Stashenko P. c-myc is required for osteoclast differentiation. J Bone Miner Res. 2002;17:763–773. doi: 10.1359/jbmr.2002.17.5.763. [DOI] [PubMed] [Google Scholar]
- Battaglino R, Fu J, Spate U, Ersoy U, Joe M, Sedaghat L, Stashenko P. Serotonin regulates osteoclast differentiation through its transporter. J Bone Miner Res. 2004;19:1420–1431. doi: 10.1359/JBMR.040606. [DOI] [PubMed] [Google Scholar]
- Collin-Osdoby P, Yu X, Zheng H, Osdoby P. RANKL-mediated osteoclast formation from murine RAW 264.7 cells. Methods Mol Med. 2003;80:153–166. doi: 10.1385/1-59259-366-6:153. [DOI] [PubMed] [Google Scholar]
- Crotti TN, Flannery M, Walsh NC, Fleming JD, Goldring SR, McHugh KP. NFATc1 regulation of the human beta(3) integrin promoter in osteoclast differentiation. Gene. 2006 doi: 10.1016/j.gene.2005.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattersley G, Chambers TJ. Generation of osteoclastic function in mouse bone marrow cultures: multinuclearity and tartrate-resistant acid phosphatase are unreliable markers for osteoclastic differentiation. Endocrinology. 1989;124:1689–1696. doi: 10.1210/endo-124-4-1689. [DOI] [PubMed] [Google Scholar]
- Hirotani H, Tuohy NA, Woo JT, Stern PH, Clipstone NA. The calcineurin/nuclear factor of activated T cells signaling pathway regulates osteoclastogenesis in RAW264.7 cells. J Biol Chem. 2004;279:13984–13992. doi: 10.1074/jbc.M213067200. [DOI] [PubMed] [Google Scholar]
- Hsu H, Lacey DL, Dunstan CR, Solovyev I, Colombero A, Timms E, Tan HL, Elliott G, Kelley MJ, Sarosi I, Wang L, Xia XZ, Elliott R, Chiu L, Black T, Scully S, Capparelli C, Morony S, Shimamoto G, Bass MB, Boyle WJ. Tumor necrosis factor receptor family member RANK mediates osteoclast differentiation and activation induced by osteoprotegerin ligand. Proc Natl Acad Sci U S A. 1999;96:3540–3545. doi: 10.1073/pnas.96.7.3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue M, Ross FP, Erdmann JM, Abu-Amer Y, Wei S, Teitelbaum SL. Tumor necrosis factor alpha regulates alpha(v)beta5 integrin expression by osteoclast precursors in vitro and in vivo. Endocrinology. 2000;141:284–290. doi: 10.1210/endo.141.1.7285. [DOI] [PubMed] [Google Scholar]
- Kong YY, Yoshida H, Sarosi I, Tan HL, Timms E, Capparelli C, Morony S, Oliveira-dos-Santos AJ, Van G, Itie A, Khoo W, Wakeham A, Dunstan CR, Lacey DL, Mak TW, Boyle WJ, Penninger JM. OPGL is a key regulator of osteoclastogenesis, lymphocyte development and lymph-node organogenesis. Nature. 1999;397:315–323. doi: 10.1038/16852. [DOI] [PubMed] [Google Scholar]
- Kurihara N, Tatsumi J, Arai F, Iwama A, Suda T. Macrophage-stimulating protein (MSP) and its receptor, RON, stimulate human osteoclast activity but not proliferation: effect of MSP distinct from that of hepatocyte growth factor. Exp Hematol. 1998;26:1080–1085. [PubMed] [Google Scholar]
- Lee SK, Goldring SR, Lorenzo J. Expression of the calcitonin receptor in bone marrow cell cultures and in bone: a specific marker of the differentiated osteoclast that is regulated by calcitonin. Endocrinology. 1995;136:4572–4581. doi: 10.1210/endo.136.10.7664679. [DOI] [PubMed] [Google Scholar]
- Lee SW, Han SI, Kim HH, Lee ZH. TAK1-dependent activation of AP-1 and c-Jun N-terminal kinase by receptor activator of NF-kappaB. J Biochem Mol Biol. 2002;35:371–376. doi: 10.5483/bmbrep.2002.35.4.371. [DOI] [PubMed] [Google Scholar]
- Li J, Sarosi I, Yan XQ, Morony S, Capparelli C, Tan HL, McCabe S, Elliott R, Scully S, Van G, Kaufman S, Juan SC, Sun Y, Tarpley J, Martin L, Christensen K, McCabe J, Kostenuik P, Hsu H, Fletcher F, Dunstan CR, Lacey DL, Boyle WJ. RANK is the intrinsic hematopoietic cell surface receptor that controls osteoclastogenesis and regulation of bone mass and calcium metabolism. Proc Natl Acad Sci U S A. 2000;97:1566–1571. doi: 10.1073/pnas.97.4.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YM, Mitsuhashi T, Wojciechowicz D, Shimizu N, Li J, Stitt A, He C, Banerjee D, Vlassara H. Molecular identity and cellular distribution of advanced glycation endproduct receptors: relationship of p60 to OST-48 and p90 to 80K-H membrane proteins. Proc Natl Acad Sci U S A. 1996;93:11047–11052. doi: 10.1073/pnas.93.20.11047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Shi Z, Silveira A, Liu J, Sawadogo M, Yang H, Feng X. Involvement of upstream stimulatory factors 1 and 2 in RANKL-induced transcription of tartrate-resistant acid phosphatase gene during osteoclast differentiation. J Biol Chem. 2003;278:20603–20611. doi: 10.1074/jbc.M212093200. [DOI] [PubMed] [Google Scholar]
- Maniatis T, Fritsch EF, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory; Cold Spring Harbor, N.Y.: 1982. [Google Scholar]
- Matsumoto M, Kogawa M, Wada S, Takayanagi H, Tsujimoto M, Katayama S, Hisatake K, Nogi Y. Essential role of p38 mitogen-activated protein kinase in cathepsin K gene expression during osteoclastogenesis through association of NFATc1 and PU.1. J Biol Chem. 2004;279:45969–45979. doi: 10.1074/jbc.M408795200. [DOI] [PubMed] [Google Scholar]
- Matsuo K, Galson DL, Zhao C, Peng L, Laplace C, Wang KZ, Bachler MA, Amano H, Aburatani H, Ishikawa H, Wagner EF. Nuclear factor of activated T-cells (NFAT) rescues osteoclastogenesis in precursors lacking c-Fos. J Biol Chem. 2004;279:26475–26480. doi: 10.1074/jbc.M313973200. [DOI] [PubMed] [Google Scholar]
- McHugh KP, Kitazawa S, Teitelbaum SL, Ross FP. Cloning and characterization of the murine beta(3) integrin gene promoter: identification of an interleukin-4 responsive element and regulation by STAT-6. J Cell Biochem. 2001;81:320–332. doi: 10.1002/1097-4644(20010501)81:2<320::aid-jcb1047>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Nakagawa N, Kinosaki M, Yamaguchi K, Shima N, Yasuda H, Yano K, Morinaga T, Higashio K. RANK is the essential signaling receptor for osteoclast differentiation factor in osteoclastogenesis. Biochem Biophys Res Commun. 1998;253:395–400. doi: 10.1006/bbrc.1998.9788. [DOI] [PubMed] [Google Scholar]
- Quinn JM, Fujikawa Y, McGee JO, Athanasou NA. Rodent osteoblast-like cells support osteoclastic differentiation of human cord blood monocytes in the presence of M-CSF and 1,25 dihydroxyvitamin D3. Int J Biochem Cell Biol. 1997;29:173–179. doi: 10.1016/s1357-2725(96)00129-x. [DOI] [PubMed] [Google Scholar]
- Quinn JM, Elliott J, Gillespie MT, Martin TJ. A combination of osteoclast differentiation factor and macrophage-colony stimulating factor is sufficient for both human and mouse osteoclast formation in vitro. Endocrinology. 1998a;139:4424–4427. doi: 10.1210/endo.139.10.6331. [DOI] [PubMed] [Google Scholar]
- Quinn JM, Neale S, Fujikawa Y, McGee JO, Athanasou NA. Human osteoclast formation from blood monocytes, peritoneal macrophages, and bone marrow cells. Calcif Tissue Int. 1998b;62:527–531. doi: 10.1007/s002239900473. [DOI] [PubMed] [Google Scholar]
- Ralph P, Nakoinz I. Antibody-dependent killing of erythrocyte and tumor targets by macrophage-related cell lines: enhancement by PPD and LPS. J Immunol. 1977;119:950–954. [PubMed] [Google Scholar]
- Raschke WC, Baird S, Ralph P, Nakoinz I. Functional macrophage cell lines transformed by Abelson leukemia virus. Cell. 1978;15:261–267. doi: 10.1016/0092-8674(78)90101-0. [DOI] [PubMed] [Google Scholar]
- Ravasi T, Wells C, Forest A, Underhill DM, Wainwright BJ, Aderem A, Grimmond S, Hume DA. Generation of diversity in the innate immune system: macrophage heterogeneity arises from gene-autonomous transcriptional probability of individual inducible genes. J Immunol. 2002;168:44–50. doi: 10.4049/jimmunol.168.1.44. [DOI] [PubMed] [Google Scholar]
- Rhee CH, Hess K, Jabbur J, Ruiz M, Yang Y, Chen S, Chenchik A, Fuller GN, Zhang W. cDNA expression array reveals heterogeneous gene expression profiles in three glioblastoma cell lines. Oncogene. 1999;18:2711–2717. doi: 10.1038/sj.onc.1202623. [DOI] [PubMed] [Google Scholar]
- Shioi A, Ross FP, Teitelbaum SL. Enrichment of generated murine osteoclasts. Calcif Tissue Int. 1994;55:387–394. doi: 10.1007/BF00299320. [DOI] [PubMed] [Google Scholar]
- Smid-Koopman E, Blok LJ, Chadha-Ajwani S, Helmerhorst TJ, Brinkmann AO, Huikeshoven FJ. Gene expression profiles of human endometrial cancer samples using a cDNA-expression array technique: assessment of an analysis method. Br J Cancer. 2000;83:246–251. doi: 10.1054/bjoc.2000.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N, Akatsu T, Udagawa N, Sasaki T, Yamaguchi A, Moseley JM, Martin TJ, Suda T. Osteoblastic cells are involved in osteoclast formation. Endocrinology. 1988;123:2600–2602. doi: 10.1210/endo-123-5-2600. [DOI] [PubMed] [Google Scholar]
- Takayanagi H, Kim S, Koga T, Nishina H, Isshiki M, Yoshida H, Saiura A, Isobe M, Yokochi T, Inoue J, Wagner EF, Mak TW, Kodama T, Taniguchi T. Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev Cell. 2002;3:889–901. doi: 10.1016/s1534-5807(02)00369-6. [DOI] [PubMed] [Google Scholar]
- Thompson CD, Frazier-Jessen MR, Rawat R, Nordan RP, Brown RT. Evaluation of methods for transient transfection of a murine macrophage cell line, RAW 264.7. Biotechniques. 1999;27 doi: 10.2144/99274rr05. [DOI] [PubMed] [Google Scholar]
- Wei S, Teitelbaum SL, Wang MW-H, Ross FP. Receptor activator of nuclear factor-κB ligand activates nuclear factor-κB in osteoclast precursors. 2001 doi: 10.1210/endo.142.3.8031. [DOI] [PubMed] [Google Scholar]
- Wong BR, Josien R, Lee SY, Vologodskaia M, Steinman RM, Choi Y. The TRAF family of signal transducers mediates NF-kappaB activation by the TRANCE receptor. J Biol Chem. 1998;273:28355–28359. doi: 10.1074/jbc.273.43.28355. [DOI] [PubMed] [Google Scholar]
- Yasuda H, Shima N, Nakagawa N, Mochizuki SI, Yano K, Fujise N, Sato Y, Goto M, Yamaguchi K, Kuriyama M, Kanno T, Murakami A, Tsuda E, Morinaga T, Higashio K. Identity of osteoclastogenesis inhibitory factor (OCIF) and osteoprotegerin (OPG): a mechanism by which OPG/OCIF inhibits osteoclastogenesis in vitro. Endocrinology. 1998;139:1329–1337. doi: 10.1210/endo.139.3.5837. [DOI] [PubMed] [Google Scholar]
- Yu X, Huang Y, Collin-Osdoby P, Osdoby P. CCR1 chemokines promote the chemotactic recruitment, RANKL development, and motility of osteoclasts and are induced by inflammatory cytokines in osteoblasts. J Bone Miner Res. 2004;19:2065–2077. doi: 10.1359/JBMR.040910. [DOI] [PubMed] [Google Scholar]
- Zhang YH, Heulsmann A, Tondravi MM, Mukherjee A, Abu-Amer Y. Tumor necrosis factor-alpha (TNF) stimulates RANKL-induced osteoclastogenesis via coupling of TNF type 1 receptor and RANK signaling pathways. J Biol Chem. 2001;276:563–568. doi: 10.1074/jbc.M008198200. [DOI] [PubMed] [Google Scholar]