Cracking the egg: molecular dynamics and evolutionary aspects of the transition from the fully grown oocyte to embryo (original) (raw)
Abstract
Fully grown oocytes (FGOs) contain all the necessary transcripts to activate molecular pathways underlying the oocyte-to-embryo transition (OET). To elucidate this critical period of development, an extensive survey of the FGO transcriptome was performed by analyzing 19,000 expressed sequence tags of the Mus musculus FGO cDNA library. Expression of 5400 genes and transposable elements is reported. For a majority of genes expressed in mouse FGOs, homologs transcribed in eggs of Xenopus laevis or Ciona intestinalis were found, pinpointing evolutionary conservation of most regulatory cascades underlying the OET in chordates. A large proportion of identified genes belongs to several gene families with oocyte-restricted expression, a likely result of lineage-specific genomic duplications. Gene loss by mutation and expression in female germline of retrotransposed genes specific to M. musculus is documented. These findings indicate rapid diversification of genes involved in female reproduction. Comparison of the FGO and two-cell embryo transcriptomes demarcated the processes important for oogenesis from those involved in OET and identified novel motifs in maternal mRNAs associated with transcript stability. Discovery of oocyte-specific eukaryotic translation initiation factor 4E distinguishes a novel system of translational regulation. These results implicate conserved pathways underlying transition from oogenesis to initiation of development and illustrate how genes acquire and lose reproductive functions during evolution, a potential mechanism for reproductive isolation.
Keywords: Eukaryotic initiation factor-4E, gene expression profiling, maternal effect gene, mRNA stability, multigene family, reproductive isolation, retroelements
Ontogenesis of sexually reproducing heterogametic organisms is widely diverse in its manifestations; however, fusion of gametes, a larger egg and smaller sperm, is a common aspect that initiates development of each new organism. The events of this earliest phase of metazoan development, the “oocyte-to-embryo transition” (OET), are uncoupled from new transcription and rely exclusively on maternal RNAs and proteins deposited during oocyte growth (Seydoux 1996). During this quiescent period, fundamental changes in nuclear function take place as the differentiated cells, egg and sperm, unite to give rise to an embryonic genome. These changes, known as “genome reprogramming,” are defined as a series of epigenetic modifications that transform the genome to the state of totipotency (Dean et al. 2003), a critical event for the successful development of an embryo.
The mechanisms and molecular pathways underlying genome reprogramming and the start of a new life remain poorly understood, largely because the functional analyses of single genes and pathways dominate in this field of developmental biology. However, alternative approaches, such as large-scale transcriptome analysis, emerge as a useful “reverse genetics” tool for extraction of essential information. In addition, these studies generate testable hypotheses about potential players that mediate developmental processes during this period (Sharov et al. 2003; Evsikov et al. 2004; Hamatani et al. 2004; Mehlmann et al. 2004; Wang et al. 2004). Another general consensus among developmental biologists is that the molecular mechanisms contributing fundamentally to oogenesis and OET in one model organism should be conserved in other species (Gilbert 2000). To test this hypothesis directly, the degree of transcriptome concordance among oocytes of different organisms would discriminate between general and species-specific molecular pathways.
The transcriptionally active, growing oocyte accumulates mRNAs essential for the OET in a stable dormant form. Transcriptional quiescence of the genome during OET necessitates post-transcriptional and post-translational mechanisms to orchestrate the multitude of processes participating in meiotic maturation, fertilization, and reprogramming of the nascent embryonic genome (Seydoux 1996; Solter et al. 2002). Post-transcriptional regulation of mRNA, consisting of transcript stability, localization, and translation, is typically mediated through short sequence motifs embedded in the 3′ untranslated regions (3′-UTRs). Large-scale analysis of mRNA abundance allows identification of cohorts of coregulated maternal transcripts during the OET. Computational assessment of 3′-UTRs within and between these groups of coregulated mRNAs for common sequence features permits identification of motifs regulating transcript stability, a necessary step in modeling and understanding of the post-transcriptional regulatory network that operates in oocytes and cleavage-stage embryos.
To address these fundamental questions efficiently, a comprehensive study of the transcripts present in the fully grown oocytes (FGOs) is essential. Here, we report the first extensive analysis of gene expression in mammalian FGOs, accomplished by sequencing of 19,000 expressed sequence tags (ESTs) from the Mus musculus FGO cDNA library. This heuristic analysis provides a unique perspective on oocyte transcriptional activity, as well as on the repertoire of gene products pivotal for oogenesis and the OET in mammals. Comprehensive analysis of gene functions, comparison with the two-cell stage mouse embryo transcriptome (Evsikov et al. 2004), and study of gene expression conserved among Ciona intestinalis eggs, Xenopus laevis oocytes, and M. musculus FGOs uncover the unique features of this cell and the major mechanisms of the OET. An extensive survey of the 3′-UTRs of the transcripts present in the oocytes reveals systematic differences between stable and unstable maternal mRNAs and exposes new clues about the properties of the translational control machinery. Taken together, these analyses bring us closer to understanding the molecular changes that drive the final stages of oogenesis, nuclear reprogramming, activation of embryonic genome, and the initiation of mammalian preimplantation development.
Results
Properties of the FGO cDNA library
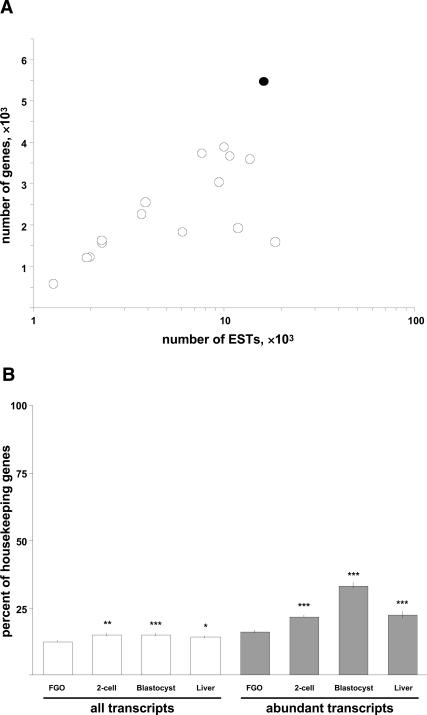
Quality of the cDNA library. Analysis of 18,790 informative 5′ ESTs from the mouse FGO library revealed expression from ∼5400 loci, 87% of which are annotated M. musculus genes (Supplemental Material; Supplementary file FGO_library_1_0.xls). The high ratio of the number of loci to the number of ESTs and comparison with other M. musculus EST libraries of oocytes and preimplantation embryos validate the excellent quality of the FGO library (Fig. 1A). As this cDNA library was not normalized, it was possible that the transcripts identified during the analysis disproportionately represent housekeeping genes. Comparison with cDNA libraries of two-cell stage embryos, blastocysts, and liver revealed that the fraction of housekeeping genes in the FGO library is strikingly low (12.6% vs. 14.2%–15.2%, ZFGO-2cell = 3.14, p < 0.01, ZFGO-blastocyst = 3.37, p < 0.001, ZFGO-liver = 2.22, p < 0.05) (Fig. 1B). Interestingly, even the proportion of housekeeping transcripts among the highest-expressed genes in the FGOs is significantly lower as well (16.1% vs. 21.9%–33.3%, ZFGO-2cell = 3.21, p < 0.001, ZFGO-blastocyst = 9.15, p < 10−6, ZFGO-liver = 3.88, p < 0.001) (Fig. 1B). These data emphasize a counterintuitive finding that housekeeping gene transcripts are actually underrepresented in the M. musculus FGO transcriptome.
FIGURE 1.
Complexity of the FGO cDNA library. (A) Numbers of genes represented by ESTs of the FGO library (black circle) and 15 cDNA libraries of oocytes and preimplantation embryos (white circles). (_X_-axis) Number of ESTs corresponding to genes in the libraries; (_Y_-axis) number of genes these ESTs represent. (B) Fraction of housekeeping genes among all (left) and abundant (right) transcripts identified in the FGOs, two-cell stage embryos, blastocysts, and liver. (*) p < 0.05; (**) p < 0.01; (***) p <0.001.
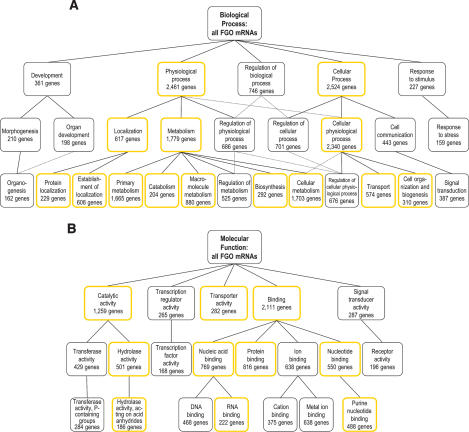
Global outlook on gene functions. In order to identify the major functional classes of the FGO-expressed genes, the VisuaL Annotation Display tool for extraction and analysis of Gene Ontology (GO) annotations for large sets of genes was used (Gene Ontology Consortium 2001; Blake et al. 2006). Considerable numbers of FGO-expressed genes fall under “development,” “regulation of biological process,” and “cell communication” categories and subgroups (Fig. 2A). With respect to molecular function, it is particularly notable that many genes lie within “transcription regulator activity,” “signal transducer activity,” “transferase activity,” and “ion-binding” groups and subgroups (Fig. 2B). Indeed, these categories associated with the FGO transcriptome are distinct from the ones typical for housekeeping genes (Supplementary Fig. S1). To resolve the functional bias of genes highly expressed in FGOs, their GO annotations were compared with the entire set of FGO genes (Supplementary Fig. S2). This analysis revealed that the highest-expressed genes in the FGO are disproportionately involved in “cell cycle” and “cell division,” and have functions of “ligase activity,” “ubiquitin-like activating enzyme activity,” and “translation regulator activity.” Similar analysis of the two-cell stage embryos and blastocysts revealed that as embryogenesis advances, the highest-expressed genes bias toward GO categories associated with housekeeping genes (Supplementary Fig. S2).
FIGURE 2.
Structure of the FGO transcriptome. Biological process (A) and molecular function (B) GO annotations common to at least 5% of FGO genes. The top three levels of GO hierarchy and the numbers of FGO genes annotated with a corresponding GO term are shown. GO terms associated with housekeeping genes are in boxes with yellow borders. Dotted connector lines indicate that a GO term is a subset of two distinct higher-level terms.
Rapid diversification of the oocyte transcriptome
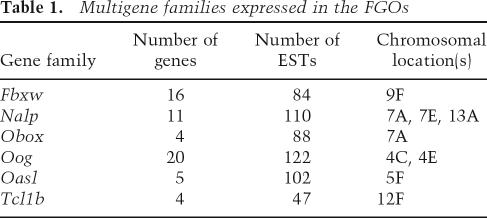
Genomic duplications of oocyte-expressed genes. A prominent feature of the FGO transcriptome is a large number of oocyte-specific protein-coding genes that fall into a handful of highly expressed gene families (Table 1; Supplementary file FGO_library_1_0.xls). Members of these families share nearly identical structure and homology at both nucleotide and protein levels. Indeed, mRNAs of some paralogs, such as members of F-box and WD-40 domain protein (Fbxw) gene families, have >98% sequence identity. Allelic polymorphisms do not cause this diversity, because the library was made from oocytes of an inbred strain C57BL/6J. These six gene families are organized as compact clusters within the M. musculus genome, implicating tandem duplications as a mechanism for their origin. Examination of syntenic regions within M. musculus and Homo sapiens genomes that harbor these genes revealed very peculiar genomic organization: Some of the families have multiple members in both genomes, while others are expanded exclusively in one species.
TABLE 1.
Multigene families expressed in the FGOs
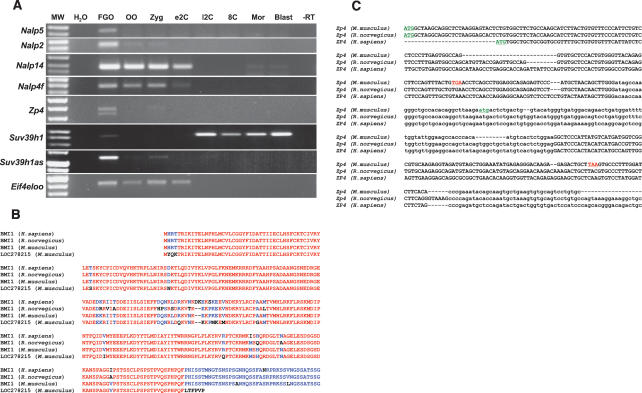
Eleven genes belong to the “NACHT, leucine-rich repeat and PYD-containing” (Nalp) family. These genes, while transcribed in oocytes, are transcriptionally silent during preimplantation development (Fig. 3A). All Nalp genes are organized in three clusters on M. musculus chromosomes 7 and 13, and each one (except for Nalp4f) has an ortholog in syntenic regions of the H. sapiens genome. The five M. musculus genes of the oligoadenylate synthetase-like (Oasl) family lack H. sapiens orthologs and exemplify intracluster duplications that occurred after the split of primates and rodents. These genes map between Oas1a and Oas3 loci of M. musculus within ancestral mammalian oligoadenylate synthetase cluster of OAS1/Oas1a, OAS3/Oas3, and OAS2/Oas2 (H. sapiens/M. musculus) genes (Mashimo et al. 2003). The genes of the oogenesin (Oog) family are also present in multiple copies within M. musculus and H. sapiens genomes. Recreation of unambiguous orthology between the M. musculus and H. sapiens Oog genes was not possible due to incompleteness of M. musculus genome assembly.
FIGURE 3.
Analysis of transcripts identified in the FGO library. (A) RT–PCR detection of Nalp family (Nalp5, Nalp2, Nalp14, Nalp4f), Eif4eloo, sense and antisense Suv39h1 (Suv39h1, Suv39h1as), and Zp4 transcripts. (FGO) Fully grown oocyte; (OO) ovulated oocyte; (Zyg) zygote; (e2C) early two-cell stage embryo; (l2C) late two-cell stage embryo; (8c) early eight-cell stage embryo; (Mor) morula; (Bl) blastocyst; (H2O) control for PCR reactions; (−RT) reverse transcriptase omitted (FGO sample); (MW) DNA size markers. (B) Alignments of LOC278215 protein with BMI1 of H. sapiens, R. norvegicus, and M. musculus. Identical amino acids are red, and residues conserved in at least half of proteins are blue. (C) Partial alignment of M. musculus, R. norvegicus, and H. sapiens Zp4 cDNA sequences. Second and fourth exons are in lowercase. Start codons are underlined in green, and stop codons in the M. musculus Zp4 are underlined in red.
Three M. musculus gene families contain only one H. sapiens ortholog. The Fbxw genes and the T-cell lymphoma breakpoint 1b (Tcl1b) genes have single H. sapiens orthologs, FBXW12 and TCL1B, respectively, in the syntenic regions of the H. sapiens genome. Examination of the oocyte-specific homeobox (Obox) genes (Rajkovic et al. 2002) and a syntenic region of H. sapiens suggests that one of the two genes, TPRX1 or CRX, underwent tandem duplications in the murine lineage, giving rise to the Obox family. Duplications of oocyte-specific genes are not limited to M. musculus. A single-copy murine gene, Rfpl4, was found to have three tandemly repeated paralogs within syntenic region of the H. sapiens genome. In summary, genome organization of these loci implies that some oocyte-specific genes highly expressed during oogenesis are subject to extensive genomic duplications, which is a recurring process in mammalian evolution.
Processed pseudogenes in the female germline. In the male germline, intronless paralogs of ancestral exon-containing genes—i.e., retrotransposed “processed pseudogenes”—encode a number of spermatogenesis-specific genes, such as Cstf2t (Dass et al. 2001) and Gk2 (Pan et al. 1999). Fourteen novel mRNAs identified in this analysis of the FGO transcriptome are transcripts of processed pseudogenes, and eight of these exhibit an oocyte-restricted expression. A case in point, the ORF of LOC278215, an intronless paralog of the Bmi1 gene, is highly similar to BMI1 protein, except for the 40 N-terminal residues that are truncated due to a single-nucleotide deletion in LOC278215 mRNA, which introduces a stop codon (Fig. 3B). Similar abundances of Bmi1 and LOC278215 (seven and five ESTs, respectively) signify comparable levels of transcription at both loci. Unlike the pseudogene-derived spermatogenesis-specific genes mentioned above, these novel loci transcribed in M. musculus oocytes (e.g., LOC278215) lack orthologs in the H. sapiens genome. This observation indicates relatively recent retrotranspositions possibly followed by acquisition of functional roles for the new processed pseudogenes in M. musculus oogenesis.
Loss of oocyte-specific genes. FGO library analysis uncovered an unusual, previously unidentified transcript, which bears a similarity to the H. sapiens and Rattus norvegicus zona pellucida 4 (ZP4) genes. This transcript was mapped to the region syntenic to the ZP4 loci in H. sapiens and R. norvegicus, with an exon–intron structure corresponding to these genes (Fig. 3C). Although Zp4 is transcribed in M. musculus oocytes (Fig. 3A), it lacks a protein product due to ORF mutations. A 19-nucleotide (nt) deletion within the first coding exon produces a frameshift and introduces a stop codon; the second exon, which bears a potential ATG start codon, carries a 2-nt frameshifting mutation as well (Fig. 3C). This intriguing example of a loss-of-gene-function in mammals adds to the list of previously identified genes that have been “lost” during evolution of specific mammalian lineages, as seen in New World monkeys (Jacobs et al. 1996; Gilad et al. 2004).
Oocytes express antisense RNAs. A large proportion of novel transcripts identified in this study most likely represent regulatory RNAs, as they do not contain ORFs longer than a few amino acids. Among these, 16 transcripts are _cis_-encoded antisense RNAs that are transcribed from the same genomic locus as the known M. musculus genes, yet in the opposite orientation (Supplementary file FGO_library_1_0.xls, sequences annotated as “antisense transcript”). These antisense transcripts have an exon–intron structure different from the conventional sense transcripts, excluding the possibility that some cDNAs were ligated in reverse orientation during library preparation. These data suggest the existence of antisense-mediated gene regulation in FGOs, an emerging concept supported by robust bioinformatics and empirical evidence (Coudert et al. 2005; Katayama et al. 2005; Loebel et al. 2005). For example, Suv39h1as, an antisense transcript of the Suv39h1 gene, apparently interferes during the OET with Suv39h1 mRNA, which becomes undetectable, using RT–PCR, during this time (Fig. 3A).
Evolutionary young retrotransposons are highly expressed in FGOs. Retrotransposable elements comprise 14% of the total ESTs in the FGO library. Among these, the most abundant are the “Mouse Transcript type A” (MTA) (Heinlein et al. 1986) long terminal repeat (LTR) retrotransposons (2419 ESTs). The next most commonly expressed repetitive elements are the RLTR10 elements (39 ESTs) and murine leukemia virus (MuLV; 11 ESTs). Interestingly, these three LTR retrotransposons are phylogenetically young, and are specific to the genus Mus (Smit 1993; Jurka et al. 2005). Transcripts of short interspersed nuclear elements and long interspersed nuclear elements are relatively rare (10 ESTs and 20 ESTs, respectively) when considering how abundant these elements are in the genome.
Comparative studies of gene expression
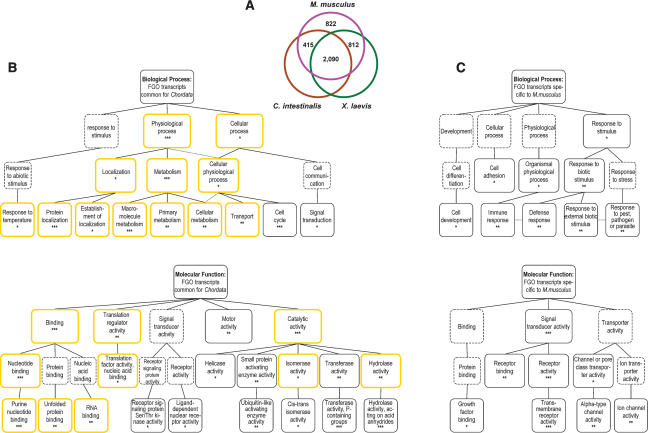
Conservation of oocyte transcriptomes in chordates. To estimate the concordance among oocyte transcriptomes of phylogenetically distant Chordata, a comparison of gene expression in M. musculus FGOs to gene expression in unfertilized eggs of X. laevis and C. intestinalis was accomplished. This survey revealed that 80% of the genes expressed in the FGOs of M. musculus are also expressed in the eggs of either X. laevis or C. intestinalis. Moreover, 50% of these genes are found in the oocyte and egg libraries of all three species (Fig. 4A). These data indicate the conservation of gene expression among evolutionary distant chordates is significant (χ2 = 537.22, p < 10−15). On one hand, this result may be an underestimate of the oocyte transcriptome conservation. Indeed, the bulk of transcripts present in the large eggs of X. laevis and C. intestinalis is thought to represent housekeeping genes, which adversely skew the representation of less abundant transcripts among sequenced ESTs. Accordingly, the proportion of transcripts encoding housekeeping proteins is significantly increased, from 12.6% among all FGO-identified genes to 20.6% in the group of common genes (cumulative hypergeometric p < 10−15) (Supplementary Fig. S3). In addition, a conservative criterion for homology identification (e < 10−15 for TBLASTX searches), while reducing the number of false positive results, may not be appropriate for genes involved in reproduction, because these are under strong selective pressure (Swanson and Vacquier 2002). On the other hand, the 822 M. musculus genes not expressed in the eggs of X. laevis and C. intestinalis may be required specifically for mammalian oogenesis. For example, none of 60 oocyte-specific genes of six families presented in Table 1 have identifiable homologs in X. laevis and C. intestinalis egg cDNA libraries (hypergeometric p < 0.05).
FIGURE 4.
Conservation of gene expression in the oocytes of Chordata. (A) Homologs of genes expressed in M. musculus FGOs are transcribed in eggs of X. laevis and C. intestinalis. (B) Genes with conserved expression are biased to specific biological processes and molecular functions. (*) p < 0.01; (**) p < 0.001; (***) p < 0.0001. Boxes with yellow borders and dotted connector lines are as in Figure 2A. Higher-level GO terms for overrepresented categories (boxes with dotted borders) are shown for reference. (C) Biological processes and molecular functions overrepresented among _M. musculus_-specific FGO-expressed genes. Presentation of data is as in B.
To determine the evolutionary hardwired molecular pathways shared among chordates, 2090 genes expressed in eggs of all three species were compared with the entire set of M. musculus FGO-expressed genes. Unexpectedly, with respect to biological process, this analysis revealed a substantial overlap with GO categories associated with housekeeping genes (Fig. 4B; Supplementary Fig. S1). However, examination of molecular function provided a different perspective. While some inclination toward “housekeeping molecular functions” of commonly expressed genes was noticeable, the overrepresentation of genes with “motor activity,” “small protein activating enzyme activity,” “transferase activity,” “helicase activity,” and two specific signal transducer activities (Fig. 4B) provides a snapshot of gene functions shared particularly by the chordates’ oocytes. Corresponding analysis of 822 genes that were found exclusively in the M. musculus FGO cDNA library (Fig. 4A) delineated processes and functions potentially specific to mammalian oocytes. Within this group, the biological processes were biased toward “cell adhesion,” “organismal physiological process,” and “response to stimulus” categories, while molecular functions slanted toward “signal transducer activity,” “channel transporter activity,” and “ion channel activity” (Fig. 4C). Overall, this broad survey established a large cohort of functional homologs in three choice models of developmental biology, and highlighted the most conserved biological mechanisms operating in the eggs of these species.
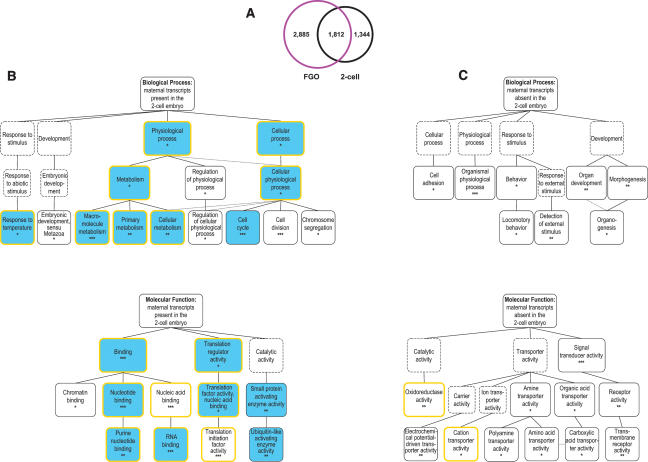
Two levels of FGO transcriptome: oogenesis versus the OET. To elucidate the molecular changes that occur during the OET, a comparison between M. musculus FGO and two-cell stage embryo transcriptomes was conducted. The two-cell stage mouse embryo contains the most stable maternal mRNAs, in addition to new transcripts that appear as the result of embryonic genome activation. Thirty-nine percent of the genes in the FGO library are also found in the previously described data set of the two-cell stage expressed genes (Fig. 5A; Evsikov et al. 2004). This set of “common” transcripts is significantly enriched, compared with the FGO or two-cell stage embryo transcriptomes for housekeeping genes (18.3% vs. 12.7% in FGO and 15.1% in two-cell, cumulative hypergeometric p < 10−15 and p < 10−10, respectively) (Supplementary Fig. S3).
FIGURE 5.
Gene expression during the OET. (A) Comparison of the FGO and two-cell stage mouse embryo transcriptomes. (B) GO categories overrepresented among genes identified in both cDNA libraries. Presentation of data is as in Figure 4B. Blue boxes indicate that the same GO terms are overrepresented among genes whose expression is conserved in the eggs of chordates. (C) GO categories overrepresented among FGO genes absent in the two-cell stage embryo. Presentation of data is as in Figure 4B.
Comparative analysis of the common genes (Fig. 5A) to all FGO-identified genes uncovered an interesting bias of these genes toward the same biological processes and molecular functions as found for the genes commonly expressed in oocytes of chordates (Fig. 5B). This can be explained, in part, by a higher-than-expected proportion of transcripts with conserved expression within this group (χ2 = 82.29, p < 10−15). Importantly, this finding underscores that evolutionary conservation of gene expression is driven primarily by the constraints and functional requirements of the OET rather than oogenesis. Indeed, analysis of genes expressed in the FGO but absent in the two-cell stage embryo (i.e., presumably required for oogenesis) to all FGO-expressed genes failed to reveal any overlapping overrepresented biological processes and molecular functions within the set of genes whose expression is conserved in chordates (Figs. 4B, 5C). Moreover, processes such as “cell adhesion” and “organismal physiological process,” and functions such as “signal transducer activity,” “receptor activity,” and “transmembrane receptor activity” are overrepresented among this group of genes as well as in genes lacking homologs in the eggs of X. laevis or C. intestinalis (Figs. 4C, 5C), most likely due to significant overlap between these two subsets of genes (χ2 = 54.28 p < 10−11). Interestingly, the bias of genes required for oogenesis toward such functions as “amine transporter activity,”“organic acid transporter activity,” and “signal transducer activity” (Fig. 5C) effectively pinpointed the known pathways that are effectors of oocyte growth and oocyte–somatic cell interactions. Arguably, these results outline a dual nature of the FGO transcriptome that comprises two functional subsets, one consisting of mRNAs required for oogenesis and the other, evolutionarily conserved one, maternal transcripts required for the OET.
Post-transcriptional regulation of maternal mRNAs
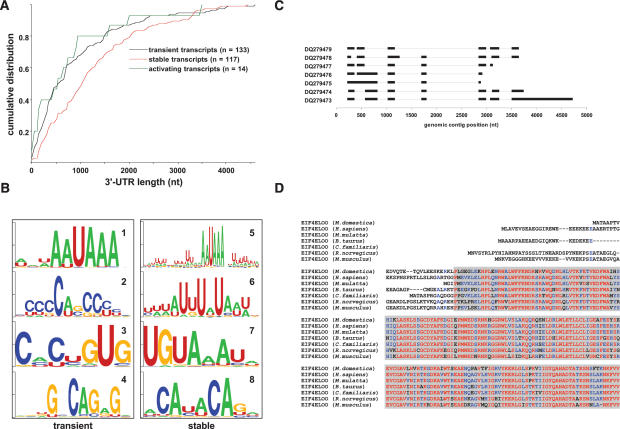
Properties of the 3_′_-UTRs. To discover novel regulatory motifs in maternal transcripts, computational analyses of mRNAs sharing similar expression patterns during the OET were performed. Comparison between the FGO and two-cell-stage mouse embryo transcriptomes generated three classes of coregulated mRNAs, defined as “stable,” “transient,” and “activating” transcripts. The set of activating mRNAs, which are absent in the FGOs but abundant in the two-cell stage embryo, was limited (n = 14), and this group was excluded from further analysis.
Surprisingly, two sets of maternal transcripts differed in the size of the 3′-UTRs (Fig. 6A), which are significantly longer in stable mRNAs than in transient ones (Kolmogorov-Smirnov test, p < 0.001). This finding is consistent with previous results on mRNA stability of genes with multiple 3′-processing sites, such as Spin, during the OET (Oh et al. 2000). Moreover, 3′-UTR sequences of the transient and stable mRNAs had significantly different nucleotide content (43.9% of guanines or cytosines in transient transcripts vs. 41.1% in stable ones, Z = 15.0, p < 10−6), caused primarily by a higher proportion of cytosines in the UTRs of transient, and uracils in the UTRs of stable maternal transcripts.
FIGURE 6.
Translational regulation of maternal mRNAs. (A) Cumulative distribution of 3′-UTR lengths for transient, stable, and activating transcripts. (B) Sequence logo representation of motif consensi identified by reciprocal analysis in the transient (motifs 1–4) and stable (motifs 5–8) mRNAs. (C) Exon–intron structure for Eif4eloo transcript variants identified by their GenBank accession numbers (all transcripts but DQ279473 are incomplete at the 3′ end). (D) Alignment for a major isoform of the M. musculus EIF4ELOO with predicted proteins from Monodelphis domestica, H. sapiens, Macaca mulatta, Bos taurus, Canis familiaris, and R. norvegicus. Color coding is as in Figure 3B. Gray shading is the IF4E/CDC33 domain.
Two analyses utilizing the expectation maximization algorithm were undertaken to compare the stable and transient maternal mRNAs. In the “reciprocal” analysis, 3′-UTRs of transient transcripts were used as a background set for the stable transcripts and vice versa (Fig. 6B). For “independent” analysis, the background sets were produced by random shuffling, with preservation of dinucleotide frequencies, of sequences within each set of UTRs (Supplementary Fig. S4A). Strikingly, in stable maternal mRNAs the nuclear polyadenylation signals (AAUAAA) occur in a larger sequence context regardless of the type of analysis (Fig. 6B, motif 5; Supplementary Fig. S4A, motif 13). In contrast, polyadenylation signals are essentially context independent in transient mRNAs (Fig. 6B, motif 1; Supplementary Fig. S4A, motif 9). Two motifs identified among stable transcripts represent a cytoplasmic polyadenylation element (McGrew et al. 1989), CPE (Fig. 6B, motif 6), and a Pumilio-binding element (White et al. 2001), PBE (Fig. 6B, motif 7; Supplementary Fig. S4A, motif 16).
To test the hypothesis that regulation of transcript stability during the OET depends on multiple factors, the co-occurrences of identified motifs within mRNAs were calculated. Within both stable and transient mRNAs, the CPE and PBE motifs were found in the same transcripts at a higher-than-expected proportion (χ2 stable = 12.88, p < 0.001; χ2 transient = 9.91, p < 0.01). Moreover, CPE motifs significantly overlapped with motif 5 in stable transcripts (χ2 = 4.98 p < 0.05). No evidence for co-occurrence or mutual exclusivity was found for any other motif combinations. A separate analysis of motifs identified above revealed that while the proportion of transient mRNAs containing motifs 3 and 4 (Fig. 6B) is higher than in the stable set, the difference is not significant (Z = 1.04, p = 0.29 and Z = 1.18, p = 0.237, respectively) (Supplementary Fig. S4B). This study also confirmed the significant overrepresentation of motifs 5, 6, and 7, but not motif 8, among stable mRNAs compared with the transient set (Z = 2.742, p < 0.01, Z = 2.604, p < 0.01, Z = 3.057, p < 0.01, and Z = 1.355, p = 0.175, respectively) (Supplementary Fig. S4B). Taken together, these data signify the prominence of motifs that define the stability of mRNA during the OET and indicate the “dual control” of some stable transcripts. Independent measures of motif significance suggest that transient transcripts lack specific sequence features that would explain their instability. As such, this result supports the hypothesis of default destabilization of obsolete maternal mRNAs in mammalian oocytes and early embryos.
Identification of a novel 5_′_-cap-binding protein, EIF4ELOO. Generally, translational repression of mRNAs in eggs of various species is conferred by interactions, either direct or via intermediate proteins, of repressive factors bound to the 3′-UTRs with the proteins of the eukaryotic translation initiation factor 4E (EIF4E) family bound to the 5′-cap of the transcripts (von der Haar et al. 2004; Richter and Sonenberg 2005). Heuristic analysis of the FGO cDNA library revealed a novel gene that was named Eif4eloo (eukaryotic translation initiation factor 4E-like, oocyte-specific). Seventy-six Eif4eloo ESTs encompass seven mRNA variants that arise predominantly as a result of alternative splicing at the 5′ end of the gene (Fig. 6C). The most abundant Eif4eloo transcript encodes a 244-amino-acid protein similar to EIF4E, and to a lesser extent, EIF4E2 and EIF4E3 proteins of M. musculus. Using RT–PCR methods, Eif4eloo mRNA is not detected beyond the early two-cell stage (Fig. 3A), indicating its maternal origin in the embryos. All surveyed mammalian genomes contain orthologs of Eif4eloo (Fig. 6D). Interestingly, the N termini of EIF4ELOO proteins are poorly conserved even among closely related mammals (i.e., M. musculus, R. norvegicus), correlating with the “ambiguous” splicing of mRNA. Discovery of this new gene creates a new perspective for dissecting the mechanisms controlling translation during OET in mammals.
Discussion
Sequencing of cDNA libraries and microarray analysis of gene expression are two approaches proven successful for large-scale transcriptome analysis of the M. musculus oocytes and preimplantation embryos (Sharov et al. 2003; Evsikov et al. 2004; Hamatani et al. 2004; Wang et al. 2004; Pan et al. 2005). The principles of gene expression analysis underlying these methods are quite different. In the case of microarrays, the study of a transcriptome is restricted to only those genes represented on the array, whereas, in the case of cDNA libraries, the transcriptome is defined by the number of sequenced clones. Therefore, while microarray techniques can validate (and, if different samples or stages are surveyed, quantify) expression of a predefined gene set, EST analysis a discovery-oriented approach to transcriptome studies.
The FGO is the only stage that embodies all the transcripts necessary for the OET. Later stages, such as the ovulated oocyte, fertilized zygote, and two-cell stage embryo, contain merely a fraction of this initial transcriptome. Indeed, the two-cell stage mouse embryo retains just 30% of the poly(A) RNA of the FGO (Piko and Clegg 1982; Clegg and Piko 1983). For that reason, transcriptome diversity through the OET dramatically decreases as well (Hamatani et al. 2004; Wang et al. 2004). This is the first large-scale EST-based study of transcripts present in the FGOs that provide the broad snapshot of the FGO transcriptome and advance the explorations of the molecular mechanisms underlying the OET. The catalog of gene expression in the M. musculus FGOs is an open access resource for researchers interested in mammalian oogenesis and the initiation of development (Supplementary file FGO_library_1_0.xls). Cross-species analysis of gene expression in oocytes distinguished functionally conserved genes and pathways among chordates. Finally, identification of novel, distinctive features in the 3′-UTRs of maternal mRNAs with differential stability is a step toward deciphering the mechanisms of translational control during this transcriptionally silent period of development.
FGO transcriptome and the functional genomics of the OET
Application of the knowledge obtained with one experimental model to understand other similar systems is widely used in developmental biology (Gilbert 2000). For example, discovery of CPE-dependent mRNA translational regulation in X. laevis oocytes was a prerequisite that triggered investigations for this mechanism in M. musculus (Gebauer et al. 1994; Gebauer and Richter 1996; Tay et al. 2000, 2003). Recent evidence suggests that complex phenomena, such as genome remodeling to totipotency, are regulated by well-conserved pathways (Byrne et al. 2003). In this respect, exploration of gene expression in oocytes among various model organisms becomes an essential tool to detect similar pathways. More importantly, a discovery-oriented approach is especially critical when incomplete or partial data from different models are available. In silico studies of evolutionarily conserved gene expression in the eggs of Chordata is a novel approach that identified the major conserved processes “encoded” in the oocyte transcriptome and enumerated potentially critical players of the OET. While “housekeeping GO annotations” predominate among the overrepresented GO categories for genes expressed in eggs of X. laevis, C. intestinalis, and M. musculus, the housekeeping fraction among these conserved genes is only 1.5 times higher than in entire FGO transcriptome (Fig. 4B; Supplementary Fig. S3). The likely cause of this deceptive discrepancy stems from near complete coverage of housekeeping genes in the GO database and deficiency in annotations for all other genes. Therefore, completeness of the GO database is an important consideration for interpretation of GO results. Yet, our findings demonstrate that categorizing genes with conserved expression using GO annotations is a valid method to identify commonalities (and differences) among oocytes of phylogenetically distant species.
Comparison of the FGO and two-cell transcriptomes adds an insightful second dimension by pulling out important processes for both oocyte growth and the OET (Fig. 5). The outstanding abundance of “cell communication” and “response to stimulus” genes in the FGO transcriptome (Fig. 2) and their scarcity in the two-cell embryo transcriptome (Fig. 5C; Evsikov et al. 2004) document the oocyte–granulosa cell cross-talk essential for oogenesis (Eppig 2001; Matzuk et al. 2002) and pinpoint the molecular pathways involved in these communications, such as G-protein-coupled receptor pathways (GPCRs). Indeed, the biological importance of GPCRs during the final stages of mouse oocyte growth has been demonstrated recently (Mehlmann et al. 2004; Freudzon et al. 2005; Ledent et al. 2005). During the OET and preimplantation development, the embryo becomes essentially autonomous and, therefore, elimination of these pathways may be compulsory. Indeed, by the two-cell stage, the major pathways regulated by maternal mRNAs are targeted protein degradation, translational control, and chromatin remodeling (Fig. 5B; Evsikov et al. 2004; Hamatani et al. 2004). Among the novel findings that exemplify the strength of our heuristic EST-based approach is an implicit change in energy metabolism between oocytes and early embryos. Examination of the FGO genes absent in the two-cell stage embryo revealed significant overrepresentation of genes involved in amino acid transport and fatty acid metabolism. Indeed, all 11 amino acid transporters and 25 of 31 fatty acid metabolism genes expressed in the FGO are absent in the two-cell stage embryo (Fig. 5C). These data imply a change in nutrients utilized by cleavage-stage embryos versus oocytes. Involvement of lipid metabolism in the physiology of oogenesis and OET is an attractive novel hypothesis, as previous research in the field focused on the roles of glucose, lactate, and pyruvate during oocyte maturation and preimplantation development (Biggers et al. 1967; Downs et al. 2002; Summers and Biggers 2003).
Stability and translation of maternal mRNA
The key regulatory networks of the OET depend on strict temporal control of maternal mRNA expression (Seydoux 1996; Solter et al. 2002). According to current models of translational control, certain events, such as phosphorylation of mRNA-bound regulatory proteins, launch a cascade that results in activation of maternal transcripts (Richter and Sonenberg 2005). For example, during oocyte growth, genes containing a functional CPE in their 3′-UTRs are initially processed as normal transcripts, but these mRNAs are deadenylated in the ooplasm, leaving just a few adenosines (Huarte et al. 1992). Inappropriate translation is prevented by CPE-binding protein (CPEB), a protein most probably conserved in all metazoans. CPEB associates with Maskin, and this complex is bound to the 5′-cap mRNA-binding protein eIF4E. The resulting multiprotein complex is capable of rendering a dormant transcript. Studies of X. laevis oocyte maturation revealed that phosphorylation of CPEB reinitiates polyadenylation via breakdown of the complex, followed by recruitment of poly(A)-binding protein and displacement of Maskin with eIF-4G protein. These events restore dormant mRNAs to an active “translational state” (Cao and Richter 2002; Mendez et al. 2000a, b). A very similar mechanism operates during the maturation of M. musculus oocytes (Hodgman et al. 2001; Tay et al. 2000, 2003). The library-based discovery of the novel oocyte-specific Eif4eloo gene is a significant revision to the model of translational control in mammalian oocytes. Interestingly, the predominant poly(A)-binding protein in M. musculus oocytes identified during the analysis of the FGO library, ePABP2, is also encoded by an oocyte-specific gene, Pabpnl1 (Seli et al. 2005).
Given the abundance of RNA-binding protein-encoding genes expressed in the FGO (Fig. 2) and evidence for translational control mechanisms across model organisms, it is logical to assume a broad diversity of regulatory interactions between mRNAs and their cognate proteins during the OET in mammals. The detection of 3′-UTR motifs using computational analysis produced valuable insights into the features associated with transcript stability in mammalian oocytes and cleavage-stage embryos, a necessary first step toward identification and characterization of mRNA–protein interactions. The strategy chosen to discover common features within the 3′-UTRs between stable and transient mRNAs exposed the known effectors of RNA stability, such as CPE and PBE. Functionality, or binding partners, of other identified motifs may be deduced from the studies of other model organisms. For example, cytosine-rich sequences interact with PCBP2 protein in X. laevis embryos (Paillard et al. 2000), and CUG-containing motifs may be the targets of CUGBP1 and CUGBP2 (Barreau et al. 2006). Maternal transcripts of Pcbp2, Cugbp1, and Cugbp2 genes are abundant in the mouse FGOs, suggesting these proteins and motifs are likely candidates for regulatory control of mRNAs during the OET. Finally, mRNA stability may be controlled by multiple mechanisms. For example, disinhibition of PUM2 repression of Ringo/Speedy mRNA leads to CPEB activation in X. laevis oocytes (Padmanabhan and Richter 2006) and pinpoints an intricate link between two systems of translational regulation. The significant co-occurrence of CPEs and PBEs in maternal transcripts suggests that these two mechanisms may act on the same targets and operate in parallel during the OET, as evidenced, for example, by interaction of CPEB and PUM1 and their coregulation of cyclin B1 mRNA during X. laevis oocyte maturation (Nakahata et al. 2001, 2003). Finally, a failure to identify any unambiguous and robust “instability motifs” among FGO mRNAs indicates that mRNA degradation may be context independent and occurs by default in mammalian oocytes.
Genetic variation and the oocyte transcriptome
Maternal effect genes: abundant or redundant? The comprehensive survey of gene expression in mouse oocytes has uncovered unique properties of these cells. Judging by the profiles of gene expression in the M. musculus EST database, one-tenth of genes identified in the FGO library are expressed exclusively in oocytes. The observation that a large number of such genes belong to a few gene families (Table 1) might be, to a certain extent, a functional analogy of developmental gene amplification occurring in other species (Claycomb and Orr-Weaver 2005). However, while the similarity between individual members is striking, it appears that the genes within these families cannot functionally substitute for each other. Indeed, targeted deletions of individual family members suggest that each of these genes may be pivotal for the OET. In mice, the Mater (Nalp5) gene, a member of the Nalp family, was one of the first identified mammalian maternal effect genes; i.e., it encodes mRNA required for successful development of a fertilized oocyte (Tong et al. 2000a). Targeted deletion of one of the Oasl genes, Oasl1d, causes reduced fertility in females but does not affect males (Yan et al. 2005), demonstrating that members of this family are maternal effect genes. Tcl1, a gene whose duplication gave rise to TCL1B/Tcl1b loci in H. sapiens and M. musculus, is essential for female fertility and viability of cleavage stage embryos as well (Narducci et al. 2002). Finally, the seminal member of a large Oog family, Oog1, encodes a nuclear protein abundant in the zygote and two-cell stage mouse embryos (Minami et al. 2003), and Rfpl4 encodes a highly expressed oocyte-specific E3 ubiquitin ligase (Suzumori et al. 2003). These observations imply important functions for Oog and Rflp4 genes during this developmental period.
These data underscore the principle of genomic duplications as an underlying mechanism of expansion of oocyte-specific genes. A complication for interpretations is that currently, little to nothing is known about the biological functions for the encoded proteins. For example, the Nalp genes share similarity with ribonuclease inhibitors and potentially may act as protectors for maternal RNAs (Tong et al. 2000b). Mater, and other members of the Nalp family, may be vital for OET by acting in similar pathways, or being differentially translated, or having different targets. However, function is only inferred by sequence similarity and must be tested directly in vivo. Proteins encoded by the Fbxw gene family contain an F-box domain at the N termini and may function as receptors that recruit different substrates to the Skp1–Cullin–F-box protein ubiquitin ligase complexes (Cardozo and Pagano 2004). Indeed, the ubiquitin–proteasome protein degradation pathway is a large component of the FGO transcriptome (Fig. 2; Supplementary Fig. S2) essential for the OET (Josefsberg et al. 2000; Solter et al. 2004). Currently, it is not clear if Fbxw genes are functionally redundant or if different members of this family have diverse protein targets. Mammalian oligoadenylate synthetases are central to the innate immune defense mechanism against double-stranded RNA viruses (Castelli et al. 1998). However, the distinguishing (and confusing) feature of all Oasl genes is a lack of 2′,5′-OAS enzymatic activity (Kakuta et al. 2002). By and large, the oocyte-restricted expression of a number of “functionally hypervariable” loci may reflect the natural variation of gene expression in the oocytes that promote the processes of gametic selection in female germline and compatibility selection in zygotes.
Gene expression in FGO: speciation in action? Analysis of the FGO transcriptome gives hints about the underlying processes of biological evolution, specifically for gametic selection and reproductive isolation. For instance, it has been established that genes associated with reproduction are subjected to strong selective pressure (Swanson et al. 2001; Swanson and Vacquier 2002). A general explanation for this phenomenon is that it serves as a primary mechanism attaining reproductive isolation during speciation. Indeed, the zona pellucida genes, which serve as sperm receptors during fertilization, are among the most rapidly evolving genes in mammals. As exemplified by Zp4, the novel zona pellucida gene identified in this study, fast-paced molecular evolution ultimately led to its functional elimination in M. musculus. By extrapolation, gene families, such as Oasl and Tcl1, may reflect the other side of the same process: New genes arise by duplication and acquire a unique function, which facilitates reproductive isolation, thereby providing a substrate for natural selection to act upon and result in speciation.
Antisense RNAs and LTR retrotransposons: regulatory molecules or escapees from degradation? Analysis of the FGO library revealed a number of transcripts with no evidence of an ORF. Some of these may play a regulatory role in post-transcriptional control of distinct genes, such as the highly expressed antisense transcript of the Suv39h1 locus that apparently modulates stability of mRNA encoding histone methyltransferase SUV39H1 (Bultman and Magnuson 2000). The functional role, if any, of others is elusive. A number of these non-protein-coding transcripts are clearly processed from premRNAs, and their genomic structure encompasses an exon–intron organization. The presence of such transcripts, and especially the transcript of the Zp4 locus described here, prompts the notion that the mechanisms of nonsense-mediated mRNA decay, which are responsible for rapid degradation of incorrectly spliced, truncated, or other aberrant mRNAs (Conti and Izaurralde 2005), may be inactive in the FGOs.
Another case of highly expressed transcripts with elusive functions is LTR retrotransposons. Retrotransposons must be expressed in the germline for their successful propagation in the genetic pool of host species. Indeed, this notion is supported by the observation that oocytes (as well as cleavage-stage embryos) transcribe the phylogenetically youngest and therefore more active LTR retrotransposons, such as MTA or MuERV-L (Evsikov et al. 2004). A number of roles that retrotransposons may play in mammalian oocytes and cleavage-stage embryos have been recently discussed (Evsikov et al. 2004; Peaston et al. 2004). Typically, transcriptional activity of retrotransposons inversely correlates with methylation of their genomic loci (Yoder et al. 1997). Thus, this atypical high activity, limited to a handful of repetitive elements present within the mouse genome, may indicate that specific retrotransposons transcribed in oocytes are hypomethylated during oocyte growth. However, it was previously shown that transcription profiles of LTR retrotransposons do not perfectly coincide with global changes to cytosine methylation during the OET (Kim et al. 2004; Peaston et al. 2004). Therefore, factors other than DNA methylation may affect this peculiar pattern of retrotransposon transcriptional activity in oocytes.
Conclusion
The large-scale survey of the oocyte transcriptional activity revealed a delicate balance between novelty and conservation, two major forces that shape the evolution of life. Two-level analysis of the transcriptome pinpointed that the molecules and, by extrapolation, the mechanisms of the OET tend to be well conserved among phylogenetically distant taxa. Unexpectedly, the fraction of the FGO transcriptome that is unique to M. musculus harbors most maternal effect genes identified to date in mammals. The strong evidence for rapid diversification of critical mammalian oocyte-specific genes prompts us to suggest that the functions of evolutionary novel loci expressed in the oocytes are shaped by the forces of gametic and compatibility selection that target the processes unique to mammals, or even individual species. This hypothesis is a strong foundation for the studies of molecular mechanisms of reproductive isolation and, consequently, the processes of speciation.
Materials and methods
Collection of oocytes and embryos
To construct the FGO cDNA library, C57BL6/J female mice were used. Fully grown, germinal vesicle-intact cumulus-enclosed oocytes were collected as described (Eppig and O'Brien 1996). Cumulus cells were removed from the oocytes by pipetting; oocytes with abnormal morphology were discarded. Zona pellucida was removed by 5-min incubation of oocytes in MEM supplemented with 0.5% proteinase K. The oocytes were washed and processed for RNA isolation. For RT–PCR assays, the mice were F1 hybrid B6D2F1/J. Hormonal stimulation, mating, and collection of oocytes and embryos were performed as described (Oh et al. 2000). Batches of oocytes or embryos (55 per batch) were harvested, processed as described (Evsikov et al. 2004), snap-frozen in liquid nitrogen, and stored at −80°C until use.
cDNA library construction and sequencing
Total RNA was isolated from 12,000 FGOs. The cDNA library was prepared as described (Rothstein et al. 1992) using NotI(dT) primer and SalI linker, size-selected for 500 base pairs (bp), and cloned into the SalI/NotI sites of pSPORT1 vector (Invitrogen). The FGO cDNA library (Eppig/Hampl oocyte library, I.M.A.G.E. ID: 1182) is representative (>9 × 105 cfu/μL), with an average insert size of 1500 nt. Library arraying and single-pass sequencing from the 5′ ends of inserts was performed by GATC Biotech AG. Sequencing produced high-quality ESTs with an average length of 635 nt (range 85–980 nt).
Preparation of cDNA for RT–PCR and detection of selected transcripts
RNA from oocytes and embryos was isolated using the PicoPure RNA Isolation Kit (Arcturus). DNase-treated RNA was used for cDNA first-strand synthesis using SuperScript III (Invitrogen). Absence of genomic DNA contamination was confirmed by PCR of the Catnb1 intronic region. PCR of mt-Atp6 was used as a constitutive control. Optimization of PCR conditions for each primer pair was performed using ovarian cDNA and FailSafe PCR system (Epicentre). Primer sequences are provided in the Supplemental Material. PCRs were conducted on at least two independent sets of cDNA. Thirty-five amplification cycles were routinely performed, with the amount of cDNA template corresponding to two oocytes or embryos.
Bioinformatics and computational analyses
Procedures for the analysis of ESTs, comparison of gene expression, and computational analysis of the 3′-UTRs are described in the Supplemental Material.
GenBank accession numbers
The seven transcripts of M. musculus oocyte-specific Eif4eloo are DQ279473–DQ279479. The M. musculus Zp4 transcript is DQ279480. ESTs of Eppig/Hampl oocyte library are DV644482–DV663271.
Acknowledgments
We thank Drs. Caralina M. de Evsikova and Jason P. Affourtit for critical reading and helpful suggestions on the manuscript. This work was supported by The Lalor Foundation (A.V.E.), US NSF grant EPS-0132384, US PHS NIH CORE grant CA34196, grants HD037102 (B.B.K.), HD021970 (J.J.E. and A.H.), NIH/ NCRR INBRE Maine contract P20 RR16463 (J.H.G.), and the Max Planck Society (D.S.).
Footnotes
References
- Barreau, C., Paillard, L., Mereau, A., Osborne, H.B. Mammalian CELF/Bruno-like RNA-binding proteins: Molecular characteristics and biological functions. Biochimie. 2006;88:515–525. doi: 10.1016/j.biochi.2005.10.011. [DOI] [PubMed] [Google Scholar]
- Biggers, J.D., Whittingham, D.G., Donahue, R.P. The pattern of energy metabolism in the mouse oocyte and zygote. Proc. Natl. Acad. Sci. 1967;58:560–567. doi: 10.1073/pnas.58.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake, J.A., Eppig, J.T., Bult, C.J., Kadin, J.A., Richardson, , J.E. and Mouse Genome Database Group. The Mouse Genome Database (MGD): Updates and enhancements. Nucleic Acids Res. 2006;34:D562–D567. doi: 10.1093/nar/gkj085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultman, S., Magnuson, T. Molecular and genetic analysis of the mouse homolog of the Drosophila suppressor of position-effect variegation 3–9 gene. Mamm. Genome. 2000;11:251–254. doi: 10.1007/s003350010049. [DOI] [PubMed] [Google Scholar]
- Byrne, J.A., Simonsson, S., Western, P.S., Gurdon, J.B. Nuclei of adult mammalian somatic cells are directly reprogrammed to oct-4 stem cell gene expression by amphibian oocytes. Curr. Biol. 2003;13:1206–1213. doi: 10.1016/s0960-9822(03)00462-7. [DOI] [PubMed] [Google Scholar]
- Cao, Q., Richter, J.D. Dissolution of the maskin–eIF4E complex by cytoplasmic polyadenylation and poly(A)-binding protein controls cyclin B1 mRNA translation and oocyte maturation. EMBO J. 2002;21:3852–3862. doi: 10.1093/emboj/cdf353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardozo, T., Pagano, M. The SCF ubiquitin ligase: Insights into a molecular machine. Nat. Rev. Mol. Cell Biol. 2004;5:739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- Castelli, J., Wood, K.A., Youle, R.J. The 2-5A system in viral infection and apoptosis. Biomed. Pharmacother. 1998;52:386–390. doi: 10.1016/s0753-3322(99)80006-7. [DOI] [PubMed] [Google Scholar]
- Claycomb, J.M., Orr-Weaver, T.L. Developmental gene amplification: Insights into DNA replication and gene expression. Trends Genet. 2005;21:149–162. doi: 10.1016/j.tig.2005.01.009. [DOI] [PubMed] [Google Scholar]
- Clegg, K.B., Piko, L. Poly(A) length, cytoplasmic adenylation and synthesis of poly(A)+RNA in early mouse embryos. Dev. Biol. 1983;95:331–341. doi: 10.1016/0012-1606(83)90034-9. [DOI] [PubMed] [Google Scholar]
- Conti, E., Izaurralde, E. Nonsense-mediated mRNA decay: Molecular insights and mechanistic variations across species. Curr. Opin. Cell Biol. 2005;17:316–325. doi: 10.1016/j.ceb.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Coudert, A.E., Pibouin, L., Vi-Fane, B., Thomas, B.L., Macdougall, M., Choudhury, A., Robert, B., Sharpe, P.T., Berdal, A., Lezot, F. Expression and regulation of the Msx1 natural antisense transcript during development. Nucleic Acids Res. 2005;33:5208–5218. doi: 10.1093/nar/gki831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dass, B., McMahon, K.W., Jenkins, N.A., Gilbert, D.J., Copeland, N.G., MacDonald, C.C. The gene for a variant form of the polyadenylation protein CstF-64 is on chromosome 19 and is expressed in pachytene spermatocytes in mice. J. Biol. Chem. 2001;276:8044–8050. doi: 10.1074/jbc.M009091200. [DOI] [PubMed] [Google Scholar]
- Dean, W., Santos, F., Reik, W. Epigenetic reprogramming in early mammalian development and following somatic nuclear transfer. Semin. Cell Dev. Biol. 2003;14:93–100. doi: 10.1016/s1084-9521(02)00141-6. [DOI] [PubMed] [Google Scholar]
- Downs, S.M., Humpherson, P.G., Leese, H.J. Pyruvate utilization by mouse oocytes is influenced by meiotic status and the cumulus oophorus. Mol. Reprod. Dev. 2002;62:113–123. doi: 10.1002/mrd.10067. [DOI] [PubMed] [Google Scholar]
- Eppig,, J.J. Oocyte control of ovarian follicular development and function in mammals. Reproduction. 2001;122:829–838. doi: 10.1530/rep.0.1220829. [DOI] [PubMed] [Google Scholar]
- Eppig, J.J., O'Brien, M.J. Development in vitro of mouse oocytes from primordial follicles. Biol. Reprod. 1996;54:197–207. doi: 10.1095/biolreprod54.1.197. [DOI] [PubMed] [Google Scholar]
- Evsikov, A.V., de Vries, W.N., Peaston, A.E., Radford, E.E., Fancher, K.S., Chen, F.H., Blake, J.A., Bult, C.J., Latham, K.E., Solter, D., et al. Systems biology of the 2-cell mouse embryo. Cytogenet. Genome Res. 2004;105:240–250. doi: 10.1159/000078195. [DOI] [PubMed] [Google Scholar]
- Freudzon, L., Norris, R.P., Hand, A.R., Tanaka, S., Saeki, Y., Jones, T.L., Rasenick, M.M., Berlot, C.H., Mehlmann, L.M., Jaffe, L.A. Regulation of meiotic prophase arrest in mouse oocytes by GPR3, a constitutive activator of the Gs G protein. J. Cell Biol. 2005;171:255–265. doi: 10.1083/jcb.200506194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer, F., Richter, J.D. Mouse cytoplasmic polyadenylylation element binding protein: An evolutionarily conserved protein that interacts with the cytoplasmic polyadenylylation elements of c-mos mRNA. Proc. Natl. Acad. Sci. 1996;93:14602–14607. doi: 10.1073/pnas.93.25.14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer, F., Xu, W., Cooper, G.M., Richter, J.D. Translational control by cytoplasmic polyadenylation of cmos mRNA is necessary for oocyte maturation in the mouse. EMBO J. 1994;13:5712–5720. doi: 10.1002/j.1460-2075.1994.tb06909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gene Ontology Consortium. Creating the gene ontology resource: Design and implementation. Genome Res. 2001;11:1425–1433. doi: 10.1101/gr.180801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilad, Y., Wiebe, V., Przeworski, M., Lancet, D., Pääbo, S. Loss of olfactory receptor genes coincides with the acquisition of full trichromatic vision in primates. PLoS Biol. 2004;2:120–125. doi: 10.1371/journal.pbio.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert,, S.F. Developmental biology. 6th ed. Sinauer Associates; Sunderland, MA: 2000. [Google Scholar]
- Hamatani, T., Carter, M.G., Sharov, A.A., Ko, M.S. Dynamics of global gene expression changes during mouse preimplantation development. Dev. Cell. 2004;6:117–131. doi: 10.1016/s1534-5807(03)00373-3. [DOI] [PubMed] [Google Scholar]
- Heinlein, U.A., Lange-Sablitzky, R., Schaal, H., Wille, W. Molecular characterization of the MT-family of dispersed middle-repetitive DNA in rodent genomes. Nucleic Acids Res. 1986;14:6403–6416. doi: 10.1093/nar/14.16.6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgman, R., Tay, J., Mendez, R., Richter, J.D. CPEB phosphorylation and cytoplasmic polyadenylation are catalyzed by the kinase IAK1/Eg2 in maturing mouse oocytes. Development. 2001;128:2815–2822. doi: 10.1242/dev.128.14.2815. [DOI] [PubMed] [Google Scholar]
- Huarte, J., Stutz, A., O'Connell, M.L., Gubler, P., Belin, D., Darrow, A.L., Strickland, S., Vassalli, J.D. Transient translational silencing by reversible mRNA deadenylation. Cell. 1992;69:1021–1030. doi: 10.1016/0092-8674(92)90620-r. [DOI] [PubMed] [Google Scholar]
- Jacobs, G.H., Neitz, M., Neitz, J. Mutations in S-cone pigment genes and the absence of colour vision in two species of nocturnal primate. Proc. Biol. Sci. 1996;263:705–710. doi: 10.1098/rspb.1996.0105. [DOI] [PubMed] [Google Scholar]
- Josefsberg, L.B., Galiani, D., Dantes, A., Amsterdam, A., Dekel, N. The proteasome is involved in the first metaphase-to-anaphase transition of meiosis in rat oocytes. Biol. Reprod. 2000;62:1270–1277. doi: 10.1095/biolreprod62.5.1270. [DOI] [PubMed] [Google Scholar]
- Jurka, J., Kapitonov, V.V., Pavlicek, A., Klonowski, P., Kohany, O., Walichiewicz, J. Repbase update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- Kakuta, S., Shibata, S., Iwakura, Y. Genomic structure of the mouse 2′, 5′-oligoadenylate synthetase gene family. J. Interferon Cytokine Res. 2002;22:981–993. doi: 10.1089/10799900260286696. [DOI] [PubMed] [Google Scholar]
- Katayama, S., Tomaru, Y., Kasukawa, T., Waki, K., Nakanishi, M., Nakamura, M., Nishida, H., Yap, C.C., Suzuki, M., Kawai, J., et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- Kim, S.H., Kang, Y.K., Koo, D.B., Kang, M.J., Moon, S.J., Lee, K.K., Han, Y.M. Differential DNA methylation reprogramming of various repetitive sequences in mouse preimplantation embryos. Biochem. Biophys. Res. Commun. 2004;324:58–63. doi: 10.1016/j.bbrc.2004.09.023. [DOI] [PubMed] [Google Scholar]
- Ledent, C., Demeestere, I., Blum, D., Petermans, J., Hamalainen, T., Smits, G., Vassart, G. Premature ovarian aging in mice deficient for Gpr3. Proc. Natl. Acad. Sci. 2005;102:8922–8926. doi: 10.1073/pnas.0503840102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loebel, D.A., Tsoi, B., Wong, N., Tam, P.P. A conserved noncoding intronic transcript at the mouse Dnm3 locus. Genomics. 2005;85:782–789. doi: 10.1016/j.ygeno.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Mashimo, T., Glaser, P., Lucas, M., Simon-Chazottes, D., Ceccaldi, P.E., Montagutelli, X., Despres, P., Guenet, J.L. Structural and functional genomics and evolutionary relationships in the cluster of genes encoding murine 2′, 5′-oligoadenylate synthetases. Genomics. 2003;82:537–552. doi: 10.1016/s0888-7543(03)00176-9. [DOI] [PubMed] [Google Scholar]
- Matzuk, M.M., Burns, K.H., Viveiros, M.M., Eppig, J.J. Intercellular communication in the mammalian ovary: Oocytes carry the conversation. Science. 2002;296:2178–2180. doi: 10.1126/science.1071965. [DOI] [PubMed] [Google Scholar]
- McGrew, L.L., Dworkin-Rastl, E., Dworkin, M.B., Richter, J.D. Poly(A) elongation during Xenopus oocyte maturation is required for translational recruitment and is mediated by a short sequence element. Genes & Dev. 1989;3:803–815. doi: 10.1101/gad.3.6.803. [DOI] [PubMed] [Google Scholar]
- Mehlmann, L.M., Saeki, Y., Tanaka, S., Brennan, T.J., Evsikov, A.V., Pendola, F.L., Knowles, B.B., Eppig, J.J., Jaffe, L.A. The Gs-linked receptor GPR3 maintains meiotic arrest in mammalian oocytes. Science. 2004;306:1947–1950. doi: 10.1126/science.1103974. [DOI] [PubMed] [Google Scholar]
- Mendez, R., Hake, L.E., Andresson, T., Littlepage, L.E., Ruderman, J.V., Richter, J.D. Phosphorylation of CPE binding factor by Eg2 regulates translation of c-mos mRNA. Nature. 2000a;404:302–307. doi: 10.1038/35005126. [DOI] [PubMed] [Google Scholar]
- Mendez, R., Murthy, K.G., Ryan, K., Manley, J.L., Richter, J.D. Phosphorylation of CPEB by Eg2 mediates the recruitment of CPSF into an active cytoplasmic polyadenylation complex. Mol. Cell. 2000b;6:1253–1259. doi: 10.1016/s1097-2765(00)00121-0. [DOI] [PubMed] [Google Scholar]
- Minami, N., Aizawa, A., Ihara, R., Miyamoto, M., Ohashi, A., Imai, H. Oogenesin is a novel mouse protein expressed in oocytes and early cleavage-stage embryos. Biol. Reprod. 2003;69:1736–1742. doi: 10.1095/biolreprod.103.018051. [DOI] [PubMed] [Google Scholar]
- Nakahata, S., Katsu, Y., Mita, K., Inoue, K., Nagahama, Y., Yamashita, M. Biochemical identification of Xenopus Pumilio as a sequence-specific cyclin B1 mRNA-binding protein that physically interacts with a Nanos homolog, Xcat-2, and a cytoplasmic polyadenylation element-binding protein. J. Biol. Chem. 2001;276:20945–20953. doi: 10.1074/jbc.M010528200. [DOI] [PubMed] [Google Scholar]
- Nakahata, S., Kotani, T., Mita, K., Kawasaki, T., Katsu, Y., Nagahama, Y., Yamashita, M. Involvement of Xenopus Pumilio in the translational regulation that is specific to cyclin B1 mRNA during oocyte maturation. Mech. Dev. 2003;120:865–880. doi: 10.1016/s0925-4773(03)00160-6. [DOI] [PubMed] [Google Scholar]
- Narducci, M.G., Fiorenza, M.T., Kang, S.M., Bevilacqua, A., Di Giacomo, M., Remotti, D., Picchio, M.C., Fidanza, V., Cooper, M.D., Croce, C.M., et al. TCL1 participates in early embryonic development and is overexpressed in human seminomas. Proc. Natl. Acad. Sci. 2002;99:11712–11717. doi: 10.1073/pnas.182412399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, B., Hwang, S., McLaughlin, J., Solter, D., Knowles, B.B. Timely translation during the mouse oocyte-to-embryo transition. Development. 2000;127:3795–3803. doi: 10.1242/dev.127.17.3795. [DOI] [PubMed] [Google Scholar]
- Padmanabhan, K., Richter, J.D. Regulated Pumilio-2 binding controls RINGO/Spy mRNA translation and CPEB activation. Genes & Dev. 2006;20:199–209. doi: 10.1101/gad.1383106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paillard, L., Maniey, D., Lachaume, P., Legagneux, V., Osborne, H.B. Identification of a C-rich element as a novel cytoplasmic polyadenylation element in Xenopus embryos. Mech. Dev. 2000;93:117–125. doi: 10.1016/s0925-4773(00)00279-3. [DOI] [PubMed] [Google Scholar]
- Pan, Y., Decker, W.K., Huq, A.H., Craigen, W.J. Retrotransposition of glycerol kinase-related genes from the X chromosome to autosomes: Functional and evolutionary aspects. Genomics. 1999;59:282–290. doi: 10.1006/geno.1999.5874. [DOI] [PubMed] [Google Scholar]
- Pan, H., O'Brien, M.J., Wigglesworth, K., Eppig, J.J., Schultz, R.M. Transcript profiling during mouse oocyte development and the effect of gonadotropin priming and development in vitro. Dev. Biol. 2005;286:493–506. doi: 10.1016/j.ydbio.2005.08.023. [DOI] [PubMed] [Google Scholar]
- Peaston, A.E., Evsikov, A.V., Graber, J.H., de Vries, W.N., Holbrook, A.E., Solter, D., Knowles, B.B. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev. Cell. 2004;7:597–606. doi: 10.1016/j.devcel.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Piko, L., Clegg, K.B. Quantitative changes in total RNA, total poly(A), and ribosomes in early mouse embryos. Dev. Biol. 1982;89:362–378. doi: 10.1016/0012-1606(82)90325-6. [DOI] [PubMed] [Google Scholar]
- Rajkovic, A., Yan, C., Yan, W., Klysik, M., Matzuk, M.M. Obox, a family of homeobox genes preferentially expressed in germ cells. Genomics. 2002;79:711–717. doi: 10.1006/geno.2002.6759. [DOI] [PubMed] [Google Scholar]
- Richter, J.D., Sonenberg, N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433:477–480. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- Rothstein, J.L., Johnson, D., DeLoia, J.A., Skowronski, J., Solter, D., Knowles, B. Gene expression during preim-plantation mouse development. Genes & Dev. 1992;6:1190–1201. doi: 10.1101/gad.6.7.1190. [DOI] [PubMed] [Google Scholar]
- Seli, E., Lalioti, M.D., Flaherty, S.M., Sakkas, D., Terzi, N., Steitz, J.A. An embryonic poly(A)-binding protein (ePAB) is expressed in mouse oocytes and early preimplantation embryos. Proc. Natl. Acad. Sci. 2005;102:367–372. doi: 10.1073/pnas.0408378102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux,, G. Mechanisms of translational control in early development. Curr. Opin. Genet. Dev. 1996;6:555–561. doi: 10.1016/s0959-437x(96)80083-9. [DOI] [PubMed] [Google Scholar]
- Sharov, A.A., Piao, Y., Matoba, R., Dudekula, D.B., Qian, Y., VanBuren, V., Falco, G., Martin, P.R., Stagg, C.A., Bassey, U.C., et al. Transcriptome analysis of mouse stem cells and early embryos. PLoS Biol. 2003;1:410–419. doi: 10.1371/journal.pbio.0000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit,, A.F. Identification of a new, abundant superfamily of mammalian LTR-transposons. Nucleic Acids Res. 1993;21:1863–1872. doi: 10.1093/nar/21.8.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solter, D., deVries, W.N., Peaston, A., Evsikov, A., Knowles, B.B. Morphogenesis and organogenesis. In: Tamm P., Rossant J., editors. Fertilization and activation of the embryonic genome. Mouse development. Academic Press; New York: 2002. pp. 5–19. [Google Scholar]
- Solter, D., Hiiragi, T., Evsikov, A.V., Moyer, J., de Vries, W.N., Peaston, A.E., Knowles, B.B. Epigenetic mechanisms in early mammalian development. Cold Spring Harb. Symp. Quant. Biol. 2004;69:11–17. doi: 10.1101/sqb.2004.69.11. [DOI] [PubMed] [Google Scholar]
- Summers, M.C., Biggers, J.D. Chemically defined media and the culture of mammalian preimplantation embryos: Historical perspective and current issues. Hum. Reprod. Update. 2003;9:557–582. doi: 10.1093/humupd/dmg039. [DOI] [PubMed] [Google Scholar]
- Suzumori, N., Burns, K.H., Yan, W., Matzuk, M.M. RFPL4 interacts with oocyte proteins of the ubiquitin–proteasome degradation pathway. Proc. Natl. Acad. Sci. 2003;100:550–555. doi: 10.1073/pnas.0234474100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson, W.J., Vacquier, V.D. The rapid evolution of reproductive proteins. Nat. Rev. Genet. 2002;3:137–144. doi: 10.1038/nrg733. [DOI] [PubMed] [Google Scholar]
- Swanson, W.J., Yang, Z., Wolfner, M.F., Aquadro, C.F. Positive Darwinian selection drives the evolution of several female reproductive proteins in mammals. Proc. Natl. Acad. Sci. 2001;98:2509–2514. doi: 10.1073/pnas.051605998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay, J., Hodgman, R., Richter, J.D. The control of cyclin B1 mRNA translation during mouse oocyte maturation. Dev. Biol. 2000;221:1–9. doi: 10.1006/dbio.2000.9669. [DOI] [PubMed] [Google Scholar]
- Tay, J., Hodgman, R., Sarkissian, M., Richter, J.D. Regulated CPEB phosphorylation during meiotic progression suggests a mechanism for temporal control of maternal mRNA translation. Genes & Dev. 2003;17:1457–1462. doi: 10.1101/gad.1071403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong, Z.B., Gold, L., Pfeifer, K.E., Dorward, H., Lee, E., Bondy, C.A., Dean, J., Nelson, L.M. Mater, a maternal effect gene required for early embryonic development in mice. Nat. Genet. 2000a;26:267–268. doi: 10.1038/81547. [DOI] [PubMed] [Google Scholar]
- Tong, Z.B., Nelson, L.M., Dean, J. Mater encodes a maternal protein in mice with a leucine-rich repeat domain homologous to porcine ribonuclease inhibitor. Mamm. Genome. 2000b;11:281–287. doi: 10.1007/s003350010053. [DOI] [PubMed] [Google Scholar]
- von der Haar, T., Gross, J.D., Wagner, G., McCarthy, J.E. The mRNA cap-binding protein eIF4E in post-transcriptional gene expression. Nat. Struct. Mol. Biol. 2004;11:503–511. doi: 10.1038/nsmb779. [DOI] [PubMed] [Google Scholar]
- Wang, Q.T., Piotrowska, K., Ciemerych, M.A., Milenkovic, L., Scott, M.P., Davis, R.W., Zernicka-Goetz, M. A genome-wide study of gene activity reveals developmental signaling pathways in the preimplantation mouse embryo. Dev. Cell. 2004;6:133–144. doi: 10.1016/s1534-5807(03)00404-0. [DOI] [PubMed] [Google Scholar]
- White, E.K., Moore-Jarrett, T., Ruley, H.E. PUM2, a novel murine puf protein, and its consensus RNA-binding site. RNA. 2001;7:1855–1866. [PMC free article] [PubMed] [Google Scholar]
- Yan, W., Ma, L., Stein, P., Pangas, S.A., Burns, K.H., Bai, Y., Schultz, R.M., Matzuk, M.M. Mice deficient in oocyte-specific oligoadenylate synthetase-like protein OAS1D display reduced fertility. Mol. Cell. Biol. 2005;25:4615–4624. doi: 10.1128/MCB.25.11.4615-4624.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder, J.A., Walsh, C.P., Bestor, T.H. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]