GB Virus B Disrupts RIG-I Signaling by NS3/4A-Mediated Cleavage of the Adaptor Protein MAVS (original) (raw)
Abstract
Understanding the mechanisms of hepatitis C virus (HCV) pathogenesis and persistence has been hampered by the lack of small, convenient animal models. GB virus B (GBV-B) is phylogenetically the closest related virus to HCV. It causes generally acute and occasionally chronic hepatitis in small primates and is used as a surrogate model for HCV. It is not known, however, whether GBV-B has evolved strategies to circumvent host innate defenses similar to those of HCV, a property that may contribute to HCV persistence in vivo. We show here in cultured tamarin hepatocytes that GBV-B NS3/4A protease, but not a related catalytically inactive mutant, effectively blocks innate intracellular antiviral responses signaled through the RNA helicase, retinoic acid-inducible gene I (RIG-I), an essential sensor molecule that initiates host defenses against many RNA viruses, including HCV. GBV-B NS3/4A protease specifically cleaves mitochondrial antiviral signaling protein (MAVS; also known as IPS-1/Cardif/VISA) and dislodges it from mitochondria, thereby disrupting its function as a RIG-I adaptor and blocking downstream activation of both interferon regulatory factor 3 and nuclear factor kappa B. MAVS cleavage and abrogation of virus-induced interferon responses were also observed in Huh7 cells supporting autonomous replication of subgenomic GBV-B RNAs. Our data indicate that, as in the case of HCV, GBV-B has evolved to utilize its major protease to disrupt RIG-I signaling and impede innate antiviral defenses. These data provide further support for the use of GBV-B infection in small primates as an accurate surrogate model for deciphering virus-host interactions in hepacivirus pathogenesis.
Chronic hepatitis C virus (HCV) infection affects millions of people worldwide and poses a major threat to human health (42). However, efforts to understand HCV pathogenesis and identify specific HCV antivirals to supplement or substitute for current interferon (IFN)-based therapies have been impeded by the lack of a robust, fully permissive tissue culture system and the absence of small, convenient animal models of HCV infection. While the former problem has been partially solved recently by the development of systems allowing productive HCV infection in human hepatoma cells (25, 45, 49, 51), the chimpanzee remains the only well-validated animal model that is susceptible to HCV infection. Several drawbacks, however, including availability, ethical considerations, and extraordinary cost, limit use of the chimpanzee for modeling HCV infection and confirming the activity of candidate antivirals.
HCV is a positive-sense, single-stranded RNA virus that belongs to the Flaviviridae family, which comprises three genera, Flavivirus, Pestivirus, and Hepacivirus (33). GBV-B is also classified within the Flaviviridae, as a tentative species within the genus Hepacivirus, and is phylogenetically most closely related to HCV, sharing about 28% amino acid homology (1, 29). GBV-B was initially isolated from small New World monkeys (tamarins) inoculated with the serum of a surgeon who contracted hepatitis of unknown origin in the 1960s (29, 43). Although its natural host remains unknown, GBV-B is hepatotropic and causes generally acute and occasionally chronic hepatitis in tamarins and marmosets, with pathological features that are similar to those of hepatitis C in humans (1, 19, 27). In addition, NS3/4A, the major protease of GBV-B, shares substrate specificity with HCV NS3/4A in vitro (5, 37). GBV-B has thus been proposed as a surrogate model for the study of HCV pathogenesis and used for the evaluation of HCV antivirals, including protease-targeting drugs (3). However, the mechanisms by which HCV and GBV-B are able to cause chronic infections in their respective hosts are not well understood. HCV is known to circumvent both innate and adaptive immune responses, which, in turn, may facilitate viral persistence (13).
One of the most immediate responses to viral infections in mammalian cells is the rapid induction of type I IFNs (IFN-α and -β), which directly inhibit virus replication and also help orchestrate subsequent adaptive immune responses (38). This innate, early antiviral response is initiated through cellular recognition of viral products by the membrane-bound Toll-like receptors (TLRs) and/or the recently identified caspase recruitment domain (CARD)-containing cytoplasmic RNA helicases, retinoic acid-inducible gene I (RIG-I), and/or melanoma differentiation-associated gene 5 (MDA5) (39). While single-stranded viral RNA is sensed by TLR7/8 in plasmacytoid dendritic cells (7), double-stranded (ds) RNA, a replication intermediate produced during RNA virus infections, is specifically recognized by TLR3 and/or RIG-I in hepatocytes (20), the major target cell type that HCV and GBV-B infect in vivo. Upon binding to dsRNA, TLR3 and RIG-I (also MDA5) recruit their respective adaptors, Toll-interleukin-1 (IL-1) receptor homology domain containing adaptor inducing IFN-β (TRIF) and MAVS (also known as IPS-1/Cardif/VISA), that relay the signal to downstream kinases, leading to activation of IFN regulatory factor 3 (IRF-3) and nuclear factor kappa B (NF-κB), transcription factors that coordinately regulate type I IFN synthesis (17, 28, 31, 39, 40, 47, 48). HCV effectively targets two adaptor proteins, TRIF and MAVS, for proteolysis by its NS3/4A serine protease and thereby disrupts host IFN induction initiated through either the TLR3 or RIG-I pathway (21, 23, 26, 28). It is not known whether GBV-B has evolved a similar ability to counteract host defenses. However, given the similarities in the NS3/4A serine proteases of HCV and GBV-B, we sought to determine whether GBV-B NS3/4A also targets the IFN-inducing pathways for inhibition. This is an intriguing question, given that GBV-B seems to have less propensity than HCV to establish persistent infections in vivo. We focused on investigating the effect of GBV-B NS3/4A on the RIG-I signaling pathway in this study, as RIG-I has recently been shown to contribute to host responses to HCV, as well as to several flaviviruses, including Japanese encephalitis virus and Dengue virus 2 (6, 16, 44). Our results demonstrate that the GBV-B NS3/4A protease closely mimics that of HCV in that it disrupts RIG-I signaling by cleaving MAVS and dislodging it from the mitochondrial membrane, a localization essential for its signaling function.
MATERIALS AND METHODS
Cells.
Human embryo kidney (HEK) 293, HEC1B, human hepatoma Huh7, Huh7.5 (kindly provided by Charles Rice), and simian virus 40 (SV40) T antigen-immortalized tamarin hepatocyte cell lines TH1-5s and TH1-14s were cultured by conventional techniques. TH1 cells were established by procedures described briefly as follows. Primary tamarin hepatocytes were transfected with pSV3neo that expresses both large T and small T antigens from the SV40 early promoters. Cell lines were isolated by the gradual increase of G418 in the medium until distinct proliferating foci were visible, and then clonal cell lines were picked and expanded. The hepatocyte nature of the cell lines was confirmed based on expression of hepatocyte-specific proteins. UNS3-4A-24 cells, which conditionally express HCV NS3/4A (genotype 1a) under tetracycline regulation (kindly provided by Darius Moradpour), were cultured as described elsewhere (46). cB76/Huh7 cells (kindly provided by Cinzia Traboni) were derived from Huh7 cells harboring a GBV-B replicon that has been eliminated by prior IFN-α treatment (8) and proved more permissive to GBV-B RNA replication. cB76 cells were transfected with in vitro-transcribed RNA representing a dicistronic subgenomic GBV-B replicon derived from a modified molecular clone of GBV-B (27) in which five nucleotide substitutions were introduced (L. Warter et al., unpublished data). Two representative G418-resistant cell clones were selected for this study, RepGBNeo5m #15 and #28, and were cultured in the presence of 0.25 mg/ml G418. IFN-cured cells were prepared by culturing RepGBNeo5m-containing cells in the presence of 100 IU/ml of IFN-α2b and in the absence of G418 for 2 weeks. The absence of GBV-B RNA after IFN treatment was confirmed by quantitative reverse transcription-PCR (RT-PCR). HeLa Tet-Off cells were maintained as indicated by the provider (Clontech).
Plasmids.
All plasmids were generated by conventional PCR techniques except for those provided by other contributors.
To construct the tetracycline (Tet)-regulated expression plasmid for GBV-B NS3/4A, we modified the original pTRE2 plasmid (Clontech) to include a selectable marker gene that allows stable selection of transfected cells with blasticidin. The resulting plasmid, designated pTRE2Bla, was generated by inserting a fragment containing the SV40 promoter and origin-blasticidin-SV40 early polyadenylation sequence that was amplified from pcDNA6 V5-HisB (Invitrogen) using primers SV40 Xho(+) (CTTCACTCGAGTGTGTCAGTTAGGGTGTGGAAAG) and SV40 Xho pA(-) (GTAAACTCGAGGCAGTGAAAAAAATGCT) into XhoI-digested, dephosphorylated pTRE2 vector.
To construct a GBV-B NS3/4A expression plasmid, cDNA encoding GBV-B NS3-4A was amplified by PCR from a GBV-B infectious clone (27) using primers GBB NS3 Kpn (cggtaccatGGCACCTTTTACGCTGCAG; lowercase letters, introduced sequences; capital letters, viral sequences) and GBB 4A (-) (gggaatgaattattaACACTCCTCCACGATTTCTTC; lowercase letters, introduced sequences; capital letters, viral sequences). The PCR fragment was first cloned into pSTblue (Novagen) and subsequently transferred to pTRE2Bla to generate pTRE2Bla-GBpro, which expresses GBV-B NS3/4A under the Tet-regulated promoter. To generate pcDNA6-GBpro, the GBV-B NS3-4A fragment was released from pTRE2Bla-GBpro and ligated into pcDNA6-V5-HisB (Invitrogen) that was digested with NheI and BamHI. The catalytically inactive mutant of the GBV-B NS3/4A construct, pcDNA6-GBpro-S139A, was generated by QuikChange site-directed mutagenesis using pcDNA6-GBpro as a template.
To construct pcDNA6-BVDVpro and pcDNA6-YFVpro, cDNA encoding the bovine viral diarrhea virus (BVDV) NADL NS3-4A proteins was amplified by PCR from a plasmid containing a copy of the BVDV NADL genome (kindly provided by Ilya Frolov) with primers BVDV-NS3 (cgctagctctagaccatGGGGCCTGCCGTGTGTAAGAAG; lowercase letters, introduced sequences; capital letters, viral sequences) and BVDV-NS4A (tttctcgagaagcttaCAGTTCTTTCAGTTCAGTCTCTG; lowercase letters, introduced sequences; capital letters, viral sequences); cDNA encoding the Yellow Fever virus (YFV) 17D NS2B-NS3 fragment was amplified by PCR from a plasmid template containing a fragment of YFV 17D (provided by Ilya Frolov) using primers YF NS2B (gctagctctagaccatGAGTATCCCAGTGAATGAGGCAC; lowercase letters, introduced sequences; capital letters, viral sequences) and YF NS3 (gagctcgagtcgacttaCCTCCTACCTTCAGCAAACTTA; lowercase letters, introduced sequences; capital letters, viral sequences). The BVDV NS3-4A and YFV NS2B-3 fragments were initially cloned using the pTOPO TA PCR cloning kit (Invitrogen) and subsequently released with XbaI and XhoI restriction enzymes and ligated into pcDNA6/V5-HisB (Invitrogen) digested with NheI and XhoI restriction sites, to result in the plasmids, pcDNA6-BVDVpro and pcDNA6-YFVpro, respectively. To facilitate detection, we also constructed pcDNA6-BVDVpro-V5, in which BVDV NS3-4A fragment was placed in frame with a C-terminal V5 epi-tag.
The cDNA encoding MAVS/IPS-1/Cardif/VISA was amplified from Huh7 total RNA using primers MAVS NdeI Start+, cgtattcatATGCCGTTTGCTGAAGAC (lowercase letters, introduced sequences; capital letters, viral sequences), and MAVS XhoI stop-, catatctcgagttaGTGCAGACGCCGCCGGTAC (lowercase letters, introduced sequences; capital letters, viral sequences), and cloned into pCMVScript (Stratagene) to generate pCMVScript-MAVS. pEFtak-FlagIPS-1 was a gift from Michael Gale. N-terminally green fluorescent protein (GFP)-tagged MAVS (GFP-MAVS) was generated by inserting Flag-MAVS in frame with GFP into pEGFP-C1 (Clontech). We made the C283R,C508R mutants of Flag-MAVS by QuikChange site-directed mutagenesis (Stratagene). pCDNA6-FlagRIG-I was constructed by inserting an N-terminal Flag-tagged RIG-I cDNA fragment into pCDNA6-V5/HisB. pcDNA6-NS3/4A, which encodes HCV-N NS3/4A, has been described elsewhere (21). The reporter plasmids pIFN-β-Luc, pISG56-Luc, PRDII-Luc, and (PRDIII-I)4-Luc were kindly provided by Rongtuan Lin, Michael Gale, and Christina Ehrhardt, respectively. pCMVβgal (Clontech) or pRL-TK (Promega) was used to normalize transfection efficiencies. The following plasmids were also used in this study: pEF-Bos(+), pEF-Bos Flag-RIG-I, pEF-Bos Flag-RIG-I KA, pEF-Bos RIG-I C, and pEF-Bos Flag-N-RIG (gifts from Takashi Fujita); pcDNA3-FlagTBK1 and pcDNA3-FlagIKKɛ (gifts from Kate Fitzgerald); IRF-3 5D (a gift from Rongtuan Lin).
Plasmid DNAs were transfected into cells using Fugene 6 (Roche) for HEK293, Huh7, and TH1 cells or Lipofectamine 2000 (Invitrogen) for HEC1B and HeLa cells following the manufacturer's instructions.
Establishment of a HeLa cell line with conditional expression of GBV-B NS3/4A.
HeLa Tet-Off cells (Clontech) were transfected with pTRE2Bla-GBpro and double-selected in complete medium supplemented with 100 μg/ml G418, 1 μg/ml blasticidin, and 2 μg/ml tetracycline. Three weeks later, individual cell colonies were selected, expanded, and examined for GBV-B NS3 expression by indirect immunofluorescence staining using a rabbit antiserum against GBV-B NS3 (kindly provided by Bruce Malcolm) after being cultured in the absence of Tet for 4 days. A cell clone, designated HeLa GBpro-10, allowed tight regulation of GBV-B NS3/4A expression under Tet control and was selected for further characterization.
SenV infection.
Where indicated, cells were infected with 100 hemagglutinin units (HAU)/ml of Sendai virus (SenV; Cantell strain; Charles River Laboratory) for 16 h before harvest for luciferase/β-galactosidase (β-Gal) reporter assays and/or immunoblot analysis as previously described (12, 20).
In vitro RNA transcription.
Five micrograms of an infectious GBV-B cDNA clone (pGBB-2) (27) was digested overnight with XhoI in a 10-μl reaction mixture. One microgram of this linear DNA was used to program an in vitro transcription reaction by using a T7 MEGAscript kit (Ambion, Austin, TX), followed by DNase I treatment to remove the template DNA. The resulting GBV-B RNA product was extracted once with acid-phenol-chloroform and once with chloroform. Following precipitation in 2-propanol and washing in 70% ethanol, the GBV-B RNA was resuspended in nuclease- and endotoxin-free water, quantified, examined by nondenaturing agarose gel electrophoresis for quality, and stored at −70°C in small aliquots. For treatment of cells, GBV-B RNA was complexed with DMRIE-C (Invitrogen) and loaded onto cells in serum-free Opti-MEM for 4 h. The RNA-DMRIE-C complex was removed, and cells were refed with complete medium for an additional 16 h before lysis for reporter gene assay or Western blot analysis. Mock-treated cells were manipulated similarly except that the RNA was omitted during transfection.
Reporter gene assay.
Cells (5 × 104 to 105 cells per well in 24-well plates) were transfected with reporter plasmids (100 ng), pCMVβgal (100 ng), and the indicated amount of an expression vector. Where indicated, cells were mock treated, transfected with poly(I-C) or GBV-B RNA, or challenged with SenV at 24 h posttransfection and then subsequently lysed and assayed for luciferase and β-galactosidase activities. For comparisons, luciferase activity was normalized to β-galactosidase activity. In some experiments, Renilla luciferase was used as an internal control and a dual-luciferase assay was performed to calculate relative luciferase activity. Data are expressed as mean relative luciferase activity plus the standard deviation for one representative experiment carried out in triplicate, typically from a minimum of three separate experiments. The induction of promoter activity was calculated by dividing the relative luciferase activity of stimulated cells by that of mock-treated cells.
Indirect immunofluorescence staining.
Cells grown in chamber slides were fixed, permeabilized, blocked, and subsequently immunostained with GBV-B NS3 and/or IRF-3 antiserum as described elsewhere (12, 22). Cells were examined under a Zeiss 510 META confocal microscope in the UTMB Optimal Imaging Core or under a Leica Widefield ApoTome Coolsnap inverted microscope at the Plate-forme d'Imagerie Dynamique (Institut Pasteur).
Isolation of crude mitochondria.
Cells expressing or not expressing GBV-B NS3/4A were scraped, resuspended in HIM buffer (200 mM mannitol, 70 mM sucrose, 1 mM EGTA, 10 mM HEPES, pH 7.5), and homogenized by 20 strokes in a tight-fitting Dounce homogenizer. Cell lysates were centrifuged at 500 × g for 10 min to pellet nuclei and unbroken cells, and the supernatant was subjected to centrifugation at 10,000 × g for 10 min to collect crude mitochondria. The mitochondrial pellet was resuspended in HIM buffer and used immediately for immunoblot analysis or aliquoted and stored at −80°C.
Immunoblot analysis.
Cellular extracts were prepared, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to nitrocellulose membranes, and subjected to immunoblot analysis as described previously (12, 20). The following monoclonal (MAb) or polyclonal (PAb) antibodies were used: anti-FLAG M2 and antiactin MAbs (Sigma); anti-GFP MAb (Roche); anti-V5 MAb (Invitrogen); anti-HCV NS3 MAb (Vector Labs); rabbit anti-GBV-B NS3 PAb (a gift from Bruce Malcolm); rabbit anti-YFV and anti-BVDV PAbs (gifts from Charles Rice); rabbit anti-IRF-3 PAb (a gift from Michael David); rabbit anti-MAVS PAb (a gift from Zhijian Chen); anti-OxPhos complex I 39-kDa subunit (CI39) MAb (Molecular Probes); rabbit anti-ISG56 PAb (a gift from Ganes Sen); rabbit anti-ISG15 PAb (a gift from Arthur Haas); rabbit anti-Sendai virus PAb (a gift from Ilkka Julkunen); peroxidase-conjugated secondary anti-rabbit and anti-mouse PAbs (Southern Biotechnology). Protein bands were visualized using ECL Plus Western blotting detection reagents (Amersham), followed by exposure to Kodak Biomax film.
Quantitative real-time PCR.
mRNAs for IFN-β and IL-6 and18S rRNA were quantified in total cellular RNA extracts using commercially available primers and TaqMan probes (Applied Biosystems) by RT-PCR at the UTMB Sealy Center for Cancer Biology Real-Time PCR core. The relative abundance of each target was obtained by normalization with endogenous 18S RNA.
RESULTS
GBV-B NS3/4A protease blocks viral activation of the IFN-β promoter.
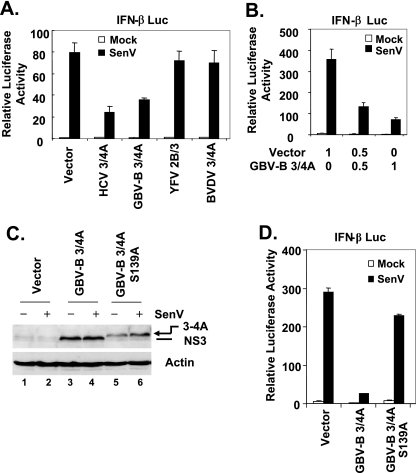
RIG-I signaling contributes to host defenses against many RNA viruses and is known to be disrupted by the HCV NS3/4A protease (11). Given the fact that the NS3/4A protease of GBV-B shares substrate specificity with that of HCV (5), we explored the effect of GBV-B NS3/4A expression on induction of IFN-β promoter activity following infection with SenV, which specifically triggers a RIG-I-dependent signaling pathway in epithelial cells, fibroblasts, and hepatocytes (20, 50). In HEK293 cells, ectopic expression of GBV-B NS3/4A, like HCV NS3/4A, significantly inhibited SenV-induced activation of the IFN-β promoter, causing a ∼60% reduction in IFN-β promoter activity when compared with empty vector-transfected cells (Fig. 1A). Inhibition of the virus-induced IFN response was specific to HCV and GBV-B NS3/4A proteases and not observed with serine proteases of other members of the Flaviviridae family, NS2B/3 of YFV, a flavivirus, and NS3/4A of BVDV, a pestivirus (35) (Fig. 1A). Therefore, the disruption of virus-induced IFN responses appears to be specific to the serine proteases of hepaciviruses, including GBV-B.
FIG. 1.
GBV-B NS3/4A protease, but not YFV NS2B/3 or BVDV NS3/4A, inhibits SenV-induced activation of the IFN-β promoter. A. HEK293 cells were cotransfected with pIFN-β-Luc and pCMVβgal plasmids and plasmids encoding HCV NS3/4A, GBV-B NS3/4A, YFV NS2B/3, or BVDV NS3/4A, or a control vector. Twenty-four hours later, cells were either mock infected (empty bars) or infected with SenV at 100 HAU/ml for 16 h (solid bars) prior to lysis for both luciferase and β-galactosidase assays. Bars show relative luciferase activity normalized to β-galactosidase activity, i.e., IFN-β promoter activity. B. IFN-β promoter activity in tamarin hepatocytes (TH1-5s) transfected with the indicated amounts (in micrograms) of GBV-B NS3/4A-expressing plasmid supplemented with a control vector, to keep the total amount of DNA transfected constant, and then mock infected or infected with SenV for 16 h. C. Immunoblot analysis of TH1-14s cells transiently transfected with plasmids encoding WT GBV-B NS3/4A or an active site mutant, S139A, and then mock infected or infected with SenV. Note that cells transfected with the S139A mutant plasmid expressed unprocessed NS3-4A precursor (arrow). D. IFN-β promoter activity in tamarin hepatocytes (TH1-14s) transfected with plasmid DNAs expressing GBV-B NS3/4A, the S139A mutant GBV-B NS3/4A, or with a control vector and mock infected or infected with SenV for 16 h.
As GBV-B infects small New World monkeys, such as tamarins and marmosets, but not chimpanzees (4, 19), we sought to determine whether the inhibition of the virus-induced IFN response that we observed in HEK293 cells also occurred in tamarin hepatocytes expressing the GBV-B protease. This is important, as the tamarin homolog of the human substrate putatively cleaved by NS3/4A in HEK293 cells might differ in its ability to be cleaved by the protease. We thus ectopically expressed the protease in two T-antigen-immortalized tamarin hepatocyte cell lines (TH1-5s and -14s) and challenged these cells with SenV. These results confirmed that GBV-B NS3/4A blocks viral induction of the IFN-β promoter in a dose-dependent manner in tamarin cells (Fig. 1B; results in TH1-5S are shown). Furthermore, we observed a similar inhibitory effect of GBV-B NS3/4A on viral activation of the interferon-stimulated gene 56 (ISG56) promoter, a well-characterized IRF-3 target (14), in TH1-5s cells (data not shown).
Like the HCV serine protease, GBV-B NS3/4A is a bifunctional molecule that contains an N-terminal serine protease activity (along with the NS4A cofactor) and a C-terminal RNA helicase domain (36, 52). To investigate whether the protease activity mediated the IFN blockade, as in HCV, we introduced a point mutation (S139A) within the active site of the GBV-B NS3 serine protease and determined the ability of this mutant NS3/4A to block SenV-induced IFN responses. The S139A mutant GBV-B NS3/4A was catalytically inactive, as it was expressed as an unprocessed NS3-4A precursor in transfected cells due to the inability to mediate the _cis_-cleavage between NS3 and NS4A (Fig. 1C, lanes 5 and 6). We found that the S139A mutant GBV-B NS3/4A lost the ability to inhibit SenV-induced up-regulation of the IFN-β promoter (Fig. 1D), indicating that GBV-B NS3/4A blocks virus-induced IFN responses by cleaving a cellular substrate(s).
GBV-B NS3/4A protease inhibits virus-induced activation of both IRF-3 and NF-κB through the RIG-I signaling pathway.
Transcriptional induction of the IFN-β promoter requires coordinate activation of IRF-3 and NF-κB transcription factors that specifically bind to the PRDIII-I and PRDII elements of the promoter, respectively (38). To determine the mechanism by which GBV-B NS3/4A inhibits viral activation of the IFN-β promoter, we investigated the effect of GBV-B NS3/4A on SenV-induced activation of synthetic promoters driven by the IRF-3-dependent PRDIII-I and NF-κB-dependent PRDII elements of the human IFN-β promoter, respectively. We found that the expression of wild-type (WT) GBV-B NS3/4A, but not the S139A mutant, effectively blocked SenV-induced activation of both PRDIII-I (Fig. 2A, left panel) and PRDII (Fig. 2A, right panel) promoters in tamarin hepatocytes (TH1-14s cells). Activation of both PRDIII-I and PRDII promoters was also significantly reduced when ectopic expression of the constitutive active CARD of RIG-I (N-RIG) was used in place of SenV infection to trigger the signaling (Fig. 2B). These results confirm that GBV-B NS3/4A, but not the related inactive S139A mutant, inhibits RIG-I signaling to both IRF-3 and NF-κB. The NS3/4A protease of GBV-B thus disrupts vial activation of the IFN-β promoter in a fashion similar to NS3/4A of HCV (12).
FIG. 2.
GBV-B NS3/4A protease disrupts virus or constitutive active RIG-I CARD (N-RIG)-induced activation of both IRF-3 and NF-κB. A. TH1-14s cells were cotransfected with plasmids encoding luciferase under the control of either an IRF-3-dependent PRDIII-I promoter (four-time repeat of the PRDIII-I element, left) or an NF-κB-dependent PRDII promoter (right), pCMVβgal, and plasmids encoding HCV NS3/4A, GBV-B NS3/4A, or GBV-B NS3/4A S139A or control vector. Twenty-four hours later, cells were either mock infected or infected with SenV at 100 HAU/ml for 16 h prior to lysis for both luciferase and β-galactosidase assays. B. Constitutively active RIG-I CARD (N-RIG)-induced PRDIII-I (left) and PRDII (right) promoter activities in HEC1B cells cotransfected similarly to those represented in panel A. In these experiments, cotransfection of an N-RIG-encoding plasmid or empty vector was substituted for SenV infection or mock infection, respectively, and cells were harvested at 24 h posttransfection.
To better characterize the mechanisms of GBV-B NS3/4A blockade on RIG-I signaling, we developed a Tet-regulated HeLa cell line, designated GBpro-10, which conditionally expresses GBV-B NS3/4A under the control of the Tet-Off promoter. In GBpro-10 cells, expression of GBV-B NS3/4A was tightly regulated by Tet and only occurred when Tet was removed from the culture medium (Fig. 3B, GBV-B NS3 panel, compare lanes 1 and 2 with lanes 3 and 4). Activation of IRF-3 requires its specific C-terminal phosphorylation and is accompanied by its nuclear translocation (15)—these events are blocked in cells expressing HCV NS3/4A (12). Similarly, we found that induction of GBV-B NS3/4A expression in GBpro-10 cells blocked SenV-induced IRF-3 phosphorylation (Fig. 3B, IRF-3 panel, compare lanes 2 and 4) and nuclear translocation (Fig. 3A), as well as upregulation of ISG56 (Fig. 3B, ISG56 panel, compare lanes 2 and 4).
FIG. 3.
GBV-B NS3/4A disrupts the endogenous IFN response to virus infection in HeLa GBpro-10 cells with tetracycline-regulated, conditional expression of GBV-B NS3/4A. A. Confocal microscopy of IRF-3 subcellular localization in HeLa GBpro-10 cells repressed (+tet) or induced (-tet) for GBV-B NS3/4A expression and mock infected (left) or infected with SenV (right) for 16 h. Nuclei were counterstained with 4′,6′-diamidino-2- phenylindole. B. HeLa GBpro-10 cells were cultured to repress or induce GBV-B NS3/4A expression for 3 days, followed by mock infection or infection with 100 HAU/ml of SenV for 16 h prior to immunoblot analysis of whole-cell extracts for IRF-3, ISG56, SenV, or GBV-B NS3. The arrow in the IRF-3 panel indicates the hyperphosphorylated form of IRF-3 (IRF3-P). A nonspecific band (*) detected by anti-ISG56 antiserum indicates equal loading. C. Real-time RT-PCR analysis of IFN-β (left) and IL-6 (right) mRNA transcripts in HeLa GBpro-10 cells repressed or induced for GBV-B NS3/4A expression and mock infected or infected with SenV. mRNA abundance was normalized to cellular 18S rRNA.
To further validate our findings, we conducted real-time PCR monitoring of IFN-β mRNA in SenV-challenged GBpro-10 cells, induced or repressed for GBV-B NS3/4A expression. As shown in Fig. 3C, SenV-induced up-regulation of IFN-β was almost ablated when GBV-B NS3/4A was induced (Fig. 3C, left panel). In addition, SenV-induced expression of IL-6, an NF-κB-dependent cytokine (34), was also ablated in GBV-B NS3/4A-expressing cells (Fig. 3C, right panel). Therefore, GBV-B NS3/4A, like the HCV protease, targets a proximal point in the RIG-I signaling pathway, prior to bifurcation towards IRF-3 and NF-κB.
GBV-B NS3/4A protease specifically targets the RIG-I adaptor protein, MAVS/IPS-1/Cardif/VISA, for proteolysis.
To explore the mechanism by which GBV-B NS3/4A protease disrupts RIG-I signaling, we determined whether known signaling proteins in the RIG-I pathway were substrates for GBV-B NS3/4A. The noncanonical IκB kinases, TBK1 and IKKɛ, mediate virus-activated IRF-3 phosphorylation (10, 41). Although the underlying mechanisms remain to be determined, these kinases are linked to the RIG-I dsRNA sensor by an adaptor protein, MAVS (also known as IPS-1, Cardif, or VISA), which was recently shown to be a substrate for the HCV NS3/4A protease (23, 24, 26, 28, 41). We found that RIG-I, TBK1, and IKKɛ were not cleaved by GBV-B NS3/4A when ectopically expressed in GBpro-10 cells (data not shown). In contrast, the two MAVS-immunoreactive species (with apparent molecular masses of approximately 75 and 60 kDa, respectively) were cleaved at a single site by GBV-B NS3/4A in GBpro-10 cells, with a pattern similar to that observed in UNS3-4A-24 cells expressing HCV NS3/4A (Fig. 4A). The 60-kDa MAVS species is likely an internally initiated translation product that shares a C terminus with the full-length MAVS, as it was detected only by MAVS antiserum and not with an anti-Flag antibody in immunblot assays when expressed from an N-terminal Flag-tagged MAVS vector (data not shown).
FIG. 4.
GBV-B NS3/4A protease targets MAVS/IPS-1/Cardif/VISA for proteolysis. A. Immunoblot analysis of endogenous MAVS in UNS3-4A-24 cells (left) and HeLa GBpro-10 cells (right) with (-tet) or without (+tet) induction of HCV and GBV-B NS3/4A, respectively. Open circles indicate intact MAVS, while solid circles indicate cleaved MAVS. B. TH1-14s cells were mock transfected (lane 4), transfected with a plasmid encoding MAVS (lane 1), or cotransfected with plasmids encoding MAVS and either GBV-B NS3/4A (lane 2) or S139A mutant GBV-B NS3/4A (lane 3) for 24 h prior to harvest for immunoblot analysis of whole-cell extracts with MAVS antiserum (upper panel) or GBV-B NS3 antibodies (lower panel). C. HeLa GBpro-10 cells induced or repressed for GBV-B NS3/4A expression were transfected with WT or the indicated mutants of N-terminally Flag-tagged MAVS. Forty-eight hours later, MAVS protein was detected with anti-Flag antibody by immunoblot analysis of whole-cell extracts. D. Activation of the IFN-β promoter by ectopic expression of IKKɛ, IRF-3 5D, or WT or C508R mutant MAVS in HEK293 cells in the absence (empty bars) or presence (solid bars) of ectopic coexpression of GBV-B NS3/4A.
As the anti-human MAVS antiserum did not detect the tamarin MAVS homolog (Fig. 4B, lane 4) in whole-cell exacts, we tried several approaches to obtain specific evidence of cleavage of tamarin MAVS by GBV-B NS3/4A. First, we tried to fractionate tamarin hepatocytes with ectopic expression of WT or S139A mutant GBV-B protease into mitochondrial and cytosolic fractions, as MAVS is known to be enriched in mitochondria while redistributed into cytosolic fractions from cells expressing HCV NS3/4A protease (23, 24, 26, 40). However, we were unable to detect tamarin MAVS in these fractionated cellular extracts with the anti-human MAVS antiserum, irrespective of expression of the GBV-B protease (data not shown). We were also unsuccessful in multiple attempts at PCR cloning of tamarin MAVS using degenerate primers based on an alignment of the nucleotide sequences of known MAVS proteins (human, chimpanzee, rhesus monkey, mouse, and rat). Thus, direct evidence of cleavage of the tamarin homolog of MAVS is lacking. However, the GBV-B NS3/4A protease, but not its catalytically inactive S139A mutant, was capable of cleaving human MAVS when ectopically expressed in tamarin hepatocytes (Fig. 4B, compare lanes 2 and 3). More importantly, it was capable of disrupting interferon signaling through the endogenous protein in these cells (Fig. 1B and 2A).
Both HCV and GBV-B NS3/4A cleave their respective viral polyproteins in trans at Cys/Ser or Cys/Ala dipeptides, although either protease can tolerate some amino acid substitutions at the P1 (Ser or Ala) position (35). HCV NS3/4A cleaves MAVS between Cys508 and His509, and the cleavage is blocked by an Arg substitution at Cys508 (23, 26, 28). To determine whether GBV-B NS3/4A also cleaves human MAVS at Cys508, we transfected expression vectors encoding WT or mutant forms of MAVS with an Arg substitution at Cys283 or Cys508 into GBpro-10 cells that were repressed or induced for GBV-B NS3/4A expression. We found that the C508R mutant MAVS was no longer cleavable by GBV-B NS3/4A, whereas the C283R mutant MAVS was still cleaved, in a manner similar to that of WT MAVS (Fig. 4C). Therefore, GBV-B NS3/4A also cleaves human MAVS at Cys508.
Consistent with the fact that cleavage of MAVS by GBV-B NS3/4A is responsible for the inhibition of RIG-I signaling, the GBV-B NS3/4A blockade of RIG-I signaling occurs downstream of RIG-I (Fig. 2B) and upstream of IKKɛ and IRF-3 (Fig. 4D) and TBK1 (data not shown). Furthermore, the protease-resistant C508R mutant MAVS, but not its WT counterpart, induced a similar level of IFN-β promoter activation in GBV-B NS3/4A-expressing cells and nonexpressing cells (Fig. 4D).
Cleavage of MAVS by GBV-B NS3/4A causes its redistribution from mitochondrial membrane to cytosol.
MAVS contains a hydrophobic transmembrane domain (TM) near its C terminus (aa 514 to 535) that anchors it to the mitochondrial outer membrane. Although poorly understood, the mitochondrial localization is essential for the function of MAVS (40). As GBV-B NS3/4A cleaves MAVS at a site located immediately upstream of its TM, we sought to determine whether GBV-B NS3/4A protease cleavage releases MAVS from the mitochondrial outer membrane as expected. To visualize MAVS, we ectopically expressed an N-terminally GFP-tagged MAVS (GFP-MAVS) in HeLa GBpro-10 cells that were induced or repressed for GBV-B NS3/4A expression. We stained mitochondria with Mitotracker Red, a specific mitochondria dye, and examined the subcellular localization of GFP-MAVS by confocal microscopy. As shown in Fig. 5A, GFP-MAVS almost exclusively colocalized with Mitotracker Red in cells that do not express GBV-B NS3-4A, with a punctuate pattern predominantly in the perinuclear region. In contrast, GFP-MAVS mitochondrial localization was no longer observed in cells induced for GBV-B NS3/4A expression. Instead, GFP-MAVS fluorescence became diffuse and distributed evenly in the cytoplasm. Parallel immunoblot analysis confirmed that GFP-MAVS was cleaved in cells induced to express GBV-B NS3/4A (Fig. 5B). To confirm that the endogenous MAVS was cleaved off mitochondria, we isolated crude mitochondrial fractions from GBpro-10 cells induced or repressed for GBV-B NS3/4A expression. As shown in Fig. 5C, the MAVS-immunoreactive products were present abundantly in the mitochondrial fraction in cells without GBV-NS3/4A expression. However, they were absent in mitochondrial fractions isolated from cells induced to express GBV-B NS3/4A. In contrast, similar amounts of CI39, a mitochondrial complex I 39-kDa protein, were present in mitochondrial fractions of GBpro-10 cells, regardless of GBV-B NS3/4A expression status, demonstrating the integrity of our mitochondria isolation. Therefore, as in the case of the HCV serine protease (23, 26), GBV-B NS3/4A also cleaves MAVS off mitochondria and dislodges it to the cytosol, where it fails to signal.
FIG. 5.
Cleavage of MAVS/IPS-1/Cardif/VISA by GBV-B NS3/4A causes its redistribution from mitochondria to the cytosol. A. HeLa GBpro-10 cells induced or repressed for GBV-B NS3/4A expression were transfected with a vector encoding N-terminally GFP-tagged MAVS (GFP-MAVS) and analyzed by confocal microscopy 24 h later. Mitochondria were stained with Mitotracker Red. B. Immunoblot analysis of GFP-MAVS (using GFP antibody) and GBV-B NS3 in whole-cell extracts of HeLa GBpro-10 cells treated as for panel A. Open circles indicate intact GFP-MAVS, while solid circles indicate cleaved GFP-MAVS. C. Immunoblot analysis of MAVS (using anti-MAVS antiserum), a mitochondrial inner membrane protein, complex I 39-kDa subunit (CI39), and GBV-B NS3 in crude mitochondrial fractions isolated from HeLa GBpro-10 cells induced (-tet) or repressed (+tet) for GBV-B NS3/4A expression.
MAVS is cleaved in cells actively replicating GBV-B subgenomic replicons.
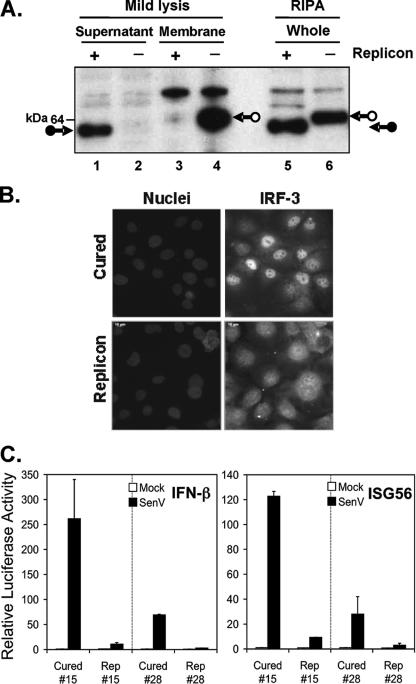
To determine whether MAVS is cleaved during GBV-B viral replication, we conducted immunoblot analysis of endogenous MAVS in Huh7 cells supporting the autonomous replication of GBV-B subgenomic RNAs that express GBV-B nonstructural proteins NS3 through NS5B as a membrane-bound replication complex. In parallel, MAVS status was examined in IFN-cured counterparts of these cell lines. We found that the MAVS-immunoreactive product present in whole-cell extracts (lysed using RIPA buffer) from replicon-bearing cells migrated with an apparently lower molecular mass than that from IFN-cured cells, consistent with MAVS cleavage by GBV-B NS3/4A (Fig. 6A, compare lanes 5 and 6). In addition, under mild lysis conditions using the reporter lysis buffer (Promega), full-length MAVS was only present in cured cells and was recovered from the cell pellet (which contains membrane-bound proteins) (Fig. 6A, compare lanes 4 and 3), whereas cleaved MAVS was only present in replicon-bearing cells and was recovered from the lysate supernatant (which contains cytosolic proteins) (Fig. 6A, compare lanes 1 and 2). Consistent with the fact that MAVS was cleaved in GBV-B replicon cells, SenV-induced IRF-3 nuclear translocation (Fig. 6B) and activation of IFN-β and ISG56 promoters (Fig. 6C) were blocked in two individually selected GBV-B replicon cell lines, while they were restored in their IFN-cured counterparts.
FIG. 6.
The virus-induced IFN response is abrogated in cells supporting the autonomous replication of GBV-B subgenomic RNAs due to MAVS cleavage. A. MAVS is cleaved in cells containing GBV-B replicons. In IFN-cured cells (− replicon lanes), an intact MAVS-immunoreactive product (arrow with open circle) was found in whole-cell extracts prepared in RIPA buffer, as well as in membranes collected after mild lysis using reporter lysis buffer. In cells containing the GBV-B replicon RNA (RepGBNeo5m#15), a protein with a slightly lower molecular mass (arrow with solid circle) was found in RIPA-lysed extracts, as well as in the supernatant from mildly lysed cells, consistent with NS3/4A cleavage and redistribution of MAVS from mitochondria to cytosol. B. IRF-3 subcellular localization monitored 16 h after SenV infection by immunofluorescence was shown to be predominantly nuclear in cured cells but mostly distributed throughout the cytoplasm in cells containing GBV-B replicon (RepGBNeo5m#15) or in noninfected cells (not shown). Nuclei were counterstained with 4′6′-diamidino-2-phenylindole (DAPI). C. cB76 cell lines containing GBV-B replicon RepGBNeo5m or corresponding IFN-cured cells were cotransfected with pIFN-β-Luc (left) or pISG56-Luc (right) and pCMV-βGal plasmids. Twenty-four hours later, cells were mock infected (empty bars) or infected with SenV at 100 HAU/ml (solid bars) for 16 h prior to lysis for a reporter assay of promoter activities.
RIG-I mediates host IFN response to GBV-B RNA.
Previous data indicate that RIG-I is an essential cytoplasmic sensor for recognition of HCV RNA and subsequent initiation of IFN responses in hepatoma cells (44). Given the similarities between GBV-B and HCV and the fact that GBV-B NS3/4A protease cleaves the RIG-I adaptor MAVS, we sought to determine whether GBV-B RNA also triggers host IFN response through RIG-I. In vector-transfected Huh7 cells, introduction of GBV-B genomic RNA activated the ISG56 promoter by twofold, similar to the level of induction by transfected poly(I-C), which activates the RIG-I pathway in these cells (44). This induction, however, was ablated in cells expressing a dominant negative mutant form of RIG-I, RIG-I C, which encodes the helicase domain of RIG-I (50) (Fig. 7A). In contrast, ectopic expression of WT RIG-I weakly activated the basal level of the ISG56 promoter and strongly potentiated its induction in response to transfected GBV-B RNA or poly(I-C) (Fig. 7A). To further confirm that RIG-I mediates the host response to GBV-B RNA, we ectopically expressed WT RIG-I or the RIG-I KA mutant (which contains an inactivating point mutation at the Walker-type ATP binding site) (50) in Huh7.5 cells and determined the induction of ISG56 expression following transfection of GBV-B RNA or, as a control, SenV challenge (Fig. 7B). Huh7.5 cells were selected for these experiments as they express a T55I mutant RIG-I and, as a result, respond to SenV or intracellular dsRNA only when a functional RIG-I molecule is reconstituted (44). As shown in Fig. 7B, both GBV-B RNA and SenV failed to induce ISG56 expression in Huh7.5 cells transfected with an empty vector (compare lanes 2 and 3 with lane 1). Ectopic expression of WT RIG-I (lanes 4 through 6), but not the KA mutant (lanes 7 through 9), restored ISG56 induction in response to GBV-B RNA stimulation or SenV infection (compare lanes 5 and 6 with lane 4 and lanes 8 and 9 with lane 7, respectively). Taken together, these data strongly suggest that RIG-I plays an essential role in the cellular IFN response to GBV-B RNA.
FIG. 7.
RIG-I is an intracellular sensor of GBV-B RNA. A. Huh7 cells in 48-well plates were cotransfected with ISG56-Luc and pCMVβgal (50 ng of each), along with 200 ng of vector, RIG-I, or RIG-I C. At 24 h later, cells were mock transfected (DMRIE-C only) or transfected with 2 μg of in vitro-transcribed GBV-B genomic RNA (GBB RNA) or poly(I-C) (pIC) for 20 h before harvest for the reporter gene assay. Relative luciferase activity represents ISG56 promoter activity. B. Huh7.5 cells grown in six-well plates were transfected with 2 μg of the empty vector (lanes 1 through 3), a Flag-tagged WT RIG-I (lanes 4 through 6), or a Flag-tagged mutant RIG-I (KA; lanes 7 through 9). At 48 h posttransfection, cells were mock treated (lanes 1, 4, and 7) or transfected with GBV-B RNA (lanes 2, 5, and 8) or infected with Sendai virus for 20 h (lanes 3, 6, and 9) before cell lysis for immunoblot analysis of ISG56, RIG-I (using an anti-Flag antibody), Sendai virus, and actin (loading control).
DISCUSSION
RIG-I is one of the major signaling pathways for activation of innate intracellular antiviral defenses against a variety of negative- and positive-strand RNA viruses, including several other flaviviruses and HCV (6, 16, 44, 50). We have expanded previous observations in this study by showing that RIG-I also contributes to innate immune recognition of GBV-B genomic RNA (Fig. 7). Furthermore, we have demonstrated that GBV-B has evolved to counteract this innate defense mechanism. GBV-B NS3/4A proteolytically cleaves human MAVS/IPS-1/Cardif/VISA, the RIG-I adaptor protein, and dislodges it from the mitochondrial outer membrane, a localization that is essential for MAVS function (40). Importantly, we demonstrated this upon both transient and inducible expression of GBV-B NS3/4A as an isolated protein in cells of human origin, in hepatocyte cell lines derived from tamarins (Fig. 4 and 5), and in human hepatoma cells, supporting the autonomous replication of a GBV-B subgenomic replicon that expresses GBV-B NS3/4A in the context of a membrane-bound replication complex (Fig. 6). GBV-B thus closely mimics HCV (12, 23, 26) in interfering with RIG-I signaling. This suggests in turn that this interference with host innate antiviral responses may play an important role during the establishment of both HCV and GBV-B infections. These data thus provide additional, strong support for the use of the GBV-B model as a surrogate for HCV.
This conclusion is further substantiated by the fact that the ability to cleave MAVS appears specific for GBV-B and HCV NS3/4A proteases, as serine proteases from a flavivirus, YFV (NS2B/3), and a pestivirus, BVDV (NS3/4A), do not inhibit RIG-I signaling (Fig. 1A). This is consistent with the differences between natural substrate specificities of flavivirus and pestivirus serine proteases and that of HCV and GBV-B NS3/4A proteases. While HCV and GBV-B NS3/4A cleave at Cys/Ser, Ala, or Gly dipeptides, serine proteases from pestiviruses and flaviviruses target distinct sequences, namely, Leu/Ser or Ala and Arg/Ser or Gly, respectively (35). Furthermore, several residues flanking the HCV and GBV-B NS3/4A cleavage site in human MAVS that may contribute to substrate recognition (e.g., at the P6 position) are identical to those found around cleavage sites in GBV-B and HCV polyproteins (Fig. 8), but not in flavivirus and pestivirus polyproteins (not shown). This further validates the value of GBV-B as a surrogate model for HCV over other viruses of the Flaviviridae family, as these two viruses appear to have adopted similar strategies to disrupt host antiviral defenses.
FIG. 8.
Alignment of peptide junctions present in viral polyproteins of GBV-B and HCV and cleaved in trans by their respective serine protease with junctions present within MAVS from various species that are cleaved (or predicted to be cleaved) by GBV-B NS3/4A. Residues that flank MAVS cleavage sites by GBV-B NS3/4A and align with natural viral substrates are underlined. Note that the MAVS proteins from various species are different in length. Therefore the positions of (predicted) GBV-B NS3/4A cleavage sites are different.
GBV-B replicates in small New World primates such as tamarins and marmosets (19). However, as sequence information about a tamarin/marmoset homolog of human MAVS is not available and our attempts to detect and clone tamarin MAVS were unsuccessful, we could not directly evaluate the susceptibility of the tamarin or marmoset MAVS homologs to GBV-B NS3/4A cleavage. Nonetheless, given that GBV-B NS3/4A cleaves human MAVS and targets a proximal point in the RIG-I signaling pathway in tamarin hepatocytes prior to the bifurcation towards IRF-3 and NF-κB (Fig. 2 and 4), it is reasonable to postulate that the tamarin homolog of MAVS is cleaved by GBV-B NS3/4A protease. This is also consistent with the conservation of the NS3/4A cleavage site in MAVS proteins derived from human, chimpanzee, rhesus monkey, mouse, and rat sources (Fig. 8), despite the MAVS proteins being of different lengths across these species. Future molecular cloning of tamarin MAVS would help to confirm this hypothesis as well as to allow us to better understand the cleavage kinetics of MAVS by GBV-B NS3/4A.
Given these newly discovered properties of HCV and GBV-B NS3/4A proteases in counteracting host antiviral defenses, small-molecule inhibitors of NS3/4A may theoretically possess dual efficacy in treating HCV and GBV-B infections. They may both impede viral polyprotein processing essential for viral replication and also relieve the viral blockade of host innate immune responses (12). This could explain the striking potency of HCV protease inhibitors in recent clinical trials (18). In addition, such a dual inhibition mechanism may also be the reason a _trans_-lactam HCV protease inhibitor, GW0014X, demonstrated potent antiviral activity in a GBV-B-infected marmoset model (3). Further in vivo investigations in GBV-B-infected tamarins or marmosets would be helpful to validate the dual action of protease inhibitors.
The ability of HCV to inactivate critical, early innate antiviral defenses via its NS3/4A serine protease has been postulated to contribute to its persistence in the infected host. Despite the fact that GBV-B has evolved a strategy similar to that employed by HCV, resulting in the disruption of RIG-I signaling, there are important differences in the outcomes of HCV and GBV-B infections. Although GBV-B is capable of causing persistent infections (27, 30), this is a rare event compared to the high frequency of chronic HCV infections in chimpanzees and humans (1). While it remains speculative whether disruption of RIG-I signaling through cleavage of MAVS contributes significantly to HCV persistence, it is interesting to see this phenomenon mirrored in GBV-B, a virus that rarely causes chronic infections. However, it is important to recognize that the efficiency and kinetics of MAVS cleavage by the NS3/4A proteases of these two viruses may differ in vivo. The HCV protease also cleaves TRIF and suppresses TLR3 signaling (21), and this might also contribute to the extraordinary ability of HCV to counteract host defenses and establish persistence. Given the similarities in the substrate specificities of the NS3/4A proteases of HCV and GBV-B (Fig. 8), it will be interesting in future studies to determine whether GBV-B NS3/4A also cleaves TRIF.
At present, we do not know why GBV-B infections in tamarins are associated with more robust viral replication and higher-titer viremia (often 108 to 109 genome equivalents/ml) than HCV infections in chimpanzees and humans. It is possible that this robust replication of GBV-B predisposes to the triggering of strong adaptive immune responses, such as cytotoxic T cells, which are important for HCV clearance (2) and are implicated in the control of GBV-B by the fact that persistent infections can be induced by T-cell immunosuppression (19). The results obtained with GBV-B in this study, and with other viruses, e.g., rhinovirus 14 (32) and hepatitis A virus (9), which do inhibit cellular IFN responses in vitro yet do not persist in humans, suggest that mechanisms other than MAVS cleavage are likely to contribute to HCV persistence in vivo and may be modulated differently in the course of GBV-B and HCV infections.
Acknowledgments
This work was supported by NIH grants R21-DA018054 (K.L.), U19-AI40035 and R24-RR15081 (S.M.L.), RO1-AA012863 (S.A.W.), and RO1-AI049574 (R.E.L.), a grant from the French National Agency for Research on AIDS and Viral Hepatitis, ANRS (A.M.), and the John Mitchell Hemophilia of Georgia Liver Scholar Award from the American Liver Foundation (K.L.). K.L. is a Cain Foundation Investigator in Innate Immunity.
We thank Michael Gale, Takashi Fujita, Ilya Frolov, Katherine Fitzgerald, Christina Ehrhardt, and Rongtuan Lin for providing plasmids, Bruce Malcolm, Zhijian Chen, Charles Rice, Ganes Sen, Ilkka Julkunen, Michael David, and Arthur Haas for providing antibodies, Charles Rice for providing the Huh7.5 cells, Darius Moradpour for providing the UNS3-4A-24 cells, and Cinzia Traboni, IRBM, Italy, for providing the parental cB76/Huh7 cells. We thank Lisette Cohen for helpful discussions, Mardelle Susman for critical review of the manuscript, the UTMB Optimal Imaging Core and the Plate-Forme d'Imagerie Dynamique for assistance with confocal microscopy, and the UTMB Sealy Center for Cancer Biology Real Time PCR core for assistance with real-time RT-PCR assays.
Footnotes
▿
Published ahead of print on 8 November 2006.
REFERENCES
- 1.Beames, B., D. Chavez, and R. E. Lanford. 2001. GB virus B as a model for hepatitis C virus. ILAR J. 42**:**152-160. [DOI] [PubMed] [Google Scholar]
- 2.Bowen, D. G., and C. M. Walker. 2005. Adaptive immune responses in acute and chronic hepatitis C virus infection. Nature 436**:**946-952. [DOI] [PubMed] [Google Scholar]
- 3.Bright, H., A. R. Carroll, P. A. Watts, and R. J. Fenton. 2004. Development of a GB virus B marmoset model and its validation with a novel series of hepatitis C virus NS3 protease inhibitors. J. Virol. 78**:**2062-2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bukh, J., C. L. Apgar, S. Govindarajan, and R. H. Purcell. 2001. Host range studies of GB virus-B hepatitis agent, the closest relative of hepatitis C virus, in New World monkeys and chimpanzees. J. Med. Virol. 65**:**694-697. [DOI] [PubMed] [Google Scholar]
- 5.Butkiewicz, N., N. Yao, W. Zhong, J. Wright-Minogue, P. Ingravallo, R. Zhang, J. Durkin, D. N. Standring, B. M. Baroudy, D. V. Sangar, S. M. Lemon, J. Y. Lau, and Z. Hong. 2000. Virus-specific cofactor requirement and chimeric hepatitis C virus/GB virus B nonstructural protein 3. J. Virol. 74**:**4291-4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, T. H., C. L. Liao, and Y. L. Lin. 2006. Flavivirus induces interferon-beta gene expression through a pathway involving RIG-I-dependent IRF-3 and PI3K-dependent NF-κB activation. Microbes Infect. 8**:**157-171. [DOI] [PubMed] [Google Scholar]
- 7.Crozat, K., and B. Beutler. 2004. TLR7: a new sensor of viral infection. Proc. Natl. Acad. Sci. USA 101**:**6835-6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Tomassi, A., M. Pizzuti, R. Graziani, A. Sbardellati, S. Altamura, G. Paonessa, and C. Traboni. 2002. Cell clones selected from the Huh7 human hepatoma cell line support efficient replication of a subgenomic GB virus B replicon. J. Virol. 76**:**7736-7746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fensterl, V., D. Grotheer, I. Berk, S. Schlemminger, A. Vallbracht, and A. Dotzauer. 2005. Hepatitis A virus suppresses RIG-I-mediated IRF-3 activation to block induction of beta interferon. J. Virol. 79**:**10968-10977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fitzgerald, K. A., S. M. McWhirter, K. L. Faia, D. C. Rowe, E. Latz, D. T. Golenbock, A. J. Coyle, S. M. Liao, and T. Maniatis. 2003. IKKɛ and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4**:**491-496. [DOI] [PubMed] [Google Scholar]
- 11.Foy, E., K. Li, R. Sumpter, Jr., Y. M. Loo, C. L. Johnson, C. Wang, P. M. Fish, M. Yoneyama, T. Fujita, S. M. Lemon, and M. Gale, Jr. 2005. Control of antiviral defenses through hepatitis C virus disruption of retinoic acid-inducible gene-I signaling. Proc. Natl. Acad. Sci. USA. 102**:**2986-2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foy, E., K. Li, C. Wang, R. Sumpter, Jr., M. Ikeda, S. M. Lemon, and M. Gale, Jr. 2003. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science 300**:**1145-1148. [DOI] [PubMed] [Google Scholar]
- 13.Gale, M., Jr., and E. M. Foy. 2005. Evasion of intracellular host defence by hepatitis C virus. Nature 436**:**939-945. [DOI] [PubMed] [Google Scholar]
- 14.Grandvaux, N., M. J. Servant, B. tenOever, G. C. Sen, S. Balachandran, G. N. Barber, R. Lin, and J. Hiscott. 2002. Transcriptional profiling of interferon regulatory factor 3 target genes: direct involvement in the regulation of interferon-stimulated genes. J. Virol. 76**:**5532-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hiscott, J., N. Grandvaux, S. Sharma, B. R. Tenoever, M. J. Servant, and R. Lin. 2003. Convergence of the NF-κB and interferon signaling pathways in the regulation of antiviral defense and apoptosis. Ann. N. Y. Acad. Sci. 1010**:**237-248. [DOI] [PubMed] [Google Scholar]
- 16.Kato, H., O. Takeuchi, S. Sato, M. Yoneyama, M. Yamamoto, K. Matsui, S. Uematsu, A. Jung, T. Kawai, K. J. Ishii, O. Yamaguchi, K. Otsu, T. Tsujimura, C. S. Koh, E. S. C. Reis, Y. Matsuura, T. Fujita, and S. Akira. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441**:**101-105. [DOI] [PubMed] [Google Scholar]
- 17.Kawai, T., K. Takahashi, S. Sato, C. Coban, H. Kumar, H. Kato, K. J. Ishii, O. Takeuchi, and S. Akira. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6**:**981-988. [DOI] [PubMed] [Google Scholar]
- 18.Lamarre, D., P. C. Anderson, M. Bailey, P. Beaulieu, G. Bolger, P. Bonneau, M. Bos, D. R. Cameron, M. Cartier, M. G. Cordingley, A. M. Faucher, N. Goudreau, S. H. Kawai, G. Kukolj, L. Lagace, S. R. LaPlante, H. Narjes, M. A. Poupart, J. Rancourt, R. E. Sentjens, R. St George, B. Simoneau, G. Steinmann, D. Thibeault, Y. S. Tsantrizos, S. M. Weldon, C. L. Yong, and M. Llinas-Brunet. 2003. An NS3 protease inhibitor with antiviral effects in humans infected with hepatitis C virus. Nature 426**:**186-189. [DOI] [PubMed] [Google Scholar]
- 19.Lanford, R. E., D. Chavez, L. Notvall, and K. M. Brasky. 2003. Comparison of tamarins and marmosets as hosts for GBV-B infections and the effect of immunosuppression on duration of viremia. Virology 311**:**72-80. [DOI] [PubMed] [Google Scholar]
- 20.Li, K., Z. Chen, N. Kato, M. Gale, Jr., and S. M. Lemon. 2005. Distinct poly(I-C) and virus-activated signaling pathways leading to interferon-beta production in hepatocytes. J. Biol. Chem. 280**:**16739-16747. [DOI] [PubMed] [Google Scholar]
- 21.Li, K., E. Foy, J. C. Ferreon, M. Nakamura, A. C. Ferreon, M. Ikeda, S. C. Ray, M. Gale, Jr., and S. M. Lemon. 2005. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc. Natl. Acad. Sci. USA 102**:**2992-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, K., T. Prow, S. M. Lemon, and M. R. Beard. 2002. Cellular response to conditional expression of hepatitis C virus core protein in Huh7 cultured human hepatoma cells. Hepatology 35**:**1237-1246. [DOI] [PubMed] [Google Scholar]
- 23.Li, X. D., L. Sun, R. B. Seth, G. Pineda, and Z. J. Chen. 2005. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc. Natl. Acad. Sci. USA 102**:**17717-17722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin, R., J. Lacoste, P. Nakhaei, Q. Sun, L. Yang, S. Paz, P. Wilkinson, I. Julkunen, D. Vitour, E. Meurs, and J. Hiscott. 2006. Dissociation of a MAVS/IPS-1/VISA/Cardif-IKKɛ molecular complex from the mitochondrial outer membrane by hepatitis C virus NS3-4A proteolytic cleavage. J. Virol. 80**:**6072-6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309**:**623-626. [DOI] [PubMed] [Google Scholar]
- 26.Loo, Y. M., D. M. Owen, K. Li, A. K. Erickson, C. L. Johnson, P. M. Fish, D. S. Carney, T. Wang, H. Ishida, M. Yoneyama, T. Fujita, T. Saito, W. M. Lee, C. H. Hagedorn, D. T. Lau, S. A. Weinman, S. M. Lemon, and M. Gale, Jr. 2006. Viral and therapeutic control of IFN-beta promoter stimulator 1 during hepatitis C virus infection. Proc. Natl. Acad. Sci. USA 103**:**6001-6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin, A., F. Bodola, D. V. Sangar, K. Goettge, V. Popov, R. Rijnbrand, R. E. Lanford, and S. M. Lemon. 2003. Chronic hepatitis associated with GB virus B persistence in a tamarin after intrahepatic inoculation of synthetic viral RNA. Proc. Natl. Acad. Sci. USA 100**:**9962-9967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meylan, E., J. Curran, K. Hofmann, D. Moradpour, M. Binder, R. Bartenschlager, and J. Tschopp. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437**:**1167-1172. [DOI] [PubMed] [Google Scholar]
- 29.Muerhoff, A. S., T. P. Leary, J. N. Simons, T. J. Pilot-Matias, G. J. Dawson, J. C. Erker, M. L. Chalmers, G. G. Schlauder, S. M. Desai, and I. K. Mushahwar. 1995. Genomic organization of GB viruses A and B: two new members of the Flaviviridae associated with GB agent hepatitis. J. Virol. 69**:**5621-5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nam, J. H., K. Faulk, R. E. Engle, S. Govindarajan, M. St. Claire, and J. Bukh. 2004. In vivo analysis of the 3′ untranslated region of GB virus B after in vitro mutagenesis of an infectious cDNA clone: persistent infection in a transfected tamarin. J. Virol. 78**:**9389-9399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oshiumi, H., M. Matsumoto, K. Funami, T. Akazawa, and T. Seya. 2003. TICAM-1, an adaptor molecule that participates in Toll-like receptor 3-mediated interferon-beta induction. Nat. Immunol. 4**:**161-167. [DOI] [PubMed] [Google Scholar]
- 32.Peng, T., S. Kotla, R. E. Bumgarner, and K. E. Gustin. 2006. Human rhinovirus attenuates the type I interferon response by disrupting activation of interferon regulatory factor 3. J. Virol. 80**:**5021-5031. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Rice, C. M. 1996. Flaviviridae: the viruses and their replication, p. 931-959. In B. N. Fields and D. M. Knipe, Fields virology, 3rd ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 34.Richmond, A. 2002. NF-kappa B, chemokine gene transcription and tumour growth. Nat. Rev. Immunol. 2**:**664-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ryan, M. D., S. Monaghan, and M. Flint. 1998. Virus-encoded proteinases of the Flaviviridae. J. Gen. Virol. 79**:**947-959. [DOI] [PubMed] [Google Scholar]
- 36.Sbardellati, A., E. Scarselli, V. Amati, S. Falcinelli, A. S. Kekule, and C. Traboni. 2000. Processing of GB virus B non-structural proteins in cultured cells requires both NS3 protease and NS4A cofactor. J. Gen. Virol. 81**:**2183-2188. [DOI] [PubMed] [Google Scholar]
- 37.Scarselli, E., A. Urbani, A. Sbardellati, L. Tomei, R. De Francesco, and C. Traboni. 1997. GB virus B and hepatitis C virus NS3 serine proteases share substrate specificity. J. Virol. 71**:**4985-4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sen, G. C. 2001. Viruses and interferons. Annu. Rev. Microbiol. 55**:**255-281. [DOI] [PubMed] [Google Scholar]
- 39.Seth, R. B., L. Sun, and Z. J. Chen. 2006. Antiviral innate immunity pathways. Cell Res. 16**:**141-147. [DOI] [PubMed] [Google Scholar]
- 40.Seth, R. B., L. Sun, C. K. Ea, and Z. J. Chen. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF3. Cell 122**:**669-682. [DOI] [PubMed] [Google Scholar]
- 41.Sharma, S., B. R. tenOever, N. Grandvaux, G. P. Zhou, R. Lin, and J. Hiscott. 2003. Triggering the interferon antiviral response through an IKK-related pathway. Science 300**:**1148-1151. [DOI] [PubMed] [Google Scholar]
- 42.Shepard, C. W., L. Finelli, and M. J. Alter. 2005. Global epidemiology of hepatitis C virus infection. Lancet Infect. Dis. 5**:**558-567. [DOI] [PubMed] [Google Scholar]
- 43.Simons, J. N., T. J. Pilot-Matias, T. P. Leary, G. J. Dawson, S. M. Desai, G. G. Schlauder, A. S. Muerhoff, J. C. Erker, S. L. Buijk, M. L. Chalmers, et al. 1995. Identification of two flavivirus-like genomes in the GB hepatitis agent. Proc. Natl. Acad. Sci. USA 92**:**3401-3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sumpter, R., Jr., Y. M. Loo, E. Foy, K. Li, M. Yoneyama, T. Fujita, S. M. Lemon, and M. Gale, Jr. 2005. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J. Virol. 79**:**2689-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11**:**791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wolk, B., D. Sansonno, H. G. Krausslich, F. Dammacco, C. M. Rice, H. E. Blum, and D. Moradpour. 2000. Subcellular localization, stability, and trans-cleavage competence of the hepatitis C virus NS3-NS4A complex expressed in tetracycline-regulated cell lines. J. Virol. 74**:**2293-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu, L. G., Y. Y. Wang, K. J. Han, L. Y. Li, Z. Zhai, and H. B. Shu. 2005. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell 19**:**727-740. [DOI] [PubMed] [Google Scholar]
- 48.Yamamoto, M., S. Sato, K. Mori, K. Hoshino, O. Takeuchi, K. Takeda, and S. Akira. 2002. Cutting edge: a novel Toll/IL-1 receptor domain-containing adapter that preferentially activates the IFN-beta promoter in the Toll-like receptor signaling. J. Immunol. 169**:**6668-6672. [DOI] [PubMed] [Google Scholar]
- 49.Yi, M., R. A. Villanueva, D. L. Thomas, T. Wakita, and S. M. Lemon. 2006. Production of infectious genotype 1a hepatitis C virus (Hutchinson strain) in cultured human hepatoma cells. Proc. Natl. Acad. Sci. USA 103**:**2310-2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoneyama, M., M. Kikuchi, T. Natsukawa, N. Shinobu, T. Imaizumi, M. Miyagishi, K. Taira, S. Akira, and T. Fujita. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5**:**730-737. [DOI] [PubMed] [Google Scholar]
- 51.Zhong, J., P. Gastaminza, G. Cheng, S. Kapadia, T. Kato, D. R. Burton, S. F. Wieland, S. L. Uprichard, T. Wakita, and F. V. Chisari. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. USA 102**:**9294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhong, W., P. Ingravallo, J. Wright-Minogue, A. Skelton, A. S. Uss, R. Chase, N. Yao, J. Y. Lau, and Z. Hong. 1999. Nucleoside triphosphatase and RNA helicase activities associated with GB virus B nonstructural protein 3. Virology 261**:**216-226. [DOI] [PubMed] [Google Scholar]