Structure of human MRG15 chromo domain and its binding to Lys36-methylated histone H3 (original) (raw)
Abstract
Human MRG15 is a transcription factor that plays a vital role in embryonic development, cell proliferation and cellular senescence. It comprises a putative chromo domain in the N-terminal part that has been shown to participate in chromatin remodeling and transcription regulation. We report here the crystal structure of human MRG15 chromo domain at 2.2 Å resolution. The MRG15 chromo domain consists of a β-barrel and a long α-helix and assumes a structure more similar to the Drosophila MOF chromo barrel domain than the typical HP1/Pc chromo domains. The β-barrel core contains a hydrophobic pocket formed by three conserved aromatic residues Tyr26, Tyr46 and Trp49 as a potential binding site for a modified residue of histone tail. However, the binding groove for the histone tail seen in the HP1/Pc chromo domains is pre-occupied by an extra β-strand. In vitro binding assay results indicate that the MRG15 chromo domain can bind to methylated Lys36, but not methylated Lys4, Lys9 and Lys27 of histone H3. These data together suggest that the MRG15 chromo domain may function as an adaptor module which can bind to a modified histone H3 in a mode different from that of the HP1/Pc chromo domains.
INTRODUCTION
MORF4 (mortality factor on chromosome 4), MRG15 (MORF4-related gene on chromosome 15) and MRGX (MORF4-related gene on chromosome X) are members of the MRG protein family that were first identified as transcription factors involved in cellular senescence (1,2). Among those MRG proteins, MRG15 (a 37 kDa protein consisting of 323 amino acid residues) is of particular interest because it is expressed in a wide variety of human tissues and its homologues have been identified in many other eukaryotes (2,3). In addition to its involvement in cellular senescence, MRG15 is found to be crucial in embryonic development and cell proliferation. Knockout of MRG15 in mice is embryonic lethal and exhibits developmental delay (4). Cell biological and biochemical studies have shown that MRG15 is most likely to function in chromatin remodeling and transcriptional regulation through participation in two nucleoprotein complexes, MAF1 and MAF2 (MRG15-associated factors 1 and 2, respectively) (5). The C-terminal part of MRG15 has interactions with the tumor suppressor protein retinoblastoma (Rb) and a novel nuclear protein PAM14 (protein associated with MRG15 of 14 kDa) in MAF1 (6). It is also involved in interactions with the HDAC (histone deacetylase) containing transcriptional corepressor mSin3A and the plant homeodomain zinc finger protein Pf1 (7). The N-terminal part of MRG15 interacts with hMOF (human male absent on first) in MAF2 (6). In addition, MRG15 is associated with a mammalian TRRAP/Tip60 HAT (histone acetyltransferase) complex through protein MRGBP (MRG15/MRGX-binding protein) (8). Several MRG15 homologues in other species, such as MRG1 in Caenorhabditis elegans, MSL3 (male-specific lethal protein 3) in Drosophila, Eaf3p (Esa1p-associated factor 3 protein) in Saccharomyces cerevisiae, Alp13 (altered polarity protein 13) in fission yeast, are also found to be part of multi-subunit HAT/HDAC complexes that are involved in transcriptional regulation through chromatin remodeling (9–17). However, the exact functions of MRG15 and its homologues in these complexes and the underlying molecular mechanism(s) are unknown.
Human MRG15 consists of a putative chromo domain (the N-terminal residues 1–85) and a conserved MRG domain (the C-terminal residues 151–323) which are linked together by a flexible region (residues 86–150) (2). The MRG domain is highly conserved among all MRG proteins and the crystal structure of the MRG domain of human MRG15 has recently been determined (18,19). It assumes a fold consisting of mainly α-helices and appears to function as an adaptor module to interact with other proteins in nuclear protein complexes. Site-directed mutagenesis studies indicate that several hydrophobic residues form a shallow hydrophobic pocket to interact with the N-terminal region of PAM14. The exact function of the chromo domain of MRG15 is not yet well understood. However, the conservation of the chromo domain in many MRG15 homologues underscores its functional importance. Previous biochemical and structural studies have shown that chromo and chromo-like domains (such as Tudor and PWWP domains) are involved in recognition and interaction with histones or other proteins containing modified residues (such as methylated lysines or arginines) in nucleoprotein complexes (such as HAT and HDAC complexes), and play important roles in chromatin remodeling that leads to transcriptional activation or repression of a large number of genes [for reviews see (20–23)]. Chromatin-binding proteins HP1 (heterochromatin-binding protein 1) and Pc (Polycomb) chromo domains bind to methylated Lys9 and Lys27 of histone H3 (H3K9 and H3K27), respectively (24–29). The highly related HP1 chromo shadow domain can interact with numerous proteins containing a PXVXL motif (30–33). Many of these interactions play an important role in directing heterochromatin formation and/or gene silencing (21,30,34). The yeast Eaf3p chromo domain binds to methylated Lys4 and Lys36 of histone H3 (H3K4 and H3K36) and this interaction links histone deacetylation to phosphorylation of the RNA polymerase II C-terminal domain and thus the transcriptional elongation (14–16). The human CHD1 (chromo-ATPase/helicase DNA-binding protein 1) double chromo domains cooperate together to bind to methylated H3K4 (35). A number of Tudor, PWWP and other chromo-like domains have also been shown to bind to methylated N-terminal tails of histones or other proteins (36–40).
To explore its biological function, we determined the crystal structure of the chromo domain of human MRG15 at 2.2 Å resolution, which assumes a structure more similar to the Drosophila MOF (dMOF) chromo barrel domain than the typical HP1/Pc chromo domain. Using in vitro binding assay, we found that the MRG15 chromo domain can bind to methylated H3K36. The structural and biochemical data together suggest that the MRG15 chromo domain may function as an adaptor module to interact with a modified histone in a mode different from that of the HP1/Pc chromo domains.
MATERIALS AND METHODS
Protein expression and purification
The cDNA encoding the chromo domain of human MRG15 (residues 1–90) was cloned into a modified pET-3D-His expression vector (Novagen) which adds a His6 tag at the N-terminus. The plasmid was transformed into Escherichia coli strain BL21(DE3) (Novagen) and the transformed bacterial cells were cultured at 37°C in Luria–Bertani medium containing 0.1 mg/ml ampicillin. Protein expression was induced by adding IPTG into the medium to a final concentration of 1 mM. The cells were harvested by centrifugation at 5000 g for 10 min at 4 °C, resuspended in a lysis buffer (50 mM Tris–HCl, pH 8.0, 500 mM NaCl, 2 mM β-ME and 1 mM PMSF), and then lysed on ice by sonication. The recombinant protein was purified first with affinity chromatography using a Ni-NTA superflow column (Qiagen) and further with gel filtration using a Superdex G75 HiLoad 26/60 column (Amersham). The target protein was concentrated to ∼20 mg/ml in a buffer (50 mM Tris–HCl, pH 8.0 and 50 mM NaCl) by ultra-filtration for further structural and biochemical studies. To obtain Se-Met substituted protein suitable for structural determination, a mutant MRG15 chromo domain containing mutations I44M and L76M was generated. The Se-Met substituted mutant protein was expressed in E.coli strain B834(DE3) (Novagen) and purified using the same methods as for the native protein.
In vitro binding assay
To explore the potential binding of the MRG15 chromo domain with histone, we performed in vitro binding assay of the chromo domain with the histone mixture from calf thymus. The cNDAs encoding the human MRG15 chromo domain (residues 1–90), the yeast Eaf3p chromo domain (residues 1–113) and the mouse HP1α (residues 1–191), respectively, were cloned into the pGEX-4T1 expression vector (Amersham) which adds a GST tag at the N-terminus. The GST-fused proteins were expressed and purified using standard methods. An aliquot of 20 μg of the target protein was first incubated with 100 μl of 25% Glutathione–Sepharose bead slurry (Amersham) for 1 h. After removal of the solution, the bead was mixed with 100 μg of BSA protein in 1 ml of buffer A (10 mM Na2HPO4, 2.7 mM KCl, 1.8 mM KH2PO4, 140 mM NaCl, pH 7.4, 1 mM PMSF and 2 mM DTT) and the mixture was incubated at 4°C for 1 h to prevent non-specific binding. Then, 100 μg of the histone mixture from calf thymus (H9250; Sigma) was added and incubated at 4°C for 2 h. After centrifugation, the bead was washed with the binding buffer twice and then with buffer B (20 mM Tris–HCl, pH 8.0, 1 mM PMSF, 2 mM DTT, 1 mM EDTA, 1% NP-40 and 0.5 M NaCl) four times. The bound protein was eluted with loading buffer and analyzed by SDS–PAGE with Coomassie blue staining. The GST protein was used as a negative control and the mouse HP1α and the yeast Eaf3p chromo domain as positive controls. To identify which specific modification of histone the MRG15 chromo domain binds with, the eluted sample was electrophoresed and transferred onto PVDF membrane (Bio-Rad) and then probed with specific histone antibodies (Upstate Biotechnology): anti-H3K9me2 (dilution 1:2000), anti-H3K27me2 (dilution 1:1000), anti-H3K36me2 (dilution 1:2000) and anti-H3K4me3 (dilution 1:1000). HRP-conjugated secondary antibody (Chemicon) and ECL-advance western blotting kit (Amersham) were used for exposure detection.
To further confirm the interaction between the MRG15 chromo domain and the methylated Lys36 of histone H3, in vitro binding assay was carried out between the MRG15 chromo domain and di- and tri-methylated H3K36 peptides (H3K36me2/3, residues 28–44, synthesized by Sigma-Aldrich) and the bound peptide was visualized by peptide dot blot analysis. Specifically, ∼20 μg of the N-terminal His-tagged MRG15 chromo domain was first bound to Ni-NTA superflow bead slurry (Qiagen) and then incubated with an excess amount of the H3K36me2/3 peptide (protein/peptide molar ratio of 1:3) for 2 h. After extensive washing with buffer A, the protein with bound peptide was eluted with 30 μl of buffer C (20 mM Tris–HCl, pH 8.0 and 300 mM imidazole). An aliquot of 2 μl of the eluting solution was spotted onto PVDF membrane, probed with the anti-H3K36me2 (dilution 1:2000; Upstate) or anti-H3K36me3 (dilution 1:500; Abcam) antibody and visualized using the same method as described above.
Crystallization and diffraction data collection
Crystallization was performed using hanging drop vapor diffusion method. Crystals of both native and Se-Met mutant MRG15 chromo domains were grown at 4°C in drops containing equal volumes (2 μl) of the protein solution (20 mg/ml) and the reservoir solution (14% PEG3350 and 0.2 M KNO3) to approximate dimensions of 0.2 × 0.2 × 0.1 mm3 in 2 weeks. The diffraction data were collected from flash-cooled crystals at 100K. The single anomalous dispersion (SAD) data were collected to 3.0 Å resolution at CHESS (Cornell High Energy Synchrotron Source) beamline F2 and the native data to 2.2 Å resolution at CHESS beamline F1. The diffraction data were processed, integrated and scaled together with HKL2000 (41). Crystals of the Se-Met MRG15 chromo domain belong to space group _R_32 and crystals of the native protein belong to space group _C_2. The statistics of the diffraction data are summarized in Table 1.
Table 1.
Summary of diffraction data and structure refinement statistics
| Peak | Native | |
|---|---|---|
| Statistics of diffraction data | ||
| Wavelength (Å) | 0.9793 | 0.9124 |
| Resolution range (Å)a | 30.0–3.00 | 50.0–2.20 |
| (3.11–3.00) | (2.28–2.20) | |
| Space group | R32 | C2 |
| Cell parameters | ||
| a (Å) | 79.2 | 109.5 |
| b (Å) | 79.2 | 80.3 |
| c (Å) | 84.8 | 81.3 |
| β (°) | 123.9 | |
| Observed reflections | 43 456 | 108 140 |
| Unique reflections [I/σ(I) > 0] | 2144 | 29 581 |
| Mosaicity | 0.43 | 0.47 |
| Average redundancy | 20.3 (17.9) | 3.7 (3.2) |
| Average I/σ(I) | 29.2 (17.2) | 15.8 (3.8) |
| Completeness (%) | 99.4 (100) | 99.8 (99.6) |
| _R_merge(%)b | 8.2 (20.1) | 3.2 (22.7) |
| Statistics of refinement and model | ||
| Number of reflections [_F_o > 0σ(_F_o)] | ||
| Working set | 28 096 | |
| Free R set | 1485 | |
| _R_-factor (%)c | 22.4 | |
| Free _R_-factor (%) | 27.3 | |
| Number of residues | 483 | |
| Number of water molecules | 457 | |
| Average _B_-factor of all atoms (Å2) | 52.7 | |
| RMS bond lengths (Å) | 0.014 | |
| RMS bond angles (°) | 1.52 | |
| Luzzati atomic positional error (Å) | 0.31 | |
| Ramachandran plot (%) | ||
| Most favored regions | 92.1 | |
| Allowed regions | 6.7 | |
| Generously allowed regions | 1.2 | |
| Disallowed regions | 0 |
Structure determination and refinement
The structure of the MRG15 chromo domain was solved using the SAD method implemented in the program SOLVE(42). The SAD results revealed two Se-Met sites in an asymmetric unit corresponding to the two Met residues introduced by mutagenesis. The SAD phases were improved by statistical density modification including solvent flattening and histogram matching using the program RESOLVE (43), increasing the overall figure-of-merit from 0.45 to 0.75 at 3.0 Å resolution. RESOLVE automatically built a partial model of 52 polyalanines out of 90 residues and a complete model was manually built using the program O (44). This model was used as the search model to determine the initial phases for the 2.2 Å resolution native dataset by the molecular replacement method implemented in the program CNS (45). Structure refinement was carried out against the native dataset using CNS. There are six molecules in an asymmetric unit. Therefore, 6-fold non-crystallographic symmetry (NCS) restrains were used at the early stage of refinement, but were released after the structure refinement converged. The final structure refinement was accomplished with the maximum-likelihood algorithm implemented in the program REFMAC5 (46). The statistics of the structure refinement and the quality of structure models are summarized in Table 1.
RESULTS AND DISCUSSION
Structure of the MRG15 chromo domain
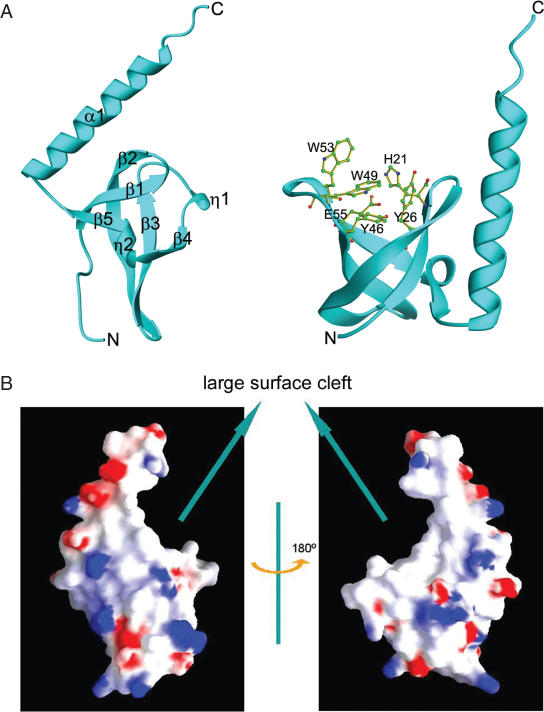
The structure of the MRG15 chromo domain was solved using the SAD method and the structure refinement converged to an _R_-factor of 22.4% and a free _R_-factor of 27.3% to 2.2 Å resolution (Table 1). There are six MRG15 chromo domain molecules in the asymmetric unit and structural comparison shows no substantial difference between the six molecules, except variation of disordered residues at the N- (usually 5–6 residues) and C-termini (usually 0–6 residues). The structure of the MRG15 chromo domain is composed of a twisted β-barrel and a C-terminal long α-helix (Figure 1A). The β-barrel core consists of five β-strands (β1–β5); β-strands β3, β4 and β5 are intercalated by two short 310 helices (η1 and η2). Highly conserved residues Tyr26, Tyr46 and Trp49 form a hydrophobic pocket at one end of the β-barrel which appears to be a potential binding site for a modified lysine of histone (see Discussion) (Figure 1A). The long α-helix folds along one side of the β-barrel. Residues Leu70, Lys72 and Leu76 of the α-helix form hydrophobic interactions with Leu18 and Phe20 of β1, Leu24 and Leu25 of β2, and Leu63 of β5. The side chain Nδ2 of Asn69 forms a hydrogen bond with the main-chain carbonyl of Lys64 (3.0 Å), and the side chain Nɛ2 of Gln73 forms a hydrogen bond with the carbonyl of Leu25 (2.7 Å). These interactions dictate the position and orientation of the α-helix relative to the β-barrel, forming a large cleft (Figure 1B).
Figure 1.
Structure of human MRG15 chromo domain. (A) Overall structure. Left panel: secondary structure elements. Right panel: structure of the potential binding pocket for a modified residue. Residues forming the pocket are shown with side chains. (B) Electrostatic surface of the MRG15 chromo domain. The β-barrel core and the C-terminal α-helix form a large surface cleft.
Comparison with other chromo and chromo-like domains
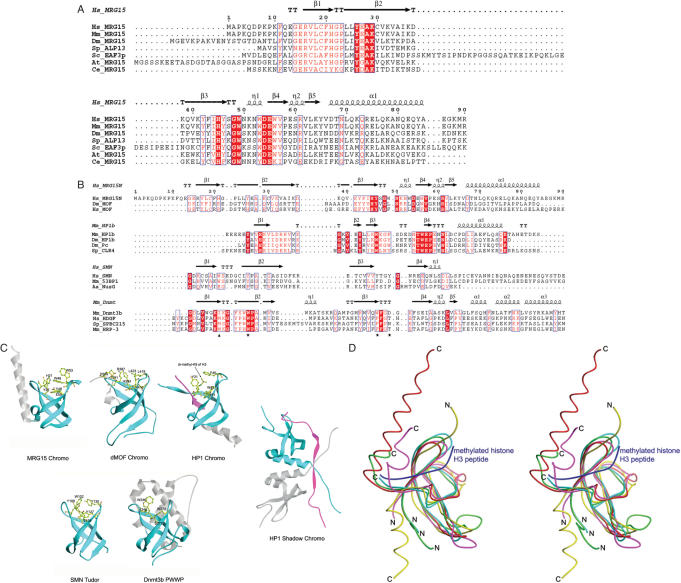
Structural comparison indicates that the overall structure of the MRG15 chromo domain is more similar to that of the dMOF chromo barrel domain (47) than that of the prototype HP1/Pc chromo domains (24–29) and the HP1 chromo shadow domain (31–33) (Figure 2). Superposition of the β-barrel core of the MRG15 chromo domain with that of other chromo domains reveals an RMSD of 1.6 Å with the HP1 chromo domain (1GUW, 41 Cα atoms), an RMSD of 2.2 Å with the HP1 chromo shadow domain (1S4Z, 35 Cα atoms) and an RMSD of 1.4 Å with the dMOF chromo barrel domain (2BUD, 49 Cα atoms). The HP1/Pc chromo domains act as adaptor modules to bind methylated H3K9 or H3K27 in chromatin remodeling and transcriptional regulation (24–29). The histone tail is bound between two β-strands of the β-barrel and the methylated lysine is bound at a hydrophobic pocket formed by three conserved aromatic residues (Tyr21, Trp42 and Phe45 in mouse HP1 or Tyr26, Trp47 and Trp50 in Drosophila Pc). Unlike the HP1/Pc chromo domains, the HP1 chromo shadow domain forms a homodimer as the functional unit (31–33). Though the chromo shadow domain shares a similar β-barrel core with the chromo domain, it binds a PXVXL motif containing peptide in a completely different manner in which the bound peptide is sandwiched between the extended β-strands from the C-terminus of the two subunits (33). Different from the HP1/Pc chromo domains in which the C-terminal region forms an α-helix flanking the β-barrel core, the C-terminal part of the MRG15 chromo domain forms a long α-helix against the β-barrel core in the opposite direction (Figure 2C and D). The C-terminal part of the dMOF chromo barrel domain occupies a position similar to that of the MRG15 chromo domain but forms a short α-helix and an extended coil. Similar to the dMOF chromo barrel domain, the β-barrel of the MRG15 chromo domain is in general similar to that of the HP1/Pc chromo domains but contains an extra β-strand (β1) which occupies the histone tail binding groove seen in the HP1/Pc chromo domains (Figure 2C and D). Though the MRG15 chromo domain also contains a hydrophobic pocket formed by conserved residues Tyr26, Tyr46 and Trp49, the binding site for methylated lysine is partially occupied by the side chain of His21 (equivalent to Arg387 of dMOF) which would have steric conflict with the side chain of methylated Lys9 in the HP1 chromo domain complex with the H3K9 peptide. Based on the structural similarities of the MRG15 and dMOF chromo domains and their common differences with the HP1/Pc chromo domains and the chromo shadow domain, we suggest that the MRG15 chromo domain should also be called a chromo barrel domain.
Figure 2.
Comparison of the MRG15 chromo domain with representative chromo and chromo-like domains. (A) Sequence comparison of the chromo domains between MRG15 and its homologues in other species. Hs, Homo sapiens; Mm, Mus musculus; Dm, Drosophila melanogaster; Sp, Schizosaccharomyces pombe; Sc, Saccharomyces cerevisiae; At, Arabidopsis thaliana; and Ce, Caenorhabditis elegans. Strictly conserved residues are highlighted in shaded red boxes and conserved residues in open red boxes. The secondary structure of the MRG15 chromo domain is placed on top of the alignment. (B) Structure-based sequence alignment of the MRG15 chromo domain with representative chromo, Tudor and PWWP domains. The secondary structure for the first member of each group is placed on top of the alignment. Dm_MOF: the dMOF chromo barrel domain, PDB code 2BUD; Mm_MOF: the mouse MOF chromo barrel domain, PDB code 1WGS; Mm_HP1b: the mouse HP1β chromo domain, PDB code 1GUW; Dm_HP1: the Dm HP1 chromo domain, PDB code 1KNA; Dm_Pc: the Dm Pc chromo domain, PDB code 1PDQ; Sp_CLR4: the Sp CLR4 chromo domain, PDB code 1G6Z; Hs_SMN: the human SMN Tudor domain, PDB code 1G5V; Mm_53BP1: the mouse 53BP1 Tudor domain, PDB code 1XNI; Aa_NusG: the Aquifex aeolicus transcription factor NusG Tudor domain, PDB code 1M1G; Mm_Dnmt3b: the mouse Dnmt3b PWWP domain, PDB code 1KHC; Hs_HDGF: the human HDGF domain of hepatoma-derived growth factor (HDGF)-related protein, PDB code 1RIO; Sp_SPBC215: the Sp protein SPBC215 PWWP domain, PDB code 1H3Z; Mm_HRP: the PWWP domain of mouse HDGF-related protein 3, PDB code 1N27. The stars indicate conserved residues that form the hydrophobic pocket in the HP1/Pc chromo domains and the triangle indicates the residue that occupies in part the hydrophobic pocket. (C) Structural comparison of the MRG15 chromo domain with the dMOF and HP1 chromo domain, the HP1 chromo shadow domain, the SMN Tudor domain and the Dnmt3b PWWP domain. Residues forming the hydrophobic pocket are shown with side chains and the bound peptides in the HP1 chromo domain complex and the HP1 chromo shadow domain complex are shown in magenta. (D) Superposition of the MRG15 chromo domain (red), the dMOF chromo barrel domain (magenta), the HP1 chromo domain (yellow), the SMN Tudor domain (cyan) and the Dnmt3b PWWP domain (green). The bound histone H3 peptide in complex with the HP1 chromo domain is shown in blue.
In addition to the chromo domain, several chromo-like domains involved in chromatin function have been identified, including Tudor domain (36–39) and PWWP domain (48). Structural comparisons of these protein modules (chromo, Tudor and PWWP domains) indicate that although these domains have very low sequence similarity (<20% identity) they share a strong structural similarity to a conserved β-barrel core and a hydrophobic pocket (Figure 2), suggesting that they may be evolved from the same ancestor and have similar biological functions (22,47). Nevertheless, there are notable structural differences between these domains (Figure 2C and D). The HP1/Pc chromo domains have a four-stranded β-barrel; the histone peptide is sandwiched between two β-strands and the modified residue is embedded in a hydrophobic pocket formed by three aromatic residues. On the other hand, the chromo barrel domains of MRG15 and dMOF, the SMN Tudor domain and the Dnmt3b PWWP domain have a five-stranded β-barrel; the putative binding groove for the histone tail is occupied by an extra N-terminal β-strand (β1) and the binding pocket for the modified residue is occupied in part by a residue of β1 (His21 of MRG15, Arg387 of dMOF, Trp102 of the SMN Tudor and Ile240 of the Dnmt3b PWWP). Moreover, the three aromatic residues forming the hydrophobic pocket are less conserved (Figure 2B). These structural differences might imply that these domains may use different modes to recognize and bind their protein substrates.
The MRG15 chromo domain can bind to methylated H3K36
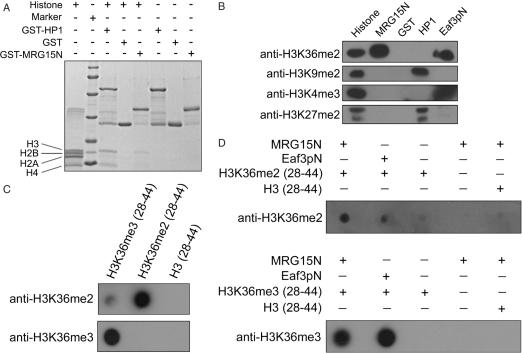
Chromo domains and chromo-like domains have been shown to be involved in the binding of modified histones or other proteins in nucleoprotein complexes (such as HAT and HDAC complexes) and play important functional roles in chromatin remodeling [for reviews see (20–23)]. The evident structural similarity of the MRG15 chromo domain to the other chromo domains implies that it may function as a module to interact with a modified histone. Since the chromo domain of Eaf3p, the yeast homolog of human MRG15 was shown to bind directly with methylated H3K4 and H3K36 (14,16), a similar function for the MRG15 chromo domain is possible. To pursue this notion, we first examined whether the MRG15 chromo domain can bind to any specific histone. The GST pull-down assay results showed that the GST-fused MRG15 chromo domain can bind with histone H3 from the calf thymus histone mixture and the binding affinity appears to be relatively weaker than that of the GST-fused HP1 (Figure 3A). To identify which modified residue(s) of histone H3 is involved in the binding, the GST pull-down histone sample was further analyzed using anti-histone antibodies targeting specifically at several commonly modified residues of histone H3, including methylated Lys4, Lys9, Lys27 and Lys36 (Figure 3B). Consistent with the literature, the mouse HP1α can bind to methylated H3K9 and H3K27, and the yeast Eaf3p chromo domain can bind to methylated H3K36 and H3K4. Moreover, our results clearly show that the MRG15 chromo domain can bind to methylated H3K36, but not methlyated H3K4, H3K9 and H3K27. To verify the interaction between the MRG15 chromo domain and the methylated H3K36, we carried out in vitro binding assay between the MRG15 chromo domain and di- and tri-methylated H3K36 peptides (H3K36me2/3, residues 28–44) (Figure 3D). The results indicate that the MRG15 chromo domain can bind to both H3K36me2 and H3K36me3 peptides, but not the unmethylated H3K36 peptide.
Figure 3.
In vitro binding assays showing that the MRG15 chromo domain can bind to methylated H3K36. (A) GST pull-down assays of the GST-fused MRG15 chromo domain (MRG15N) and the mouse HP1α with the calf thymus histone mixture. The protein samples were analyzed by SDS–PAGE with Coomassie blue staining. The results clearly show that the MRG15 chromo domain can bind to histone H3. (B) Western blot analysis of the GST pull-down samples of the MRG15 chromo domain (MRG15N), the mouse HP1α and the yeast Eaf3p chromo domain (Eaf3pN) with the calf thymus histone mixture. The mouse HP1α binds to H3K9me2 and H3K27me2. The yeast Eaf3p chromo domain binds to H3K36me2 and H3K4me3. The MRG15 chromo domain binds to H3K36me2. GST was used as a negative control. (C) Control experiments showing the specificity of the anti-H3K36me2/3 antibodies. The anti-H3K36me2 antibody has a high specificity with the H3K36me2 peptide and a weak cross-reaction with the H3K36me3 peptide, but no reaction with the unmethylated H3K36 peptide. The anti-H3K36me3 antibody recognizes only the H3K36me3 peptide. (D) In vitro binding assays of the N-terminal His-tagged MRG15 chromo domain with the H3K36me2 and H3K36me3 peptides. The yeast Eaf3p chromo domain (Eaf3pN) was used as a positive control. The results show that the MRG15 chromo domain can bind to both H3K36me2 and H3K36me3 peptides, but not the unmethylated H3K36 peptide.
Structural comparison of the MRG15 chromo domain with the HP1 chromo domain indicate that three conserved aromatic residues Tyr26, Tyr46 and Trp49 of the MRG15 chromo domain form a hydrophobic pocket, corresponding to that formed by residues Tyr21, Trp42 and Phe45 of mouseHP1α which is the binding site of methylated H3K9. Previous biochemical data have also shown that mutations of two aromatic residues Tyr81 and Trp84 in the corresponding hydrophobic pocket of the Eaf3p chromo domain (equivalent to Trp42 and Phe45 of mouse HP1α, respectively) disrupt the binding of Eaf3p with methylated H3K36 (16). Thus, it is possible that the hydrophobic pocket formed by Tyr26, Tyr46 and Trp49 of the MRG15 chromo domain might also be the potential binding site for the methylated Lys36 of histone H3. On the other hand, since the MRG15 chromo domain has an extra β-strand occupying the histone binding groove seen in the HP1/Pc chromo domains, it seems very likely that the MRG15 chromo domain might use other surface cleft or depression to bind the methylated H3K36. Besides the canonical peptide binding mode found in the HP1/Pc chromo domains, different substrate binding modes have been observed in other chromo domains (33,35). The crystal structure of the MRG15 chromo domain in complex with a histone peptide and further biochemical studies will eventually resolve the exact substrate binding mode of the MRG15 chromo domain and possibly other chromo barrel domains.
Acknowledgments
We are grateful to other members of our group for helpful discussion. This work was supported by NSFC grant (30570379) and MOST grants (2004CB720102 and 2006CB806501). Funding to pay the Open Access publication charges for this article was provided by MOST.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bertram M.J., Berube N.G., Hang-Swanson X., Ran Q., Leung J.K., Bryce S., Spurgers K., Bick R.J., Baldini A., Ning Y., et al. Identification of a gene that reverses the immortal phenotype of a subset of cells and is a member of a novel family of transcription factor-like genes. Mol. Cell. Biol. 1999;19:1479–1485. doi: 10.1128/mcb.19.2.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bertram M.J., Pereira-Smith O.M. Conservation of the MORF4 related gene family: identification of a new chromo domain subfamily and novel protein motif. Gene. 2001;266:111–121. doi: 10.1016/s0378-1119(01)00372-9. [DOI] [PubMed] [Google Scholar]
- 3.Marin I., Baker B.S. Origin and evolution of the regulatory gene male-specific lethal-3. Mol. Biol. Evol. 2000;17:1240–1250. doi: 10.1093/oxfordjournals.molbev.a026407. [DOI] [PubMed] [Google Scholar]
- 4.Tominaga K., Kirtane B., Jackson J.G., Ikeno Y., Ikeda T., Hawks C., Smith J.R., Matzuk M.M., Pereira-Smith O.M. MRG15 regulates embryonic development and cell proliferation. Mol. Cell. Biol. 2005;25:2924–2937. doi: 10.1128/MCB.25.8.2924-2937.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pardo P.S., Leung J.K., Lucchesi J.C., Pereira-Smith O.M. MRG15, a novel chromodomain protein, is present in two distinct multiprotein complexes involved in transcriptional activation. J. Biol. Chem. 2002;277:50860–50866. doi: 10.1074/jbc.M203839200. [DOI] [PubMed] [Google Scholar]
- 6.Leung J.K., Berube N., Venable S., Ahmed S., Timchenko N., Pereira-Smith O.M. MRG15 activates the B-myb promoter through formation of a nuclear complex with the retinoblastoma protein and the novel protein PAM14. J. Biol. Chem. 2001;276:39171–39178. doi: 10.1074/jbc.M103435200. [DOI] [PubMed] [Google Scholar]
- 7.Yochum G.S., Ayer D.E. Role for the mortality factors MORF4, MRGX, and MRG15 in transcriptional repression via associations with Pf1, mSin3A, and Transducin-Like Enhancer of Split. Mol. Cell. Biol. 2002;22:7868–7876. doi: 10.1128/MCB.22.22.7868-7876.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai Y., Jin J., Tomomori-Sato C., Sato S., Sorokina I., Parmely T.J., Conaway R.C., Conaway J.W. Identification of new subunits of the multiprotein mammalian TRRAP/TIP60-containing histone acetyltransferase complex. J. Biol. Chem. 2003;278:42733–42736. doi: 10.1074/jbc.C300389200. [DOI] [PubMed] [Google Scholar]
- 9.Fujita M., Takasaki T., Nakajima N., Kawano T., Shimura Y., Sakamoto H. MRG-1, a mortality factor-related chromodomain protein, is required maternally for primordial germ cells to initiate mitotic proliferation in C. elegans. Mech. Dev. 2002;114:61–69. doi: 10.1016/s0925-4773(02)00058-8. [DOI] [PubMed] [Google Scholar]
- 10.Gorman M., Franke A., Baker B.S. Molecular characterization of the male-specific lethal-3 gene and investigations of the regulation of dosage compensation in Drosophila. Development. 1995;121:463–475. doi: 10.1242/dev.121.2.463. [DOI] [PubMed] [Google Scholar]
- 11.Smith E.R., Pannuti A., Gu W., Steurnagel A., Cook R.G., Allis C.D., Lucchesi J.C. The Drosophila MSL complex acetylates histone H4 at lysine 16, a chromatin modification linked to dosage compensation. Mol. Cell. Biol. 2000;20:312–318. doi: 10.1128/mcb.20.1.312-318.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morales V., Regnard C., Izzo A., Vetter I., Becker P.B. The MRG domain mediates the functional integration of MSL3 into the dosage compensation complex. Mol. Cell. Biol. 2005;25:5947–5954. doi: 10.1128/MCB.25.14.5947-5954.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reid J.L., Moqtaderi Z., Struhl K. Eaf3 regulates the global pattern of histone acetylation in Saccharomyces cerevisiae. Mol. Cell. Biol. 2004;24:757–764. doi: 10.1128/MCB.24.2.757-764.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carrozza M.J., Li B., Florens L., Suganuma T., Swanson S.K., Lee K.K., Shia W.J., Anderson S., Yates J., Washburn M.P., et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 15.Joshi A.A., Struhl K. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation Pol II elongation. Mol. Cell. 2005;20:971–978. doi: 10.1016/j.molcel.2005.11.021. [DOI] [PubMed] [Google Scholar]
- 16.Keogh M.C., Kurdistani S.K., Morris S.A., Ahn S.H., Podolny V., Collins S.R., Schuldiner M., Chin K., Punna T., Thompson N.J., et al. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 17.Nakayama J., Xiao G., Noma K., Malikzay A., Bjerling P., Ekwall K., Kobayashi R., Grewal S.I. Alp13, an MRG family protein, is a component of fission yeast Clr6 histone deacetylase required for genomic integrity. EMBO J. 2003;22:2776–2787. doi: 10.1093/emboj/cdg248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bowman B.R., Moure C.M., Kirtane B.M., Welschhans R.L., Tominaga K., Pereira-Smith O.M., Quiocho F.A. Multipurpose MRG domain involved in cell senescence and proliferation exhibits structural homology to a DNA-interacting domain. Structure. 2006;14:151–158. doi: 10.1016/j.str.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 19.Zhang P., Zhao J., Wang B., Du J., Lu Y., Chen J., Ding J. The MRG domain of human MRG15 uses a shallow hydrophobic pocket to interact with the N-terminal region of PAM14. Protein Sci. 2006;15:2423–2434. doi: 10.1110/ps.062397806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cavalli G., Paro R. Chromo-domain proteins: linking chromatin structure to epigenetic regulation. Curr. Opin. Cell. Biol. 1998;10:354–360. doi: 10.1016/s0955-0674(98)80011-2. [DOI] [PubMed] [Google Scholar]
- 21.Jones D.O., Cowell I.G., Singh P.B. Mammalian chromodomain proteins: their role in genome organisation and expression. Bioessays. 2000;22:124–137. doi: 10.1002/(SICI)1521-1878(200002)22:2<124::AID-BIES4>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 22.Maurer-Stroh S., Dickens N.J., Hughes-Davies L., Kouzarides T., Eisenhaber F., Ponting C.P. The Tudor domain ‘Royal Family’: Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem. Sci. 2003;28:69–74. doi: 10.1016/S0968-0004(03)00004-5. [DOI] [PubMed] [Google Scholar]
- 23.Brehm A., Tufteland K.R., Aasland R., Becker P.B. The many colours of chromodomains. Bioessays. 2004;26:133–140. doi: 10.1002/bies.10392. [DOI] [PubMed] [Google Scholar]
- 24.Bannister A.J., Zegerman P., Partridge J.F., Miska E.A., Thomas J.O., Allshire R.C., Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 25.Lachner M., O'Carroll D., Rea S., Mechtler K., Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 26.Jacobs S.A., Khorasanizadeh S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002;295:2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- 27.Nielsen P.R., Nietlispach D., Mott H.R., Callaghan J., Bannister A., Kouzarides T., Murzin A.G., Murzina N.V., Laue E.D. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature. 2002;416:103–107. doi: 10.1038/nature722. [DOI] [PubMed] [Google Scholar]
- 28.Fischle W., Wang Y., Jacobs S.A., Kim Y., Allis C.D., Khorasanizadeh S. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 2003;17:1870–1881. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Min J., Zhang Y., Xu R.M. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys27. Genes Dev. 2003;17:1823–1828. doi: 10.1101/gad.269603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y., Kirschmann D.A., Wallrath L.L. Does heterochromatin protein 1 always follow code? Proc Natl Acad. Sci. USA. 2002;99:16462–16469. doi: 10.1073/pnas.162371699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cowieson N.P., Partridge J.F., Allshire R.C., McLaughlin P.L. Dimerisation of a chromo shadow domain and distinctions from the chromodomain as revelaed by structural analysis. Curr. Biol. 2000;10:517–525. doi: 10.1016/s0960-9822(00)00467-x. [DOI] [PubMed] [Google Scholar]
- 32.Brasher S.V., Smith B.O., Fogh R.H., Nietlispach D., Thiru A., Nielsen P.R., Broadhurst R.W., Ball L.J., Murzina N.V., Laue E.D. The structure of mouse HP1 suggests a unique mode of single peptide recognition by the shadow chromo domain dimer. EMBO J. 2000;19:1587–1597. doi: 10.1093/emboj/19.7.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thiru A., Nietlispach D., Mott H.R., Okuwaki M., Lyon D., Nielsen P.R., Hirshberg M., Verreault A., Murzina N.V., Laue E.D. Structural basis of HP1/PXVXL motif peptide interactions and HP1 localisation to heterochromatin. EMBO J. 2004;23:489–499. doi: 10.1038/sj.emboj.7600088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cao R., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R.S., Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 35.Flanagan J.F., Mi L.Z., Chruszcz M., Cymborowski M., Clines K.L., Kim Y., Minor W., Rastinejad F., Khorasanizadeh S. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature. 2005;438:1181–1185. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- 36.Selenko P., Sprangers R., Stier G., Buhler D., Fischer U., Sattler M. SMN tudor domain structure and its interaction with the Sm proteins. Nature Struct. Biol. 2001;8:27–31. doi: 10.1038/83014. [DOI] [PubMed] [Google Scholar]
- 37.Sprangers R., Groves M.R., Sinning I., Sattler M. High-resolution X-ray and NMR structures of the SMN Tudor domain: conformational variation in the binding site for symmetrically dimethylated arginine residues. J. Mol. Biol. 2003;327:507–520. doi: 10.1016/s0022-2836(03)00148-7. [DOI] [PubMed] [Google Scholar]
- 38.Huyen Y., Zgheib O., Ditullio R.A.J., Gorgoulis V.G., Zacharatos P., Petty T.J., Sheston E.A., Mellert H.S., Stavridi E.S., Halazonetis T.D. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 2004;432:406–411. doi: 10.1038/nature03114. [DOI] [PubMed] [Google Scholar]
- 39.Cote J., Richard S. Tudor domains bind symmetrical dimethylated arginines. J. Biol. Chem. 2005;280:28476–28483. doi: 10.1074/jbc.M414328200. [DOI] [PubMed] [Google Scholar]
- 40.Kim J., Daniel J., Espejo A., Lake A., Krishna M., Xia L., Zhang Y., Bedford M. Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep. 2006;7:397–403. doi: 10.1038/sj.embor.7400625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Meth. Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 42.Terwilliger T.C., Berendzen J. Evaluation of macromolecular electron-density map quality using the correlation of local r.m.s. density. Acta Crystallogr. D. 1999;55:1872–1877. doi: 10.1107/S090744499901029X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Terwilliger T.C. Maximum-likelihood density modification. Acta Crystallogr. D. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jones T.A., Zou J.Y., Cowan S.W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 45.Brunger A.T., Adams P.D., Clore G.M., DeLano W.L., Gros P., Grosse-Kunstleve R.W., Jiang J.S., Kuszewski J., Nilges M., Pannu N.S., et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 46.Murshudov G.N., Vagin A.A., Dodson E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 47.Nielsen P.R., Nietlispach D., Buscaino A., Warner R.J., Akhtar A., Murzin A.G., Murzina N.V., Laue E.D. Structure of the chromo barrel domain from the MOF acetyl-transferase. J. Biol. Chem. 2005;280:32326–32331. doi: 10.1074/jbc.M501347200. [DOI] [PubMed] [Google Scholar]
- 48.Qiu C., Sawada K., Zhang X., Cheng X. The PWWP domain of mammalian DNA methyltransferase Dnmt3b defines a new family of DNA-binding folds. Nature Struct. Biol. 2002;9:217–224. doi: 10.1038/nsb759. [DOI] [PMC free article] [PubMed] [Google Scholar]