Mutations in STRA6 Cause a Broad Spectrum of Malformations Including Anophthalmia, Congenital Heart Defects, Diaphragmatic Hernia, Alveolar Capillary Dysplasia, Lung Hypoplasia, and Mental Retardation (original) (raw)
Abstract
We observed two unrelated consanguineous families with malformation syndromes sharing anophthalmia and distinct eyebrows as common signs, but differing for alveolar capillary dysplasia or complex congenital heart defect in one and diaphragmatic hernia in the other family. Homozygosity mapping revealed linkage to a common locus on chromosome 15, and pathogenic homozygous mutations were identified in STRA6, a member of a large group of “stimulated by retinoic acid” genes encoding novel transmembrane proteins, transcription factors, and secreted signaling molecules or proteins of largely unknown function. Subsequently, homozygous STRA6 mutations were also demonstrated in 3 of 13 patients chosen on the basis of significant phenotypic overlap to the original cases. While a homozygous deletion generating a premature stop codon (p.G50AfsX22) led to absence of the immunoreactive protein in patient’s fibroblast culture, structural analysis of three missense mutations (P90L, P293L, and T321P) suggested significant effects on the geometry of the loops connecting the transmembrane helices of STRA6. Two further variations in the C-terminus (T644M and R655C) alter specific functional sites, an SH2-binding motif and a phosphorylation site, respectively. STRA6 mutations thus define a pleiotropic malformation syndrome representing the first human phenotype associated with mutations in a gene from the “STRA” group.
Clinical anophthalmia (AO) is the complete absence of the eye and may be the most severe end of a clinical spectrum of ocular malformations including microphthalmia (MO), which is a small eye usually defined in terms of corneal diameter or axial length.1 Estimates of the birth prevalence of anophthalmia and microphthalmia from well-maintained population-based registers are 14 and 3 per 100,000 births, respectively.1 Autosomal recessive origin is likely in ∼10% of cases.2 CHX10 mutations have been shown to underlie autosomal recessive isolated clinical anophthalmia in two families3 and microphthalmia with cataract and abnormalities of the iris,4 whereas syndromic autosomal dominant anophthalmia has been associated with mutations in SOX2.5 According to a national study on microphthalmia, anophthalmia, and coloboma, 33% of cases had one or more associated major malformations, and 21% had learning disabilities.1 As only a few of the latter syndromal cases had features of recognized entities, it was assumed that several new syndrome diagnoses are yet to be delineated in this group.
To increase the understanding of syndromic anophthalmia, we performed positional cloning in two unrelated consanguineous families with new, apparently not yet reported conditions, including clinical anophthalmia and variable malformations of the lung, the heart, and the diaphragm, as well as mental retardation. This work was performed as part of our research study addressing the genetics of mental retardation, which was approved by the research ethics committee of the University of Erlangen-Nuremberg.
The proband in family 1 (IV:2 in fig. 1_A_) was a female infant born at 33 wk of gestation following a pregnancy during which bilateral anophthalmia had been diagnosed by ultrasound scan at 16 wk of gestation. She had normal intrauterine growth, with a birth length of 49 cm (97th percentile), weight 2,035 g (50th percentile), and head circumference 32 cm (75th percentile). In the perinatal period, bilateral clinical anophthalmia was confirmed. Additional malformations were noted at that time: right-sided pelvic kidney, circulatory nonrelevant pulmonic valve stenosis, and persistent ductus arteriosus, which was surgically closed at the age of 3 wk. When assessed at the age of 2 mo, growth was normal, with the crown-to-heel length (50 cm) and weight (3,200 g) at the age of 2 mo corresponding to the 10th–25th percentile for prematurely born girls, whereas head circumference (35 cm) corresponded to the 25th–50th percentile. She had mild facial dysmorphism, with marked blepharophimosis with an unusual trichoglyphic pattern of the eyebrows, which were broad, flaring, and only upward growing (fig. 2_E_). She had a broad nasal bridge, micrognathia, and large, low-set ears (fig. 2_A_ and 2_B_). Cerebral magnetic resonance imaging (MRI) showed no abnormality of brain structure and showed visible optic nerves and chiasm. Mechanical ventilatory support was required from birth, because of persistent respiratory insufficiency. Chest CT at the age of 6 wk showed no evidence of pulmonary malformations, lymphangiectasia, or interstitial lung disease. Open lung biopsy performed at the age of 2.5 mo revealed a reduced number of alveolar units and pulmonary capillary vessels with thickening of the interalveolar septa, as well as medial thickening of small pulmonary arteries with muscularization, which are the key features of alveolar capillary dysplasia (MIM 235680) (fig. 2_G_) without misalignment of lung vessels. She did not show any psychomotor development and, despite high-dose steroid treatment, she was extubated for only 13 d before dying at the age of 6 mo from respiratory insufficiency. The family history was significant. The parents are first cousins of Turkish origin. The paternal uncle of the proband, who was also married to his half-cousin, had a daughter with bilateral anophthalmia, who died at the age of 2 d from a complex cyanotic congenital heart defect with atresia of the pulmonary artery and single ventricle (IV:4 in fig. 1_A_). She had normal intrauterine growth, with a birth length of 51 cm (25th–50th percentile), weight 3,240 g (25th–50th percentile), and head circumference 36 cm (75th percentile). Karyotype, metabolic screen, and cerebral and renal ultrasound all showed normal results. Parents consented to postmortem examination of thoracic organs, which confirmed the single ventricle with atresia of the pulmonary artery.
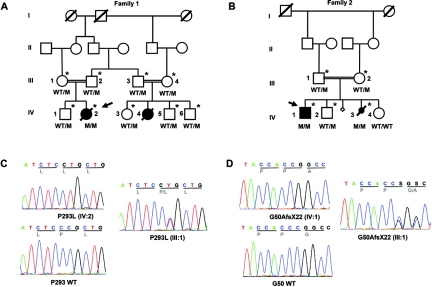
Figure 1. .
A and B, Pedigrees of the two families and segregation of the respective mutation (M). Arrows indicate the probands. Individuals marked with asterisks participated in the genomewide linkage analysis. C and D, Examples of electropherograms of STRA6 mutations in families 1 (C) and 2 (D). WT = Wild type.
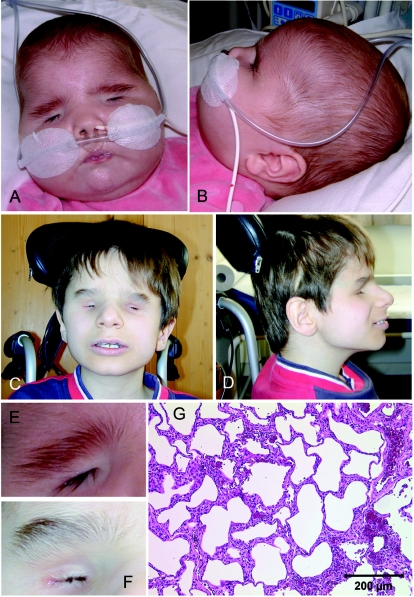
Figure 2. .
A–D, Frontal and lateral views of patient IV:2 of family 1 at age 6 mo (A and B) and patient IV:1 of family 2 at age 13 years (C and D). Note similar mild dysmorphism with broad, flaring, and only upward-growing eyebrows; broad nasal bridge; large, low-set ears; and receding chin. E and F, Close-up of right eyebrows of IV:2 of family 1 (E) and IV:1 of family 2 (F). G, Hematoxylin-eosin staining of lung biopsy, showing deficiency in the number of alveolar units and pulmonary capillary vessels with thickening of the interalveolar septa.
The proband in family 2 (IV:1 in fig. 1_B_) was a 14-year-old boy with bilateral clinical anophthalmia, diaphragmatic hernia, and profound mental retardation, with a performance IQ estimated to be <20. He was the eldest son of a healthy consanguineous couple of Turkish origin. When assessed at the age of 13 years 3 mo, he had severe short stature (height 123 cm [−4.47 SD]; weight 21 kg [BMI −2.95]) with relative preservation of head growth (occipitofrontal circumference 51 cm [3rd percentile]). He had no speech and had no obvious receptive language skills. Although he used a wheelchair, he was able to take a few steps when supported. He had both an atrial and a ventricular septal defect, which did not require any therapy. Cerebral MRI performed at the age of 4 years showed a structurally normal brain, apart from absent optic nerves. He had mild facial dysmorphism with severe blepharophimosis and an unusual trichoglyphic pattern of both eyebrows similar to that seen in family 1 (fig. 2_F_). He had a broad nasal bridge, micrognathia, and large, low-set ears (fig. 2_C_ and 2_D_). He had a healthy brother and sister. His mother had had one termination of pregnancy because of perceived high risk and another pregnancy that was terminated at 23 wk gestation after a diagnosis of bilateral anophthalmia and severe diaphragmatic hernia on antenatal ultrasound scan (IV:3). The fetus showed mild facial dysmorphism similar to that of the probands in this family and family 1. Parents did not consent to postmortem examination but agreed to skin biopsy for fibroblast culture.
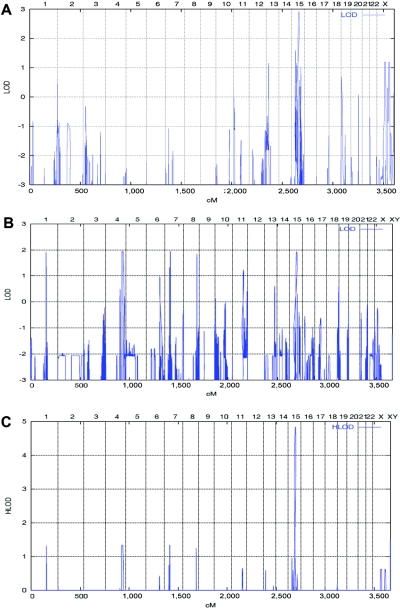
To identify the underlying disease genes, a genomewide linkage scan was performed using the Affymetrix GeneChip Human Mapping 10K SNP array Xba142 (version 2.0) and both affected and unaffected individuals from both families. The sex of each sample was verified by counting heterozygous SNPs on the X chromosome. Relationship errors were evaluated with the help of the program Graphical Relationship Representation.6 The program PedCheck was applied to detect Mendelian errors,7 and data for SNPs with such errors were removed from the data set. Non-Mendelian errors were identified by use of the program MERLIN,8 and unlikely genotypes for related samples were deleted. LOD score calculations were performed using the Allegro program,9 under the assumption of autosomal recessive inheritance with full penetrance. The parametric analysis unveiled the expected maximum multipoint LOD score of 2.9 on chromosome 15 (q23-25.1) in family 1 for a region of ∼15 cM (fig. 3_A_) and multiple possible loci in family 2, including the same locus on chromosome 15 with the expected maximum LOD score of 1.9 (fig. 3_B_). Parametric and nonparametric linkage analysis of both families together revealed a single maximum LOD score of 4.8 for the region 15q23-25.1, which was also achieved when allowing for locus heterogeneity (HLOD) (fig. 3_C_). Haplotypes were reconstructed with ALLEGRO and were presented graphically with HaploPainter.10 This latter program also reveals informative SNP markers as points of recombination between parental haplotypes. All data handling was performed using the graphical user interface ALOHOMORA.11 Haplotype reconstruction showed homozygosity for two different alleles at the same 12-Mb region between markers SNP_A-1511966 (rs1822829) and SNP_A-1509050 (rs1077965) in affected children from both families (fig. 4).
Figure 3. .
Graphic summaries of multipoint linkage analyses in families 1 and 2, performed using the Allegro program under the assumption of autosomal recessive inheritance with full penetrance. A, Genomewide multipoint LOD scores of family 1, including genotypes from individuals III:1–4, IV:1–3, and IV:5–6 (fig. 1_A_), identifying a single locus on chromosome 15 with the expected maximum LOD score of 2.9. B, Genomewide multipoint LOD scores of family 2, including genotypes from individuals III:1–2 and IV:1–3 (fig. 1_B_), identifying multiple loci on chromosomes 1, 4, 7, 8, and 15 with the expected maximum LOD score of 1.9. C, Summary LOD scores including both families allowing also for locus heterogeneity (HLOD) revealing a single locus on chromosome 15 with a maximum LOD and HLOD of 4.8.
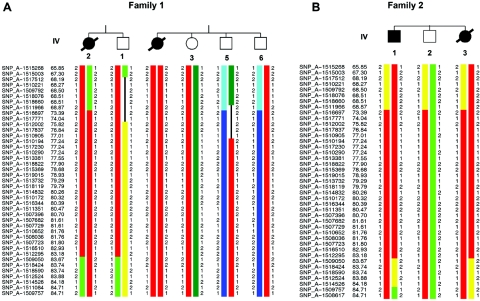
Figure 4. .
Haplotype analyses on chromosome 15q23-25.1, showing homozygous markers linked to the disease locus in family 1 (A) and family 2 (B). According to the NCBI human genome overview page, build 35.1, the flanking markers SNP_A-1511966 (rs1822829) and SNP_A-1509050 (rs1077965) span a region of ∼12 Mb between 65.21 and 77.85 Mb from pter.
This critical region contained >280 known and predicted genes annotated in the UCSC database. After evaluation of the available data on developmental expression and function of each of the genes, we selected two for mutational analysis. UACA (encoding uveal autoantigen with coiled-coil domains and ankyrin repeats protein) was selected because it plays an important role in the regulation of stress-induced apoptosis and because it is expressed in heart and in choroid, retina, and eye muscles, among other tissues, according to the SwissProt protein database. STRA6 (FLJ12541; Stra for “stimulated by retinoic acid”) was chosen because of its involvement in the retinoic acid pathway and its therefore likely role in morphogenesis, as well as reported expression in mouse tissues corresponding to affected organs in our patients.
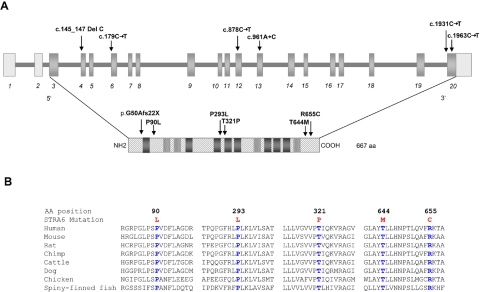
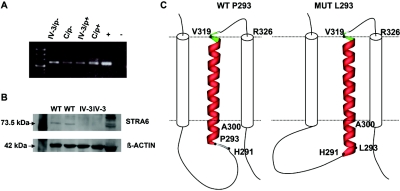
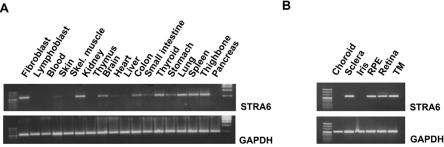
Sequencing of UACA revealed no mutation in the probands of family 1 (IV:2) and 2 (IV:1). Sequence analysis of all 20 exons (fig. 5_A_) and intronic flanking regions of STRA6 revealed single-nucleotide variants in both families. A homozygous missense mutation in exon 12, c.878C→T (P293L), was found in the affected child IV:2 of family 1 (fig. 1_C_). A homozygous frameshift mutation leading to a premature stop codon (c.145_147delC; p.G50AfsX22) in exon 4 was demonstrated in both affected children of family 2 (fig. 1_D_). Both mutations were shown to cosegregate with the disease phenotype in the respective families (fig. 1_A_ and 1_B_). After the identification of STRA6 mutations in these families, the mutational analysis was extended using a cohort of 13 unrelated white patients selected on the basis of having a severe eye malformation and malformations of the diaphragm or one of the latter associated with malformations of the lungs or heart (table 1). This analysis led to the identification of four further homozygous amino acid changes (P90L, T321P, T644M, and R655C) in STRA6 in three patients (tables 1 and 2 and fig. 5_A_). Parental heterozygosity for the respective mutation was proved in families for which parental samples were available (MWS1-EE and MWS4-BE). Patient MWS6-BK carries two of these missense mutations (P90L and T321P), both homozygously, but no other family members were available for further analysis. None of the mutations were found in a panel of 190 healthy, adult white control individuals. Further support for the pathogenicity of the missense mutations was provided by the demonstration that each of the substituted amino acids was evolutionarily conserved, which was done using multiple sequence alignments by ClustalW (fig. 5_B_). RT-PCR analysis of RNA isolated from cultured fibroblasts of the affected aborted fetus (IV:3) from family 2, which harbored the premature stop mutation, showed detectable levels of the mutated STRA6 transcript with and without puromycin treatment (fig. 6_A_). Western blot analysis of protein extracted from these fibroblasts was performed using a rabbit polyclonal antibody raised to the C-terminal region of STRA612 (kindly supplied by Pierre Chambon). This showed absence of immunoreactive protein in the patient’s fibroblasts (fig. 6_B_). RT-PCR on normal adult human mRNAs from different tissues confirmed the broad expression pattern previously found in mice (fig. 7_A_). Detailed analysis of distinct parts of an adult human eye revealed expression in sclera, retina, retinal pigment epithelium, and trabecular mashwork but not in choroid and iris (fig. 7_B_).
Figure 5. .
Location and conservation of mutated residues. A, Location of mutations along STRA6 genomic and protein schematic representation, respectively. Transmembrane helices predicted by all three algorithms used are shown as dark gray boxes; helices predicted only by one or two of the algorithms are shown as dotted light gray boxes. B, ClustalW multiple sequence alignments of human STRA6 regions to orthologues encompassing mutated residues. All these residues are evolutionary conserved. The respective substituted amino acid is highlighted in blue.
Table 1. .
Overview of Phenotype in Patients Investigated for STRA6 Mutations[Note]
| Patient | Mutation | Parental Consanguinity | Eye | Lung | Diaphragm | Heart | Palate | Kidney | Uterus | Age at Death | Other |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Fam1-IV:2 | P293L | + | b AO | ACD | − | PSt, PDA | − | Pelvic | − | 6 mo | PTB 33 weeks, DD |
| Fam1-IV:4 | NA | + | b AO | − | − | CHD,PA | − | − | − | 2 d | |
| Fam2-IV:1 | p.G50AfsX22 | + | b AO | − | CDH | ASD, VSD | − | − | − | Alive at age 14 years, profound MR, SOS | |
| Fam2-IV:3 | p.G50AfsX22 | + | b AO | − | CDH | − | − | − | − | TOP | |
| MWS1-EE | R655C | + | b AO | Hypo | DE | 3 mo | Inguinal hernia, severe hypotonia, failure to thrive | ||||
| Brother | NA | + | b AO | − | − | TAC-IV, RAA, PDA, PA | − | − | − | 22 mo | SOS |
| MWS4-BE | T644M | − | b AO | Hypo | CDH | b hydronephrosis | Alive at age 3 mo | ||||
| Brother | NA | − | ? | Hypo, unilo | − | TOF, PDA | − | Horseshoe | − | 1 d | Undescended testes, hypoplastic renal arteries |
| Sister | NA | − | b AO | Hypo, unilo | − | PDA, CoA | − | − | Dysplasia | 1 d | |
| MWS6-BK | P90L, T321P | + | b AO | Hypo | CDH, DE | PDA | Hypo | Bicornuate | 1 d | PTB 36 wk, Meckel diverticle | |
| MWS2-FA | − | + | b Col | − | CDH | − | − | − | − | Skin patches, brittle hair | |
| MWS3-KH | − | − | b MO | − | CDH | − | − | − | − | MO: extreme | |
| MWS5-LR | − | − | Col | − | CDH | − | − | − | − | ||
| RHP006.070 | − | − | b MO | − | b DE | − | − | − | − | MO: extreme, MR | |
| GM23728 | − | − | b MO | Hypo, unilo | DE, hypo | Hypo Pa, CoA | Dysplastic | Neonatal | Abnormal cornea and iris | ||
| AvdW22260 -Twin1 | − | − | − | Hypo | CDH | − | − | − | − | 1 d | PTB (28 wk) |
| Twin 2 | − | − | − | Hypo | CDH | − | CP | − | − | 1 d | PTB (28 wk) |
| AS20861-FF264 | − | − | ri MO | − | CDH | − | − | − | − | MO: max. diameter 9 mm at age 13 mo with internal, dense calcification within the globe, and a larger, inferiorly located benign cyst measuring 2.5 cm; DD (11 mo level at age 13 mo) | |
| CD50396 | − | − | b AO | Hypo | DE | VSD | CP | − | Hypo, bicornuate | 1 d | Matthew-Woods syndrome, hypoplastic spleen, hypoplastic alae nasi |
| PM22479 | − | + | − | − | CDH | − | − | − | − | Neonatal | Suspected Donnai-Barrow syndrome (MIM 222448), large omphalocele, hypoplasia of corpus callosum, enlarged ventricles, extreme hypertelomerism |
| Brother | NA | + | − | − | CDH | ASD | b CLP | − | − | Neonatal | Suspected Donnai-Barrow syndrome (MIM 222448), hypoplasia of corpus callosum, enlarged ventricles, extreme hypertelomerism |
| PB-E03_053 | − | − | b MO | − | CDH | − | − | − | − | − | MO: severe, b inguinal hernia, sparse hair, brachycephaly, MR, spasticity, alive at 10 years |
Table 2. .
Overview of STRA6 Mutations
| Alterationa | |||
|---|---|---|---|
| Patient | Exon | Genomic | Protein |
| Fam1-IV:2 | 12 | c.878C→T | P293L |
| Fam2-IV:1 | 4 | c.145-147delC | p.G50AfsX22 |
| Fam2-IV:3 | 4 | c.145-147delC | p.G50AfsX22 |
| MWS1-EE | 20 | c.1963C→T | R655C |
| MWS4-BE | 20 | c.1931C→T | T644M |
| MWS6-BK | 6, 13 | c.269C→T, c.961A→C | P90L, T321P |
Figure 6. .
Characterization of mutations. A, STRA6 RT-PCR on cultured fibroblast cells from the affected fetus (IV:3, family 2) and from a healthy individual (C) grown in the absence and the presence of puromycin (p−/p+) as inhibitor of translation, respectively, showing no evidence for early nonsense-mediated mRNA decay; (+) positive control, (−) negative control. B, Western blot analysis of cultured fibroblast protein cell extract showing a STRA6 protein band in two human healthy control lanes (indicated as “WT”) but not in the two lanes with the homozygous mutant c.145_147delC (from IV:3, family 2). C, Model showing the effect of the P293L mutation (right) on the STRA6 structure in comparison with the wild type (left). The membrane is indicated by a dotted line. The transmembrane helix (A300-V319) that is proximal to the site of the mutation is shown in ribbon presentation, and the two adjacent helices are indicated by cylinders. In the wild type, residue P293 represents the N-terminal cap of a helix starting at L294, whereas the L293 present in the mutant allows an N-terminal extension of the helix, now starting at H291 and changing the topology of the respective loop. Therefore, the P293L mutation is predicted to cause an extension of the helix by three residues, thus affecting the structure and orientation of the respective loop. Moreover, it is important to note that V319 is also the N-terminal residue of an extremely short loop (maximum predicted length between aa 319 and aa 326), in which a second mutation (T321P) has been identified.
Figure 7. .
Expression pattern of STRA6 determined by RT-PCR in normal adult human (A) and eye (B) tissues. RPE = Retinal pigment epithelium; TM = Trabecular meshwork.
Both the function and the tertiary structure of STRA6 peptide are unknown. To explain the potential effects of the missense mutations, we performed secondary structure analysis by use of three different prediction programs (TMHMM,13 TMpred,14 and TopPred,15), which suggested that STRA6 has between 8 and 12 transmembrane helices. Accordingly, three missense mutations (P90L, P293L, and T321P) are predicted to be located in loops connecting these transmembrane helices, whereas two missense mutations (T644M and R655C) are located in the evolutionary conserved C-terminal region of the protein. Analysis of the secondary structure outside the transmembrane segments was performed using a consensus secondary prediction from the NPS@ server.16 This approach suggested that the mutations P293L and P90L increase the extent of helical structures in STRA6, resulting in an extension of the transmembrane helix (P293L) (fig. 6_C_) and the formation of a novel helix within a loop (P90L). As a consequence, the topology of the loops is altered, and numerous conserved residues are brought into a different orientation with respect to the membrane. Amino acid T321 was also predicted to be located in an extremely short loop (maximum length comprising residues 319–326) connecting two transmembrane helices. As a consequence of the T321P mutation, a rigid diproline-motif (P320–P321) is generated that is probably incompatible with the sterically demanding topology of this tight loop (fig. 6_C_). Thus, mutants P90L and T321P, observed homozygously in the same patient, are both expected to have significant effects on the secondary and tertiary structure of the loops in STRA6.
In contrast, mutations T644M and R655C are predicted by the ELM program17 to impair functional sites at the C-terminal region. T644M alters the STAT5 Src Homology 2 (SH2) domain binding motif, YTLL, which triggers the JAK2/STAT5 signaling cascade. STAT5 and related members of the STAT family are activated in different tissues by means of a series of ligands and are involved in interferon signaling, development of the mammary gland, response to growth hormone, and embryogenesis.18 R655C alters the invariant arginine of the R-X-[ST] consensus sequence representing a phosphorylation site of proteinkinase A19 and therefore probably alters the successive signal transduction pathways.
The phenotype of the three patients detected on the follow-up mutation analysis shows remarkable overlap with that of the initial cases, with bilateral clinical anophthalmia and normal birth measurements as consistent features. The healthy parents of patient MWS1-EE are distantly related through a common great-great-grandparent. After a pregnancy remarkable for polyhydramnion, MWS1-EE was born at term with normal measurements (weight 3,130 g [10th–25th percentile], length 50 cm [10th–25th percentile], and head circumference 35 cm [25th–50th percentile]). Because of respiratory insufficiency, ventilatory support was needed for 4 d. The boy showed bilateral anophthalmia, left-sided diaphragmatic eventration, and right-sided inguinal hernia. He had severe hypotonia, poor feeding, and almost no weight gain until he died at the age of 3 mo. No postmortem examination was performed. The older brother of MWS1-EE also showed bilateral anophthalmia, with only remains of nervi optici detected at autopsy. In addition, he had truncus arteriosus communis type IV with right-sided aorta, lack of pulmonary arteries, and lung supply by bronchial arteries. He died from a thrombosis of the bronchial arterial branches at the age of 22 mo. Although his birth measurements at 36 wk gestation were normal (weight 2,300 g [10th percentile], length 46 cm [10th–25th percentile], and head circumference 33 cm [25th–50th percentile]), he had short stature at autopsy (length 78 cm [−3.26 SD] and head circumference 48 cm [25th–50th percentile]). He was not able to walk but could feed himself and spoke in short sentences and thus showed no evidence of mental retardation. Patient MWS4-BE is a female infant who is the third child of seemingly unrelated healthy parents. She was born at 38 wk gestation, weighing 3,180 g (50th percentile), with length of 47 cm (25th percentile), and a head circumference of 34 cm (50th–75th percentile). At birth, she was noted to have bilateral anophthalmia, with a right-sided diaphragmatic hernia, pulmonary hypoplasia, and bilateral hydronephrosis. She was last assessed at the age of 3 mo. The family history was significant. The first child was male and was born at 40 wk gestation, weighing 3,200 g (10th–25th percentile) with a length of 49.5 cm (10th percentile). This infant died at 24 h of age. At postmortem examination, pulmonary hypoplasia, unilobular left lung, tetralogy of Fallot, patent ductus arteriosus, undescended testes, horseshoe kidney, and hypoplastic renal arteries were noticed. No mention was made of the eyes in the postmortem report or neonatal record. The second child was female and was born at 39 wk gestation, weighing 3,160 g (25th–50th percentile). Bilateral anophthalmia was noted at birth. This child also died in the first 24 h of life. At postmortem examination, pulmonary hypoplasia with unilobular lungs was noted, in addition to the severe eye malformation. Patent ductus arteriosus, coarctation of the aorta, and uterine dysplasia were also reported. Unfortunately, no samples were available from the first two children. Patient MWS6-BK was the offspring of a marriage between Pakistani first cousins. The mother had two midtrimester miscarriages. The girl was born at 36 wk gestation, with normal measurements (weight 2,750 g [25th–50th percentile], length 47 cm [25th percentile], and head circumference 33 cm [25th–50th percentile]. She died at age 7 h from respiratory insufficiency with rapid deterioration. Autopsy showed absent eyes with small palpebral fissures, left-sided diaphragmatic hernia, right-sided diaphragmatic eventration, bilateral severe lung hypoplasia, structural normal heart with wide-open ductus arteriosus, bicornuate uterus, small kidneys (6 and 5.7 g) with prominent but microscopically normal adrenals (7.9 g), Meckel diverticle, and prominent labia minora. Karyotyping revealed homozygosity for the common pericentromeric inversion of chromosome 9. Skeletal survey revealed no abnormalities.
The phenotype of our patients with STRA6 mutations shows overlap with that of the sib pair reported as having “Matthew-Wood syndrome” (MIM 601186).20 However, one of our patients, CD50396, who had the full clinical picture described in this family, including cleft palate, hypoplastic alae nasi, hypoplastic spleen, and bicornuate uterus, did not show a STRA6 mutation. This patient may still harbor a STRA6 mutation outside the analyzed region or may indicate locus heterogeneity for this disorder. Nevertheless, there may be subtle differences in the facial dysmorphisms—in particular, the shape of the nose and eyebrows, the involvement of the palate, and the large head size relative to body length—between patients with STRA6 mutations and those with Matthew-Wood syndrome, but for these to be considered significant, we will need to analyze further cases. The combination of microphthalmia, pulmonary hypoplasia, and diaphragmatic hernia has been described in five patients with sporadic disease,21 all of whom had intrauterine growth retardation, a feature not seen in our patients with STRA6 mutations. The combination of severe microphthalmia, Fallot tetralogy, and diaphragmatic hernia has been reported in a child with de novo balanced chromosomal translocation (1;15)(q41;q21.2),22 but the breakpoint on chromosome 15 is located several bands proximal to the STRA6 locus. These sporadic cases and cases of Matthew-Wood syndrome may not be associated with STRA6 mutations but are likely to be caused by a gene from the same pathway.
STRA6 belongs to a novel group of retinoic acid (RA)–inducible genes that are likely to be direct targets of the retinoid receptors, such as RXRα and RARγ.23 STRA6 is predicted to encode a highly hydrophobic transmembrane protein, which does not display similarities with other known integral membrane proteins. Immunohistochemical and RNA in situ hybridization analyses revealed site- and stage-specific expression of Stra6 during murine embryonic development. In placenta, Stra6 is prominently expressed in the region of exchanges between maternal and embryonic circulations (i.e., yolk sac and labyrinthine zone of mature placenta). During embryogenesis, Stra6 is strongly expressed in the periocular mesenchyme, in the developing eyes, in respiratory mesenchymes, and in respiratory/bronchial epithelium, as well as in the developing CNS (meninges, cranial ganglia, choroid plexi, and brain microvasculature) and in different embryonic gut derivatives (the epithelium of the pharyngeal pouches, mesenchyme of the esophagus, stomach, intestine, and rectum). In adult organs, it is strongly expressed at the level of blood-organ barriers. These barriers are made of epithelial and endothelial cells connected by tight junctions and play a role in limiting passive diffusion of chemicals, as well as in the facilitative transport of many nutrients. The high expression of Stra6 in 16 of these different barriers, together with its localization in the basal region of the Sertoli cell plasma membrane, suggested that it may function as a component of an as-yet-unidentified transport machinery.12,23
The RA signaling pathway is complex and incompletely understood. RA appears to directly regulate >500 proteins.24 RA signaling is known to be important in normal lung and alveolar development.25 The developing diaphragm,26,27 heart,28 and CNS24 also strongly depend on proteins associated with the metabolism and binding of retinoids. It is interesting that the homozygous null mutation was compatible with life (family 2), whereas the patients with missense mutations all died of respiratory insufficiency (family 1: MWS1-EE, MWS4-BE, and MWS6-BK). The assembly of an altered protein into the membrane thus may be more deleterious than the total absence of the protein. Alveolar dysplasia and pulmonary hypoplasia may be the result of an impaired RA signal transduction resulting in abnormal levels of elastin production, which is essential for alveolization.25
Congenital diaphragmatic hernia (CDH) is an important component of the phenotype in the STRA6 mutation-positive cases, and, in this malformation, the etiology is unclear in most isolated and syndromal cases. CDH has an incidence of 1 in 3,000 births and a high mortality rate of ∼33%–58%.29 Many cytogenetic anomalies have been described in CDH involving almost every chromosome but most commonly chromosome 15.30 A 5-Mb critical interval has been defined molecularly on 15q26.1-q26.2,31 but no causal mutations in individual genes within this region have been identified in CDH cases.32 The only gene that has been implicated in diaphragmatic and lung development in humans is FOG2, or ZFPM2 (8q23),33 which was found mutated in a single deceased child with bilateral pulmonary hypoplasia and abnormally muscularized diaphragm but also in some patients with Fallot tetralogy.34 Alveolar capillary dysplasia is a rare but probably underdiagnosed cause of persistent pulmonary hypertension of the neonate, usually resulting in death within 4–6 wk after birth.35 It will be interesting to investigate isolated cases for STRA6 mutations.
In summary, we show that homozygous mutations in STRA6 cause a pleiotropic, multisystem malformation syndrome characterized by bilateral anophthalmia, mild facial dysmorphism, normal intrauterine growth, early lethality in most cases, and a variety of malformations of the lungs, diaphragm, heart, and urogenital system. Profound mental retardation and short stature with relatively large head were present in the one patient with long-term survival. This STRA6 anophthalmia syndrome shows phenotypic overlap with Matthew-Wood syndrome, which may be distinct, however, in facial dysmorphism and involvement of the palate. Very preliminary genotype-phenotype correlation suggests that missense mutations may be associated with early lethality due to respiratory insufficiency, whereas truncating mutation may be compatible with long-term survival and normal lung function.
Further studies are necessary to elucidate whether STRA6 and related genes from the RA pathway are also involved in the pathogenesis of other types of syndromal or isolated anophthalmia, diaphragmatic hernia, and alveolar capillary dysplasia.
_Note added in proof.—_STRA6 was just identified as the long-sought membrane receptor for retinol binding protein that mediates cellular uptake of vitamin A.36
Acknowledgments
We thank the family members for their kind participation, Claudia Preller for excellent technical assistance, and Renate Ulmer for cultivation of the fibroblast cell line. We also thank Dr. Kathy Williamson for help with samples transfer and Dr. Jose Martinez for sending samples and clinical details. This work was supported in part by grant C2 from the SFB539 (A. Reis) and RA 833/7-1 (A. Rauch), funded by the Deutsche Forschungsgemeinschaft (DFG), and by the German Federal Ministry of Science and Education through the National Genome Research Network (grant 01GR0416 to G.N., C.B., and P.N.).
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- ClustalW, http://www.ebi.ac.uk/clustalw/
- ELM, The Eukaryotic Linear Motif Resource for Functional Sites in Proteins, http://elm.eu.org/
- SwissProt, http://ca.expasy.org/sprot/ (for reference sequence Q8TB21_Human)
- National Center for Biotechnology Information (NCBI), http://www.ncbi.nlm.nih.gov/
- NPS@ Consensus Secondary Structure Prediction, http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_seccons.html
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/
- TMHMM, http://www.cbs.dtu.dk/services/TMHMM-2.0/ (for prediction of transmembrane helices in proteins)
- TMpred, http://www.ch.embnet.org/software/TMPRED_form.html (for prediction of transmembrane helices in proteins)
- TopPred, http://bioweb.pasteur.fr/seqanal/interfaces/toppred.html (for prediction of transmembrane helices in proteins)
- University of California Santa Cruz (UCSC) Genome Browser, http://genome.ucsc.edu/cgi-bin/hgTracks (reference sequences NT_010194 and NM_022369)
References
- 1.Morrison D, FitzPatrick D, Hanson I, Williamson K, van Heyningen V, Fleck B, Jones I, Chalmers J, Campbell H (2002) National study of microphthalmia, anophthalmia, and coloboma (MAC) in Scotland: investigation of genetic aetiology. J Med Genet 39:16–22 10.1136/jmg.39.1.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vogt G, Puho E, Czeizel AE (2005) A population-based case-control study of isolated anophthalmia and microphthalmia. Eur J Epidemiol 20:939–946 10.1007/s10654-005-2382-z [DOI] [PubMed] [Google Scholar]
- 3.Bar-Yosef U, Abuelaish I, Harel T, Hendler N, Ofir R, Birk OS (2004) CHX10 mutations cause non-syndromic microphthalmia/ anophthalmia in Arab and Jewish kindreds. Hum Genet 115:302–309 10.1007/s00439-004-1154-2 [DOI] [PubMed] [Google Scholar]
- 4.Ferda Percin E, Ploder LA, Yu JJ, Arici K, Horsford DJ, Rutherford A, Bapat B, Cox DW, Duncan AM, Kalnins VI, et al (2000) Human microphthalmia associated with mutations in the retinal homeobox gene CHX10. Nat Genet 25:397–401 10.1038/78071 [DOI] [PubMed] [Google Scholar]
- 5.Fantes J, Ragge NK, Lynch SA, McGill NI, Collin JR, Howard-Peebles PN, Hayward C, Vivian AJ, Williamson K, van Heyningen V, et al (2003) Mutations in SOX2 cause anophthalmia. Nat Genet 33:461–463 10.1038/ng1120 [DOI] [PubMed] [Google Scholar]
- 6.Abecasis GR, Cherny SS, Cookson WO, Cardon LR (2001) GRR: graphical representation of relationship errors. Bioinformatics 17:742–743 10.1093/bioinformatics/17.8.742 [DOI] [PubMed] [Google Scholar]
- 7.O’Connell JR, Weeks DE (1998) PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abecasis GR, Cherny SS, Cookson WO, Cardon LR (2002) Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30:97–101 10.1038/ng786 [DOI] [PubMed] [Google Scholar]
- 9.Gudbjartsson DF, Jonasson K, Frigge ML, Kong A (2000) Allegro, a new computer program for multipoint linkage analysis. Nat Genet 25:12–13 10.1038/75514 [DOI] [PubMed] [Google Scholar]
- 10.Thiele H, Nurnberg P (2005) HaploPainter: a tool for drawing pedigrees with complex haplotypes. Bioinformatics 21:1730–1732 10.1093/bioinformatics/bth488 [DOI] [PubMed] [Google Scholar]
- 11.Ruschendorf F, Nurnberg P (2005) ALOHOMORA: a tool for linkage analysis using 10K SNP array data. Bioinformatics 21:2123–2125 10.1093/bioinformatics/bti264 [DOI] [PubMed] [Google Scholar]
- 12.Sapin V, Bouillet P, Oulad-Abdelghani M, Dastugue B, Chambon P, Dolle P (2000) Differential expression of retinoic acid-inducible (Stra) genes during mouse placentation. Mech Dev 92:295–299 10.1016/S0925-4773(00)00241-0 [DOI] [PubMed] [Google Scholar]
- 13.Krogh A, Larsson B, von Heijne G, Sonnhammer EL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- 14.Reddy EP, Korapati A, Chaturvedi P, Rane S (2000) IL-3 signaling and the role of Src kinases, JAKs and STATs: a covert liaison unveiled. Oncogene 19:2532–2547 10.1038/sj.onc.1203594 [DOI] [PubMed] [Google Scholar]
- 15.Claros MG, von Heijne G (1994) TopPred II: an improved software for membrane protein structure predictions. Comput Appl Biosci 10:685–6867704669 [Google Scholar]
- 16.Combet C, Blanchet C, Geourjon C, Deleage G (2000) NPS@: network protein sequence analysis. Trends Biochem Sci 25:147–150 10.1016/S0968-0004(99)01540-6 [DOI] [PubMed] [Google Scholar]
- 17.Puntervoll P, Linding R, Gemund C, Chabanis-Davidson S, Mattingsdal M, Cameron S, Martin DM, Ausiello G, Brannetti B, Costantini A, et al (2003) ELM server: a new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Res 31:3625–3630 10.1093/nar/gkg545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Calo V, Migliavacca M, Bazan V, Macaluso M, Buscemi M, Gebbia N, Russo A (2003) STAT proteins: from normal control of cellular events to tumorigenesis. J Cell Physiol 197:157–168 10.1002/jcp.10364 [DOI] [PubMed] [Google Scholar]
- 19.Shabb JB (2001) Physiological substrates of cAMP-dependent protein kinase. Chem Rev 101:2381–2411 10.1021/cr000236l [DOI] [PubMed] [Google Scholar]
- 20.Seller MJ, Davis TB, Fear CN, Flinter FA, Ellis I, Gibson AG (1996) Two sibs with anophthalmia and pulmonary hypoplasia (the Matthew-Wood syndrome). Am J Med Genet 62:227–229 [DOI] [PubMed] [Google Scholar]
- 21.Lee SYR, Shiu YK, Ng WF, Chow CB (2006) Another patient with pulmonary hypoplasia, microphthalmia and diaphragmatic hernia. Clin Dysmorphol 15:43–44 10.1097/01.mcd.0000181604.78252.29 [DOI] [PubMed] [Google Scholar]
- 22.Smith SA, Martin KE, Dodd KL, Young ID (1994) Severe microphthalmia, diaphragmatic hernia and Fallot’s tetralogy associated with a chromosome 1;15 translocation. Clin Dysmorphol 3:287–291 [PubMed] [Google Scholar]
- 23.Bouillet P, Sapin V, Chazaud C, Messaddeq N, Decimo D, Dolle P, Chambon P (1997) Developmental expression pattern of Stra6, a retinoic acid-responsive gene encoding a new type of membrane protein. Mech Dev 63:173–186 10.1016/S0925-4773(97)00039-7 [DOI] [PubMed] [Google Scholar]
- 24.Blomhoff R, Blomhoff HK (2006) Overview of retinoid metabolism and function. J Neurobiol 66:606–630 10.1002/neu.20242 [DOI] [PubMed] [Google Scholar]
- 25.Kumar VH, Lakshminrusimha S, Abiad MT, Chess PR, Ryan RM (2005) Growth factors in lung development. Adv Clin Chem 40:261–316 [DOI] [PubMed] [Google Scholar]
- 26.Greer JJ, Babiuk RP, Thebaud B (2003) Etiology of congenital diaphragmatic hernia: the retinoid hypothesis. Pediatr Res 53:726–730 10.1203/01.PDR.0000062660.12769.E6 [DOI] [PubMed] [Google Scholar]
- 27.Gallot D, Marceau G, Coste K, Hadden H, Robert-Gnansia E, Laurichesse H, Dechelotte PJ, Labbe A, Dastugue B, Lemery D, et al (2005) Congenital diaphragmatic hernia: a retinoid-signaling pathway disruption during lung development? Birth Defects Res A Clin Mol Teratol 73:523–531 10.1002/bdra.20151 [DOI] [PubMed] [Google Scholar]
- 28.Zile MH (2004) Vitamin A requirement for early cardiovascular morphogenesis specification in the vertebrate embryo: insights from the avian embryo. Exp Biol Med (Maywood) 229:598–606 [DOI] [PubMed] [Google Scholar]
- 29.Langham MR Jr, Kays DW, Ledbetter DJ, Frentzen B, Sanford LL, Richards DS (1996) Congenital diaphragmatic hernia: epidemiology and outcome. Clin Perinatol 23:671–688 [PubMed] [Google Scholar]
- 30.Lurie IW (2003) Where to look for the genes related to diaphragmatic hernia? Genet Couns 14:75–93 [PubMed] [Google Scholar]
- 31.Klaassens M, van Dooren M, Eussen HJ, Douben H, den Dekker AT, Lee C, Donahoe PK, Galjaard RJ, Goemaere N, de Krijger RR, et al (2005) Congenital diaphragmatic hernia and chromosome 15q26: determination of a candidate region by use of fluorescent in situ hybridization and array-based comparative genomic hybridization. Am J Hum Genet 76:877–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Slavotinek AM, Moshrefi A, Davis R, Leeth E, Schaeffer GB, Burchard GE, Shaw GM, James B, Ptacek L, Pennacchio LA (2006) Array comparative genomic hybridization in patients with congenital diaphragmatic hernia: mapping of four CDH-critical regions and sequencing of candidate genes at 15q26.1-15q26.2. Eur J Hum Genet 14:999–1008 10.1038/sj.ejhg.5201652 [DOI] [PubMed] [Google Scholar]
- 33.Ackerman KG, Herron BJ, Vargas SO, Huang H, Tevosian SG, Kochilas L, Rao C, Pober BR, Babiuk RP, Epstein JA, et al (2005) Fog2 is required for normal diaphragm and lung development in mice and humans. PLoS Genet 1:58–65 10.1371/journal.pgen.0010010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clark KL, Yutzey KE, Benson DW (2006) Transcription factors and congenital heart defects. Annu Rev Physiol 68:97–121 10.1146/annurev.physiol.68.040104.113828 [DOI] [PubMed] [Google Scholar]
- 35.Michalsky MP, Arca MJ, Groenman F, Hammond S, Tibboel D, Caniano DA (2005) Alveolar capillary dysplasia: a logical approach to a fatal disease. J Pediatr Surg 40:1100–1105 10.1016/j.jpedsurg.2005.03.067 [DOI] [PubMed] [Google Scholar]
- 36.Kawaguchi R, Yu J, Honda J, Hu J, Whitelegge J, Ping P, Wiita P, Bok D, Sun H (2007) A membrane receptor for retinol binding protein mediates cellular uptake of vitamin A. Science (http://www.sciencemag.org/cgi/rapidpdf/1136244v1.pdf) (electronically published ahead of print January 25, 2007; accessed January 29, 2007) [DOI] [PubMed] [Google Scholar]