Identification, activation, and selective in vivo ablation of mouse NK cells via NKp46 (original) (raw)
Abstract
Natural killer (NK) cells contribute to a variety of innate immune responses to viruses, tumors and allogeneic cells. However, our understanding of NK cell biology is severely limited by the lack of consensus phenotypic definition of these cells across species, by the lack of specific marker to visualize them in situ, and by the lack of a genetic model where NK cells may be selectively ablated. NKp46/CD335 is an Ig-like superfamily cell surface receptor involved in human NK cell activation. In addition to human, we show here that NKp46 is expressed by NK cells in all mouse strains analyzed, as well as in three common monkey species, prompting a unifying phenotypic definition of NK cells across species based on NKp46 cell surface expression. Mouse NKp46 triggers NK cell effector function and allows the detection of NK cells in situ. NKp46 expression parallels cell engagement into NK differentiation programs because it is detected on all NK cells from the immature CD122+NK1.1+DX5− stage and on a minute fraction of NK-like T cells, but not on CD1d-restricted NKT cells. Moreover, human NKp46 promoter drives NK cell selective expression both in vitro and in vivo. Using NKp46 promoter, we generated transgenic mice expressing EGFP and the diphtheria toxin (DT) receptor in NK cells. DT injection in these mice leads to a complete and selective NK cell ablation. This model paves a way for the in vivo characterization and preclinical assessment of NK cell biological function.
Keywords: genetic models, innate immunity
Natural killer (NK) cells are large granular lymphocytes that belong to the innate immune system (1). NK cells are present in lymphoid organs as well as in nonlymphoid peripheral tissues (2). They are involved in defense mechanisms against several types of microbial infections and tumors (3). They also have a role in shaping adaptive immune responses and in the control of placental development (4, 5). Strategies are emerging to apply NK cells as therapeutic agents against a broad range of malignancies (6, 7). However, several NK cell features have limited our understanding of their biological function.
First, NK cells share phenotypic properties with various T cell populations such as CD1d-restricted NKT cells, γδ T cells, and discrete subsets of antigen-experienced CD122+CD8+ T cells (8). NK cell identification by flow cytometry thus requires other markers to exclude T cells.
Second, there is no consensus phenotypic definition of NK cells across species. In humans, NK cells are defined as CD56+CD3− lymphocytes. In NK cells, CD56 corresponds to the 140-kDa isoform of the neural cell adhesion molecule (N-CAM). Yet, N-CAM is also expressed by T cell subsets, muscle cells, and neurons, but it is not expressed by murine NK cells (9). In the rat, NK cells express the activating receptor NKR-P1A, but this molecule is also expressed by T cell subsets (10). In the mouse, the widely used PK136 antibody reacts with NK1.1, an epitope shared by the activating receptor NKR-P1C in C57BL/6 mice and the inhibitory receptor NKR-P1B in SJL mice (11). However, NK cells from other mouse strains do not react with the anti-NK1.1 antibody, due to allelic divergence of the nkrp1b/c genes (11). In NK1.1− mouse strains, the identification of NK cells is based on the expression of the integrin subunit CD49b that is recognized by DX5 antibodies (12), despite its expression on T cell and myeloid subsets (13, 14). These various phenotypic NK cell definitions and the lack of specific NK cell markers impede the comparison of data across species.
Third, there is no genetic model where NK cells can be selectively deleted (15, 16). Transgenic mice lacking NK cells but with a normal T/NKT cell compartment have been reported (17). The cause of NK cell ablation in these mice is unknown, but is linked to the expression of the ubiquitous transcription factor ATF2, raising the possibility of other defects yet to be further investigated (18). In vivo NK cell depletion has thus so far relied on anti-asialo-GM1 or anti-NK1.1 depleting antibodies. However, the expression of these markers outside of the NK cell compartment has hampered the interpretation of results obtained with these protocols.
Natural cytotoxicity receptors (NCRs) include the NKp30, NKp44, and NKp46 molecules, and are Ig-like transmembrane glycoproteins (19). Their transmembrane regions contain positively charged amino acids allowing association with the ITAM-bearing polypeptides CD3ζ and FcRγ for NKp46 and NKp30 or KARAP/DAP12 for NKp44 (20). NCRs are involved in the recognition of tumor targets, but the cellular ligands recognized by the NCRs are still elusive (20). NKp46 (product of the NCR1 gene) has also been shown to recognize the hemagglutinin of influenza virus and the hemagglutinin-neuraminidase of Sendai virus (21). NKP46 is conserved between human and mouse, whereas no mouse orthologue of NKP44/NCR2 has been found (22), and mouse Nkp30/Ncr3 is a pseudogene in Mus musculus (23).
Previous studies have shown that NKp46 expression was restricted to CD56+CD3−HLA-DR− lymphocytes in human peripheral blood mononuclear cells (24). Here, we show that NKp46 cell surface expression defines NK cells in human, in all strains of mice tested, and in three species of monkey, leading to the proposal that NKp46 is a unifying marker for NK cells across mammalian species. Moreover, NK cells can be unambiguously visualized in situ and selectively activated via NKp46. Finally, we identified a 400-bp NKP46 promoter that drives NK cell-specific gene expression, allowing the generation of the first transgenic mouse model of conditional and selective NK cell ablation in vivo.
Results
Cell Surface Expression of NKp46 Defines NK Cells Across Species.
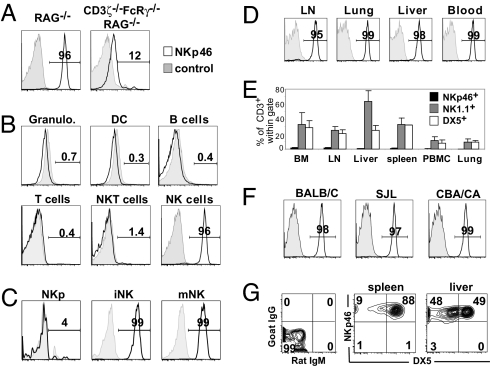
NKp46 expression was previously shown to be virtually restricted to CD56+CD3−HLA-DR− cells in humans (24). Here, we further show that, unlike CD56, NKp46 was not expressed by γδ T cells or CD1d-restricted Vα24+ T cells that represent minor fractions of the human blood CD3+ population [supporting information (SI) Fig. 6]. Thus, NKp46 cell surface expression defined human peripheral blood NK cells in normal individuals. These results prompted us to test whether NKp46 can also be used as a phenotypic marker of mouse NK cells, using either monoclonal (29A1.4) or affinity-purified polyclonal antibodies raised against a chimeric mouse NKp46-Fc fusion protein (SI Fig. 7). NKp46 expression was detected in nearly all NK1.1+ cells from RAG−/− mice but was severely reduced when cells were isolated from CD3ζ−/−FcRγ−/−RAG−/− mice (Fig. 1A). The latter lack the CD3ζ and FcRγ adapter molecules required for cell surface expression of human NKp46 (25). These data thus suggest that both human and mouse NKp46 depend on CD3ζ and FcRγ for cell surface expression and establish the specificity of the mouse NKp46 staining performed with anti-NKp46 monoclonal and polyclonal antibodies. In C57BL/6 mice, NKp46 was not expressed in granulocytes, dendritic cells, B cells, T cells, CD1d-α-gal-cer tetramer+ NKT cells (Fig. 1B), monocytes or macrophages (data not shown). By contrast, NK cells, defined as NK1.1+CD3−, were characterized by a high and uniform expression of NKp46 (Fig. 1B). In line with previous studies in humans (26), the cell surface expression of NKp46 is initiated at the immature stage of NK cell development (CD122+NK1.1+DX5−) in the bone marrow (Fig. 1C) and subsequently remains at the same level by all NK cells isolated from all organs tested (Fig. 1D and data not shown). Thus, NKp46 cell surface expression defines NK cells in C57BL/6 mice in contrast to NK1.1 alloantigen that is expressed by a large percentage of CD3+ cells in various organs (Fig. 1E). Next, we measured NKp46 expression in NK1.1− strains of mice, where NK cell identification is the most problematic. A bright NKp46 staining was observed on DX5+CD3− cells from all strains of mice tested (BALB/C, SJL, CBA/CA, DBA/2, B6.129, NOD, NZW; Fig. 1F and data not shown). We also noticed the presence of a substantial population of NKp46+DX5−CD3− cells (data not shown). These cells were also present in C57BL/6 mice where they expressed NK1.1. They correspond to immature NK cells, enriched in young mice, especially in the liver when they represented up to 50% of NK cells (Fig. 1G) (27). Thus, NKp46 cell surface expression defines NK cells in all mouse strains in contrast to DX5/CD49b that is expressed by a large percentage of CD3+ cells in various organs (Fig. 1E) and by only a subset of NK cells (Fig. 1G). The cell surface expression of DX5 has also been reported on lung basophils, as well as on ill-defined subsets of CD3−NK1.1− splenocytes (28). Importantly, NKp46 is not expressed by bone marrow basophils, or by the small fraction of splenic NK1.1−CD3−DX5low cells that are likely of myeloid origin (data not shown).
Fig. 1.
Expression of the activating NKp46 cell surface receptor on mouse NK cells. (A) Flow cytometric measurement of NKp46 expression in gated NK1.1+ spleen cells from RAG−/− and RAG−/−CD3ζ−/−FcRγ−/− mice (C57BL/6 background). NKp46 expression (open histogram, thick line) or isotype control (gray histogram, thin line) is shown in the indicated subsets in A and in subsequent panels. (B) Indicated cell types were identified from C57BL/6 splenocytes; Granulo., granulocytes; DC, dendritic cells. (C) Flow cytometric measurement of NKp46 expression on C57BL/6 NK precursors (NKp), immature and mature NK cells, identified as described (47) as CD122+lin−NK1.1−DX5−, CD122+NK1.1+DX5−, and CD122+NK1.1+DX5+, respectively. (D) Flow cytometric measurement of NKp46 expression in gated NK1.1+ CD3− cells from C57BL/6 mouse lymph nodes (inguinal), lung, liver, and peripheral blood. (E) Comparison of the percentage of CD3+ cells within the NKp46+ (black bars), NK1.1+ (gray bars), and DX5+ subsets in various organs of C57BL/6 mice. (F) Flow cytometric measurement of NKp46 expression in gated splenic DX5+CD3− cells from the indicated strains. (G) Flow cytometric measurement of NKp46/DX5 expression in gated NK1.1+CD3− cells from the spleen or the liver (27), obtained from 5-week-old C57BL/6 mice. (Left) Isotype staining. Results in A–C and D–G show one representative set of data of three independent experiments. Results in E are presented as the mean ± SD of at least three independent experiments. Similar staining results were obtained by using either affinity purified goat anti-mouse NKp46 polyclonal antibodies and the rat anti-NKp46 29A1.4 mAb (SI Fig. 7).
Although these results indicate that NKp46 represents the most specific surface marker for mouse NK cells, <2% of NKp46+ cells also express low levels of surface CD3ε (Fig. 1E). To determine whether antigenic exposure induced NKp46 expression on T cells, we infected C57BL/6 mice with mouse cytomegalovirus, which is associated with a robust CD8+ T cell response. At the peak of the response, 40–50% of CD8+ T cells displayed an activated CD43high phenotype and acquired NKG2A/C/E cell surface expression (29) (SI Fig. 8). However, all T cells in virus-infected mice remained NKp46− (SI Fig. 8). These results indicate that acute T cell stimulation does not lead to the cell surface expression of NKp46. Rather, NKp46+CD3+ cells could originate from chronically activated T cells reprogrammed into NK-like cells, as recently proposed for intraepithelial T cells in celiac patients (30). Consistent with this hypothesis, these NKp46+CD3+ cells harbor a very peculiar cell surface phenotype, as they are NK1.1+Ly49+CD1d tetramer−CD4−CD8−, and 75% of them are TCRγδ+. These NKp46+ γδ T cells represent a minute fraction of γδ T cells, as 95–99% of splenic and nearly 100% of gut intraepithelial γδ T cells are NKp46− (data not shown).
NK Cells Can Be Visualized in Situ and Activated via NKp46.
Visualization of NK cells in situ has been hampered by the lack of reactivity and specificity of NK cell markers. Therefore, we tested the reactivity of anti-NKp46 antibodies on mouse tissue sections. A bright staining was observed on spleen sections in comparison with control antibodies. This staining was specific to NKp46, as it was abrogated by the addition of NKp46-Fc recombinant protein (data not shown). Three-color immunofluorescence staining showed that NK cells were mostly localized in the splenic red pulp at steady state. Under these conditions, NK cells were also present in parafollicular (Fig. 2) and medullar (SI Fig. 9) lymph node areas. In spleen and lymph nodes, NK cells were in close proximity to CD11cbright and CD11bbright cells that include dendritic cell and macrophage subsets. NKp46 antibodies also stained NK cells in other organs such as the lung or the liver (SI Fig. 9). Thus, NKp46 staining allows the visualization of NK cells in their microenvironment.
Fig. 2.
NKp46 staining allows the in situ visualization of mouse NK cells. For C57BL/6 mouse spleen (A) or inguinal lymph node (B), 7-μm serial frozen sections were fixed with acetone and stained with anti-NKp46 antiserum (green), CD19 mAb (red), and either CD3, CD11c, or CD11b mAb (blue). Samples were analyzed by confocal microscopy. A representative picture for each group is shown in the same anatomical region for each staining. WP, white pulp; RP, red pulp; T, T cell zone. (Original magnification, ×16.)
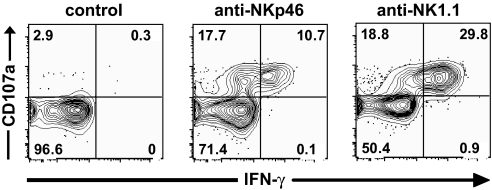
Consistent with NKp46 association with CD3ζ or FcRγ and data obtained with human NK cells (25), NKp46 antibody-mediated triggering induced freshly isolated mouse NK cells to secrete IFN-γ and to release their cytotoxic granules content, as measured by surface exposure of the lytic granule marker CD107a (31) (Fig. 3). Thus, resting NK cells may be activated through NKp46 providing means to specifically activate NK cell effector functions. These results contrast with a recent study showing that only CD16 engagement triggers freshly isolated human NK cells (32). However, the density of CD16 surface expression is much higher than that of NKp46 on human NK cells. In contrast, CD16 is expressed at low density on freshly isolated mouse NK cells and NKp46 is expressed at higher levels. Our reproducible activation of freshly isolated mouse NK cells by anti-NKp46 reagents (both 29A1.4 mAb and antiserum), thus indicate that CD16 is not the only NK cell surface receptor whose engagement leads to NK cell activation. This finding is consistent with the association of NKp46 with the same ITAM-bearing transduction polypeptides (FcRγ and/or CD3ζ) as CD16 (19).
Fig. 3.
Mouse NK cells can be activated via NKp46. C57BL/6 spleen cell suspensions were stimulated for 4 h in wells coated with the indicated antibodies, in the presence of soluble anti-CD107a antibodies. Control antibodies included mouse and goat IgG. Cells were then stained for surface DX5/CD3 and intracellular IFN-γ. Results show the expression of CD107a and IFN-γ in gated DX5+CD3− cells, and are representative of five independent experiments.
In Vivo Tagging of NK Cells via NKP46.
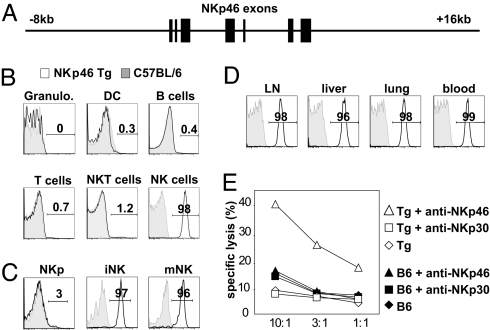
The dissection of NK cell biological functions has been complicated by the lack of selective deficiency models. Having shown that NKp46 was a specific NK cell marker, our goal was to use NKP46 regulatory sequences to create such models. To validate the feasibility of this strategy, we first generated a transgenic vector consisting of a 24-kb human genomic region located between the NKP46 adjacent genes FCAR and NALP7 (Fig. 4A). From a transgenic founder (referred to as huNKp46 Tg hereafter), offspring were obtained at Mendelian frequencies, developed normally, and were fertile. BAB281 (anti-human NKp46) mAb that do not cross-react with mouse NKp46 (SI Fig. 7) (24) were used to assess the cell surface expression of human NKp46 in these mice. Human NKp46 was not expressed on granulocytes, dendritic cells, B cells, T cells, and CD1d-α-gal-cer tetramer+ NKT cells but expressed at a high and uniform level on NK cells (Fig. 4B). Moreover, human NKp46 starts to be expressed at the immature stage of NK cell development in the bone marrow (Fig. 4C) and remains subsequently expressed at the same level by all NK cells isolated from all organs tested (Fig. 4D and data not shown). Remarkably, the pattern of human NKp46 expression in huNKp46 Tg mice was thus similar to that of endogenous mouse NKp46 molecules in parental mice. Therefore, the cell surface expression of human NKp46 defined NK cells in huNKp46 Tg mice demonstrating that NKP46 regulatory sequences could be used to drive NK-specific gene expression. NK cells in huNKp46 Tg mice exhibited normal counts, phenotype and effector function (data not shown). Importantly, redirected lysis was induced through human NKp46 (Fig. 4E), indicating that human NKp46 molecules are functional in mouse NK cells. Finally, i.v. administration of anti-human NKp46 mAb led to a nearly complete disappearance of NK cells from blood and all organs tested, 2 days after injection (SI Fig. 10). By contrast, NKT cell and γδ T cell counts were not significantly affected (SI Fig. 10), indicating that huNKp46 Tg mice can be used as a mouse model of NK cell-selective depletion.
Fig. 4.
NKP46 genomic sequence can be used to genetically tag mouse NK cells. (A) Schematic representation of the human genomic fragment used for transgenesis. NKP46 exons are shown as black bars. (B) Flow cytometric measurement of human NKp46 expression in various gated subsets from huNKp46 transgenic mice. The indicated cell types were identified as described in Methods. Granulo., granulocytes; DC, dendritic cells. Human NKp46 expression (open histogram, thick line) or isotype control (gray histogram, thin line) is shown in the indicated subsets in B and in subsequent panels. (C) Flow cytometric measurement of human NKp46 expression on NK precursors (NKp), immature and mature NK cells, identified as described in Fig. 1. (D) Flow cytometric measurement of human NKp46 expression in gated NK1.1+ CD3− cells from huNKp46 Tg mouse lymph nodes (inguinal), lung, liver and peripheral blood. (E) Redirected lysis assay of LAK cells derived from B6 (C57BL/6) or huNKp46 Tg (Tg) spleen cells against Daudi cells incubated with the indicated antibodies. The cytolytic function of LAK cells prepared from B6 and huNKp46 Tg mice were comparable. Results are representative of three experiments.
Generation of Mouse Genetic Models for the in Vivo Dissection of NK Cell Function.
Next, we sought to identify a minimal promoter from the 24-kb huNKp46 construct. DNA alignments between several mammalian species revealed a 400-bp sequence upstream of NKP46 exon 1 that is highly conserved through evolution and could thus correspond to a functionally active NKP46 promoter region (SI Fig. 11). A putative TATA box was predicted in this region upstream of the transcription start site and downstream of a series of consensus motifs for transcription factor binding sites. This 400-bp sequence was cloned into a luciferase reporter vector to examine its capacity to promote transcription in vitro. The 400-bp NKP46 sequence drove efficient luciferase expression in the NK cell line, NKL, as compared with the ubiquitous SV40 promoter (Fig. 5A). In contrast, this 400-bp NKP46 sequence was inefficient in the erythroleukemia K562 line, suggesting that the 400-bp sequence upstream of NKP46 exon 1 featured both efficient promoter activity and NK cell specificity. We thus used this 400-bp NKP46 sequence to drive the transgenic expression of a bicistronic cassette consisting of the cDNA sequences encoding for the human diphtheria toxin (DT) receptor (DTR) and EGFP (Fig. 5B). Expression of the DTR protein should confer DT sensitivity to NKp46-expressing cells leading to the selective ablation of these cells after DT administration (33). This construct, referred to as NKDTR/EGFP hereafter, was injected into fertilized mouse ovocytes. Of six founders, one displayed high transgene expression and was bred to start the NKDTR/EGFP transgenic mouse colony. Remarkably, EGFP was expressed in NK cells in these mice, confirming that the 400-bp NKP46 promoter retained the tissue-specificity of the endogenous Nkp46 regulatory sequences in vivo (Fig. 5C). DTR expression was similar to that of EGFP (data not shown). NK cells in NKDTR/EGFP transgenic mice occur at normal frequency and display a normal phenotype (data not shown). Two injections of DT at 24-h interval led to the disappearance of NK cells in mouse peripheral blood mononuclear cells (Fig. 5D), as well as in spleen, lymph nodes, bone marrow, liver, and lung (Fig. 5E). NK cell ablation persisted for at least 7 days, before a progressive repopulation of the circulating compartment. The kinetics of reconstitution of circulating NK cells is compatible with the 17-day NK cell half-life reported earlier (34). Importantly, DT injection did not affect T cell populations, including NKT cells and γδ T cells in all organs tested (Fig. 5F). To confirm the depletion of NK cells, splenocytes from control mice or NKDTR/EGFP mice treated with DT, were assayed in vitro against YAC-1 tumor cells. No cytotoxicity was detected when splenocytes were isolated from NKDTR/EGFP mice treated with DT, in contrast to control mice (Fig. 5G). These data show that DT infusion in NKDTR/EGFP mice leads to a specific depletion of NK cells in all organs, that is associated with the complete disappearance of in vitro cytotoxicity against YAC-1. As the small population of NKp46+ T cells expresses low level of NKp46 as compared with bone fide NK cells, it was not depleted using the above DT administration protocols (data not shown). The generation of NKDTR/EGFP transgenic mice thus provides an in vivo model of selective and conditional NK cell ablation.
Fig. 5.
Dissection of NKP46 regulatory sequences and generation of NKDTR/EGFP mice. (A) The putative 400-bp human NKP46 promoter was used in a dual luciferase assay in K562 or NKL cell lines. The assay was also performed with promoterless or SV40 promoter vectors for negative and positive control, respectively. (B) Schematic representation of the transgenic construct used to make NKDTR/EGFP transgenic mice. For details, see Methods. (C) Flow cytometric measurement of EGFP expression in immune cell types of NKDTR/EGFP or control mice, identified as in Fig. 1. Granulo, granulocytes. Results are representative of at least three experiments. (D) NKDTR/EGFP transgenic or control littermates were injected i.p. twice with DT within a 24-h interval. The percentage of NKp46+CD3− or CD3+ cells in the blood was measured over time after DT injections. Results are representative of at least three experiments. (E) NKDTR/EGFP (open histograms) and wild-type littermates (filled histograms) were injected with DT. Twenty-four hours later, the number of NK cells was measured in the indicated organs. Results show the mean ± SD number of NK cells in DT-treated wild-type or NKDTR/EGFP mice, expressed as the percentage of the controls (NK cell numbers in untreated wild-type mice); four mice per group. (F) NKDTR/EGFP transgenic or control mice were injected i.p. twice with DT within a 24-h interval. Forty-eight hours after DT injections, the percentage of NK cells, γδ T cells and CD1d-restricted NKT cells was measured in the peripheral blood. Representative results of three experiments are expressed as the percentage of indicated cell subsets as compared with control mice. (G) Spleen cells from mice in E were cultured o/n with IL-2 and used as effector cells in a standard 4-h Cr51 cytotoxicity assay against YAC-1 cells. Results are expressed as the mean ± SD percentage of specific lysis of YAC-1 cells with four mice in each group.
Discussion
Until now, the identification of mouse NK cells relied on NK1.1 or CD49b cell surface expression. However, these markers are not specific of NK cells. Moreover, NK1.1 and CD49b patterns of expression are not totally overlapping. In particular, subsets of NK cells do not express CD49b (Fig. 1G), which complicates the comparison of data regarding NK cells obtained in NK1.1+ and NK1.1− strains (15). By contrast, we show here that NKp46 is a specific marker for all NK cells (CD49b+ and CD49b−), in all mouse strains tested. This finding opens perspectives in the study of the role of NK cells in various conditions. For instance, there are numerous mouse disease models, either autoimmune or infectious, that rely on specific mouse strains. The use of NKp46 will thus make it possible to study of the role of NK cells in these strain-dependent disorders. In particular, the cell surface expression of NKp46 allows to detect NK cells in autoimmune prone strains such as NOD and NZW (data not shown), where the roles of NK cells remain to be precisely dissected or revisited (35, 36).
Moreover, the comparison of NK cells across species has been complicated by various phenotypic definitions of NK cells. Here, we show that NKp46 is selectively expressed by human and mouse NK cells, consistent with the original description of NKp46 in humans (24) and the recent generation of _Ncr1_gfp/gfp mice (37). In addition, we also documented the NK cell-specific expression of NKp46 in three monkey species (Baboon, Rhesus, and Cynomolgus; SI Fig. 12). Previous reports have shown that rat and bovine NK cells specifically express NKp46 (38, 39). We thus propose to unify the phenotypic definition of NK cells across mammalian species on the basis of NKp46 cell surface expression.
We show that NK cells can be visualized in situ by means of NKp46 staining on tissue sections. Our results document the presence of NK cells in the red pulp of the spleen, as well as in the paracortex and medulla of lymph nodes consistent with previous data (40–42). We further showed that NK cells in secondary lymphoid organs are in close proximity to CD11b+ and CD11c+ cells, consistent with the cross-talks between NK and DC (43), as well as NK and macrophages (44). These results provide the proof of principle that anti-NKp46 antibodies represent an efficient means for the identification of NK cells in situ. In addition, GFP-tagging in NKDTR-EGFP transgenic mice constitutes an alternative strategy of NK cell visualization.
To evaluate the role of NK cells in vivo, previous studies have classically used anti-NK1.1 antibodies to deplete NK cells in C57BL/6 mice, or anti-asialo-GM1 in other strains of mice. However, these markers are not NK cell-specific and might induce depletion of various other T cell subsets, complicating the interpretation of such experiments. Moreover, administration of antibodies may induce undesirable side effects, such as NK cell activation upon anti-NK1.1 cross-linking (45), or nonspecific effects through interactions with Fc receptors expressed on many cell types. To overcome these limitations, we took advantage of the identification of a functional NKP46 promoter to generate a mouse model of conditional NK cell ablation based on the DT/DTR system, successfully used before to ablate other cell types (33, 46). We showed that DT injection led to a complete and selective ablation of NK cells.
In conclusion, we propose a unifying phenotypic definition of NK cells across species based on the cell surface expression of NKp46. Moreover, we foresee the extensive use of NKDTR-GFP transgenic mice to delineate the role of NK cells in various conditions. Taken together, these innovative technical approaches will help to reveal the biological functions of NK cells and to develop informative preclinical models.
Methods
Mice.
All inbred mice (Charles River Laboratories, L'Arbresle, France) were purchased for use in this study. RAG1−/−, RAG1−/−CD3ζ−/−FcRγ−/−, and transgenic mice were bred in pathogen-free breeding facilities at the Centre d'Immunologie de Marseille-Luminy (Marseille, France). All of the mice used in this study were between 6 and 10 weeks of age. Experiments were conducted in accordance with institutional guidelines for animal care and use.
Flow Cytometry and Lymphocyte Preparation.
Flow cytometric analysis was done on a FACS Canto (Becton Dickinson, San Diego, CA). For all staining procedures, cells were first incubated with a buffer containing 10 μg/ml 2.4G2 antibody, 2% normal mouse serum, and 2% horse serum to prevent binding of the goat or rat antibodies to Fc receptors. Cells were subsequently stained with primary and secondary antibodies diluted in PBS 2% bovine serum. Granulocytes were identified in the bone marrow by high expression of GR1 and CD11b. Dendritic cells, B cells, TCRαβ+ T cells, TCRγδ+ T cells, and NK cells were identified in the spleen as CD11chighIA/IEhigh, CD19+, CD3+TCRβ+, CD3+TCRγδ+ cells and CD3−NK1.1+, respectively. NK precursors were identified in the bone marrow as described (47), as CD122+lin− cells, whereas immature/mature NK cells were defined as CD122+NK1.1+DX5− and CD122+NK1.1+DX5+, respectively. All antibodies were from PharMingen. CD1d-restricted NKT cells were identified by alphagalactosylceramide-loaded CD1d tetramer staining (48). NKp46 expression was detected by using a polyclonal antibody against mouse NKp46 (R & D Systems, Minneapolis, MN) or using the rat 29A1.4 mAb raised against a mouse NKp46-Fc fusion protein from R & D Systems. More information on this mAb are available in SI Text and SI Fig. 7. Secondary staining was performed with goat anti-rat IgG or donkey anti-goat IgG (Invitrogen, Carlsbad, CA). Anti-human NKp46 mAb (BAB281) was from Beckman Coulter (Fullerton, CA). Lung and liver lymphocytes were prepared as described (49).
Immunofluorescence on Tissue Sections.
Immunofluorescence was performed on 5- to 10-μm-thick serial frozen sections. Sections were fixed with acetone before staining with anti-CD3-APC, anti-CD19-PE and goat anti-mouse NKp46 followed by secondary donkey anti-goat IgG-alexa 488 (Invitrogen). Slides were analyzed by confocal microscopy (Zeiss LSM 510, Welwyn Garden City, Hertfordshire, U.K.).
NK Cell Stimulation Assay.
Anti-NK1.1 (PK136) and anti- NKp46 antibodies (antiserum or 29A1.4 mAb) were bound on plastic (96-well plates) overnight in carbonate buffer. Spleen lymphocytes isolated from RAG−/− or C57BL/6 mice were stimulated 4 h in the presence of FITC-coupled anti-CD107a antibodies and Golgi-stop (PharMingen). Cells were subsequently stained with DX5/CD3 antibodies and intracellular IFN-γ using cytofix/cytoperm kit (PharMingen) before flow cytometric analysis.
Supplementary Material
Supporting Information
Acknowledgments
We thank Claude Grégoire, Sophie Ugolini, Elena Tomasello, Ana Reynders, and C. Andrew Stewart (Centre d'Immunologie de Marseille-Luminy; CIML) for comments and advice; Mitch Kronenberg (La Jolla Institute of Allergy and Immunology, La Jolla, CA) for the generous gift of CD1d tetramers; Adrien Kissenpfennig (CIML, Plateforme RIO), Cécile Goujet (Centre National de la Recherche Scientifique, Villejuif, France), Anne Gillet, and Hervé Sanchez (CIML) for help in the generation and maintenance of the transgenic mice; the mouse functional genomics Platform of the Marseille-Nice Genopole for immunohistochemistry; Angélique Bole for help in the generation of the anti-mouse NKp46 mAb. E.V.'s laboratory is supported by European Union FP6, LSHB-CT-2004-503319-Allostem, Ligue Nationale contre le Cancer (“Equipe labellisée La Ligue”), Agence Nationale de la Recherche (“Réseau Innovation Biotechnologies” and “Microbiologie Immunologie–Maladies Emergentes”), Institut National de la Santé et de la Recherche Médicale, Centre National de la Recherche Scientifique, and Ministère de l'Enseignement Supérieur et de la Recherche. N.F. is supported by Agence Nationale de la Recherche (Réseau Innovation Biotechnologies). Innate Pharma laboratory is partly supported by LSHB-CT-2004-503319-Allostem, and Agence Nationale de la Recherche (Réseau Innovation Biotechnologies).
Abbreviations
NK
natural killer
N-CAM
neural cell adhesion module
NCR
natural cytotoxicity receptor
DT
diphtheria toxin
DTR
DT receptor.
Footnotes
Conflict of interest statement: M.B., P.A., K.C., L.G., and Y.M. are employees and shareholders of Innate-Pharma. F.R. is founder, employee, and shareholder of Innate-Pharma. E.V. and A.M. are founders and shareholders of Innate-Pharma.
This article is a PNAS direct submission.
References
- 1.Moretta A, Bottino C, Mingari MC, Biassoni R, Moretta L. Nat Immunol. 2002;3:6–8. doi: 10.1046/j.1365-3083.2002.01055.x. [DOI] [PubMed] [Google Scholar]
- 2.Degli-Esposti MA, Smyth MJ. Nat Rev Immunol. 2005;5:112–124. doi: 10.1038/nri1549. [DOI] [PubMed] [Google Scholar]
- 3.Stewart CA, Vivier E, Colonna M. Curr Top Microbiol Immunol. 2006;298:1–21. doi: 10.1007/3-540-27743-9_1. [DOI] [PubMed] [Google Scholar]
- 4.Moffett-King A. Nat Rev Immunol. 2002;2:656–663. doi: 10.1038/nri886. [DOI] [PubMed] [Google Scholar]
- 5.Raulet DH. Nat Immunol. 2004;5:996–1002. doi: 10.1038/ni1114. [DOI] [PubMed] [Google Scholar]
- 6.Smyth MJ, Hayakawa Y, Takeda K, Yagita H. Nat Rev Cancer. 2002;2:850–861. doi: 10.1038/nrc928. [DOI] [PubMed] [Google Scholar]
- 7.Ruggeri L, Capanni M, Mancusi A, Perruccio K, Burchielli E, Martelli MF, Velardi A. Int J Hematol. 2005;81:13–17. doi: 10.1532/ijh97.04172. [DOI] [PubMed] [Google Scholar]
- 8.Vivier E, Anfossi N. Nat Rev Immunol. 2004;4:190–198. doi: 10.1038/nri1306. [DOI] [PubMed] [Google Scholar]
- 9.Lanier LL, Testi T, Bindl J, Phillips JH. J Exp Med. 1989;169:2233–2238. doi: 10.1084/jem.169.6.2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ryan JC, Niemi EC, Goldfien RD, Hiserodt JC, Seaman WE. J Immunol. 1991;147:3244–3250. [PubMed] [Google Scholar]
- 11.Carlyle JR, Mesci A, Ljutic B, Belanger S, Tai LH, Rousselle E, Troke AD, Proteau MF, Makrigiannis AP. J Immunol. 2006;176:7511–7524. doi: 10.4049/jimmunol.176.12.7511. [DOI] [PubMed] [Google Scholar]
- 12.Arase H, Saito T, Phillips JH, Lanier LL. J Immunol. 2001;167:1141–1144. doi: 10.4049/jimmunol.167.3.1141. [DOI] [PubMed] [Google Scholar]
- 13.Slifka MK, Pagarigan RR, Whitton JL. J Immunol. 2000;164:2009–2015. doi: 10.4049/jimmunol.164.4.2009. [DOI] [PubMed] [Google Scholar]
- 14.Hamaguchi-Tsuru E, Nobumoto A, Hirose N, Kataoka S, Fujikawa-Adachi K, Furuya M, Tominaga A. Br J Haematol. 2004;124:819–827. doi: 10.1111/j.1365-2141.2004.04850.x. [DOI] [PubMed] [Google Scholar]
- 15.Di Santo JP. Annu Rev Immunol. 2006;24:257–286. doi: 10.1146/annurev.immunol.24.021605.090700. [DOI] [PubMed] [Google Scholar]
- 16.Blom B, Spits H. Annu Rev Immunol. 2005;24:287–320. doi: 10.1146/annurev.immunol.24.021605.090612. [DOI] [PubMed] [Google Scholar]
- 17.Kim S, Iizuka K, Aguila HL, Weissman IL, Yokoyama WM. Proc Natl Acad Sci USA. 2000;97:2731–2736. doi: 10.1073/pnas.050588297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim S, Song YJ, Higuchi DA, Kang HP, Pratt JR, Yang L, Hong CM, Poursine-Laurent J, Iizuka K, French AR, et al. Blood. 2006;107:1024–1030. doi: 10.1182/blood-2005-04-1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moretta A, Biassoni R, Bottino C, Mingari MC, Moretta L. Immunol Today. 2000;21:228–234. doi: 10.1016/s0167-5699(00)01596-6. [DOI] [PubMed] [Google Scholar]
- 20.Moretta L, Moretta A. EMBO J. 2004;23:255–259. doi: 10.1038/sj.emboj.7600019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mandelboim O, Lieberman N, Lev M, Paul L, Arnon TI, Bushkin Y, Davis DM, Strominger JL, Yewdell JW, Porgador A. Nature. 2001;409:1055–1060. doi: 10.1038/35059110. [DOI] [PubMed] [Google Scholar]
- 22.Moretta A, Bottino C, Vitale M, Pende D, Cantoni C, Mingari MC, Biassoni R, Moretta L. Annu Rev Immunol. 2001;19:197–223. doi: 10.1146/annurev.immunol.19.1.197. [DOI] [PubMed] [Google Scholar]
- 23.Hollyoake M, Campbell RD, Aguado B. Mol Biol Evol. 2005;22:1661–1672. doi: 10.1093/molbev/msi162. [DOI] [PubMed] [Google Scholar]
- 24.Sivori S, Vitale M, Morelli L, Sanseverino L, Augugliaro R, Bottino C, Moretta L, Moretta A. J Exp Med. 1997;186:1129–1136. doi: 10.1084/jem.186.7.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pessino A, Sivori S, Bottino C, Malaspina A, Morelli L, Moretta L, Biassoni R, Moretta A. J Exp Med. 1998;188:953–960. doi: 10.1084/jem.188.5.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sivori S, Falco M, Marcenaro E, Parolini S, Biassoni R, Bottino C, Moretta L, Moretta A. Proc Natl Acad Sci USA. 2002;99:4526–4531. doi: 10.1073/pnas.072065999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takeda K, Cretney E, Hayakawa Y, Ota T, Akiba H, Ogasawara K, Yagita H, Kinoshita K, Okumura K, Smyth MJ. Blood. 2005;105:2082–2089. doi: 10.1182/blood-2004-08-3262. [DOI] [PubMed] [Google Scholar]
- 28.Voehringer D, Shinkai K, Locksley RM. Immunity. 2004;20:267–277. doi: 10.1016/s1074-7613(04)00026-3. [DOI] [PubMed] [Google Scholar]
- 29.Robbins SH, Terrizzi SC, Sydora BC, Mikayama T, Brossay L. J Immunol. 2003;170:5876–5885. doi: 10.4049/jimmunol.170.12.5876. [DOI] [PubMed] [Google Scholar]
- 30.Meresse B, Curran SA, Ciszewski C, Orbelyan G, Setty M, Bhagat G, Lee L, Tretiakova M, Semrad C, Kistner E, et al. J Exp Med. 2006;203:1343–1345. doi: 10.1084/jem.20060028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rubio V, Stuge TB, Singh N, Betts MR, Weber JS, Roederer M, Lee PP. Nat Med. 2003;9:1377–1382. doi: 10.1038/nm942. [DOI] [PubMed] [Google Scholar]
- 32.Bryceson YT, March ME, Ljunggren HG, Long EO. Blood. 2005;107:159–166. doi: 10.1182/blood-2005-04-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jung S, Unutmaz D, Wong P, Sano G, De los Santos K, Sparwasser T, Wu S, Vuthoori S, Ko K, Zavala F, et al. Immunity. 2002;17:211–220. doi: 10.1016/s1074-7613(02)00365-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jamieson AM, Isnard P, Dorfman JR, Coles MC, Raulet DH. J Immunol. 2004;172:864–870. doi: 10.4049/jimmunol.172.2.864. [DOI] [PubMed] [Google Scholar]
- 35.Poirot L, Benoist C, Mathis D. Proc Natl Acad Sci USA. 2004;101:8102–8107. doi: 10.1073/pnas.0402065101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johansson S, Berg L, Hall H, Hoglund P. Trends Immunol. 2005;26:613–618. doi: 10.1016/j.it.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 37.Gazit R, Gruda R, Elboim M, Arnon TI, Katz G, Achdout H, Hanna J, Qimron U, Landau G, Greenbaum E, et al. Nat Immunol. 2006;7:517–523. doi: 10.1038/ni1322. [DOI] [PubMed] [Google Scholar]
- 38.Storset AK, Kulberg S, Berg I, Boysen P, Hope JC, Dissen E. Eur J Immunol. 2004;34:669–676. doi: 10.1002/eji.200324504. [DOI] [PubMed] [Google Scholar]
- 39.Westgaard IH, Berg SF, Vaage JT, Wang LL, Yokoyama WM, Dissen E, Fossum S. J Leukoc Biol. 2004;76:1200–1206. doi: 10.1189/jlb.0903428. [DOI] [PubMed] [Google Scholar]
- 40.Andrews DW, Farrell HE, Densley EH, Scalzo AA, Shellam GR, Degli-Esposti MA. J Immunol. 2001;166:1796–1802. doi: 10.4049/jimmunol.166.3.1796. [DOI] [PubMed] [Google Scholar]
- 41.Bajenoff M, Breart B, Huang AY, Qi H, Cazareth J, Braud VM, Germain RN, Glaichenhaus N. J Exp Med. 2006;203:619–631. doi: 10.1084/jem.20051474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dokun AO, Chu DT, Yang L, Bendelac AS, Yokoyama WM. J Immunol. 2001;167:5286–5293. doi: 10.4049/jimmunol.167.9.5286. [DOI] [PubMed] [Google Scholar]
- 43.Walzer T, Dalod M, Robbins SH, Zitvogel L, Vivier E. Blood. 2005;106:2252–2258. doi: 10.1182/blood-2005-03-1154. [DOI] [PubMed] [Google Scholar]
- 44.Baratin M, Roetynck S, Lepolard C, Falk C, Sawadogo S, Uematsu S, Akira S, Ryffel B, Tiraby JG, Alexopoulou L, et al. Proc Natl Acad Sci USA. 2005;102:14747–14752. doi: 10.1073/pnas.0507355102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arase N, Arase H, Park SY, Ohno H, Ra C, Saito T. J Exp Med. 1997;186:1957–1963. doi: 10.1084/jem.186.12.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kissenpfennig A, Henri S, Dubois B, Laplace-Builhe C, Perrin P, Romani N, Tripp CH, Douillard P, Leserman L, Kaiserlian D, et al. Immunity. 2005;22:643–654. doi: 10.1016/j.immuni.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 47.Vosshenrich CA, Samson-Villeger SI, Di Santo JP. Curr Opin Immunol. 2005;17:151–158. doi: 10.1016/j.coi.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 48.Matsuda JL, Gapin L, Fazilleau N, Warren K, Naidenko OV, Kronenberg M. Proc Natl Acad Sci USA. 2001;98:12636–12641. doi: 10.1073/pnas.221445298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Masopust D, Vezys V, Usherwood EJ, Cauley LS, Olson S, Marzo AL, Ward RL, Woodland DL, Lefrancois L. J Immunol. 2004;172:4875–4882. doi: 10.4049/jimmunol.172.8.4875. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information