Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster (original) (raw)
Abstract
The nuage is an electron-dense perinuclear structure that is known to be a hallmark of animal germ-line cells. Although the conservation of the nuage throughout evolution accentuates its essentiality, its role(s) and the exact mechanism(s) by which it functions in the germ line still remain unknown. Here, we report a nuage component, Krimper (KRIMP), in Drosophila melanogaster and show that it ensures the repression of the selfish genetic elements in the female germ line. The Krimp loss-of-function allele exhibited female sterility, defects in karyosome formation and oocyte polarity, and precocious osk translation. These phenotypes are commonly observed in the other nuage component mutants, vasa (vas) and maelstrom (mael), and the RNA-silencing component mutants, spindle-E (spn-E) and aubergine (aub), suggesting a shared underlying defect that uses RNA silencing. Moreover, we demonstrated that the localization of the nuage components depends on both SPN-E and AUB and that the selfish genetic elements were derepressed to different extents in the nuage component mutants, as well as in aub and armitage (armi) mutants. In the nuage component mutants, vas, krimp, and mael, the levels of roo, I-element, and HeT-A repeat-associated small interfering RNAs were greatly reduced. Hence, our data suggest that the nuage functions as a specialized center that protects the genome in the germ-line cells via gene regulation mediated by repeat-associated small interfering RNAs.
Keywords: Krimper, repeat-associated small interfering RNA
Germ-line cells in numerous animals are characterized by the presence of an electron-dense unique organelle called the nuage (which means “cloud” in French), also known as the chromatoid body in mouse or perinuclear P granules in Caenorhabditis elegans (reviewed in ref. 1). The nuage forms an amorphous and fibrous structure that is localized to the cytoplasmic face of the nuclear envelope when visualized by electron microscopy. In some cells, the nuage is large enough to be visible by light microscopy. Although the nuage was reported half a century ago, its role(s) and the mechanism(s) by which it functions in the animal germ line remain unknown.
The ovary of Drosophila melanogaster has proven to be an excellent system in which to study the composition and function(s) of the nuage. In Drosophila, Vasa (VAS) (2), Aubergine (AUB) (3), and Maelstrom (MAEL) (4) are reported to be nuage components. VAS is a well conserved DEAD box RNA helicase expressed in animal germ-line cells (2) and is reported to localize to the nuage in other organisms, such as mouse, planarians, and C. elegans (5–7). MAEL is also shown to localize to the nuage in mouse (8), thereby implying a conserved role for it in the germ line throughout evolution. The Drosophila germ line is characterized by the presence of a specialized region, known as the pole plasm, at the posterior end of the embryo. During early embryogenesis, most of the pole plasm is incorporated into the pole cells (or primordial germ-line cells). The pole cells invaginate into the embryo and migrate through the midgut epithelium to form the gonads with the somatic gonadal cells. The nuage can be observed as early as the formation of the primordial germ-line cells, and it persists in all germ-line cells until the oocyte is determined (9, 10). During the remaining stages of oogenesis, the nuage can be found in all nurse cells but not in the oocyte.
The possibility that the nuage uses RNA silencing as a gene-regulation mechanism was proposed on the basis of the observations that one of the argonaute family proteins, AUB, localizes to the nuage (11) and that another RNA-silencing component, Spindle-E (SPN-E), governs the perinuclear localization of two nuage components, VAS and MAEL (4, 12). Furthermore, in C. elegans, it was shown that mutations in the retinoblastoma pathway components cause somatic cells to enhance RNA interference (RNAi) and elaborate perinuclear P granules (i.e., nuage), suggesting that the nuage is an essential player of the germ line in regulating gene expression by RNAi (13). Here we report a nuage component, Krimper (KRIMP), whose loss-of-function allele, as well as vas and mael mutants, exhibits a defect in the processing of the selfish genetic elements roo, I-element, and HeT-A into repeat-associated small interfering RNAs (rasiRNAs). The derepression of long interspersed nuclear elements (LINEs), such as HeT-A and I-element, is also observed in the nuage component mutants krimp and mael and RNA-silencing component mutants aub and armitage (armi). Hence, we provide evidence that the nuage ensures the repression of the selfish genetic element gene expression, possibly via the production of rasiRNAs.
Results and Discussion
KRIMP Localizes to the Nuage.
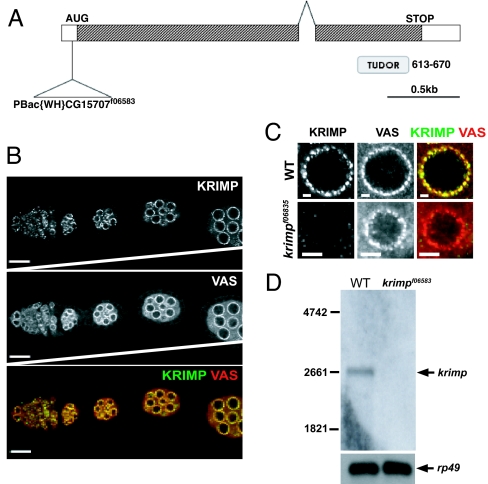
By comparing the expression profiles of isolated tumor germ-line stem cells (GSCs) induced by the loss of bag-of-marbles expression or the overexpression of decapentaplegic to those of the somatic cells, CG15707 (hereby known as Krimper) was identified as one of the potential candidate genes that is highly expressed in the GSCs (14). Krimper (Krimp) encodes a protein predicted to contain a tudor domain (Fig. 1A). Tudor domain-containing proteins, such as the Drosophila Tudor (TUD), SPN-E, mouse RNF17, and mouse tudor repeat 1 (MTR-1), are reported to play essential roles in both the female and male germ lines (15–18). Although krimp was identified as a highly expressed mRNA in the GSCs from the microarray analysis, immunostaining of KRIMP indicates a wide expression in germ-line cells, including the differentiating germ cells in the germarium and egg chambers (Fig. 1B). KRIMP seems to localize to perinuclear foci reminiscent of the nuage (Fig. 1B). In fact, costaining of KRIMP with a well known nuage component, VAS, shows an overlap of virtually all KRIMP and VAS foci in the nuage (Fig. 1C). Unlike VAS, which is both a nuage and pole plasm component (19, 20), KRIMP is detected only in perinuclear foci and not in the pole plasm (Fig. 1B).
Fig. 1.
KRIMP is a nuage component. (A) Gene structure of krimp is shown. krimp contains two exons and is predicted to contain a tudor domain. A piggyBac insertion, f06583, 35 bp upstream of the krimp ORF results in female sterility. (B) KRIMP localizes to the perinuclear regions of the germ-line cells in the Drosophila ovary. Ovaries were immunostained with anti-KRIMP (shown in green) and anti-VAS (shown in red). KRIMP perinuclear foci colocalize with VAS foci. All ovarioles are orientated with the anterior to the left. (Scale bars: 20 μm.) (C) Closer view of a nurse cell nucleus confirms the colocalization. (Scale bars: 4 μm.) (D) _krimp_f06583 is a loss-of-function allele. Northern analysis indicates the expected transcript size of ≈2.5 kb in the WT ovaries. No detectable transcript is observed in krimp mutant ovaries.
_krimp_f06583 Is a Loss-of-Function Allele and Exhibits Meiosis and Oocyte-Patterning Defects.
A piggyBac transposable element inserted 35 bp upstream of the Krimp ORF was identified as a possible krimp mutant allele, whereas females homozygous for _krimp_f06583 were sterile. Northern blotting analysis revealed the absence of the 2.5-kb krimp transcript in the mutant ovary (Fig. 1D), indicating that _krimp_f06583 is a loss-of-function allele. Moreover, immunostaining of _krimp_f06583 ovary with anti-KRIMP also indicated the loss of perinuclear foci (Fig. 1C). To confirm that _krimp_f06583 is a suitable mutant allele for the characterization of the krimp phenotypes, we placed this allele over an available deletion that uncovers krimp genomic region Df(2R)Exel6063. Transheterozygotes _krimp_f06583/Df(2R)Exel6063 exhibited female sterility and an extent of loss in KRIMP perinuclear staining similar to that exhibited by homozygous _krimp_f06583 (data not shown). Hence, _krimp_f06583 was used as a loss-of-function allele to characterize krimp phenotypes in this work.
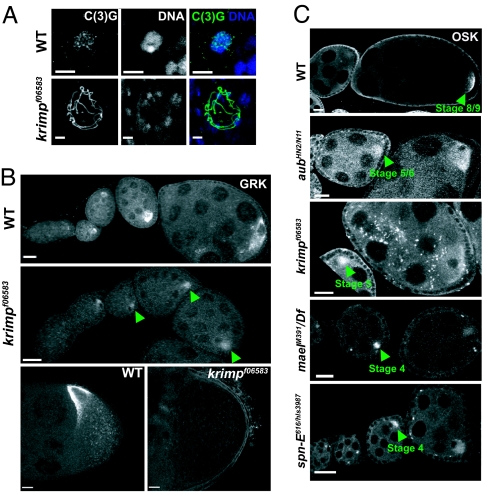
In _krimp_f06583 mutant ovary, the progression of oogenesis was compromised, and degeneration of the egg chambers was observed from stage 8 onward (data not shown). A closer examination of _krimp_f06583 revealed meiotic progression and oocyte polarity defects that are commonly seen in the nuage component mutants vas and mael (4, 21) and the RNA-silencing component mutant spn-E (22). krimp mutant oocyte nucleus failed to form a compact karyosome, and the synaptonemal marker C(3)G remained chromosomal (Fig. 2A). This is in contrast to the WT oocyte nucleus, which compacts into a karyosome by stage 3, when C(3)G dissociates to become extrachromosomal (23). Examination of a dorsal/ventral marker, Gurken (GRK), indicated a loss of dorsal/ventral polarity in krimp mutant oocytes. The level of GRK expression was markedly reduced in 100% (n = 30) of the mutant ovarioles, and its localization to the anterior-dorsal corner of the oocyte was affected in 93% (n = 61) of mutant egg chambers in stage 8 and onward (Fig. 2B) (24). Last, we saw the precocious translation of osk mRNA in 80% (n = 55) of krimp mutant ovarioles (Fig. 2C). In the WT, osk mRNA is transcribed at the onset of oogenesis, but translation is initiated only at stage 9 (Fig. 2C) (25). This is consistent with the osk silencing defects reported previously for armi, mael, aub, and spn-E mutants (Fig. 2C) (26). Taking all our observations together, our nuage component, KRIMP, shares similarities in at least two or more phenotypes with the other nuage component mutants, vas and mael, and RNA-silencing component mutants, armi, aub, and spn-E. This suggests a common underlying defect that uses RNA silencing.
Fig. 2.
krimp mutants exhibit defective karyosome formation and oocyte polarity. (A) Immunostaining with a synaptonemal complex marker, C(3)G (shown in green), shows that the _krimp_f06583 oocyte nucleus fails to compact into a karyosome (shown in blue). (Scale bars: 5 μm.) (B) Ovary staining with anti-GRK. The level of GRK expression is down-regulated (indicated by green arrowheads), and its dorsal-anterior localization is disrupted in stage-8 egg chamber. (Scale bars: 20 μm.) (C) Ovary staining with anti-OSK. In WT, osk is translated at stages 7–9. Precocious translation of osk mRNA is observed in the nuage component mutants, including spn-E mutants, as indicated by green arrowheads. (Scale bars: 20 μm.)
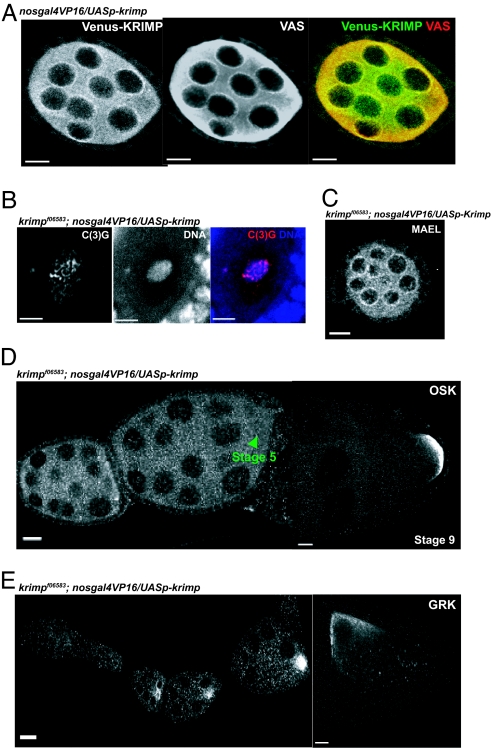
We generated transgenic flies that expressed a Venus yellow fluorescent protein (YFP)-tagged version of KRIMP protein under the control of a UASp promoter. By crossing the flies harboring the Venus-tagged KRIMP to flies that expressed the nosgal4VP16 transgene, Venus-tagged KRIMP protein was visualized as perinuclear foci that colocalized with endogenous VAS perinuclear foci, therefore paralleling the localization of endogenous KRIMP protein (Figs. 1B and 3A). However, when compared with endogenous KRIMP expression, more cytoplasmic Venus-tagged KRIMP was observed (Figs. 1B and 3A), suggesting that the Venus tag affected KRIMP localization to the nuage slightly. Although nosgal4VP16 overexpressed the UASp-venus-krimp transgene, we did not see any gain-of-function phenotypes even in the presence of functional KRIMP. The UASp-venus-krimp transgene driven by nosgal4VP16 could fully rescue the female sterility defect in krimp mutants, with the compaction of the oocyte nucleus into a karyosome by stage 6 in 100% (n = 44) of the egg chambers (Fig. 3B), an accurate repression of osk translation in 100% (n = 24) of the ovarioles (Fig. 3D), a normal GRK expression in 100% (n = 23) of the ovarioles, and the localization of GRK to the anterior-dorsal corner of the oocyte in 97% (n = 31) of stage 8 egg chambers (Fig. 3E). This indicates that the fusion protein was fully functional and that KRIMP localization to the nuage is essential for proper meiosis and oocyte polarity specification. Hence, all of the observed phenotypes in _krimp_f06583 mutant ovaries were a result of the loss of CG15707 gene functions.
Fig. 3.
The UASp-venus(YFP)-krimp transgene fully rescues krimp mutant defects. Transgenic flies carrying the venus-krimp transgene under the UASp promoter were crossed with flies harboring the nosgal4VP16 transgene to drive the expression of Venus-tagged KRIMP in the ovary. (A) Immunostaining with anti-GFP (shown in green) and anti-VAS (shown in red) indicates that Venus-tagged KRIMP protein localizes to the perinuclear regions of the germ-line cells, which colocalize with VAS perinuclear foci. (Scale bars: 10 μm.) (B) By crossing flies harboring the venus-Krimp transgene into the _krimp_f06583 mutant background, the oocyte nucleus compacts into a karyosome (shown in blue), and C(3)G (shown in red) becomes chromosomal. (Scale bars: 5 μm.) (C) MAEL, whose localization depends on KRIMP, localizes to the perinuclear regions of the germ-line cells. (Scale bar: 10 μm.) (D) osk mRNA is translationally repressed normally. (Scale bars: 20 μm.) (E) GRK expression is comparable with the WT ovariole, and the protein localizes to the anterior-dorsal region of the stage-8 egg chamber. (Scale bars: 20 μm.)
Hierarchical Relationship of the Nuage Components.
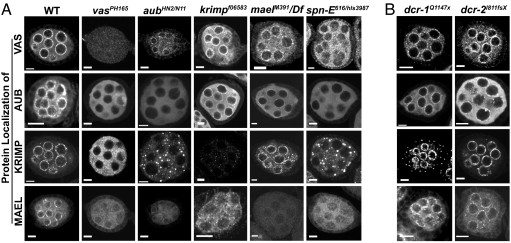
Previous work (4) has suggested that the nuage assembly reflects a hierarchical relationship among the known components. Localization of AUB to the nuage depends on VAS, and MAEL localization depends on VAS and AUB (Fig. 4A) (4). To examine whether KRIMP participates in this hierarchical assembly of the nuage, we examined KRIMP localization in vas, aub, and mael mutants. KRIMP localization was affected in _vas_PH165 and _aub_HN2/N11 mutants but unaffected by the loss of MAEL (Fig. 4A). We also showed that MAEL localization depended on KRIMP, as well as on VAS and AUB (Fig. 4A) (4). Moreover, MAEL perinuclear localization was rescued in 100% (n = 30) of the ovarioles in the presence of a UASp-venus-krimp transgene that was driven by nosgal4VP16 in a krimp mutant background (Fig. 3C). Hence, VAS seems to function as a recruiting “platform” that lies in the perinuclear region and facilitates the recruitment of subsequent nuage components. This suggests the possibility that these examined components have comparable activities in a common molecular machine; alternatively, it suggests that they have distinct functions that are carried out by using a common platform, the nuage.
Fig. 4.
Nuage foci are mislocalized in the germ-line-specific RNA-silencing component mutants. Ovaries from different mutant flies were immunostained for the nuage components. Homozygous mutant alleles or allelic combinations were used for all of the mutants, except for dcr-1, where clonal analysis was used. (A) Localization of the nuage components at the perinuclear regions of the germ-line cells reflects a hierarchical assembly. All of the nuage components, VAS, AUB, KRIMP, and MAEL, depend on SPN-E to localize normally to the perinuclear regions. AUB, KRIMP, and MAEL depend on VAS for proper localization; KRIMP and MAEL depend on VAS and AUB to localize to the nuage; MAEL depends on VAS, AUB, and KRIMP to localize normally. (B) Nuage localization is unaffected in the conventional dicing enzyme mutants, dcr-1 and dcr-2. (Scale bars: 10 μm.)
Nuage Components Are Mislocalized in Germ-Line-Specific RNA-Silencing Component Mutants.
In recent years, evidence has accumulated suggesting that RNAi or similar mechanisms function as a gene-regulation mechanism in the Drosophila germ line. For example, AUB and SPN-E have been implicated as key components that relate RNA silencing to the regulation of polarity during ovary development (26). To examine the dependence of the nuage components on the germ-line-specific RNA-silencing components, we investigated their localization in both aub and spn-E mutants. As described earlier (4), the localization of both KRIMP and MAEL was affected in aub mutants (Fig. 4A). In the spn-E mutant, we also saw mislocalization of all of the nuage components, VAS, AUB, KRIMP, and MAEL (Fig. 4A). We also noticed that VAS localization depends partially on proper AUB localization. Although VAS foci were apparent in aub mutants, cytoplasmic VAS was visibly more abundant than in the WT (Fig. 4A). Hence, we did not rule out the possibility of the existence of a feedback mechanism between VAS and AUB.
Vagin et al. (27) have also shown that Drosophila Dicer 1 (DCR-1) and Dicer 2 (DCR-2) are not involved in the processing of rasiRNAs or the repression of selfish genetic elements, suggesting that they are not germ-line-specific RNAi machinery. To test the effect of Dicers on nuage localization, we examined the localization of VAS, AUB, KRIMP, and MAEL in either dcr-1 or dcr-2 mutants. We found that all of the nuage components localized normally in both mutants (Fig. 4B). These data imply a clear distinction in the use of RNA-silencing machinery between the germ line and the soma. The nuage, a unique feature of the animal germ line, may be a key in the regulation of germ-line-specific gene expression. Germ-line-specific RNA-silencing components SPN-E and AUB seem to act upstream of the nuage, where they regulate the localization of nuage components.
Some Selfish Genetic Elements Are Derepressed in Nuage Component Mutants.
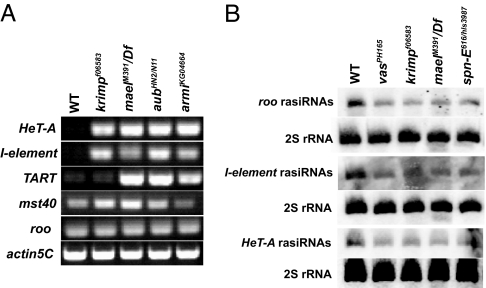
To examine the nuage's role in regulating gene expression in the germ-line cells, we analyzed the expression of some selfish genetic elements using semiquantitative RT-PCR with RNA extracted from mutant ovaries (Fig. 5A). The selfish genetic elements examined include the repetitive LINEs such as HeT-A, TART, and I-element (28), a tandem repeat that lies near the β-heterochromatin mst40 (29), and a euchromatic long terminal repeat (LTR) retrotransposon, roo (30). Interestingly, HeT-A and I-element were derepressed in krimp and mael mutants (Fig. 5A), similar to what has been reported previously in vas, aub, and armi mutants (27, 31, 32). TART was derepressed only in mael and aub mutants, whereas mst40 derepression was only observed in krimp and mael mutants (Fig. 5A). This may indicate differences in element specificity among the nuage components. Alternatively, this difference may have resulted from the differences among the oogenesis progression defects among the different nuage component mutants. All of the semiquantitative RT-PCR results were confirmed by using real-time RT-PCR [supporting information (SI) Fig. 6].
Fig. 5.
LINEs are derepressed, and the production of rasiRNAs is defective in the nuage component mutants. (A) The expression of the selfish genetic elements was examined by using semiquantitative RT-PCR. HeT-A and I-element were derepressed in the nuage component mutants, krimp and mael, and RNA-silencing component mutants, aub and armi. TART was derepressed in mael, aub, and armi mutants. mst40 was derepressed in krimp and mael mutants. roo, belonging to the euchromatic LTR family, was not derepressed. (B) PAGE northern analysis of roo, I-element, and HeT-A rasiRNAs. The production of roo, I-element, and HeT-A rasiRNAs was reduced in the nuage component mutants, as well as the RNA-silencing component mutants.
Surprisingly, we did not detect derepression of roo in any of the nuage component mutants, including armi mutants (Fig. 5A and SI Fig. 6_B_). Our data differ from previous work wherein Vagin et al. (27) reported slight derepression of roo in armi and spn-E mutants. This minor derepression, when compared with the other selfish genetic elements, is seemingly inconsequential. Moreover, we confirmed our roo results with real-time RT-PCR. The LINE family elements, HeT-A, I-element, and TART, as well as mst40, are located predominantly in the heterochromatin regions of the chromosome, whereas roo is primarily a euchromatic retrotransposon (28, 30). Hence, the different effects of the nuage component mutants on the selfish genetic elements imply a role for the nuage in maintaining the silenced state of the heterochromatin.
Because Tudor (TUD) is reported to be a component of both the pole plasm and nuage (33), we also examined the repression of HeT-A, I-element, and TART transcripts in a loss-of-function tud allele, _tud_1 (34). All of the examined transcripts seemed to be repressed to an extent similar to that in the WT (SI Fig. 7), indicating that tud is not involved in the repression of retroelements. The nuage may, thus, serve as a “station” where TUD undertakes different role(s) in the germ line.
Production of rasiRNAs Is Reduced in the Nuage Component Mutants.
A previous report has linked the up-regulation of selfish genetic elements in aub and armi mutants to the failure in rasiRNA production (27). To determine whether the production of rasiRNAs is compromised in the nuage component mutants, we analyzed the levels of roo, I-element, and HeT-A rasiRNAs. Interestingly, all of the examined rasiRNAs were reduced in vas, krimp, and mael mutants, and the RNA-silencing component mutant, spn-E (Fig. 5B). This suggests a role of the nuage in regulating the production of rasiRNAs. Although roo, I-element, and HeT-A rasiRNAs were reduced in the nuage component mutants, only I-element and HeT-A, which belong to the LINE family, were significantly derepressed (Fig. 5A and SI Fig. 6). This implies that the expression of roo may be regulated by mechanism(s) other than rasiRNA-mediated silencing. Alternatively, the number of roo rasiRNAs that is produced in the nuage component mutants may be sufficient to ensure repression. However, we cannot rule out the possibility that the reduction of other LTR rasiRNAs in the nuage component mutants may affect the repression of their respective transcripts.
Conclusion
The highly conserved nature of the nuage throughout evolution strongly implies its importance and essentiality in the germ line. Here, we suggest a function for the nuage, to repress the selfish genetic elements by regulating the production of rasiRNAs. Drosophila nuage component, KRIMP, and the germ-line-specific RNA-silencing component, SPN-E, contain tudor domains that were previously implied to associate with the methylated peptides of histones H3 and H4 (35). In addition, Drosophila MAEL shuttles between the nucleus and cytoplasm (4), and mouse MAEL associates with the chromatin remodeler, SNF5/INI1 (8). Therefore, the nuage may regulate the chromatin state to repress unfavorable gene expression in the germ-line cells. Moreover, our work suggests that the nuage functions to maintain genome stability by repressing the expression of the selfish genetic elements, such as HeT-A and I-element. In fact, it has been shown that the repression of telomeric retroelements in Drosophila germ-line cells is required to control telomere length (31, 32). Protection of the genome is essential to govern high fidelity, and this is critical in the case of the germ line, which gives rise to the next generation.
Homologues of KRIMP can be identified in the Drosophilidae family, including the melanogaster group and others, such as D. virilis and D. grimshawi (SI Fig. 8). Although no close orthologues of KRIMP are found in the higher vertebrates, two mouse tudor domain proteins, RNF17 and MTR-1, are reported to localize to the chromatoid body (16, 18). Hence, RNF17 and MTR-1 may potentially be functional homologues of KRIMP, which may have evolved to repress a unique set of retroelements. On the other hand, mouse VAS and MAEL are reported to localize to the chromatoid body (7, 8), implying their conserved functions beyond species. In fact, the argonaute proteins, MIWI, AGO2, and AGO3, are also localized to the chromatoid body (36), suggesting that a similar RNA-silencing pathway is also used to regulate gene expression in higher vertebrates. Furthermore, Kotaja et al. (36) also reported the concentration of the microRNA (miRNA), Let-7a, and the processing enzyme, Dicer 1, at the chromatoid body in mouse, implying that the miRNA precursors are processed at the perinuclear site. Thus, gene regulation involving the miRNA pathway may also play a role in germ-line development. This may account for the precocious osk translation phenomena that was observed in both the nuage and RNA-silencing component mutants (Fig. 2C) because a potential miR-280 binding site is present at the 3′ UTR of osk mRNA (26). Further experiments will be needed to address the role of the nuage for osk translational repression via miR-280 and/or other regulatory mechanisms. The question remains open of whether the germ line, besides using a small RNA-silencing mechanism, also uses other form(s) of gene regulation mechanism(s) to regulate germ-line-specific gene expression.
Materials and Methods
Fly Strains and Transgenes.
For ovary staining and molecular work, yw was used as a WT control. The mutant alleles and allelic combinations used in this work were _vas_PH165 (21), _krimp_f06583 (Bloomington Drosophila Stock Center at Indiana University, Bloomington, IN), _dcr-2_L811fsX (37), _armi_KG04664 (Bloomington Drosophila Stock Center), _krimp_f06583/Df(2R)Exel6063 (Bloomington Drosophila Stock Center), _aub_HN2/N11 (38, 39), _mael_M391/Df(3L)79E-F (40, 41), and _spn-E_616/hls3987 (17, 22). The original _krimp_f06583 mutant line from the Bloomington Drosophila Stock Center exhibited male sterility due to a background mutation, and it was cleaned up before the use of this allele for all of the experiments described here. Because homozygous _dcr-1_Q1147x is lethal, mitotic clones were generated by subjecting 2-day-old female flies (hsFLP/eyFLP;_FRT82BUbi-GFP/FRT82Bdcr-1_Q1147x) to heat-shock at 37°C for 1.5 h. The treated flies were then aged for 7–10 days before immunostaining.
Transgenic flies harboring the UASp-venus-krimp transgene were generated as follows: The transgene was constructed by cloning the full-length _krimp_-coding sequence, amplified with the primers _krimpFL_-FW and _krimpFL_-RV (see SI Table 1), into pENTR/D-TOPO (Invitrogen, Carlsbad, CA) and then recombining it downstream of pPVW (The Drosophila Gateway Vector Collection, Carnegie Institution of Washington, Baltimore, MD; www.ciwemb.edu/labs/murphy/Gateway%20vectors.html) in accordance with the manufacturer's instruction manual. The resulting plasmid was injected into yw embryos by using standard methods (42). Transgenic flies carrying UASp-venus-krimp were crossed to flies harboring the nosgal4VP16 transgene to drive Venus-tagged KRIMP expression in the female germ line (43). The rescue experiment of krimp mutant defects was performed by crossing w; krimp/CyO; nosgal4VP16 into w; krimp/CyO; UASp-venus-krimp.
Antibody Generation.
KRIMP antibody was raised against a portion of KRIMP fused to glutathione _S_-transferase (GST). Krimp antigen sequence, corresponding to amino acids 163–306, was amplified with an EST clone, RE66405, by using the primers _krimpAg_-FW and _krimpAg_-RV (SI Table 1) cloned into pENTR/D-TOPO (Invitrogen) and recombined into pDEST15 GST and pDEST17 His (Invitrogen), respectively, in accordance with the manufacturer's instruction manual. KRIMP GST fusion protein was purified by using Glutathione Sepharose High Performance (Amersham Biosciences, Piscataway, NJ) and then sent to Zymed Laboratories (South San Francisco, CA) for antibody generation in rabbits. Anti-KRIMP rabbit polyclonal antibodies were affinity-purified by using KRIMP His fusion protein (purified by using Ni Sepharose High Performance from Amersham Biosciences) conjugated to a HiTrap NHS-activated HP affinity column (Amersham Biosciences).
Immunostaining.
Ovaries were dissected, stained, and mounted as described (44), with a change of the fixation time to 5 min for better preservation of the nuage structure. The following antibodies were used: anti-KRIMP rabbit polyclonal (1:10,000), anti-VAS rat polyclonal (1:200), anti-OSK rat polyclonal (1:100) (gifts from Paul Lasko, McGill University, Montréal, QC, Canada), anti-AUB rat polyclonal (1:100) (a gift from Hong Han, McGill University, Montréal, QC, Canada), anti-C(3)G (1:500) guinea pig polyclonal (23), anti-GRK mouse monoclonal 1D12 (1:10) (45), anti-MAEL rabbit polyclonal (1:200) (4), and anti-GFP rabbit polyclonal (1:1,000) (Torrey Pines Biolabs, Houston, TX). Alexa Fluor 488-, 555-, or 633-conjugated goat anti-mouse, anti-rat, anti-rabbit, or anti-guinea pig IgG secondary antibodies (1:400) (Molecular Probes/Invitrogen, Carlsbad, CA) were used. Images were acquired by using either a Leica (Wetzlar, Germany) TCS-NT or Carl Zeiss (Oberkochen, Germany) LSM 510 confocal microscope.
Northern Blotting.
Total RNA was extracted from dissected ovaries by using TRIzol Reagent (Invitrogen). Total RNA was used directly for PAGE northern rasiRNAs and semiquantitative and real-time RT-PCR. For northern analysis of krimp transcripts, Poly(A) RNA was purified from yw and _krimp_f06583 total RNA by using the Oligotex mRNA mini kit (Qiagen, Valencia, CA).
For northern analysis of krimp and rp49 transcripts, DIG DNA probes were synthesized by PCR with 10× DIG DNA labeling mix (Roche Applied Science, Indianapolis, IN) by using the EST clone, RE66405, and cloned rp49 as templates, respectively. The primer sets _krimp_-FW and _krimp_-RV and T7 and T3 (SI Table 1) were used for making krimp and rp49 probes, respectively. Two-hundred nanograms of yw and _krimp_f06583 Poly(A) RNA were loaded and separated on a formaldehyde/Mops 1% agarose gel, transferred onto a Hybond-N+ nylon membrane (Amersham Biosciences), and then cross-linked. Hybridization was carried out in DIG Easy Hyb buffer (Roche Applied Science) containing DIG DNA probes, in accordance with the manufacturer's instruction manual. The signals were visualized on Kodak (Rochester, NY) BioMax MS film.
For PAGE northern analysis of the rasiRNAs, RNA probes were synthesized from the linearized templates by in vitro transcription using either T7 or SP6 RNA polymerases (Roche Applied Science) in the presence of [γ-32P]UTP (3,000 Ci/mmol, 10 mCi/ml; 1 Ci = 37 GBq). Templates were _pGEM-T_-harboring PCR product amplified with primer sets corresponding to roo, I-element, and HeT-A (SI Table 1). Total RNA (10 or 20 μg) from the ovaries was separated on a 15% polyacrylamide/8 M urea denaturing gel, transferred onto a Hybond-N+ nylon membrane (Amersham Biosciences), and cross-linked. Hybridization was performed at 62°C in Church buffer [0.25 M sodium phosphate, pH 7.2/1 mM EDTA, pH 8.0/7% (wt/vol) SDS/1% (wt/vol) BSA] containing radio-labeled sense roo, I-element, or HeT-A RNA probes. The membranes were analyzed by using a Typhoon 9200 scanner (Amersham Biosciences).
Two-Step RT-PCR.
Total RNA (1 μg) extracted from ovaries was treated with DNaseI (Roche Applied Science). Reverse transcription was carried out by using Oligo(dT)15 and SuperScript III RT (Invitrogen). A mock reaction without reverse transcriptase was also prepared for each RNA sample. The newly synthesized cDNAs, after treatment with RNaseH (Stratagene, La Jolla, CA), were first normalized and checked for genomic DNA contamination by performing PCR with actin5C primers. PCR was subsequently performed by using 1 μl per cDNA sample/reaction with the primer sets corresponding to HeT-A, TART, I-element, mst40, and roo. All primer sequences can be found in SI Table 1.
Supplementary Material
Supporting Information
Acknowledgments
We thank P. Lasko (McGill University, Montréal, QC, Canada), H. Ruohola-Baker (University of Washington, Seattle, WA), A. Nakamura [RIKEN, Center for Developmental Biology (Kobe, Japan)], P. M. MacDonald (University of Texas, Austin, TX), D. St. Johnston (The Wellcome Trust and Cancer Research Campaign/Institute of Cancer and Developmental Biology/University of Cambridge, Cambridge, England), and the Bloomington Drosophila Stock Center at Indiana University for the fly stocks and antibodies; especially H. Han for the unpublished anti-AUB antibody; S. Cohen, A. Spradling, and F. Berger for critical readings of the manuscript; our laboratory members, J. Honda, S. Y. Chen, L. H. Tao, and V. Patil; and L. Orban's laboratory members, X. G. Wang, W. C. Liew, K. W. Chang, and R. C. Bartfai for support in laboratory work. A.K.L. is supported by the Singapore Millennium Foundation program.
Abbreviations
rasiRNA
repeat-associated small interfering RNA
LINE
long interspersed nuclear element
GSC
germ-line stem cell.
Footnotes
The authors declare no conflict of interest.
References
- 1.Eddy EM. Int Rev Cytol. 1975;43:229–281. doi: 10.1016/s0074-7696(08)60070-4. [DOI] [PubMed] [Google Scholar]
- 2.Liang L, Diehl-Jones W, Lasko P. Development (Cambridge, UK) 1994;120:1201–1211. doi: 10.1242/dev.120.5.1201. [DOI] [PubMed] [Google Scholar]
- 3.Harris AN, Macdonald PM. Development (Cambridge, UK) 2001;128:2823–2832. doi: 10.1242/dev.128.14.2823. [DOI] [PubMed] [Google Scholar]
- 4.Findley SD, Tamanaha M, Clegg NJ, Ruohola-Baker H. Development (Cambridge, UK) 2003;130:859–871. doi: 10.1242/dev.00310. [DOI] [PubMed] [Google Scholar]
- 5.Kuznicki KA, Smith PA, Leung-Chiu WMA, Estevez AO, Scott HC, Bennett KL. Development (Cambridge, UK) 2000;127:2907–2916. doi: 10.1242/dev.127.13.2907. [DOI] [PubMed] [Google Scholar]
- 6.Shibata N, Umesono Y, Orii H, Sakurai T, Watanabe K, Agata K. Dev Biol. 1999;206:73–87. doi: 10.1006/dbio.1998.9130. [DOI] [PubMed] [Google Scholar]
- 7.Toyooka Y, Tsunekawa N, Takahashi Y, Matsui Y, Satoh M, Noce T. Mech Dev. 2000;93:139–149. doi: 10.1016/s0925-4773(00)00283-5. [DOI] [PubMed] [Google Scholar]
- 8.Costa Y, Speed RM, Gautler P, Semple CA, Maratou K, Turner JMA, Cooke HJ. Hum Mol Genet. 2006;15:2324–2334. doi: 10.1093/hmg/ddl158. [DOI] [PubMed] [Google Scholar]
- 9.Mahowald AP. J Exp Zool. 1968;167:237–262. doi: 10.1002/jez.1401670211. [DOI] [PubMed] [Google Scholar]
- 10.Mahowald AP. J Exp Zool. 1971;176:329–344. doi: 10.1002/jez.1401760308. [DOI] [PubMed] [Google Scholar]
- 11.Snee MJ, Macdonald PM. J Cell Sci. 2004;117:2109–2120. doi: 10.1242/jcs.01059. [DOI] [PubMed] [Google Scholar]
- 12.Kennerdell JR, Yamaguchi S, Carthew RW. Genes Dev. 2002;16:1884–1889. doi: 10.1101/gad.990802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang D, Kennedy S, Conte D, Jr, Kim J, Gabel HW, Kamath RS, Mello CC, Ruvkun G. Nature. 2005;436:593–597. doi: 10.1038/nature04010. [DOI] [PubMed] [Google Scholar]
- 14.Kai T, Williams D, Spradling A. Dev Biol. 2005;283:486–503. doi: 10.1016/j.ydbio.2005.04.018. [DOI] [PubMed] [Google Scholar]
- 15.Boswell RE, Mahowald AP. Cell. 1985;43:97–104. doi: 10.1016/0092-8674(85)90015-7. [DOI] [PubMed] [Google Scholar]
- 16.Chuma S, Hiyoshi M, Yamamoto A, Hosokawa M, Takamura K, Nakasuji N. Mech Dev. 2003;120:979–990. doi: 10.1016/s0925-4773(03)00181-3. [DOI] [PubMed] [Google Scholar]
- 17.Gillespie DE, Berg CA. Genes Dev. 1995;9:2495–2508. doi: 10.1101/gad.9.20.2495. [DOI] [PubMed] [Google Scholar]
- 18.Pan J, Goodheart M, Chuma S, Nakatsuji N, Page DC, Wang PJ. Development (Cambridge, UK) 2005;132:4029–4039. doi: 10.1242/dev.02003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hay B, Ackerman L, Barbel S, Jan LY, Jan LN. Development (Cambridge, UK) 1988;103:625–640. doi: 10.1242/dev.103.4.625. [DOI] [PubMed] [Google Scholar]
- 20.Lasko P, Ashburner M. Genes Dev. 1990;4:905–921. doi: 10.1101/gad.4.6.905. [DOI] [PubMed] [Google Scholar]
- 21.Styhler S, Nakamura A, Swan A, Suter B, Lasko P. Development (Cambridge, UK) 1998;125:1569–1578. doi: 10.1242/dev.125.9.1569. [DOI] [PubMed] [Google Scholar]
- 22.Gonzalez-Reyes A, Elliott H, Johnston D., St Development (Cambridge, UK) 1997;124:4927–4937. doi: 10.1242/dev.124.24.4927. [DOI] [PubMed] [Google Scholar]
- 23.Page SL, Hawley RS. Genes Dev. 2001;15:3130–3143. doi: 10.1101/gad.935001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neuman-Silberberg FS, Schupbach T. Mech Dev. 1996;59:105–113. doi: 10.1016/0925-4773(96)00567-9. [DOI] [PubMed] [Google Scholar]
- 25.Johnston D., St . In: The Development of Drosophila melanogaster. Bate M, Martinez-Arias A, editors. Vol 1. Plainview, NY: Cold Spring Harbor Lab Press; 1993. pp. 325–363. [Google Scholar]
- 26.Cook HA, Koppetsch BS, Wu J, Theurkauf WE. Cell. 2004;116:817–829. doi: 10.1016/s0092-8674(04)00250-8. [DOI] [PubMed] [Google Scholar]
- 27.Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- 28.Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, Gaasterland T, Meyer J, Tuschl T. Dev Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- 29.Steven RH, Russel KK. Chromosoma. 1994;103:63–72. [Google Scholar]
- 30.Bowen NJ, McDonald JF. Genome Res. 2001;11:1527–1540. doi: 10.1101/gr.164201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Savitsky M, Kwon D, Georgiev P, Kalmykova A, Gvozdev V. Genes Dev. 2006;20:345–354. doi: 10.1101/gad.370206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vagin VV, Klenov MS, Kalmykova A, Stolyarenko AD, Kotelnikov RN, Gvozdev V. RNA Biol. 2004;1:54–58. [PubMed] [Google Scholar]
- 33.Bardsley A, McDonald K, Boswell RE. Development (Cambridge, UK) 1993;119:207–219. doi: 10.1242/dev.119.1.207. [DOI] [PubMed] [Google Scholar]
- 34.Arkov AL, Wang JS, Ramos A, Lehmann R. Development (Cambridge, UK) 2006;133:4053–4062. doi: 10.1242/dev.02572. [DOI] [PubMed] [Google Scholar]
- 35.Kim J, Daniel J, Espejo A, Lake A, Krishna M, Xia L, Zhang Y, Bedford MT. EMBO Rep. 2006;7:397–403. doi: 10.1038/sj.embor.7400625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kotaja N, Bhattacharyya SN, Jaskiewicz L, Kimmins S, Parvinen M, Filipowicz W, Sassone-Corsi P. Proc Natl Acad Sci USA. 2006;103:2647–2652. doi: 10.1073/pnas.0509333103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hatfield SD, Shcherbata HR, Fischer KA, Nakahara K, Carthew RW, Ruohola-Baker H. Nature. 2005;435:974–978. doi: 10.1038/nature03816. [DOI] [PubMed] [Google Scholar]
- 38.Schupbach T, Wieschaus E. Genetics. 1991;129:1119–1136. doi: 10.1093/genetics/129.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson JE, Connell JE, Macdonald PM. Development (Cambridge, UK) 1996;122:1631–1639. doi: 10.1242/dev.122.5.1631. [DOI] [PubMed] [Google Scholar]
- 40.Clegg NJ, Frost DM, Larkin MK, Subrahmanyan L, Bryant Z, Ruohola-Baker H. Development (Cambridge, UK) 1997;124:4661–4671. doi: 10.1242/dev.124.22.4661. [DOI] [PubMed] [Google Scholar]
- 41.Hartenstein V, Jan YN. Roux's Arch Dev Biol. 1992;201:194–220. doi: 10.1007/BF00188752. [DOI] [PubMed] [Google Scholar]
- 42.Spradling A, Rubin GM. Science. 1982;218:341–347. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
- 43.Van Doren M, Williamson AL, Lehmann R. Curr Biol. 1998;8:243–246. doi: 10.1016/s0960-9822(98)70091-0. [DOI] [PubMed] [Google Scholar]
- 44.Kai T, Spradling A. Proc Natl Acad Sci USA. 2003;100:4633–4638. doi: 10.1073/pnas.0830856100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Queenan AM, Barcelo G, van Buskirk C, Schupbach T. Mech Dev. 1999;89:35–42. doi: 10.1016/s0925-4773(99)00196-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information