Pan-S replication patterns and chromosomal domains defined by genome-tiling arrays of ENCODE genomic areas (original) (raw)
Abstract
In eukaryotes, accurate control of replication time is required for the efficient completion of S phase and maintenance of genome stability. We present a high-resolution genome-tiling array-based profile of replication timing for ∼1% of the human genome studied by The ENCODE Project Consortium. Twenty percent of the investigated segments replicate asynchronously (pan-S). These areas are rich in genes and CpG islands, features they share with early-replicating loci. Interphase FISH showed that pan-S replication is a consequence of interallelic variation in replication time and is not an artifact derived from a specific cell cycle synchronization method or from aneuploidy. The interallelic variation in replication time is likely due to interallelic variation in chromatin environment, because while the early- or late-replicating areas were exclusively enriched in activating or repressing histone modifications, respectively, the pan-S areas had both types of histone modification. The replication profile of the chromosomes identified contiguous chromosomal segments of hundreds of kilobases separated by smaller segments where the replication time underwent an acute transition. Close examination of one such segment demonstrated that the delay of replication time was accompanied by a decrease in level of gene expression and appearance of repressive chromatin marks, suggesting that the transition segments are boundary elements separating chromosomal domains with different chromatin environments.
Although all the DNA in a eukaryotic cell replicates during the S phase of cell cycle, there is a great variability in the actual point in S phase when a given chromosomal segment replicates. Segments are known to reproducibly replicate early or late in S phase, and it is generally believed that this is determined by the time at which the origins in a segment fire. All origins of replication are licensed with MCM proteins by the time S phase begins (Bell and Dutta 2002), and yet, once conditions in the cell change to favor the firing of the origins, all origins do not fire at the same time. In situ labeling techniques and other methods have led to some general principles determining the time of replication of a segment in S phase (for review, see MacAlpine and Bell 2005). Early-replicating segments are generally enriched in euchromatin, while late-replicating segments are enriched in heterochromatin. Some loci that are selectively expressed in specialized cells (e.g., immunoglobulin, beta-globin, or neural-associated genes) show a change in time of replication from late-S phase in undifferentiated, nonexpressing cells to early-S phase after differentiation (Simon et al. 2001; Zhou et al. 2002; Perry et al. 2004). The correspondence between the activation of chromatin at differentiation-induced genes with the advancement in replication time also suggests that the chromatin environment dictates time of replication (Bickmore and Carothers 1995; Rountree et al. 2000; Demeret et al. 2001).
The completion of many genomic sequences and the advent of genome-tiling microarrays provided an opportunity to correlate gene expression or chromatin structure with time of replication at a much finer resolution. DNA replicated at specific intervals in S phase were hybridized to genome-tiling microarrays to determine the exact time in S phase when specific genes replicate. Early experiments in model organisms like Saccharomyces cerevisiae and Drosophila melanogaster confirmed many of the principles outlined above (Raghuraman et al. 2001; Schubeler et al. 2002; MacAlpine et al. 2004).
Extending this method of analysis to human cells, specifically to Chromosomes 21 and 22, we confirmed that similar principles dictate time of replication in human chromosomes (Jeon et al. 2005). We made the surprising observation that almost 60% of the chromosomal probes studied gave a replication signal at multiple times in S phase, described as a pan-S-phase pattern of replication. While asynchrony of replication between alleles of a given gene would give rise to a pan-S-phase pattern of replication, it seemed highly unlikely to us that 60% of the human chromosomes would show such asynchrony. In addition, it was unclear whether the pan-S-phase replication was an artifact of cells losing their synchrony of progression through the cell cycle, of the thymidine-aphidicolin method of cell cycle synchronization, or of the aneuploidy inherent in HeLa cells.
The ENCODE region encompasses 44 segments covering ∼1% of the human genome on which multiple groups are applying different techniques to find the best methods to annotate the human genome (The ENCODE Project Consortium 2004). We measured the replication time for this region and used the data to improve our method of computing the replication profile of chromosomal segments. The improvements in our algorithm decreased the pan-S replication pattern to ∼20% of the segments interrogated. We confirmed the prediction of pan-S replication by an independent method of assessing replication time: interphase FISH. The results demonstrate that pan-S-phase replication is a real pattern of replication that cannot be explained by artifacts derived from microarray platform, methods of cell cycle synchronization, or aneuploidy of cells. Instead, pan-S-phase replication is a reflection of asynchrony of replication between alleles in a given cell, suggesting that differences in the chromatin environment of two alleles can be seen in up to 20% of the human genome in some cells.
Finally, using the high-definition temporal profile of replication over the ENCODE areas, we identified adjoining chromosomal segments of a few hundred kilobases each with differing times of replication. Hypothesizing that these areas are “replication domains,” we demonstrate for one such region that the adjoining domains have different levels of gene expression and activating and repressing marks on histones. We believe that the replication domains correspond to chromosomal domains separated by boundary elements.
Results
Replication timing analyses of 1% genome using synchronized HeLa cells
HeLa cells were synchronized at G1/S by thymidine-aphidicolin block. After release from the block, cells were pulsed with bromodeoxyuridine (BrdU) at every 2-h interval of S-phase and genomic DNA isolated. In all, five time intervals (0–2, 2–4, 4–6, 6–8, and 8–10 h) representing 10 h of the entire S phase were collected. The BrdU-incorporated heavy/light (H/L) DNA was purified using a CsCl density gradient as described earlier (Jeon et al. 2005). Purified DNA from each time interval was hybridized to the high-density genome-tiling Affymetrix array comprising unique 25-mer oligonucleotides in the ENCODE-selected chromosomal loci covering 1% of the human genome (∼30 Mb) (see Methods for details of the ENCODE regions).
Segregation of chromosomal regions into temporally specific and pan-S replicating segments
Probes that replicated in a discrete interval in S phase were called temporally specific, while probes that replicated at multiple intervals in S phase were called temporally nonspecific. The Methods and Supplemental Table 1 contain examples of the specificity classification. For the Affymetrix ENCODE array, 26.115% of the probes were temporally nonspecific.
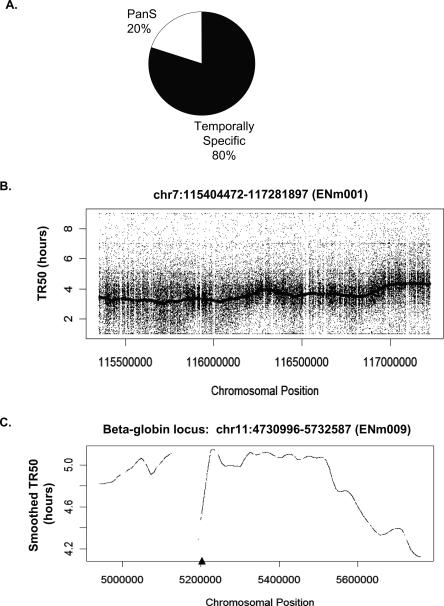
In order to classify chromosomal segments as temporally specific or asynchronously replicating (pan-S), a 10-kb sliding window was passed along the chromosome and each window defined as replicating in a pan-S manner if >60% of the probes in that window are temporally nonspecific (see Methods for details). Thus, by ensuring that the majority of contiguous probes in a given segment replicate in a temporally nonspecific manner, we eliminate artifacts from cross-hybridization or from poor probe hybridization. Since the estimated average speed of a replication fork is ∼1 kb/min, isolated segments <10 kb (<10 min) that appeared to replicate in a nonspecific manner were significantly below the resolution of the 2-h sampling method. Such segments (<0.2% of the ENCODE region) were therefore eliminated from our calculations. After these corrections, ∼20% of the ENCODE area replicated in a pan-S-phase pattern as determined by a base-pair count (Fig. 1A), while the remaining 80% shows a temporally distinct profile. Individual chromosomal segments showing these patterns are presented below.
Figure 1.
Temporal profile of replication of chromosomal segments. (A) Temporally specific versus pan-S distribution of replication for 1% of the human genome investigated in this study. (B) Raw TR50 data with a smoothed TR50 curve overlaid from the 1.9-Mb region on Chromosome 7. According to the ENCODE Consortium nomenclature, this chromosomal segment is referred to as ENm001 (http://hgwdev.cse.ucsc.edu/ENCODE/encode.hg17.html). (C) Smoothed TR50 data from the 1-Mb beta-globin locus (ENm009) on Chromosome 11. The lowest point in each valley indicates a site that is replicated before its adjoining segments and thus is likely to contain origins of replication. The gaps in the TR50 plots indicate the presence of repeats. In order to minimize cross-hybridization of oligonucleotides, repeat regions of the genome are not spotted on the tiling arrays. The triangle on the _X_-axis indicates the position of the known beta-globin origin.
Continuous TR50 profile along the length of a chromosomal segment
The time at which a temporally specific probe replicates to 50% (TR50) is calculated by summing the replication signal over the five time points and linearly interpolating the time when 50% of the total signal was reached. Supplemental Table 1 gives examples of TR50 calculation for several probes. Plotting the TR50 values for specific probes against the linear coordinate of the probe on the chromosome gives a view of the replication profile of the chromosome. Because the raw TR50 data are noisy, Lowess smoothing for all temporally specific probes in a 60-kb window was performed to ascertain the trends in the replication pattern along the length of the chromosome. Figure 1B shows the raw TR50 data and a smoothed TR50 curve for a 1.9-Mb segment of Chromosome 7 (ENm001). By averaging over relatively long segments of DNA, the smoothed curve corrects for scatter created by differences in probe hybridization efficiency or from cross-hybridization of a few errant probes and is very useful for comparing the time of replication of adjoining segments of DNA. Figure 1C shows another example of a smoothed TR50 plot from a 1-Mb region of Chromosome 11 containing the beta-globin locus. The late replication of this segment in HeLa cells agrees with previous findings that the beta-globin locus replicates late in S phase in nonerythroid cells (Epner et al. 1988; Dhar et al. 1989). The TR50 profiles for all the 43 chromosomal loci can be viewed using the UVa DNA Rep TR50 track at the http://hgwdev.cse.ucsc.edu/ENCODE/encode.hg17.html site. Temporal profiles from 12 of these regions are shown in Supplemental Figure 1.
Local minima of the TR50 curve show areas that replicate earlier than the flanking regions and thus are likely to contain origins of replication, as has been shown previously in S. cerevisiae (Raghuraman et al. 2001). Only one previously validated origin of replication lies in the ENCODE area near the large stretch of repeat sequences (chr11: 5124929–5193780) within the beta-globin locus (Kitsberg et al. 1993; Aladjem et al. 1998; Wang et al. 2004). The repeat sequences near the beta-globin gene were not represented on the microarray, causing a gap in the TR50 profile (Fig. 1C). However, the TR50 profile of the regions immediately adjoining these repeats clearly suggests that a minimum in the TR50 profile is located somewhere at or near these repeats, indicating the presence of an origin of replication at this site. Thus, the hundreds of minima in the TR50 profile are likely to be at or near origins of replication.
Segregation of temporally specific regions into early-, mid-, and late-S replicating regions
The smoothed TR50 profile suffers from a compression of the _Y_-axis values due to the smoothing operation; thus we do not get an accurate estimate of the time of replication of a given segment from the profile. We therefore processed the TR50 data to define discrete segments with early-, mid-, and late-S-phase replication in addition to the pan-S-phase replication patterns described above. A temporally specific region is classified into early, mid, or late replication based on the average TR50 of the temporally specific probes within a 10-kb window. TR50 cutoffs of 3.4 h (for early- to mid-S transition) and 3.9 h (for mid- to late-S transition) are used.
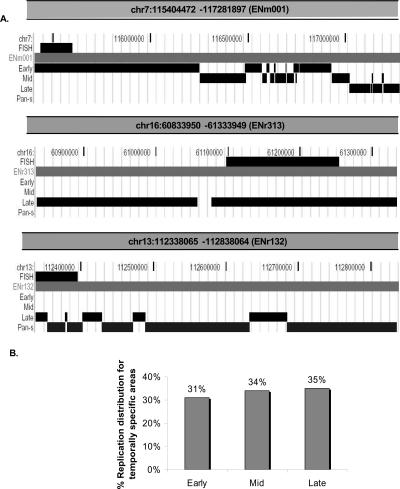
The top panel of Figure 2A shows the segregation of ENm001 after these analyses. Tracks representing segments that replicate in early-, mid-, late-, or pan-S-phase, respectively, are indicated. Since ENm001 is an early-replicating region, only a very small region shows up in the late-replication track. The general trend of the right portion of the region replicating later can be seen in the transition from a solid early-replication track into mid-replicating regions as we move left to right. The tracks are nonoverlapping at the base-pair level, and the apparent overlap in certain places is due to the low resolution of the UCSC Browser snapshot required to fit the whole region into a figure.
Figure 2.
Segregation of chromosomal segments with temporally specific and temporally nonspecific pattern of replication. (A) On the basis of TR50, temporally specific regions are further segregated and displayed as three tracks (UCSC Genome Browser), early-, mid-, or late-replicating, while chromosomal regions undergoing temporally nonspecific replication are highlighted under the pan-S track. The three panels in this figure show segregation of replication timing for three chromosomal segments. (Top panel) The 1.9-Mb region (ENm001) of Chromosome 7; (middle and bottom panels) examples of two 500-kb chromosomal segments from Chromosomes 16 (ENr313) and 13 (ENr132) that underwent late and pan-S replication, respectively. The FISH track in all the three panels refers to the chromosomal positions of BAC clones selected for the interphase FISH experiment shown in Figures 3 and 4. (B) Percent of temporally specific chromosomal segments replicating in early-, middle-, or late-S phase.
The second and third panels of Figure 2A show similar segmentation of 500-kb chromosomal regions from Chromosomes 16 (ENr313) and 13 (ENr132), respectively. ENr313 replicates late, while ENr132 shows a pan-S pattern of replication. TR50 segmentation profiles for 12 regions are shown in Supplemental Figure 1. Profiles for all the 43 regions can be viewed in the UVa DNA Rep Seg track at http://hgwdev.cse.ucsc.edu/ENCODE/encode.hg17.html site. Eighty percent of the ENCODE area replicates in a temporally specific interval (Fig. 1A). Within the specific regions, 31% segregates into early-, 34% into mid-, and 35% into late-S-phase replicating patterns (Fig. 2B).
Validation of replication time by interphase FISH
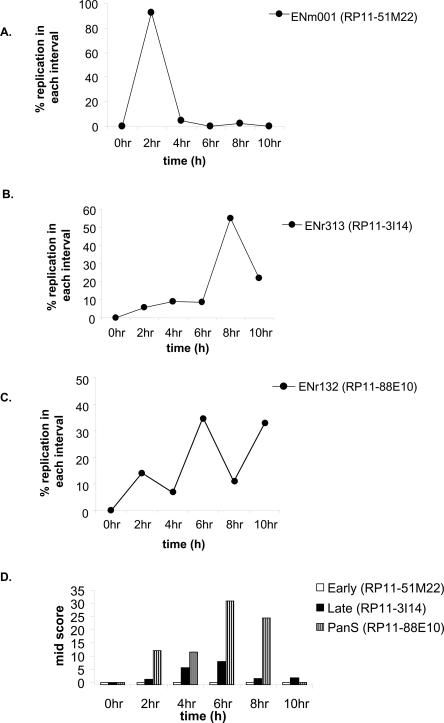
To check the temporal profile of replication generated by the microarray data, we used interphase FISH as an independent method for determining replication time. Although labor-intensive, this method has the additional advantage in that the large sizes of the probes reduce errors from poor signal strength and cross-hybridization. Ten BAC clones of 48–187 kb each (details in Supplemental Table 2) were selected to validate the microarray data for 10 segments from nine ENCODE areas: three each with early and pan-S-phase and four with late-S-phase patterns of replication. The positions of BAC clones used in Figure 3 and Figure 4 are highlighted in Figure 2A. HeLa cells were synchronized and harvested at 2-h intervals during S phase, and BAC clones were labeled and hybridized to denatured interphase nuclei. A single hybridization signal (visible as a dot under the microscope) indicates one copy of the targeted DNA. ENm001 showed 2 dots/cell in G1 (0 h) and 4 dots/cell in G2, while the remaining eight regions had 3 dots/cell in G1 and 6 dots/cell in G2 because of the aneuploidy of HeLa cells. The percent replication of a probed segment in each time interval in S phase was determined by counting the increments in dots/cell during that interval, where 100% replication means that the number of dots/cell is twice the G1 value.
Figure 3.
Interphase FISH for validating replication timing in HeLa. (A–C) Synchronously progressing HeLa cells were hybridized to fluorescence-labeled probes of BAC clone DNA RP11-51M22, RP11-3I14 (for early- and late-replicating areas, respectively) and RP11-88E10 (for pan-S pattern of replication). The chromosomal locations of these BACs are highlighted in Figure 2A. The percent replication at each interval of S phase is plotted against time in S phase. (D) The interallelic variation in replication for FISH data observed for each of the BAC clones mentioned above was determined by calculating the Mid-Score (detailed in Results and Supplemental Material).
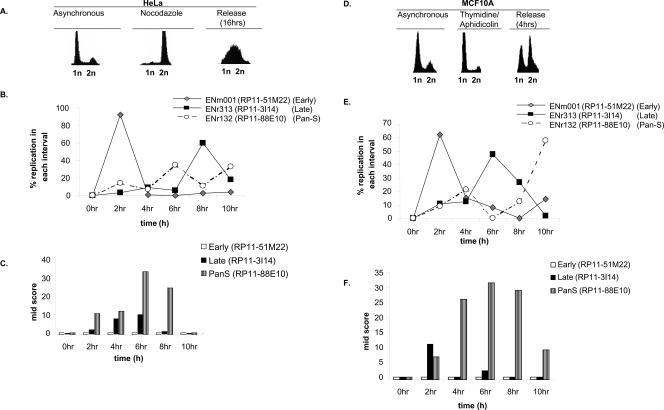
Figure 4.
Pan-S replication pattern is independent of cell synchronization method and aneuploidy. (A) HeLa cells blocked (by nocodazole) and released from mitosis followed by FACS for DNA content. (B,C) Interphase FISH was performed with HeLa cells synchronized with nocodazole and released. The _X_-axis represents time in S phase such that 0 h = 12 h post-release from the nocodazole block. The rest is as in Figure 3. (D) MCF10A cells released from a G1/S block with thymidine/aphidicolin followed by FACS for DNA content. (E,F) Interphase FISH with MCF10A cells synchronously progressing through S phase to determine the replication profile and Mid-Score with the chromosomal segments mentioned in Figure 3.
The RP11-51M22 probe shows that this region of ENm001 replicates early with the complete doubling of all signal in the first 2 h of S phase (Fig. 3A). For a late-replicating region, RP11-3I14 from ENr313, the increase in dot number was maximum in the last 4 h of S phase (Fig. 3B). RP11-88E10 from the ENr132 region indicated that significant replication occurred in multiple time intervals (Fig. 3C), consistent with the pan-S replication detected in the microarrays (Fig. 2A). All the 10 segments tested by FISH reproduced the microarray data for time of replication (see Supplemental Table 2 for details).
Pan-S replication is due to interallelic variation in time of replication
To ascertain whether the pan-S-phase pattern of replication was due to intercellular or interallelic variation in replication time, we calculated the percent nuclei in mid-replication. The Mid-Score for a time point is defined as the percentage of cells in mid-replication, having replicated one, but not all alleles, for a given probe. Thus cells in mid-replication will have 3 dots/cell for ENm001, and 4 or 5 dots/cell for ENr313 or ENr132. Segments that replicate synchronously in a narrow interval of S phase are expected to have a very narrow temporal window with a high Mid-Score (a more detailed explanation is in Supplemental Fig. 2). ENm001 (early replicating) had no time point with a high Mid-Score, while ENr313 (late replicating) had only two time points with a Mid-Score of 5.6% and 7.9%, indicating that all the alleles replicated in a narrow time window (Fig. 3D).
If pan-S-phase replication is due to intercellular variation in time of replication of the chromosomal segment, the two alleles in a cell will still replicate simultaneously so that the window of time when a cell is caught in mid-replication will remain short. Mid-Scores would be low or elevated for only a tightly restricted time interval (Supplemental Fig. 2). However, the ENr132 region (pan-S-phase replication) showed four time points with high Mid-Scores (i.e., 12.1, 11.4, 30.7, and 24.5) (Fig. 3D), suggesting that there was significant asynchrony in the time of replication of the alleles in a given cell. Thus the asynchrony in replication seen in the pan-S-phase pattern of replication is due to interallelic variation in replication time.
Pan-S-phase pattern of replication is not due to thymidine-aphidicolin block
We next investigated whether pan-S-phase replication was caused by the prolonged arrest in S phase that is inherent to the thymidine-aphidicolin double-block method of synchronization of cells in the cell cycle. HeLa cells were synchronized in mitosis using nocodazole and released. The time of replication was determined by interphase FISH for five regions (Fig. 4B): three that were temporally monophasic and two that had a pan-S-phase pattern of replication. The temporally specific segments still replicated in the expected time frames despite the different method of synchronization (Fig. 4B; Supplemental Table 2). Most important, both the pan-S-phase regions continued to replicate at multiple times in S phase (Fig. 4B; Supplemental Table 2), suggesting that pan-S-phase replication was not an artifact of the synchronization method. The observed asynchrony in replication was due to interallelic variation as determined by the wide time interval when the cells displayed high Mid-Scores (Fig. 4C).
Pan-S pattern of replication is not restricted to aneuploid HeLa cells
We wanted to rule out the possibility that the pan-S-phase pattern of replication is seen only in aneuploid cancer cells like HeLa. To address this, we repeated the interphase FISH experiments with MCF10A, a breast epithelial cell line derived from fibrocystic breast disease that is near diploid and nonmalignant (Fig. 4D). The area covered by probe RP11-88E10 (a region with pan-S-phase replication in HeLa cells) replicated at two time intervals (Fig. 4E). The first peak at 4 h corresponded with the time interval during which the Mid-Score increased (Fig. 4F). The Mid-Score remained high until the 10-h time interval, when the second peak of replication was observed, indicating a significant time lapse in the replication of two alleles. Therefore, pan-S-phase replication is also seen in MCF10A cells and is not unique to HeLa cells. Replication of RP11-51M22 (early) and RP11-3I14 (late) was also consistent with that seen in HeLa cells. FISH analyses for two more regions in MCF10A are detailed in Supplemental Table 2.
Correlation of TR50 profile with genome sequence features
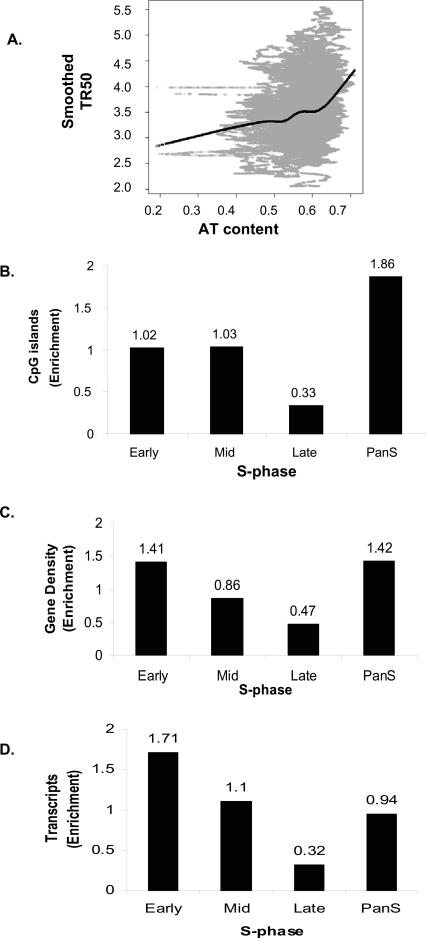
The replication timing for the 43 ENCODE regions were correlated against genome sequence features such as AT content, CpG islands, and gene density. AT content was computed using a 10-kb sliding window and plotted against the smoothed TR50 curve. A transition from low to high AT content is evident for early- to late-replicating regions (Fig. 5A). The Spearman rank correlation coefficient calculated from the plot was 0.257, suggesting a moderate positive correlation. The Pearson correlation coefficient was 0.252, also indicating a moderate positive correlation. Computation of AT content at a window size of 1 kb gave a lower correlation coefficient (0.19).
Figure 5.
Correlation between replication time and genomic features. (A) Plot of smoothed TR50 against AT content in a 10-kb sliding window. Lowess smoothed curve done at f = 0.3 (fraction of the data included in the running local fit) is overlaid in black to show the general trend. (B–D) Histograms showing distribution of (B) CpG islands, (C) gene density, and (D) transcripts (HeLa cells) against temporal segregation of replication.
DNA methylation is an important epigenetic marker (Jones and Takai 2001), with differential DNA methylation between alleles leading to monoallelic gene expression, interallelic differences in the chromatin, and asynchronous replication (Simon et al. 1999; Rountree et al. 2001; Fournier et al. 2002; Jiang et al. 2004; Fuks 2005). Since the Mid-Score calculations above suggested that the pan-S areas demonstrated interallelic differences in replication, we wondered whether the pan-S replicating segments were enriched in CpG islands and thus potentially susceptible to regulation by differential DNA methylation. Indeed, the pan-S-phase regions showed the maximum enrichment (1.86) of CpG islands (Fig. 5B).
We next compared the replication time of a segment with its gene density. In the chromosomal areas where replication was temporally specific, a threefold higher enrichment of genes was found in regions replicating early compared to those replicating late (Fig. 5C). Interestingly, the pan-S-phase regions had gene content (enrichment = 1.42) (Fig. 5C), comparable to early-replicating chromosomal segments (enrichment = 1.41), consistent with the idea that these regions could have replicated early if not for interallelic variation in chromatin structure that resulted in a subset of the alleles replicating late and producing a pan-S-phase pattern of replication.
Early-replicating regions are highly transcribed
Active transcription of genes is associated with euchromatin and may be expected to correlate with early replication. Total RNA was prepared from logarithmically growing HeLa cells and hybridized to an Affymetrix HG-U133 Plus 2.0 array to measure the level of expression of genes in different chromosomal segments. Early-replicating segments have 5.34-fold higher transcription over the late-replicating regions (Fig. 5D). The pan-S regions had an intermediate level of gene expression, consistent with the idea that all alleles of the genes in these segments are not in favorable chromatin and are not uniformly well expressed.
TR50 profile on one chromosomal segment defines chromosomal domains
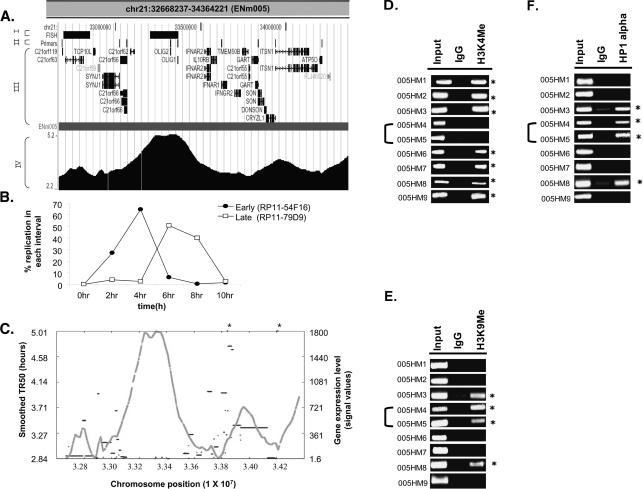
The global correlations described above are consistent with the hypothesis that early-replicating regions are usually gene dense and contain actively transcribed genes. The fine resolution of replication profile possible with the genome-tiling arrays allowed us to closely examine how such correlations hold up across contiguous stretches of chromosomes. Intriguingly, the TR50 profile of some regions revealed the presence of neighboring chromosomal segments with acute transitions in replication time. For example, in ENm005, an ∼366-kb (Chr21: 33119705–33486048) late-replicating stretch was bracketed by two early-replicating areas (Fig. 6A). Dual color interphase FISH was performed to confirm the transition in replication time from early to late in two neighboring segments of ENm005 (Fig. 6B). BAC clones (separated by ∼355 kb) from the early (RP11-54F16) and late (RP11-79D9) replicating areas confirmed that the two DNA segments, indeed, replicated in two different intervals of S phase (Fig. 6B).
Figure 6.
Replication profile demarcates chromosomal domains. (A) UCSC Genome Browser display of a 1.7-Mb region from Chromosome 21 (ENm005). This Browser picture highlights four tracks (I–IV): (I) FISH: chromosomal location of BAC clones (RP11-54F16 and RP11-79D9, from left to right) selected for the interphase FISH experiment shown in B; (II) Primers: chromosomal locations of the primers (005HM1–9, left to right) selected for ChIP assay to ascertain the histone modifications and HP1α-binding sites shown in D_–_F; (III) RefSeq: positions of all the genes in this chromosomal segment; and (IV) the contiguous TR50 profile. (B) Dual color FISH was performed with HeLa cells synchronously progressing through S phase. RP11-54F16 (from early-replicating area on left) was labeled with spectrum red dUTP, while RP11-79D9 (from late-replicating area) was labeled with spectrum green dUTP. Dual color FISH with these two BAC clones ascertained the replication time of the two regions of Chromosome 21 set 355 kb apart. (C) Plot of smoothed TR50 (_Y_-axis on left, gray) against level of transcription of genes (_Y_-axis on right, black). The two asterisk marks represent transcripts whose transcription levels exceeded the _Y_-axis limit (i.e., 2346 and 10,010 for left and right asterisks, respectively). (D–F) ChIP-PCR assay across ENm005 region (see Supplemental Table 3 for primers). PCR was performed on DNA chromatin immunoprecipitated with antibodies against methylated histones (H3 Lys4 and H3 Lys9) and HP1α. (Input) DNA control before immunoprecipitation; (IgG) ChIP with rabbit IgG was negative control for nonspecific precipitation. Forty cycles of PCR were performed for H3 Lys4 and HP1α and 30 cycles for H3 K9 di-Me. The asterisks refer to primer pairs that gave positive ChIP signal for the indicated antibodies relative to the IgG negative control. 005HM4 and 005HM5 were from the late-replicating island in ENm005.
The replication dissimilarities between the adjoining domains correlated with dissimilarities in gene expression and gene density (Fig. 6C). The late-replicating island was both gene-poor and transcriptionally less active compared to the adjoining early-replicating chromosomal segments.
These observations suggested the existence of two chromatin environments in a contiguous stretch of a chromosome separated by some type of boundary element. Since histone modifications distinguish euchromatin from heterochromatin, we decided to confirm the existence of two chromatin environments in this locus in HeLa cells by performing a chromatin immunoprecipitation (ChIP) assay for the active and inactive chromatin marks. H3 Lys4 methylation is specific for active chromatin at active promoters (Bernstein et al. 2005). We therefore selected nine genes, two in the late-replicating region (OLIG1 and OLIG2) and seven in the adjoining early-replicating chromosomal segments (C21orf119, SYNJ1, C21orf66, IFNAR1, GART, ITSN1, and ATP5O) and designed primers to amplify unique 100–300-bp fragments from the 2-kb sequences upstream of the genes (see details for primers in Supplemental Table 3). Chromatin immunoprecipitation (ChIP) and amplification of these promoters revealed that all seven genes in the early-replicating segments were positive for H3 lysine 4 (H3K4) methylation, while the two embedded in the late-replicating environment (005HM4 and 005HM5) lacked this modification (Fig. 6D). Conversely, ChIP for markers of repressed chromatin, H3 lysine 9 (H3K9) dimethylation and association of HP1α, showed that the two promoters in the late-replicating domain were in repressed chromatin. Five out of the seven promoters in the early-replicating segments were negative for markers of repressed chromatin, while the other two were positive (Fig. 6E,F).
Therefore, the island of late-replicating DNA represents a specific chromosomal domain with all the features of heterochromatin: low gene density, low gene expression, lack of activating chromatin marks, and presence of repressive chromatin marks. The rapid transition of the features of heterochromatin in this late-replicating island to those of euchromatin in the flanking areas suggests that the chromosome may be divided into discrete domains with different chromatin features. In addition, the existence of such discrete adjoining domains with different chromatin structure suggests the presence of boundary elements that prevent the spread of euchromatin from the neighboring areas to this island of heterochromatin.
Pan-S segments contain markers for both active and repressed chromatin
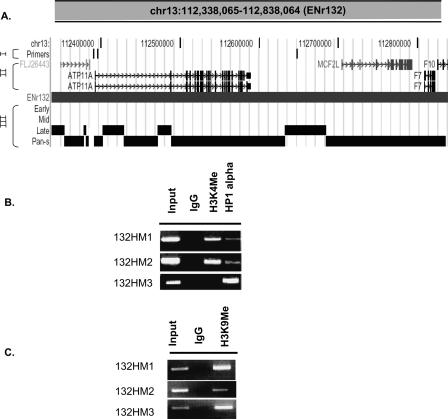
The interallelic variation in replication time observed in pan-S replicating segments predicts that one allele will be in active chromatin and another in repressed chromatin, leading us to test whether pan-S replicating segments are enriched in both types of marks. ENr132 contained extensive stretches with the pan-S replication pattern with a few interspersed segments that were exclusively late replicating. The two promoters in the pan-S replicating area, 132HM1 and 132HM2, were positive by ChIP for both the activating histone modification (H3K4 methylation) and repressive histone modification (H3K9 dimethylation) and a marker for heterochromatin (HP1) (Fig. 7). In contrast, 132HM3, from a late-S replicating segment only carried the repressed chromatin marks and not the activating histone modification. Therefore, combining the data in Figure 6 and Figure 7, three out of three late-replicating promoters were exclusively in repressed chromatin, and five out of seven early-replicating promoters were exclusively in activated chromatin. In contrast, the promoter from the pan-S replicating segment carried marks of both active and repressed chromatin, consistent with the pan-S replication pattern arising from interallelic variation in the chromatin environment.
Figure 7.
Both active and repressive chromatin marks are present in a pan-S segment. (A) UCSC Genome Browser display of a 500-kb region from Chromosome 13 (ENr132). This Browser picture highlights three tracks: (I) Primers: ChIP-PCR primers (132HM1–3, left to right) to study histone modifications and HP1α-binding sites; (II) RefSeq: positions of all the genes in this chromosomal segment; and (III) the temporal segregation of replication data. (B,C) ChIP-PCR assay across ENr132 region (see Supplemental Table 3 for primers) against methylated histones (H3 Lys4 and H3 Lys9) and HP1α (as indicated).
Discussion
Since the ENCODE project specifically selected the target 1% of the genome to be broadly representative of the whole genome based on criteria like gene density and sequence conservation, we expect that the lessons learned from these high-resolution replication time profiles can be extended to the entire genome. The pan-S-phase pattern of replication; the correlation of replication time with chromatin modifications, gene expression, and AT content; and the significance of chromosomal domains and boundary elements revealed by our studies are discussed here.
We still identify regions that replicate in multiple times in S phase in mammalian cell lines (pan-S replication pattern). Since the genome-based studies of replication in S. cerevisiae were executed only in haploid strains, they were not expected to identify regions with interallelic difference in time of replication (Raghuraman et al. 2001). Genome-based studies of replication in diploid organisms were also unsuitable to identify this pattern because of the study design (MacAlpine et al. 2004; Woodfine et al. 2004). In those studies, the time of replication was assessed by determining the ratio of DNA content for a locus in late-S (or G2) cells compared to G1 cells. In such experiments, segments showing replication in both early- and late-S phase would appear to replicate in mid-S phase, and the pan-S pattern would be missed. In contrast, the sampling of cells in multiple intervals in S phase and the use of a more sensitive method of detecting replication dependent on a positive selection for BrdU-labeled DNA enabled us to identify chromosomal segments that replicate in multiple intervals in S phase.
In this study, 20% of the studied genome appeared to replicate asynchronously, a value that is one-third that of our previous analysis on Chromosomes 21 and 22 (Jeon et al. 2005). This difference is due to an important refinement in the method of analysis in the present study. In the previous work, the hybridization data from genome-tiling arrays was analyzed by the standard Affymetrix GTRANS software to generate a track that showed when the replication signal from a given time point was significantly enriched over signal obtained from DNA replicated for the entire duration of S phase. Although this method provided an intuitive belief for replication timing, not surprisingly, replication signal was not only seen in the time period when the locus replicated but lower levels of signal were seen in adjoining time intervals. The presence of a signal in multiple time tracks led us to overestimate that nearly 60% of sequences showed a pan-S replication pattern (Supplemental Figure 3, ENm001). In contrast, in this study, we segregate probes into those that are temporally synchronous versus temporally asynchronous by quantitative criteria that take into account the spillage of replication signal into adjoining time points. In addition, only large contiguous DNA segments (≥10 kb) containing >60% of probes with asynchronous replication signals are classified as pan-S. This prevents mis-calling as pan-S short stretches where low signal strength or cross-hybridization from isolated probes give an apparent replication signal in multiple intervals in S phase. As is evident from the comparison of the two methods in one segment (Supplemental Fig. 3), the present method gives a more conservative estimate of segments that replicate at multiple times in S phase. Because microarray-based profiling of replication is a relatively new approach, we also validated the time and pattern of replication for some of the segments by a completely independent method, interphase FISH. The confirmation of all three pan-S regions as replicating asynchronously adds to the confidence that ∼20% of the chromosomal segments in HeLa cells, indeed, show this unexpected pattern of replication.
All 10 regions tested by interphase FISH (including the temporally specific regions) reproduced the time of replication estimated by the microarray-based replication profile. In addition, the time of replication for five of five tested chromosomal regions remained unaltered when a different cell cycle block method was used in HeLa cells. Interphase FISH allowed us to check the time of replication of the same five regions in another cell line, MCF10A, where we found replication times of 3/5 chromosomal segments to match that of HeLa. The differences at the other two loci are likely due to differences in the chromatin environment of these loci in the two cell lines. Since MCF10A cells are near-diploid and untransformed, the detection of pan-S-phase replication in these cells indicates that pan-S replication is not an artifact arising exclusively from the aneuploidy or the transformed state of HeLa cells. It is, of course, entirely possible that aneuploidy or cell transformation increases the fraction of the genome that shows pan-S replication.
Since FISH-based methods analyze replication in the context of individual nuclei, the Mid-Scores showed that the asynchrony in replication time was due to interallelic difference in replication. Homologous alleles usually replicate synchronously in S phase, but there are some notable exceptions to this general rule. In humans, examples of such exceptions include monoallelically expressed genes such as those imprinted depending on parent of origin (Simon et al. 1999), genes encoding olfactory receptors (Chess et al. 1994), genes on the female X-chromosome (Avner and Heard 2001; Boumil and Lee 2001), and immunoglobulin and T-cell receptor genes (Mostoslavsky et al. 2001). We will test in the future whether all the pan-S segments express all their genes monoallelically. The interallelic asynchrony in replication in the pan-S segments suggests that one allele is in euchromatin and the other in heterochromatin. Consistent with this, pan-S areas are unique in being enriched in both activating and repressive marks (Fig. 7), with the different marks residing presumably in the two different alleles.
Since the HeLa cell line is of female origin (XX), the inactivation of one of the X-chromosomes predicts that segments from the X-chromosome should replicate in a pan-S manner, unless the long passage and aneuploidy of these cells have disrupted such inactivation. There are two regions from the X-chromosome included under ENCODE (Supplemental Fig. 4). The 1.2-Mb ENm006 region had three areas of pan-S replication (126 kb, 62 kb, and 10 kb), one of which contained the Glucose-6-phosphate dehydrogenase (G6PD) gene, which is known to be transcriptionally repressed on the inactivated X-chromosome and delayed in replication compared to its active counterpart (Hansen et al. 1996). The second region, ENr324 (ChrX: 122,507,850–123,007,849), contained no pan-S replicating segments. Thus the survey of the X-chromosome fragments for pan-S replication gave mixed results. The lack of pan-S replication over the entire stretch of X-chromosome in HeLa cells could not only be due to transformation and long-term culture affecting inactivation, but also because ENm006 and ENr324 contain blocks of genes that normally escape X-chromosome inactivation, similar to many reported X-linked genes (Chang et al. 1990; Disteche 1995; Miller et al. 1995; Carrel et al. 1996; Vermeesch et al. 1997).
Correlation of gene expression with time of replication in eukaryotes has produced contradictory results. In S. cerevisiae, the expression of genes did not correlate with their time of replication in S phase. In contrast, in cultured Drosophila cells, there was a positive correlation between early replication and gene transcription (MacAlpine et al. 2004). In mammalian cells, housekeeping genes like Hprt, histones, beta-tubulin, actin, and rDNA are ubiquitously expressed and replicated in the first half of S phase. On the other hand, tissue-specific genes such as those coding for Factor IX, fibronectin, and myosin heavy chain replicate late in the cell lines not expressing them (Holmquist et al. 1982; Iqbal et al. 1984; Goldman 1988). The previous study from our laboratory on human Chromosomes 21 and 22 also showed a positive correlation between early replication and gene expression, but the results could have been improperly skewed because of atypical features of the two small chromosomes. The positive correlation between early replication and gene expression in this study is likely to be generally true throughout the genome, because it was obtained with a distributed set of segments that together are representative of the entire genome.
The association of early replication with gene expression suggests that there are consistent differences in chromatin environment between the early- and late-replicating segments. Cytological studies have shown spatial differences in nuclear staining for both activating and silencing histone modification marks, and these spatial differences in histone modification are correlated with differences in replication time (Wu et al. 2005). ChIP for histone modification marks reported here strengthens the correlation at a finer resolution: early replication and gene expression correlate with euchromatin, and conversely, late replication correlates with low gene expression and heterochromatin. These results are confirmed in a wider study that correlates our replication time data with ChIP-on-chip data for histone modifications done by the ENCODE Consortium (The ENCODE Project Consortium 2007).
Interestingly, the finer resolution offered by genome-tiling microarrays identified chromosomal segments with acute transitions in replication timing. For one particular segment (ENm005) (Fig. 6), the replication time transition was confirmed by interphase FISH and appeared to correlate with transitions in both gene expression and chromatin modifications: a late-replicating island of 355 kb had repressive chromatin marks and low gene expression. The genes OLIG1 and OLIG2 in this island are known to be expressed during development of oligodendrocyte (OL) lineage (Jakovcevski and Zecevic 2005), and thus the island is expected to become early replicating in oligodendrocytes.
Identification of transition zones separating chromosomal domains is an interesting outcome from the replication profiles. Thirty-one genes of biomedical significance (including 10 oncogenes/tumor suppressor genes on 11q and 21q) reside in or near replication-timing transition regions (Watanabe et al. 2002). The mechanism by which a boundary is maintained between euchromatin and heterochromatin around these transition zones is not understood. At the major histocompatibility complex (MHC), loci replication timing switches precisely where there is a transition in the GC% content and is associated with nuclear scaffold attachment regions (Tenzen et al. 1997). A similar transition in GC content in the neurofibromatosis 1 (NF1) gene demarcates early replicating from late-replicating chromatin, and a stalled replication fork was observed in this transition region (Schmegner et al. 2005). The sites of replication time switch identified by our method will likely lead to the identification of more such transition zones, and we are interested in determining in the future whether such zones cause replication forks to slow down or stall, whether they contain nuclear scaffold attachment regions, and whether they act as boundary elements responsible for keeping adjoining chromatin domains separate from each other.
In humans, the R and G chromosomal bands have been linked to both gene density and AT/GC content. G bands are AT-rich, while the R bands are more GC-rich. GC-rich regions are not only enriched in genes but specifically in expressed genes (Saccone et al. 1993; Caron et al. 2001; Lander et al. 2001; Versteeg et al. 2003). The moderately positive correlation between AT content and TR50 (0.26 at 10-kb, 0.19 at 1-kb resolution) suggests a trend favoring an increase in AT content as we move from early- to late-replicating chromosomal segments. This observation is also in concordance with our previous study on Chromosomes 21 and 22 (Jeon et al. 2005). The correlation increases as the computation is done at larger scales, suggesting that the influence of AT content on TR50 occurs at scales greater than tens of kilobases. Consistent with this, replication-timing studies done at 1-Mb resolution show an even stronger positive correlation with AT content (Woodfine et al. 2004).
Finally, the smoothed TR50 profile suggests locations of origins of replication at local minima and positions of replication fork termination at local maxima. Replication speed can also be estimated based on the slope of the smoothed TR50 profile at a given locus. These possibilities will be explored in our future work.
Methods
Cell culture, synchronization, and FACS analysis
HeLa and MCF10A cells were cultured as per standard growth conditions. For synchronous progression through S phase, HeLa and MCF10A cell lines were arrested by thymidine-aphidicolin block and released as described earlier (Jeon et al. 2005). For nocodazole block, HeLa cells at 60% confluency were treated with 40 ng/mL nocodazole for 10 h. This was followed by selection of cells blocked in mitosis by mitotic shake-off. These cells were washed three times with PBS and released into fresh medium for 12 h to reach the 0-h point when they enter S phase. Cells harvested at indicated time points of S phase were either used for FISH or fixed in 70% ethanol and stained with propidium iodide (PI) for FACS by standard methods.
Newly replicated DNA (H/L DNA) purification
Synchronously released cells were labeled with 100 μM BrdU for the indicated time interval; 10 ∼ 30 150-mm plates of cells were used to purify H/L DNA from each time point as described earlier (Jeon et al. 2005).
Microarray hybridization
To generate replication time profiles, ENCODE01-Forward (P/N 900543; Affymetrix) tiling arrays were used. These arrays are designed to study the pilot ENCODE regions of DNA, comprised of 30 Mb of DNA, or ∼1% of the human genome. These pilot regions were selected by a committee of the National Human Genome Research Institute (NHGRI). Half of the content on the ENCODE01 Array was manually selected by the NHGRI committee, while the remaining 50% was randomly selected (The ENCODE Project Consortium 2004). A total of 14.82 Mb of sequence constituted the manually selected regions and included 14 targets ranging in size from 500 kb to 2 Mb. The randomly selected content includes 30 500-kb regions selected based on gene density and level of nonexonic conservation.
Nonrepetitive, 25-mer oligonucleotide probe pairs (Perfect Match, PM; Mis-Match control, MM) spaced at intervals of ∼22 bp as measured from the central nucleotide were spotted on arrays. Heavy/light DNA from each time point was fragmented to 50–100 bp by DNase I digestion, end-labeled with biotin-ddATP using terminal transferase, and hybridized to the arrays as per the manufacturer’s protocol (Affymetrix). Each microarray was scanned and analyzed for signal intensities using a GeneChIP Scanner 3000 and GeneChIP Operating Software (GCOS; Affymetrix). Two biological and one technical replicates were hybridized to ascertain the reproducibility of array hybridizations. The primary data in the form of .cel files can be accessed at http://www.cs.virginia.edu/&cmt5n/Rawtimepoints/. All the primary and processed data have been generated using the hg17 (NCBI Build 35, May 2004) build of the Human genome assembly.
The replication signal for each probe was calculated as PM − MM. If the difference was negative, then the signal was assigned a value of 0. For a given probe on the array, we have five replication signals, one from each time point. Each probe is classified to be replicating either in a temporally specific or nonspecific (asynchronous) manner as follows. Probes were temporally specific if the signal in any one time point was at least twice the signal of each of the other four time points. To accommodate the possibility that a temporally specific replication signal could span two adjacent time points, probes were also called specific if the sum of any two adjacent time points was at least three times the signal of each of the other three time points. Probes that do not satisfy either of the criteria above are designated as temporally nonspecific. Supplemental Table 1 gives some examples of the specificity classification. For the Affymetrix ENCODE array, we classified 26.115% of the probes as temporally nonspecific in their pattern of replication.
For studying gene expression, RNA was extracted from logarithmically growing HeLa cells by using an RNeasy Kit (QIAGEN) and hybridized to the Human HG-U133 Plus 2.0 array (containing ∼38,500 genes) as described in the Affymetrix GeneChIP protocol (Affymetrix). Each microarray was scanned, visualized, and analyzed for the level of each individual transcript using a GeneChIP Scanner 3000 and GeneChIP Operating Software (GCOS; Affymetrix). Transcript signal was mapped against the chromosome coordinates (probe-by-probe basis) using the HG-U133A_2 Annotations, CSV provided by the manufacturer (Affymetrix).
Segregation of temporally specific and pan-S replicating segments
To segregate broad regions of replication, a sliding window of 10 kb was passed along each chromosomal segment, calculating the percentage of temporally nonspecific probes within the window. A pan-S region is begun when the percentage exceeds 60% and continues until it drops below the 60% threshold minus a given tolerance (e.g., 10% for our analysis). The tolerance is introduced in order to avoid thrashing between nonspecific and specific regions. Once the percentage drops below “threshold tolerance” (e.g., 50% for our settings of threshold and tolerance), the current pan-S region ends and a temporally specific region is started. The temporally specific region is continued until the percentage again rises above the threshold. In this manner, moving along the chromosome, broad regions of replication are segregated into temporally specific or pan-S classes.
The tolerance parameter, which helps us avoid thrashing between the two classes, introduces a directional bias into the segregation algorithm. As we move from lower chromosomal positions to higher chromosomal positions, the percentage must exceed 60% in order to begin a pan-S region. But the pan-S region does not end until the percentage drops below 50%. In order to correct for this directional bias, we perform two passes of the algorithm. One pass moves the window toward higher chromosomal positions, while the other pass moves the window toward lower chromosomal positions. Then we merge the two passes into a single segregation, which no longer suffers from a directional bias.
Interphase fluorescence in situ hybridization
Cells in S phase were harvested at indicated time points and incubated in pre-warmed 75 mM KCl solution for 15 min at 37°C to prepare nuclei. These nuclei were fixed in 3:1 (v/v) methanol/glacial acetic acid and mounted on a slide. A nick translation kit and SpectrumGreen dUTP/Spectrum Red dUTP (Vysis Inc.) were used for labeling the probe. Hybridization was carried out in a humidified chamber for 16 h at 37°C as described in the Vysis FISH protocol (Vysis Inc.). Slides were washed with 0.4× SSC/0.3% NP-40 for 2 min at 73°C and again with 2× SSC/0.1% NP-40 solution for 1 min at room temperature. Chromosomal DNA was counterstained with DAPI (VECTASHIELD Mounting Medium; Vector Labratories) and visualized with a Nikon Microphot.SA fluorescent microscope equipped with a DAPI filter, FITC, and a TRITC cube set (for Spectrum Green and Spectrum red fluors, respectively). Images were digitally obtained with a Nikon UFX-DX camera and Spot version 3.5.4 software. All the BAC clones were purchased from Children’s Hospital Oakland Research Institute.
The number of dots was visually counted in ∼100 cells at each time interval, and the number of dots/cell was calculated; 100% replication (in G2 cells) was when the increase in the number of dots/cell equaled the number of dots/cell observed in G1. After determining the dots/cell at 0, 2, 4, 6, 8, and 10 h of S phase, for each interval (e.g., 0–2 h, 2–4 h, etc.), we calculated the increase in dots/cell during that interval and converted it to the percent of replication.
Correlation of TR50 with genome features
CpG island annotations were obtained from the UCSC Genome Browser Web site (http://genome.ucsc.edu/cgi-bin/hgTrackUi?hgsid=64765488&c=chr16&g=cpgIsland). The density of CpG islands was calculated for all the chromosomal regions in each of the replication segments, that is, early-, mid-, late-, and pan-S. For the CpG islands that overlapped two temporal segments, the number of bases in the CpG island were counted and a 60% cutoff was used to assign it a specific temporal classification.
For determining gene density, we used the annotated genes under the Refseq database from the UCSC Genome Browser (http://genome.ucsc.edu/cgi-bin/hgTrackUi?hgsid= 64796361&c=chr16&g=refGene).
Enrichment in each of the replication segments (early-, mid-, late-, and pan-S) within a given data set (CpG islands, gene density, and transcripts) was calculated as follows. The number of elements from the data set whose majority base count fell into early segments was calculated. This was divided by the total number of elements in the data set to get a ratio of early-replicating elements. This ratio was divided by the ratio of early segments to all segments to give the enrichment ratio. Hence, a value of 1.0 would indicate that the data set was distributed in early segments as was expected by chance, while a value of 2.0 would indicate twice as many as expected by chance. Enrichment of the mid-, late-, and pan-S replicating regions was calculated similarly.
Chromatin immunoprecipitation
A chromatin immunoprecipitation assay was performed as per the protocol described at http://www.upstate.com with a variation in the sonication step. Samples were sonicated using a Branson microtip (3.2 mm) and Fisher Model 500 Sonic Dismembrator. Eight cycles of 15-sec pulse at 50% amplitude and 45 sec of cooling on ice were done to disrupt the cells. The antibodies used for ChIP were for identifying sites of histone 3 Lys 4 mono-, di-, and trimethylation (H3K4 Me), histone 3 Lys9 dimethylation (H3K9 di-Me), and HP1α. These antibodies were purchased from Upstate (Anti-H3K4 Me; 05-791 and H3K9 Anti-Me; 07-441) and Abcam (Anti- HP1α; ab9057). To determine the ChIP signal for H3K9 di-Me, 4 μL of ChIP DNA were first amplified in a linear range (14 cycles) using the WGA2 kit from Sigma-Aldrich and cleaned up by the QIAGEN PCR clean-up kit. Two microliters of this purified DNA was used as template for ChIP assay with primers. To rule out any amplification bias, three independent amplifications were performed and PCR with primers repeated with each of these template preparations. As a negative control, ChIP DNA from an IgG sample was amplified in a similar way. The details on primers used for ENm005 and ENr132 regions are provided in Supplemental Table 3.
Acknowledgments
This work was supported by National Institutes of Health Grant HG003157 (to A.D.). We thank members of the Dutta laboratory for helpful discussions.
Footnotes
References
- Aladjem M.I., Rodewald L.W., Kolman J.L., Wahl G.M., Rodewald L.W., Kolman J.L., Wahl G.M., Kolman J.L., Wahl G.M., Wahl G.M. Genetic dissection of a mammalian replicator in the human beta-globin locus. Science. 1998;281:1005–1009. doi: 10.1126/science.281.5379.1005. [DOI] [PubMed] [Google Scholar]
- Avner P., Heard E., Heard E. X-Chromosome inactivation: Counting, choice and initiation. Nat. Rev. Genet. 2001;2:59–67. doi: 10.1038/35047580. [DOI] [PubMed] [Google Scholar]
- Bell S.P., Dutta A., Dutta A. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- Bernstein B.E., Kamal M., Lindblad-Toh K., Bekiranov S., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., III, Gingeras T.R., Kamal M., Lindblad-Toh K., Bekiranov S., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., III, Gingeras T.R., Lindblad-Toh K., Bekiranov S., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., III, Gingeras T.R., Bekiranov S., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., III, Gingeras T.R., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., III, Gingeras T.R., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., III, Gingeras T.R., McMahon S., Karlsson E.K., Kulbokas E.J., III, Gingeras T.R., Karlsson E.K., Kulbokas E.J., III, Gingeras T.R., Kulbokas E.J., III, Gingeras T.R., Gingeras T.R., et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Bickmore W.A., Carothers A.D., Carothers A.D. Factors affecting the timing and imprinting of replication on a mammalian chromosome. J. Cell Sci. 1995;108:2801–2809. doi: 10.1242/jcs.108.8.2801. [DOI] [PubMed] [Google Scholar]
- Boumil R.M., Lee J.T., Lee J.T. Forty years of decoding the silence in X-chromosome inactivation. Hum. Mol. Genet. 2001;10:2225–2232. doi: 10.1093/hmg/10.20.2225. [DOI] [PubMed] [Google Scholar]
- Caron H., van Schaik B., van der Mee M., Baas F., Riggins G., van Sluis P., Hermus M.C., van Asperen R., Boon K., Voute P.A., van Schaik B., van der Mee M., Baas F., Riggins G., van Sluis P., Hermus M.C., van Asperen R., Boon K., Voute P.A., van der Mee M., Baas F., Riggins G., van Sluis P., Hermus M.C., van Asperen R., Boon K., Voute P.A., Baas F., Riggins G., van Sluis P., Hermus M.C., van Asperen R., Boon K., Voute P.A., Riggins G., van Sluis P., Hermus M.C., van Asperen R., Boon K., Voute P.A., van Sluis P., Hermus M.C., van Asperen R., Boon K., Voute P.A., Hermus M.C., van Asperen R., Boon K., Voute P.A., van Asperen R., Boon K., Voute P.A., Boon K., Voute P.A., Voute P.A., et al. The human transcriptome map: Clustering of highly expressed genes in chromosomal domains. Science. 2001;291:1289–1292. doi: 10.1126/science.1056794. [DOI] [PubMed] [Google Scholar]
- Carrel L., Clemson C.M., Dunn J.M., Miller A.P., Hunt P.A., Lawrence J.B., Willard H.F., Clemson C.M., Dunn J.M., Miller A.P., Hunt P.A., Lawrence J.B., Willard H.F., Dunn J.M., Miller A.P., Hunt P.A., Lawrence J.B., Willard H.F., Miller A.P., Hunt P.A., Lawrence J.B., Willard H.F., Hunt P.A., Lawrence J.B., Willard H.F., Lawrence J.B., Willard H.F., Willard H.F. X inactivation analysis and DNA methylation studies of the ubiquitin activating enzyme E1 and PCTAIRE-1 genes in human and mouse. Hum. Mol. Genet. 1996;5:391–401. doi: 10.1093/hmg/5.3.391. [DOI] [PubMed] [Google Scholar]
- Chang P.L., Mueller O.T., Lafrenie R.M., Varey P.A., Rosa N.E., Davidson R.G., Henry W.M., Shows T.B., Mueller O.T., Lafrenie R.M., Varey P.A., Rosa N.E., Davidson R.G., Henry W.M., Shows T.B., Lafrenie R.M., Varey P.A., Rosa N.E., Davidson R.G., Henry W.M., Shows T.B., Varey P.A., Rosa N.E., Davidson R.G., Henry W.M., Shows T.B., Rosa N.E., Davidson R.G., Henry W.M., Shows T.B., Davidson R.G., Henry W.M., Shows T.B., Henry W.M., Shows T.B., Shows T.B. The human arylsulfatase-C isoenzymes: Two distinct genes that escape from X inactivation. Am. J. Hum. Genet. 1990;46:729–737. [PMC free article] [PubMed] [Google Scholar]
- Chess A., Simon I., Cedar H., Axel R., Simon I., Cedar H., Axel R., Cedar H., Axel R., Axel R. Allelic inactivation regulates olfactory receptor gene expression. Cell. 1994;78:823–834. doi: 10.1016/s0092-8674(94)90562-2. [DOI] [PubMed] [Google Scholar]
- Demeret C., Vassetzky Y., Mechali M., Vassetzky Y., Mechali M., Mechali M. Chromatin remodelling and DNA replication: From nucleosomes to loop domains. Oncogene. 2001;20:3086–3093. doi: 10.1038/sj.onc.1204333. [DOI] [PubMed] [Google Scholar]
- Dhar V., Skoultchi A.I., Schildkraut C.L., Skoultchi A.I., Schildkraut C.L., Schildkraut C.L. Activation and repression of a beta-globin gene in cell hybrids is accompanied by a shift in its temporal replication. Mol. Cell. Biol. 1989;9:3524–3532. doi: 10.1128/mcb.9.8.3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disteche C.M. Escape from X inactivation in human and mouse. Trends Genet. 1995;11:17–22. doi: 10.1016/s0168-9525(00)88981-7. [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium, The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium, Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007 doi: 10.1038/nature05874. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epner E., Forrester W.C., Groudine M., Forrester W.C., Groudine M., Groudine M. Asynchronous DNA replication within the human beta-globin gene locus. Proc. Natl. Acad. Sci. 1988;85:8081–8085. doi: 10.1073/pnas.85.21.8081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier C., Goto Y., Ballestar E., Delaval K., Hever A.M., Esteller M., Feil R., Goto Y., Ballestar E., Delaval K., Hever A.M., Esteller M., Feil R., Ballestar E., Delaval K., Hever A.M., Esteller M., Feil R., Delaval K., Hever A.M., Esteller M., Feil R., Hever A.M., Esteller M., Feil R., Esteller M., Feil R., Feil R. Allele-specific histone lysine methylation marks regulatory regions at imprinted mouse genes. EMBO J. 2002;21:6560–6570. doi: 10.1093/emboj/cdf655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuks F. DNA methylation and histone modifications: Teaming up to silence genes. Curr. Opin. Genet. Dev. 2005;15:490–495. doi: 10.1016/j.gde.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Goldman M.A. The chromatin domain as a unit of gene regulation. Bioessays. 1988;9:50–55. doi: 10.1002/bies.950090204. [DOI] [PubMed] [Google Scholar]
- Hansen R.S., Canfield T.K., Fjeld A.D., Gartler S.M., Canfield T.K., Fjeld A.D., Gartler S.M., Fjeld A.D., Gartler S.M., Gartler S.M. Role of late replication timing in the silencing of X-linked genes. Hum. Mol. Genet. 1996;5:1345–1353. doi: 10.1093/hmg/5.9.1345. [DOI] [PubMed] [Google Scholar]
- Holmquist G., Gray M., Porter T., Jordan J., Gray M., Porter T., Jordan J., Porter T., Jordan J., Jordan J. Characterization of Giemsa dark- and light-band DNA. Cell. 1982;31:121–129. doi: 10.1016/0092-8674(82)90411-1. [DOI] [PubMed] [Google Scholar]
- Iqbal M.A., Plumb M., Stein J., Stein G., Schildkraut C.L., Plumb M., Stein J., Stein G., Schildkraut C.L., Stein J., Stein G., Schildkraut C.L., Stein G., Schildkraut C.L., Schildkraut C.L. Coordinate replication of members of the multigene family of core and H1 human histone genes. Proc. Natl. Acad. Sci. 1984;81:7723–7727. doi: 10.1073/pnas.81.24.7723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakovcevski I., Zecevic N., Zecevic N. Olig transcription factors are expressed in oligodendrocyte and neuronal cells in human fetal CNS. J. Neurosci. 2005;25:10064–10073. doi: 10.1523/JNEUROSCI.2324-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon Y., Bekiranov S., Karnani N., Kapranov P., Ghosh S., MacAlpine D., Lee C., Hwang D.S., Gingeras T.R., Dutta A., Bekiranov S., Karnani N., Kapranov P., Ghosh S., MacAlpine D., Lee C., Hwang D.S., Gingeras T.R., Dutta A., Karnani N., Kapranov P., Ghosh S., MacAlpine D., Lee C., Hwang D.S., Gingeras T.R., Dutta A., Kapranov P., Ghosh S., MacAlpine D., Lee C., Hwang D.S., Gingeras T.R., Dutta A., Ghosh S., MacAlpine D., Lee C., Hwang D.S., Gingeras T.R., Dutta A., MacAlpine D., Lee C., Hwang D.S., Gingeras T.R., Dutta A., Lee C., Hwang D.S., Gingeras T.R., Dutta A., Hwang D.S., Gingeras T.R., Dutta A., Gingeras T.R., Dutta A., Dutta A. Temporal profile of replication of human chromosomes. Proc. Natl. Acad. Sci. 2005;102:6419–6424. doi: 10.1073/pnas.0405088102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y.H., Bressler J., Beaudet A.L., Bressler J., Beaudet A.L., Beaudet A.L. Epigenetics and human disease. Annu. Rev. Genomics Hum. Genet. 2004;5:479–510. doi: 10.1146/annurev.genom.5.061903.180014. [DOI] [PubMed] [Google Scholar]
- Jones P.A., Takai D., Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- Kitsberg D., Selig S., Keshet I., Cedar H., Selig S., Keshet I., Cedar H., Keshet I., Cedar H., Cedar H. Replication structure of the human beta-globin gene domain. Nature. 1993;366:588–590. doi: 10.1038/366588a0. [DOI] [PubMed] [Google Scholar]
- Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Devon K., Dewar K., Doyle M., FitzHugh W., Dewar K., Doyle M., FitzHugh W., Doyle M., FitzHugh W., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- MacAlpine D.M., Bell S.P., Bell S.P. A genomic view of eukaryotic DNA replication. Chromosome Res. 2005;13:309–326. doi: 10.1007/s10577-005-1508-1. [DOI] [PubMed] [Google Scholar]
- MacAlpine D.M., Rodriguez H.K., Bell S.P., Rodriguez H.K., Bell S.P., Bell S.P. Coordination of replication and transcription along a Drosophila chromosome. Genes & Dev. 2004;18:3094–3105. doi: 10.1101/gad.1246404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller A.P., Gustashaw K., Wolff D.J., Rider S.H., Monaco A.P., Eble B., Schlessinger D., Gorski J.L., van Ommen G.J., Weissenbach J., Gustashaw K., Wolff D.J., Rider S.H., Monaco A.P., Eble B., Schlessinger D., Gorski J.L., van Ommen G.J., Weissenbach J., Wolff D.J., Rider S.H., Monaco A.P., Eble B., Schlessinger D., Gorski J.L., van Ommen G.J., Weissenbach J., Rider S.H., Monaco A.P., Eble B., Schlessinger D., Gorski J.L., van Ommen G.J., Weissenbach J., Monaco A.P., Eble B., Schlessinger D., Gorski J.L., van Ommen G.J., Weissenbach J., Eble B., Schlessinger D., Gorski J.L., van Ommen G.J., Weissenbach J., Schlessinger D., Gorski J.L., van Ommen G.J., Weissenbach J., Gorski J.L., van Ommen G.J., Weissenbach J., van Ommen G.J., Weissenbach J., Weissenbach J., et al. Three genes that escape X chromosome inactivation are clustered within a 6 Mb YAC contig and STS map in Xp11.21–p11.22. Hum. Mol. Genet. 1995;4:731–739. doi: 10.1093/hmg/4.4.731. [DOI] [PubMed] [Google Scholar]
- Mostoslavsky R., Singh N., Tenzen T., Goldmit M., Gabay C., Elizur S., Qi P., Reubinoff B.E., Chess A., Cedar H., Singh N., Tenzen T., Goldmit M., Gabay C., Elizur S., Qi P., Reubinoff B.E., Chess A., Cedar H., Tenzen T., Goldmit M., Gabay C., Elizur S., Qi P., Reubinoff B.E., Chess A., Cedar H., Goldmit M., Gabay C., Elizur S., Qi P., Reubinoff B.E., Chess A., Cedar H., Gabay C., Elizur S., Qi P., Reubinoff B.E., Chess A., Cedar H., Elizur S., Qi P., Reubinoff B.E., Chess A., Cedar H., Qi P., Reubinoff B.E., Chess A., Cedar H., Reubinoff B.E., Chess A., Cedar H., Chess A., Cedar H., Cedar H., et al. Asynchronous replication and allelic exclusion in the immune system. Nature. 2001;414:221–225. doi: 10.1038/35102606. [DOI] [PubMed] [Google Scholar]
- Perry P., Sauer S., Billon N., Richardson W.D., Spivakov M., Warnes G., Livesey F.J., Merkenschlager M., Fisher A.G., Azuara V., Sauer S., Billon N., Richardson W.D., Spivakov M., Warnes G., Livesey F.J., Merkenschlager M., Fisher A.G., Azuara V., Billon N., Richardson W.D., Spivakov M., Warnes G., Livesey F.J., Merkenschlager M., Fisher A.G., Azuara V., Richardson W.D., Spivakov M., Warnes G., Livesey F.J., Merkenschlager M., Fisher A.G., Azuara V., Spivakov M., Warnes G., Livesey F.J., Merkenschlager M., Fisher A.G., Azuara V., Warnes G., Livesey F.J., Merkenschlager M., Fisher A.G., Azuara V., Livesey F.J., Merkenschlager M., Fisher A.G., Azuara V., Merkenschlager M., Fisher A.G., Azuara V., Fisher A.G., Azuara V., Azuara V. A dynamic switch in the replication timing of key regulator genes in embryonic stem cells upon neural induction. Cell Cycle. 2004;3:1645–1650. [PubMed] [Google Scholar]
- Raghuraman M.K., Winzeler E.A., Collingwood D., Hunt S., Wodicka L., Conway A., Lockhart D.J., Davis R.W., Brewer B.J., Fangman W.L., Winzeler E.A., Collingwood D., Hunt S., Wodicka L., Conway A., Lockhart D.J., Davis R.W., Brewer B.J., Fangman W.L., Collingwood D., Hunt S., Wodicka L., Conway A., Lockhart D.J., Davis R.W., Brewer B.J., Fangman W.L., Hunt S., Wodicka L., Conway A., Lockhart D.J., Davis R.W., Brewer B.J., Fangman W.L., Wodicka L., Conway A., Lockhart D.J., Davis R.W., Brewer B.J., Fangman W.L., Conway A., Lockhart D.J., Davis R.W., Brewer B.J., Fangman W.L., Lockhart D.J., Davis R.W., Brewer B.J., Fangman W.L., Davis R.W., Brewer B.J., Fangman W.L., Brewer B.J., Fangman W.L., Fangman W.L. Replication dynamics of the yeast genome. Science. 2001;294:115–121. doi: 10.1126/science.294.5540.115. [DOI] [PubMed] [Google Scholar]
- Rountree M.R., Bachman K.E., Baylin S.B., Bachman K.E., Baylin S.B., Baylin S.B. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat. Genet. 2000;25:269–277. doi: 10.1038/77023. [DOI] [PubMed] [Google Scholar]
- Rountree M.R., Bachman K.E., Herman J.G., Baylin S.B., Bachman K.E., Herman J.G., Baylin S.B., Herman J.G., Baylin S.B., Baylin S.B. DNA methylation, chromatin inheritance, and cancer. Oncogene. 2001;20:3156–3165. doi: 10.1038/sj.onc.1204339. [DOI] [PubMed] [Google Scholar]
- Saccone S., De Sario A., Wiegant J., Raap A.K., Della Valle G., Bernardi G., De Sario A., Wiegant J., Raap A.K., Della Valle G., Bernardi G., Wiegant J., Raap A.K., Della Valle G., Bernardi G., Raap A.K., Della Valle G., Bernardi G., Della Valle G., Bernardi G., Bernardi G. Correlations between isochores and chromosomal bands in the human genome. Proc. Natl. Acad. Sci. 1993;90:11929–11933. doi: 10.1073/pnas.90.24.11929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmegner C., Berger A., Vogel W., Hameister H., Assum G., Berger A., Vogel W., Hameister H., Assum G., Vogel W., Hameister H., Assum G., Hameister H., Assum G., Assum G. An isochore transition zone in the NF1 gene region is a conserved landmark of chromosome structure and function. Genomics. 2005;86:439–445. doi: 10.1016/j.ygeno.2005.06.011. [DOI] [PubMed] [Google Scholar]
- Schubeler D., Scalzo D., Kooperberg C., van Steensel B., Delrow J., Groudine M., Scalzo D., Kooperberg C., van Steensel B., Delrow J., Groudine M., Kooperberg C., van Steensel B., Delrow J., Groudine M., van Steensel B., Delrow J., Groudine M., Delrow J., Groudine M., Groudine M. Genome-wide DNA replication profile for Drosophila melanogaster: A link between transcription and replication timing. Nat. Genet. 2002;32:438–442. doi: 10.1038/ng1005. [DOI] [PubMed] [Google Scholar]
- Simon I., Tenzen T., Reubinoff B.E., Hillman D., McCarrey J.R., Cedar H., Tenzen T., Reubinoff B.E., Hillman D., McCarrey J.R., Cedar H., Reubinoff B.E., Hillman D., McCarrey J.R., Cedar H., Hillman D., McCarrey J.R., Cedar H., McCarrey J.R., Cedar H., Cedar H. Asynchronous replication of imprinted genes is established in the gametes and maintained during development. Nature. 1999;401:929–932. doi: 10.1038/44866. [DOI] [PubMed] [Google Scholar]
- Simon I., Tenzen T., Mostoslavsky R., Fibach E., Lande L., Milot E., Gribnau J., Grosveld F., Fraser P., Cedar H., Tenzen T., Mostoslavsky R., Fibach E., Lande L., Milot E., Gribnau J., Grosveld F., Fraser P., Cedar H., Mostoslavsky R., Fibach E., Lande L., Milot E., Gribnau J., Grosveld F., Fraser P., Cedar H., Fibach E., Lande L., Milot E., Gribnau J., Grosveld F., Fraser P., Cedar H., Lande L., Milot E., Gribnau J., Grosveld F., Fraser P., Cedar H., Milot E., Gribnau J., Grosveld F., Fraser P., Cedar H., Gribnau J., Grosveld F., Fraser P., Cedar H., Grosveld F., Fraser P., Cedar H., Fraser P., Cedar H., Cedar H. Developmental regulation of DNA replication timing at the human beta globin locus. EMBO J. 2001;20:6150–6157. doi: 10.1093/emboj/20.21.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenzen T., Yamagata T., Fukagawa T., Sugaya K., Ando A., Inoko H., Gojobori T., Fujiyama A., Okumura K., Ikemura T., Yamagata T., Fukagawa T., Sugaya K., Ando A., Inoko H., Gojobori T., Fujiyama A., Okumura K., Ikemura T., Fukagawa T., Sugaya K., Ando A., Inoko H., Gojobori T., Fujiyama A., Okumura K., Ikemura T., Sugaya K., Ando A., Inoko H., Gojobori T., Fujiyama A., Okumura K., Ikemura T., Ando A., Inoko H., Gojobori T., Fujiyama A., Okumura K., Ikemura T., Inoko H., Gojobori T., Fujiyama A., Okumura K., Ikemura T., Gojobori T., Fujiyama A., Okumura K., Ikemura T., Fujiyama A., Okumura K., Ikemura T., Okumura K., Ikemura T., Ikemura T. Precise switching of DNA replication timing in the GC content transition area in the human major histocompatibility complex. Mol. Cell. Biol. 1997;17:4043–4050. doi: 10.1128/mcb.17.7.4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeesch J.R., Petit P., Kermouni A., Renauld J.C., Van Den Berghe H., Marynen P., Petit P., Kermouni A., Renauld J.C., Van Den Berghe H., Marynen P., Kermouni A., Renauld J.C., Van Den Berghe H., Marynen P., Renauld J.C., Van Den Berghe H., Marynen P., Van Den Berghe H., Marynen P., Marynen P. The IL-9 receptor gene, located in the Xq/Yq pseudoautosomal region, has an autosomal origin, escapes X inactivation and is expressed from the Y. Hum. Mol. Genet. 1997;6:1–8. doi: 10.1093/hmg/6.1.1. [DOI] [PubMed] [Google Scholar]
- Versteeg R., van Schaik B.D., van Batenburg M.F., Roos M., Monajemi R., Caron H., Bussemaker H.J., van Kampen A.H., van Schaik B.D., van Batenburg M.F., Roos M., Monajemi R., Caron H., Bussemaker H.J., van Kampen A.H., van Batenburg M.F., Roos M., Monajemi R., Caron H., Bussemaker H.J., van Kampen A.H., Roos M., Monajemi R., Caron H., Bussemaker H.J., van Kampen A.H., Monajemi R., Caron H., Bussemaker H.J., van Kampen A.H., Caron H., Bussemaker H.J., van Kampen A.H., Bussemaker H.J., van Kampen A.H., van Kampen A.H. The human transcriptome map reveals extremes in gene density, intron length, GC content, and repeat pattern for domains of highly and weakly expressed genes. Genome Res. 2003;13:1998–2004. doi: 10.1101/gr.1649303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Lin C.M., Brooks S., Cimbora D., Groudine M., Aladjem M.I., Lin C.M., Brooks S., Cimbora D., Groudine M., Aladjem M.I., Brooks S., Cimbora D., Groudine M., Aladjem M.I., Cimbora D., Groudine M., Aladjem M.I., Groudine M., Aladjem M.I., Aladjem M.I. The human beta-globin replication initiation region consists of two modular independent replicators. Mol. Cell. Biol. 2004;24:3373–3386. doi: 10.1128/MCB.24.8.3373-3386.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y., Fujiyama A., Ichiba Y., Hattori M., Yada T., Sakaki Y., Ikemura T., Fujiyama A., Ichiba Y., Hattori M., Yada T., Sakaki Y., Ikemura T., Ichiba Y., Hattori M., Yada T., Sakaki Y., Ikemura T., Hattori M., Yada T., Sakaki Y., Ikemura T., Yada T., Sakaki Y., Ikemura T., Sakaki Y., Ikemura T., Ikemura T. Chromosome-wide assessment of replication timing for human chromosomes 11q and 21q: Disease-related genes in timing-switch regions. Hum. Mol. Genet. 2002;11:13–21. doi: 10.1093/hmg/11.1.13. [DOI] [PubMed] [Google Scholar]
- Woodfine K., Fiegler H., Beare D.M., Collins J.E., McCann O.T., Young B.D., Debernardi S., Mott R., Dunham I., Carter N.P., Fiegler H., Beare D.M., Collins J.E., McCann O.T., Young B.D., Debernardi S., Mott R., Dunham I., Carter N.P., Beare D.M., Collins J.E., McCann O.T., Young B.D., Debernardi S., Mott R., Dunham I., Carter N.P., Collins J.E., McCann O.T., Young B.D., Debernardi S., Mott R., Dunham I., Carter N.P., McCann O.T., Young B.D., Debernardi S., Mott R., Dunham I., Carter N.P., Young B.D., Debernardi S., Mott R., Dunham I., Carter N.P., Debernardi S., Mott R., Dunham I., Carter N.P., Mott R., Dunham I., Carter N.P., Dunham I., Carter N.P., Carter N.P. Replication timing of the human genome. Hum. Mol. Genet. 2004;13:191–202. doi: 10.1093/hmg/ddh016. [DOI] [PubMed] [Google Scholar]
- Wu R., Terry A.V., Singh P.B., Gilbert D.M., Terry A.V., Singh P.B., Gilbert D.M., Singh P.B., Gilbert D.M., Gilbert D.M. Differential subnuclear localization and replication timing of histone H3 lysine 9 methylation states. Mol. Biol. Cell. 2005;16:2872–2881. doi: 10.1091/mbc.E04-11-0997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Ermakova O.V., Riblet R., Birshtein B.K., Schildkraut C.L., Ermakova O.V., Riblet R., Birshtein B.K., Schildkraut C.L., Riblet R., Birshtein B.K., Schildkraut C.L., Birshtein B.K., Schildkraut C.L., Schildkraut C.L. Replication and subnuclear location dynamics of the immunoglobulin heavy-chain locus in B-lineage cells. Mol. Cell. Biol. 2002;22:4876–4889. doi: 10.1128/MCB.22.13.4876-4889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]