A suite of Gateway® cloning vectors for high-throughput genetic analysis in Saccharomyces cerevisiae (original) (raw)
. Author manuscript; available in PMC: 2008 Jan 10.
Published in final edited form as: Yeast. 2007 Oct;24(10):913–919. doi: 10.1002/yea.1502
Abstract
In the post-genomic era, academic and biotechnological research is increasingly shifting its attention from single proteins to the analysis of complex protein networks. This change in experimental design requires the use of simple and experimentally tractable organisms, such as the unicellular eukaryote Saccharomyces cerevisiae, and a range of new high-throughput techniques. The Gateway® system has emerged as a powerful high-throughput cloning method that allows for the in vitro recombination of DNA with high speed, accuracy and reliability. Two Gateway®-based libraries of overexpression plasmids containing the entire complement of yeast open reading frames (ORFs) have recently been completed. In order to make use of these powerful resources, we adapted the widely used pRS series of yeast shuttle vectors for use in Gateway®-based cloning. The resulting suite of 288 yeast Gateway® vectors is based upon the two commonly used GPD and GAL1 promoter expression systems that enable expression of ORFs either constitutively or under galactose-inducible conditions. In addition, proteins of interest can be fused to a choice of frequently used N- or C- terminal tags, such as EGFP, ECFP, EYFP, Cerulean, monomeric DsRed, HA or TAP. We have made this yeast Gateway® vector kit available to the research community via the non-profit Addgene Plasmid Repository (http://www.addgene.org/yeast_gateway).
Keywords: Gateway, vector, high-throughput, cloning
Introduction
Baker’s yeast, or Saccharomyces cerevisiae, is a powerful experimental system for studying complex biological processes. Notably, many of the key cellular pathways of yeast share a high degree of similarity to those of mammalian cells. In addition, the yeast genome is very well characterized and amenable to genetic manipulation. Methods are available to rapidly overexpress or knockout almost every gene and efforts to generate a collection of haploid strains, each harboring a single mutation, as well as a collection of expression plasmids with each yeast ORF under the control of an inducible promoter have recently come to fruition (Cooper et al., [2006]; Sopko, et al., [2006]). The availability of these tools turns yeast into a robust new system for investigating, on a genome-wide scale, the mechanisms underlying many cellular processes with direct relevance to human disease and for the discovery of novel drug targets for therapeutic intervention.
A frustrating aspect of high-throughput screening is that it is often desirable to test candidate genes in multiple formats and/or combinations, but current cloning strategies usually make transferring ORFs from one vector backbone to another very time-consuming. To address this problem, we have constructed a suite of 288 yeast expression vectors compatible with the Gateway® recombination-based cloning system and describe here methods for their use in multiple applications. Our collection provides a choice of two promoters (constitutive or inducible), integrating or extra-chromosomal, high- or low-copy origins of replication, and the option for N- or C-terminal fusion to various protein affinity tags (HA, TAP) or fluorescent proteins (EGFP, ECFP, EYFP, Cerulean, DsRed). We have made these plasmids available to the research community (http://www.addgene.org/yeast_gateway) and we hope their use along with the methods described here will expedite the process of high-throughput screening and target validation.
Materials and Methods
Vector Construction
We first modified a set of 24 pRS yeast shuttle vectors (pRS303, pRS304, pRS305, pRS306, pRS413, pRS414, pRS415, pRS416, pRS423, pRS424, pRS425 and pRS426, each with a CYC1 terminator and a GPD or GAL1 promoter) to make them compatible for use with the Gateway® system. To generate this core set of pAG (Advanced Gateway) vectors, we inserted the chloramphenicol/ccdB resistance Gateway® cassette A into the single SmaI restriction site of each pRS plasmid. Proper orientation of the Gateway® cassette was confirmed by DNA sequencing. The resulting plasmids were then used to generate a set of derived vectors that allows for the expression of proteins with C-terminal tags (Please see Supplementary Table 1 for nomenclature and a complete list of pAG plasmids). The coding sequences for the various tags were amplified using the primers and templates listed in Tables 1 and 2. PCR products were cloned between the HindIII and XhoI restriction sites of pAG426GPD and pAG426GAL (EGFP, ECFP, EYFP, Cerulean, DsRed and 3HA) or between the ClaI and XhoI sites of pAG423GPD and pAG423GAL (TAP). The correct amplification and integration of tag DNA sequences was confirmed by sequencing. To complete the set of pAG plasmids the entire expression cassette was removed from the primary pAG423GPD/GAL-ccdB-tag and pAG426GPD/GAL-ccdB-tag constructs by digestion with SacI and KpnI and subcloned into the remaining pRS vector backgrounds using SacI and KpnI for linearization of the target plasmids.
Table 1.
Primers used for the amplification of tags (restriction sites are underlined).
| Primer | Sequence |
|---|---|
| cEXFP-forw-HindIII | GCA GTA CGA AGC TTA ATG GTG AGC AAG GGC GAG GAG |
| cEXFP-rev-XhoI | GA TAG TGT CTC GAG TTA CTT GTA CAG CTC GTC CAT GCC G |
| DsRed-forw-HindIII | GCA GTA CGA AGC TTA ATG GAC AAC ACC GAG GAC GTC ATC AAG |
| DsRed-rev-XhoI | GA TAG TGT CTC GAG CTA CTG GGA GCC GGA GTG GCG G |
| TAP-forw-ClaI | GCA GTA CGA TCG ATA GGT GGA CCA GGT GGT GGA ATG AAG CGA CGA TGG AAA AAG |
| TAP-rev-XhoI | GA TAG TGT CTC GAG TCA CTG TTC TTT GCT CAC CGA AG |
| 3HA-forw-HindIII | GCA GTA CGA AGC TTA GGT GGA ATG TAC CCA TAC GAT GTT CCT GAC T |
| 3HA-rev-XhoI | GA TAG TGT CTC GAG TTA GCA CTG AGC AGC GTA ATC TG |
| nEXFP-forw-XmaI | G CAG CGT TCC CGG GAC AAA ATG GTG AGC AAG GGC GAG G |
| nEXFP-rev-XhoI | GA TAG TGT CTC GAG TCC ACC ACC TGG TCC ACC CTT GTA CAG CTC GTC CAT GCC G |
Table 2.
Primer combinations and templates used for the amplification of tags.
| Tag | Primer combination | Template |
|---|---|---|
| EGFP (C-terminal) | cEXFP-forw-HindIIIcEXFP-rev-XhoI | pEGFP-1 (Clontech) |
| ECFP (C-terminal) | cEXFP-forw-HindIIIcEXFP-rev-XhoI | pECFP-1 (Clontech) |
| EYFP (C terminal) | cEXFP-forw-HindIIIcEXFP-rev-XhoI | pEYFP-1 (Clontech) |
| Cerulean (C-terminal) | cEXFP-forw-HindIIIcEXFP-rev-XhoI | pmCerulean-N1 (Rizzo and Piston, [2005]) |
| DsRed M1 (C-terminal) | DsRed-forw-HindIIIDsRed-rev-XhoI | pDsRed M1 (Clontech) |
| TAP (C-terminal) | TAP-forw-ClaITAP-rev-XhoI | pVV220 (Van Mullem, et al., [2003]) |
| 3HA (C-terminal) | 3HA-forw-HindIII3HA-rev-XhoI | pVV205 (Van Mullem, et al., [2003]) |
| EGFP (N-terminal) | nEXFP-forw-XmaInEXFP-rev-XhoI | pEGFP-1 (Clontech) |
| ECFP (N-terminal) | nEXFP-forw-XmaInEXFP-rev-XhoI | pECFP-1 (Clontech) |
| EYFP (N-terminal) | nEXFP-forw-XmaInEXFP-rev-XhoI | pEYFP-1 (Clontech) |
| Cerulean (N-terminal) | nEXFP-forw-XmaInEXFP-rev-XhoI | pmCerulean-N1 (Rizzo and Piston, [2005]) |
For the cloning of vectors for expression of N-terminal fusions, PCR-generated fragments (See Tables 1 and 2 for details) were inserted into the XmaI and XhoI sites of pRS426GPD and pRS426GAL. The resulting plasmids were cut with XhoI and incubated with mung bean nuclease to remove single-stranded overhangs. Next, the C1 Gateway® cassette was ligated to the blunted plasmids. The resulting plasmids were subsequently analyzed by sequencing to confirm the correct integration of tag sequences and Gateway® cassette. As a final step a SpeI-MluI fragment comprising the Gateway® cassette and the tag sequence was cut out and cloned between the SpeI and MluI sites of the core set of pAG plasmids.
Standard BP and LR reaction protocol
We have developed modified versions of the standard Gateway® LR and BP reactions. The smaller volumes require less enzyme, reducing the cost per reaction.
BP or LR Reaction
- 150 ng expression clone (BP) or entry clone (LR): 1 microliter
- 150 ng entry vector (BP) or destination vector (LR): 1 microliter
- 1X TE (pH 8): 2 microliters
- BP (or LR) Clonase II mix (Invitrogen): 1 microliter
Incubate at room temperature for 1 hour. Transform 2–3 microliters of the reaction into DH5α, TOP10, or other ccdB-sensitive cells. Select for transformants on LB agar plates containing 100 μg/ml ampicillin.
Experiments with α-synuclein
Entry clones containing full-length human α-synuclein with or without a stop codon were used in Gateway LR reactions to generate 426GAL-α–syn-EGFP, 426GAL-α–syn-DsRed, 426GAL-α–syn-HA, 426GAL-α–syn-TAP, and 426GAL-α–syn. These plasmids were transformed into the BY4741 yeast strain. To induce expression of the α–syn fusion proteins, yeast cells were pre-grown overnight in synthetic media containing raffinose to mid log-phase. Cells were switched to synthetic media containing galactose and grown for 6 hours. Cells were either processed for fluorescence microscopy or protein lysates prepared for immunoblotting. Antibodies used for immunoblotting were as follows: GFP (Roche, 1:1,000); HA (Roche, 1:1,000); TAP (Open Biosystems, 1:1,000). Horseradish peroxidase coupled secondary antibodies were used at 1:10,000. For growth assays, serial dilutions of transformants were grown on solid synthetic media containing either glucose (control, α-syn “off”) or galactose (α-syn “on”).
Results and Discussion
Overview of the Gateway® system of recombination-based cloning
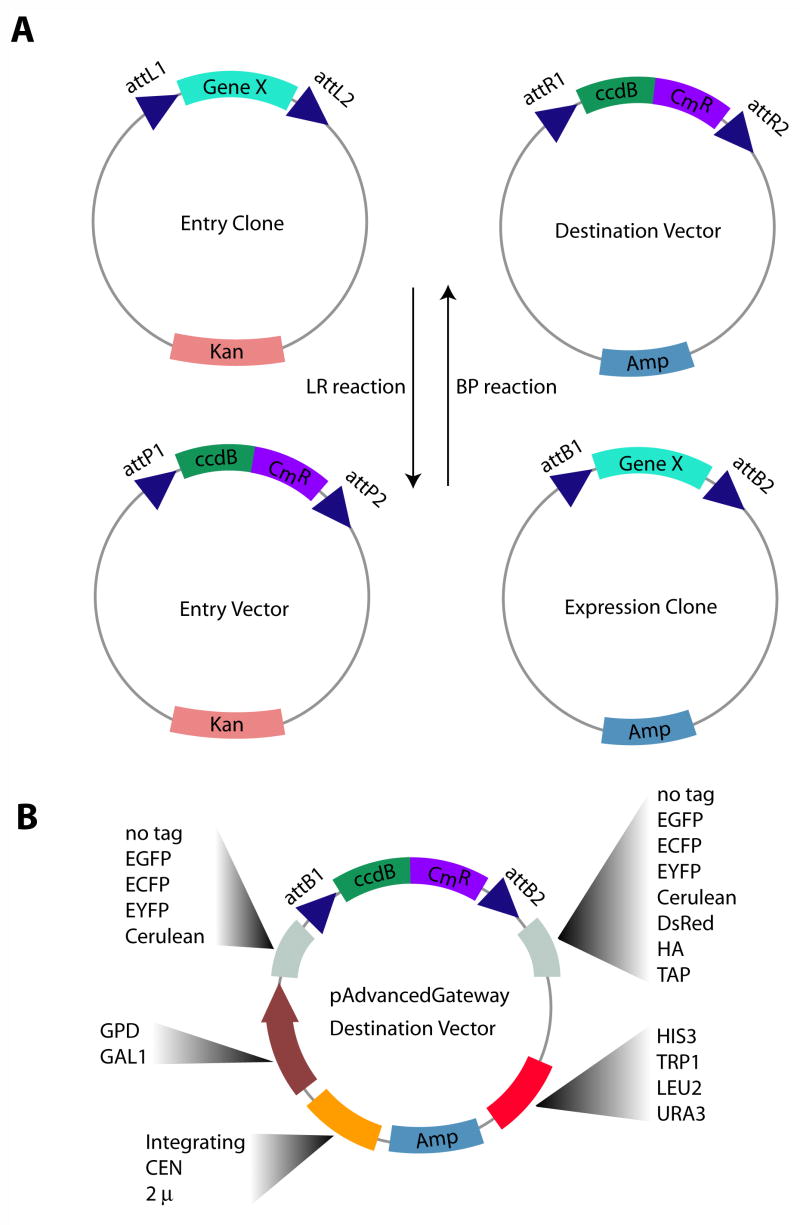
The types of plasmids and the general strategy of Gateway® cloning is diagrammed in Figure 1A. This rapid, directional, and highly efficient cloning system is based on site-specific recombination mediated by the recombination machinery of bacteriophage lambda (Landy, [1989]) and has been adapted for in vitro use in standard molecular biology protocols (Hartley, et al., [2000]; Walhout, et al., [2000]).
Figure 1. Overview of Gateway system and Generation of pAG yeast destination vectors.
A) The Gateway system of recombination-based cloning involves an LR reaction, in which an entry clone, containing a gene of interest is mixed with a destination vector, containing features of interest (e.g. promoter, protein tags, etc.). The destination vector harbors a recombination site-flanked bacterial “death” gene (ccdB) that is exchanged for the gene of interest contained in the entry clone. Transformation of E. coli that are sensitive to the ccdB effects allows for selection of expression clones. This reaction is reversible (BP reaction); expression clones can be used to re-generate entry clones. B) The set of pAdvancedGateway destination vectors allows for a choice of constitutive (GPD) or inducible (GAL1) promoters, N- or C-terminal protein tags, integrating or extra-chromosomal origins of replication, high- (2μ) or low-copy number (CEN), as well as a choice of auxotrophic markers. Caution: Since these destination vectors contain the ccdB cassette, they must be propagated in ccdB-resistant E. coli (e.g. DB3.1).
There are two main types of Gateway® reactions: LR and BP. An LR reaction consists of an “entry clone” plasmid (containing a gene of interest flanked by attL1 and attL2 sequences) harboring a kanamycin resistance cassette that is mixed with a “destination vector” (containing a bacterial death gene and chloramphenicol resistance gene flanked by attR1 and attR2 sequences) harboring an ampicillin resistance cassette. LR Clonase II enzyme mix (Invitrogen) catalyzes recombination between the recognition sites, generating an “expression clone” containing the gene of interest in the destination vector backbone. Transformation of E. coli with this reaction mixture and plating on ampicillin-containing LB agar plates allows for the specific selection of expression clones and the selective killing of bacteria containing the initial plasmid constructs. A BP reaction is essentially the opposite reaction of LR, and transfers genes from expression clones into entry vectors (isolated by selection on kanamycin-containing agar plates).
Construction and validation of a collection of Saccharomyces cerevisiae Gateway® destination vectors
We modified the pRS series of yeast shuttle vectors (Christianson, et al., [1992]; Sikorski and Hieter, [1989]) for use with the Gateway® system (Figure 1B). This involved blunt-end ligation of the ccdB/chloramphenical resistance cassette into the multiple cloning site of each of these plasmids (see supplementary Table 1 for details on the type of plasmids generated). Diagnostic restriction digest and DNA sequencing verified proper orientation of the Gateway® cassette. These initial Gateway® vectors were subsequently modified to obtain a choice of N- or C-terminal protein tags to enable visualization by fluorescence microscopy or biochemical purification. We maintained the same nomenclature as the pRS series vectors, but renamed the new Advanced Gateway® derivatives, pAG (e.g. pAG413GPD, pAG426GAL, pAG306GAL-ccdB-EGFP, pAG413GAL-EGFP-ccdB). The ccdB name portion in plasmids for N- or C-terminal fusions stands for the entire Gateway® cassette and is used in our nomenclature scheme to indicate the orientation of the Gateway® cassette relative to the tag.
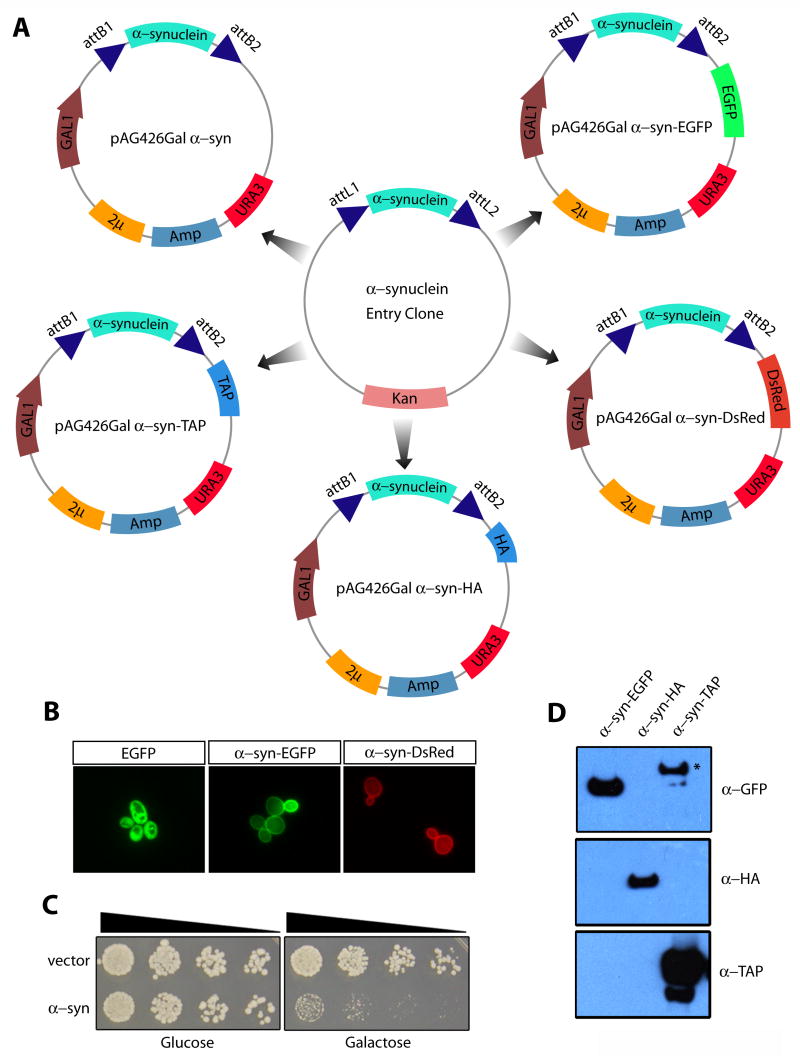
We tested the functionality of our vectors using a protein that we have been studying in our laboratory. The subcellular localization and phenotypic consequences of expressing human α-synuclein (α-syn) in yeast cells have been well characterized (Cooper et al., [2006]; Outeiro and Lindquist, [2003]). We used the Gateway® system and our collection of pAG destination vectors to rapidly generate a variety of α-syn expression constructs, which we then tested in yeast cells. In a 1 hour LR reaction we were able to generate a panel of α-syn constructs enabling toxicity studies, biochemical purification and subcellular localization (Figure 2). The generated α-syn fusion proteins behave according to previously published data indicating that the linkers (see Table 3 for linker sequences) do not interfere with proper folding and targeting of α-syn. Generating a similar set of constructs using standard molecular biology techniques would take several days and would require customized cloning strategies for each construct.
Figure 2. Practical application of Advanced Gateway vectors to experiments with α–synuclein in yeast.
A) An entry clone containing human α–synuclein (with or without stop codon) was used in 1 hour in vitro LR reactions to generate untagged, EGFP-, DsRed-, HA-, or TAP-tagged expression constructs. The Gateway-generated fluorescently tagged proteins localized properly to the plasma membrane (B), whereas EGFP alone was distributed throughout the cytoplasm; the untagged protein induced cellular toxicity (C) and immunoblot analysis detected the appropriate size tagged proteins (D). The asterisk (*) in D indicates a non-specific immunoreactive band resulting from the Protein A portion of the TAP tag.
Table 3.
Resulting linker sequences (the translated attB1 or attB2 portion is underlined).
| Primer | Sequence |
|---|---|
| EGFP, ECFP, EYFP, DsRed, Cerulean (C-terminal) | SAFLYKVVMGCRNSISSL_M_ |
| TAP (C-terminal) | SAFLYKVVMGCRNSISSLSIGGPGGG_M_ |
| 3HA (C-terminal) | SAFLYKVVMGCRNSISSLSIGG_M_ |
| EGCP, ECFP, EYFP, Cerulean (N-terminal) | GGGPGGGHQTSLYKKAE |
Concluding Remarks
In summary, we have generated a collection of 288 Saccharomyces cerevisiae Gateway® vectors enabling the rapid and efficient generation of a variety of expression constructs. All of these vectors are available to the research community via Addgene (http://www.addgene.org/yeast_gateway), individually or as an entire “kit”, and will complement the collection of existing yeast Gateway® vectors (Geiser, [2005]; Van Mullem, et al., [2003]). Furthure versions of the Gateway®-based vectors we generate will be made available through Addgene.
Acknowledgments
We are grateful to Tom DiCesare for expert graphical assistance and to Randal Halfmann for comments on the manuscript. ADG is a Lilly Fellow of the Life Sciences Research Foundation. SA is supported by a research fellowship of the Deutsche Forschungsgemeinschaft (DFG). Additional funding came from a Udall Center Grant (#NS38372).
References
- Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–22. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- Cooper AA, Gitler AD, Cashikar A, Haynes CM, Hill KJ, Bhullar B, Liu K, Xu K, Strathearn KE, Liu F, Cao S, Caldwell KA, Caldwell GA, Marsischky G, Kolodner RD, Labaer J, Rochet JC, Bonini NM, Lindquist S. Alpha-synuclein blocks ER-Golgi traffic and Rab1 rescues neuron loss in Parkinson's models. Science. 2006;313:324–8. doi: 10.1126/science.1129462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiser JR. Recombinational cloning vectors for regulated expression in Saccharomyces cerevisiae. Biotechniques. 2005;38:378–382. doi: 10.2144/05383BM06. [DOI] [PubMed] [Google Scholar]
- Hartley JL, Temple GF, Brasch MA. DNA cloning using in vitro site-specific recombination. Genome Res. 2000;10:1788–95. doi: 10.1101/gr.143000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landy A. Dynamic, structural, and regulatory aspects of lambda site-specific recombination. Annu Rev Biochem. 1989;58:913–49. doi: 10.1146/annurev.bi.58.070189.004405. [DOI] [PubMed] [Google Scholar]
- Outeiro TF, Lindquist S. Yeast cells provide insight into alpha-synuclein biology and pathobiology. Science. 2003;302:1772–5. doi: 10.1126/science.1090439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo MA, Piston DW. High-contrast imaging of fluorescent protein FRET by fluorescence polarization microscopy. Biophys J. 2005;88:L14–6. doi: 10.1529/biophysj.104.055442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopko R, Huang D, Preston N, Chua G, Papp B, Kafadar K, Snyder M, Oliver SG, Cyert M, Hughes TR, Boone C, Andrews B. Mapping pathways and phenotypes by systematic gene overexpression. Mol Cell. 2006;21:319–30. doi: 10.1016/j.molcel.2005.12.011. [DOI] [PubMed] [Google Scholar]
- Van Mullem V, Wery M, De Bolle X, Vandenhaute J. Construction of a set of Saccharomyces cerevisiae vectors designed for recombinational cloning. Yeast. 2003;20:739–46. doi: 10.1002/yea.999. [DOI] [PubMed] [Google Scholar]
- Walhout AJ, Temple GF, Brasch MA, Hartley JL, Lorson MA, van den Heuvel S, Vidal M. GATEWAY recombinational cloning: application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol. 2000;328:575–92. doi: 10.1016/s0076-6879(00)28419-x. [DOI] [PubMed] [Google Scholar]