Plant Speciation (original) (raw)
. Author manuscript; available in PMC: 2008 Jul 3.
Published in final edited form as: Science. 2007 Aug 17;317(5840):910–914. doi: 10.1126/science.1137729
Abstract
Like the formation of animal species, plant speciation is characterized by the evolution of barriers to genetic exchange between previously interbreeding populations. Prezygotic barriers, which impede mating or fertilization between species, typically contribute more to total reproductive isolation in plants than do postzygotic barriers, in which hybrid offspring are selected against. Adaptive divergence in response to ecological factors such as pollinators and habitat commonly drives the evolution of prezygotic barriers, but the evolutionary forces responsible for the development of intrinsic postzygotic barriers are virtually unknown and frequently result in polymorphism of incompatibility factors within species. Polyploid speciation, in which the entire genome is duplicated, is particularly frequent in plants, perhaps because polyploid plants often exhibit ecological differentiation, local dispersal, high fecundity, perennial life history, and self-fertilization or asexual reproduction. Finally, species richness in plants is correlated with many biological and geohistorical factors, most of which increase ecological opportunities.
Plants provide extraordinary opportunities for studying speciation. Flowering plants are especially speciose, trailing only insects in named species diversity. Much of this diversification has occurred recently, creating spectacular examples of adaptive radiation and of speciation in action (table S1). Plants are mostly sessile but vary dramatically in mating system, ploidy level, mode of dispersal, and life history, aiding efforts to understand the contribution of various ecological and evolutionary factors to speciation.
What Is a Plant Species?
The definition of a species in plants has been a major impediment to botanical studies of speciation; botanists have often expressed doubt that plant species even exist, because of frequent reports of interspecific hybrids (1) and because phenotypic variation in some plant groups does not assort readily into discrete categories (2). These concerns were amplified by claims that gene flow within many plant species was so low that populations rather than species were the most inclusive reproductive units (2, 3).
Recent work allays these concerns. Analyses of morphometric data from more than 200 plant genera indicate that discrete clusters of morphologically similar individuals occur within most sexual plant lineages, that these clusters correspond closely to groups with significant post-pollination reproductive isolation, and that interspecific hybridization is not the primary cause of poorly defined species boundaries (4). Molecular population genetic studies imply that migration rates within plant species are higher than earlier direct estimates and do not differ, on average, from those of animals (5). Theoretical (6) and empirical work further indicates that even in species with low gene flow, populations may evolve in concert through the spread of advantageous alleles (7).
Although many plant species are held together by gene flow and kept apart from other species by reproductive barriers, there are exceptions. For example, some plants reproduce without sex. These asexual taxa are composed of clonal hybrid genotypes that fill the phenotypic space between their sexual parental species (table S1). Because sexual reproduction is infrequent in such species, it is difficult to discuss their evolution in terms of sexual isolation and speciation. In contrast, self-fertilizing (selfing) species often maintain genetic and phenotypic cohesion (4) because they have higher within-species gene flow than previously hypothesized (8), and their restricted outcrossing (exchange of pollen between individuals) impedes interspecific hybridization. Reproductive isolation between species may be incomplete, however, particularly in groups that have recently undergone multiple speciation events or those that have long generation times. This incomplete isolation may result in some gene flow between groups that are otherwise well-defined species (table S1).
Reproductive Isolation
Reproductive isolation is not the proximal cause of diversification; this is the province of diversifying selection and genetic drift. However, reproductive isolation can facilitate the accumulation of genetic differences between groups of populations, thereby sharpening boundaries between them and permitting adaptive traits to move closer to their fitness optima. This does not require absolute isolation. Rather, any reduction in the effective migration rate facilitates divergence, which reduces effective migration rates even further. The resulting feedback loop, given enough time, usually leads to complete genetic isolation.
Multiple reproductive barriers isolate most plant species. These include prepollination barriers that limit the transfer of pollen from individuals of one species to stigmas of other species. Several prepollination barriers—ecogeographic, mechanical, and temporal—are found in animal species, whereas pollinator isolation is exclusively associated with plant speciation. Other barriers, such as an advantage of conspecific pollen in fertilizing eggs compared with nonconspecific pollen (conspecific pollen precedence) and the failure of nonconspecific pollen to fertilize eggs (gametic incompatibilities), act after pollination but before fertilization, resulting in postpollination, prezygotic isolation. A final set of barriers, also found in animals, act after fertilization: hybrid inviability, sterility, and the failure or reduction in successful reproduction in subsequent generations (hybrid breakdown). These postzygotic barriers may be a by-product of changes in the internal genetic environment (intrinsic isolation) or in the external ecological environment (extrinsic isolation). Current challenges are to estimate the relative contribution of different reproductive barriers in limiting gene flow among contemporary populations and to determine the order and speed with which they arose.
All else being equal, early-acting reproductive barriers will contribute more to isolation than late-acting barriers (9). For example, the production of hybrid seeds in artificial crosses and reductions in the fertility of first-generation hybrids are commonly tested in the greenhouse. Cross-compatibility data (4) reveals that hybrid fertility reduction is the slightly stronger of the two barriers. However, because hybrid seed production acts before reductions in fertility, reduced hybrid seed production actually would be expected to contribute about 75% and hybrid sterility just 25% of the total isolation caused by these two barriers.
Unfortunately, only a few studies provide comprehensive estimates of isolation between pairs of sibling species (table S1). In these, the cumulative effects of many reproductive barriers lead to almost complete isolation. Early-acting reproductive barriers such as ecogeographic, pollinator, and mating system isolation are most important (table S1 and fig. S1), whereas late-acting postzygotic barriers contribute very little to isolation. Ecogeographic isolation has long been viewed as the most important reproductive barrier in plants (10), and its preeminence has been confirmed by numerous reciprocal transplant studies showing differences in habitat preferences among closely related species or subspecies (Table 1). Pollinator and mating system isolation are less frequent, with the former arising when the focal species is numerically dominant but does not fully use the array of available animal pollinators (11).
Table 1.
Case studies of plant speciation.
| Topic | Taxa studied | Conclusions | Ref. |
|---|---|---|---|
| Reproductive isolation | Gilia capitata ssp. capitata and G. c. ssp. chamissonis | Local adaptation of interfertile subspecies to different habitats restricts successful migration and gene flow. | (48) |
| Arctic Draba | Three self-fertilizing morphological species each appears to comprise thousands of cryptic biological species. | (49) | |
| Genetics of isolation | Mimulus lewisii and M. cardinalis | Allele increasing petal carotenoid concentration reduced bee visitation by 80%; allele increasing nectar production doubled hummingbird visitation. | (50) |
| Lycopersicon hirsutum and L. pimpinellifolium | Tomato lines with resistance gene (_Cf_-2) from L. pimpinellifolium exhibit autonecrosis of mature leaves, but no autonecrosis observed when complementary gene (RC3) from S. pimpinellifolium also introduced. | (51) | |
| Hybrid and polyploid speciation | Helianthus anomalus, H. deserticola, and H. paradoxus | Three diploid species arisen via hybridization from same two parental species. Karyotypically divergent hybrids colonized extreme habitats through selection on transgressive traits (Fig. 2). | (52) |
| Brassica napus | Chromosomal rearrangement after polyploidization responsible for flowering time divergence among synthetic polyploid lineages. | (53) | |
| Factors affecting species richness | Angiosperms | Acquisition of nectar spurs in wide variety of plants correlated with increased species diversity. | (44) |
| Andean Lupinus | Most rapid species radiation in plants driven by ecological opportunities afforded by uplift of Andes. | (54) |
It is difficult to determine the order of reproductive barrier evolution. Indirect evidence from analyses of patterns of reproductive isolation suggests that prepollination barriers often arise first. For example, 19% of 1234 interspecific cross combinations (most from rapidly radiating lineages isolated by ecological barriers) failed to show evidence of cross-incompatibility or intrinsic postzygotic isolation (4). Intrinsic postzyotic barriers may arise first in polyploid species that are intersterile with their diploid progenitors but that fail to exhibit ecological differences (table S1). Likewise, intrinsic post-zygotic barriers may sometimes arise before ecological barriers (other than mating system isolation) in selfing species (Table 1).
We know surprisingly little about the speed of plant speciation, although studies of contemporary evolution imply that reproductive barriers can arise rapidly. For example, grass populations exposed to different fertilizer treatments or to mine tailings exhibit both temporal (flowering time) and habitat isolation (seeds transplanted between sites have reduced survival) (table S1). Interestingly, flowering time divergence is greatest at the boundary between habitats in both experiments, a pattern suggestive of reinforcement, where selection against unfit hybrids has enhanced prezygotic isolation. These studies of speciation in action illustrate the plausibility of reinforcement and sympatric speciation, both of which are increasingly well supported by theory (12) and empirical work (table S1).
Although individual reproductive barriers can arise rapidly, most plant species remain separated by numerous barriers, which implies that complete speciation typically requires many thousands of generations. The main exceptions to this are hybrid and polyploid speciation. Fully isolated polyploid species may arise in one or two generations, and diploid or homoploid hybrid species may achieve isolation in as few as 60 generations (13).
Genetics of Isolation
Genetic analyses provide information on the numbers and kinds of genetic changes underlying reproductive barriers, as well as on the evolutionary forces responsible for their origin. Studies of pollinator isolation have shown, for example, that major quantitative trait loci (QTLs) sometimes underlie shifts in the animals that pollinate plants (pollination syndrome) (table S1) and changes in pollinator preferences in the field (Table 1). In contrast, two studies of mating system isolation detected many smaller genetic changes (table S1). These different architectures might be explained by the fact that many intermediate pollinator syndromes are maladaptive (e.g., red flowers lacking a nectar reward are unattractive to both birds and bees) and favor larger genetic steps, whereas small increases in selfing rates may be favored if inbreeding depression costs are not prohibitive (14). Analyses of the direction of QTL effects imply that most traits contributing to prepollination isolation diverged through directional selection; as predicted for adaptive phenotypes, QTL effects for these traits are mostly in the same direction as the parental differences (15). QTL effects are predicted to vary in direction (i.e., have opposing effects) for traits not under consistent directional selection (16).
Recent genetic analyses of prezygotic and extrinsic postzygotic barriers associated with discrete habitat differences are particularly informative because many of the studies have been performed in the field. This makes it possible to estimate the strength of selection on traits and QTLs that contribute to habitat isolation. Studies have shown, for example, that the strength of selection on QTLs that contribute to habitat isolation is sufficient to permit speciation in the presence of gene flow, that hybrid inviability may arise as a by-product of habitat selection, and that interspecific hybridization can facilitate the exchange of adaptive alleles between species (table S1).
Genetic studies of postpollination, prezygotic isolation have focused on the relationship between self-incompatibility (SI) mechanisms, which enforce outcrossing in many hermaphroditic plants, and interspecific incompatibility. This interest stems from early observations that self-compatible species are more compatible in interspecific crosses than are SI species, implying that SI may contribute to both intra- and interspecific incompatibilities. This was confirmed by detection of a QTL for interspecific incompatibility that colocalizes with the SI locus, as well as observations that crosses between self-compatible species fail after transformation with a SI gene from a self-incompatible species (table S1). Diversification of genes that contribute to SI appears to result from frequency-dependent selection (17). Interestingly, other plant reproductive proteins appear to be under positive selection as well, including candidates for species-specific recognition between pollen and stigma (table S1).
Intrinsic postzygotic barriers offer special challenges to genetic analyses because the phenotypes of interest (hybrid sterility and inviability) impede genetic study and lack obvious candidate genes for functional analyses (see below, however). Intrinsic postzygotic isolation may be caused by chromosomal rearrangements and/or changes in genes (Fig. 1). Population genetic theory minimizes the importance of strongly underdominant chromosomal rearrangements (those that reduce the fitness of heterozygotes) because their negative effects on fitness should prevent them from becoming established, except in small, inbred populations. Weakly underdominant rearrangements are more easily established but contribute little to reproductive isolation. In contrast, the Bateson-Dobzhansky-Muller (BDM) model accounts for the accumulation of interspecific incompatibilities in genes without loss of fitness (Fig. 1). Briefly, as a lineage diverges, geographically isolated or neighboring allopatric populations may accumulate independent mutations. These mutations are compatible with the ancestral genotype but are incompatible when combined. BDM incompatibilities generally involve two or more loci, although it is theoretically possible for BDM incompatibilities to result from the allopatric accumulation of independent mutations at a single locus (Fig. 1).
Fig. 1.
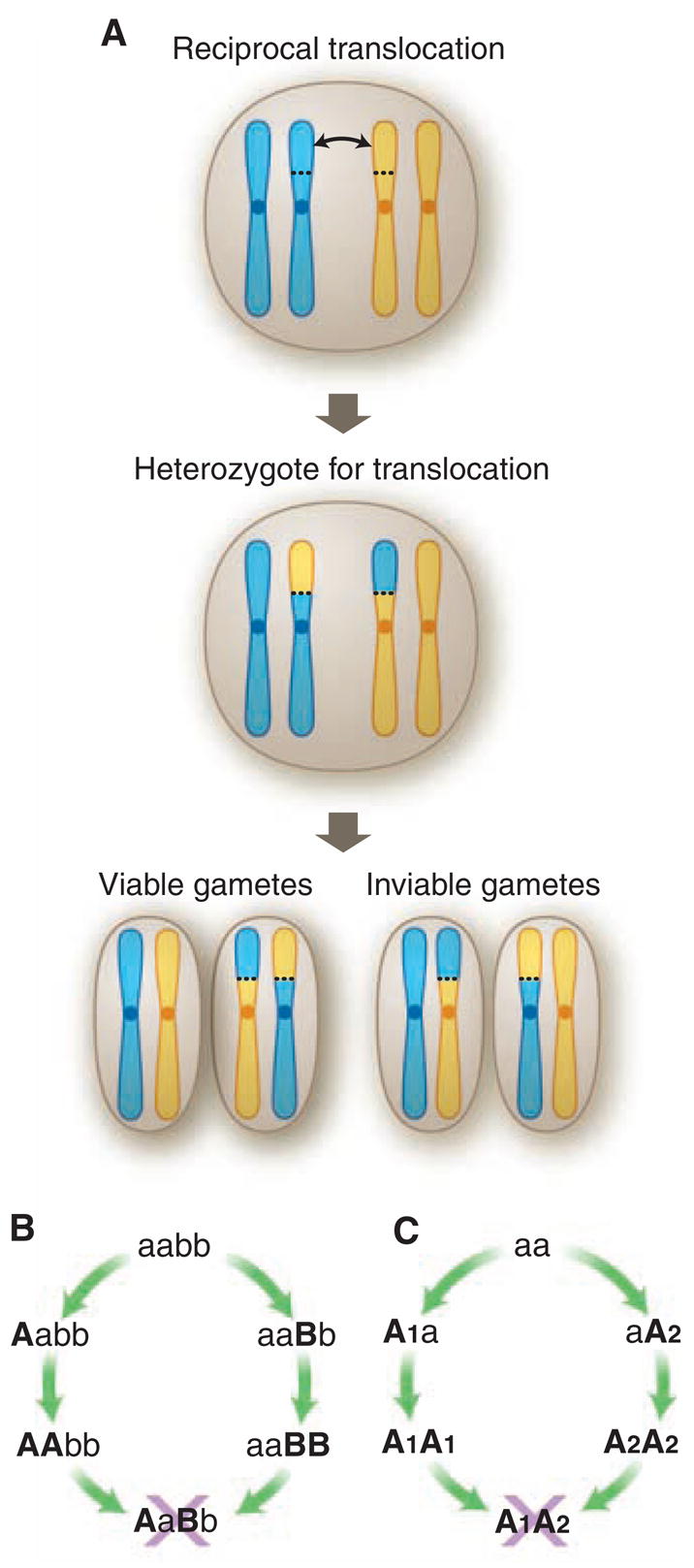
Genetics of hybrid incompatibilities. (A) Example of a typical chromosomal rearrangement in plants, showing loss of fertility in heterozygotes because 50% of gametes are unbalanced genetically and inviable. (B) Classic two-locus BDM incompatibility in which new mutations are established at alternate loci and without loss of fitness in geographically isolated populations, but which are incompatible in hybrids. (C) Single-locus BDM incompatibility in which new mutations are established at the same locus and without loss of fitness in geographically isolated populations, but which are incompatible in hybrids.
Despite theoretical doubts about their importance in speciation, chromosomal rearrangements often contribute to the sterility of hybrid plants (18, 19). Unlike Drosophila (in which hybrid sterility is mostly due to BDM incompatibilities), sterile plant hybrids often recover fertility after chromosomal doubling (18). This is expected if chromosomal rearrangements are the cause of sterility, because chromosomal doubling furnishes an exact homolog for each chromosome, whereas doubling should not affect BDM incompatibilities. Microchromosomal rearrangements such as the gain and loss of duplicate genes are more frequent than previously suspected and may lead to hybrid incompatibilities with no loss of fitness in the diverging lineages (20). Finally, hybrid sterility in plants frequently maps to chromosomal rearrangements (21), although whether the cause is chromosomal underdominance or BDM loci that have accumulated within the rearrangements is often unclear. The reduced recombination associated with chromosomal rearrangements can facilitate the accumulation of hybrid incompatibilities in these regions (19, 22) or expedite the establishment of rearrangements in the first place (23).
BDM hybrid sterility in plants may be under simple or complex genetic control. However, fewer loci contribute to hybrid sterility in plants than in Drosophila, and there appears to be no difference in the numbers of pollen (male) versus seed (female) incompatibilities, perhaps because plants largely lack differentiated sex chromosomes (24). In addition, cytoplasmic male sterility examples characterized at the molecular level (26). CMS phenotypes are rescued by nuclear-encoded, mitochondrial-targeted genes that restore fertility (Rf genes). With the exception of Rf2 from maize, all cloned Rf genes are members of the pentatricopeptide repeat gene family (PPR), an unusually large gene family in plants (441 genes in Arabidopsis) that controls organelle gene expression. Although the molecular evolution of Rf genes is unknown, they are likely to be involved in coevolutionary chases with CMS as a result of genetic conflict between cytoplasmic and nuclear genes. These evolutionary dynamics may reduce the long-term effectiveness of CMS as a species barrier, because the same evolutionary forces that cause the spread of CMS within species could facilitate the introgression of CMS and restorers across species boundaries.
BDM factors also can cause hybrid weakness or inviability. Hybrid weakness is often manifested as necrosis in developing seedlings or adult plant tissue, similar to the phenotype of pathogen attacks (27). These observations imply that hybrid weakness may result from changes in pathogen resistance genes (Table 1), which diverge in response to selection pressure exerted by pathogens. More studies are needed to determine the frequency of this mechanism for hybrid (CMS), which results from an incompatibility between the plant’s nuclear genome and its cytoplasm, is frequently reported in intra- and interspecific plant hybrids, but not in animal hybrids (25). CMS is under frequency-dependent selection in hermaphrodite-biased populations, which predominate in plants, but under strong negative selection if there are separate male and female sex morphs. CMS is caused by aberrant, frequently chimeric, mitochondrial genes in all weakness in interspecific crosses and to elucidate other mechanisms of hybrid inviability.
A final emerging difference between plants and animals (or at least Drosophila) is that most BDM incompatibilities characterized in plants are polymorphic within species (27–29) (Table 1). This is consistent with an origin of BDM incompatibilities through frequency-dependent selection, local adaptation, or drift. However, it also implies that BDM incompatibilities are rarely the cause of speciation in plants, because they correlate poorly with species boundaries and typically make small contributions to total isolation.
Hybrid and Polyploid Speciation
Although most studies of speciation focus on how lineages diverge, speciation is not always about divergence. Indeed, a substantial fraction of speciation events in plants involves the reunion of divergent genes and genomes through sexual hybridization. There are two kinds of hybrid speciation: homoploid and polyploid. Homoploid hybrid speciation refers to the origin of a new hybrid lineage without a change in chromosome number, whereas polyploid hybrid speciation involves the full duplication of a hybrid genome (allopolyploidy). Polyploids not of hybrid origin are autopolyploids.
Homoploid hybrid speciation is rarer than polyploid speciation for two reasons. First, homoploid hybrid species have strongly reduced fitness in early generation hybrids as selection eliminates incompatibilities. In contrast, polyploid species need not have low fertility during intermediate stages. Second, genome duplication protects the genetic integrity of newly derived polyploids, but no such barrier prevents homoploid hybrids from back-crossing with their parental species. In addition to these biological difficulties, homoploid hybrid species are technically challenging to detect because they often lack diagnostic features, such as a change in chromosome number. So far, there are 15 to 20 good examples in the literature (30), but more are likely to be discovered with the widespread application of genomic tools to natural plant populations (31).
Homoploid hybrid species may become reproductively isolated by rapid karyotypic evolution, ecological divergence, and spatial isolation of the new hybrid lineage. Simulation studies indicate that although strong ecological selection promotes hybrid speciation, without chromosomal or spatial isolation the hybrid population forms a steep step in a cline between the parental species (32). Karyotypic divergence and spatial isolation both reduce the probability that hybrid species will be generated but will enhance the evolutionary independence of hybrid lineages once they arise.
As hypothesized, all plant homoploid hybrid species are ecologically diverged and exhibit some degree of ecogeographic isolation, and roughly half have differing karyotypes (30). Most commonly, the hybrid species are adjacent to one or both parental species, although there are examples of long-distance dispersal as well (table S1). Some hybrids occupy habitats that are intermediate between the parental species, whereas others have colonized an extreme habit by combining QTLs with effects in the same direction from both parental species (Table 1 and Fig. 2). Homoploid hybrid species are easily recreated in the greenhouse, perhaps explaining why many are multiply derived in the wild (table S1).
Fig. 2.
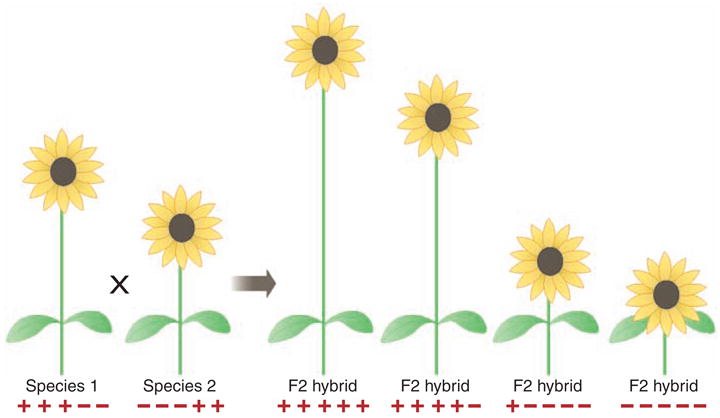
Genetic basis of transgressive segregation showing how segregating hybrids can combine plus and minus alleles from parental species, thereby generating extreme phenotypes or adaptations to extreme habitats.
In contrast to homoploid hybrid species, polyploid species are easily diagnosed because of chromosome number changes associated with genome doubling. However, the frequency of polyploid speciation remains controversial. When there are multiple polyploid species within a genus, it is difficult to determine whether there was a single transition to the new ploidal level followed by divergent speciation or whether each polyploid species arose independently. Recent model-based estimates (33) assume a single transition to a new ploidal level within a genus and provide a lower bound of the polyploid speciation rate: 2 to 4% in flowering plants and 7% in ferns. This contrasts with Stebbins’ (34) estimate of 30 to 35% for flowering plants, which assumes that all polyploid species within a genus are independently derived. Because many polyploid species are themselves multiply derived (Table 1), Stebbins’ estimate is probably closer to the true polyploid speciation rate. However, neither of these estimates (33, 34) includes intraspecific ploidal variation. At least 8 to 9% of named plant species vary in ploidal level, and this might be the tip of the iceberg (35). If each ploidal level (cytotype) is viewed as a cryptic biological species, then the contribution of polyploidy to biological species diversity is even higher than previously surmised. In addition, there has been confusion between estimates of the proportion of species that are polyploid and the rate of polyploid speciation. Analyses of the age distribution of duplicate genes in diverse flowering plants (36) indicate that essentially all may be paleopolyploids, but this should not be equated with the polyploid speciation rate.
Polyploids can arise by somatic doubling, by the fusion of unreduced gametes, and by means of a triploid bridge (Fig. 3). Unreduced gametes are common in plants and likely represent the most frequent route to polyploidy (37). However, most newly arisen polyploids fail to become established because of meiotic abnormalities and/or the paucity of appropriate mates (38). The establishment of polyploids is favored by differential niche preference, low dispersal, a selfing or asexual mating system, high fecundity, and a perennial life history (39, 40). Niche separation, low dispersal, and selfing increase the probability of successful matings during early stages of polyploid species establishment; otherwise most matings will be with the diploid progenitors (40, 41). Stochastic events due to a small number of polyploid colonizers decrease the chance of establishment, but this barrier is minimized by high fecundity and a perennial life history, which allows plants to reproduce at multiple times over their life cycle (39).
Fig. 3.
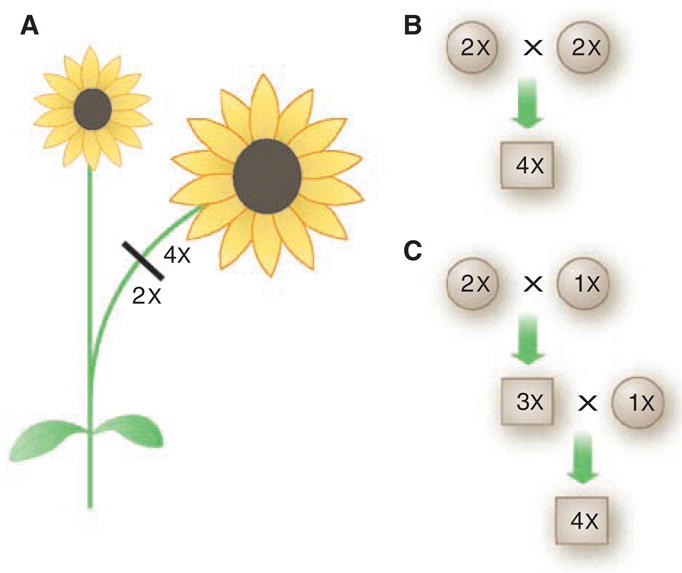
Mechanisms by which polyploids can arise. (A) Somatic doubling, in which chromosome number is doubled in vegetative tissue that gives rise to reproductive organs. (B) Fusion of unreduced gametes that are produced when cell walls fail to form in the final stage of meiosis. (C) A triploid bridge, in which unreduced and reduced gametes form triploids. If the triploids also produce unreduced gametes, the triploid gametes may fuse with reduced gametes from diploid individuals to generate stable tetraploids.
Because intraspecific matings are far more common than interspecific matings in natural populations of plants, autopolyploids must arise at a much higher rate than allopolyploids (37). However, named species are more likely to be allopolyploids (42), which implies that allopolyploids are more easily established in nature, easier to find, and/or more readily recognized by taxonomists. Establishment of allopolyploids is favored because of greater niche separation from their diploid progenitors (43), and taxonomists appear reluctant to name phenotypically cryptic autopolyploid species.
Recent attention has been given to changes in gene expression, genome content, and DNA methylation that accompany hybrid and polyploid speciation, but these genomic alterations only rarely have been linked to changes in ecology or mating system that affect polyploid establishment (Table 1). Indeed, many described genomic changes appear to be maladaptive by-products of reuniting divergent genomes. However, maladaptive changes in gene expression in first-generation interspecific hybrids may be reduced by genome doubling, and elimination of DNA sequences may help restore fertility in polyploids (table S1).
Factors Affecting Speciation or Extinction Rates
Recent advances in comparative methods have made it possible to identify biological or geohistorical factors affecting speciation or extinction rates. The most rigorous approach compares species richness of multiple sister clades that differ in the presence or absence of a given trait (44). A significant association may result from either increased speciation or reduced extinction. Traits associated with increased species richness in plants include resin canals, nectar spurs, biotic pollination, herbaceous growth form, abiotic dispersal, increased neutral evolution, bilateral symmetry of flowers, twig epiphytism, and polyploidy (45) (table S1). Many of these examples involve biotic interactions, leading to suggestions that coevolution may drive speciation in many plant groups or that niche space may be less constrained in biotic than abiotic interactions (46). The most rapid diversification rates in plants are associated with ecological opportunities created by major geological changes such as the uplift of the Andes or island formation (Table 1), which implies that mechanisms that expand niche diversity often increase species diversification (or reduce extinction). Unfortunately, the factors listed above do not fully account for the most striking trend in species richness—the negative correlation with latitude—which appears to have a pluralistic explanation (47) (table S1_)_.
Concluding Remarks
The field of plant speciation is in for an exciting decade. The wide availability of genomic tools and resources for crop and noncrop species, from green algae to mosses to angiosperms, will accelerate our understanding of the genetic and ecological bases of speciation. These resources not only will facilitate the cloning and functional characterization of genes underlying reproductive barriers but also will make it possible to quantify the effects of individual mutations or alleles on reproductive isolation or fitness in natural populations (Table 1). Likewise, the widespread application of molecular phylogenetic approaches simplifies comparative study.
We expect to see rapid progress in each of the areas highlighted in our review. For example, studies of the geography of selective sweeps should provide an objective method for evaluating the importance of different kinds of reproductive barriers and geohistorical processes in speciation. Our understanding of reproductive isolation will also be enhanced by additional field-based estimates of isolation across all life history stages. With the cloning of BDM incompatibilities in plants, the next step is molecular evolutionary studies of these genes to identify the forces that drive their evolution. Finally, comparative analyses of the effects of different kinds of reproductive barriers on species richness should allow us to determine whether reproductive barriers themselves increase speciation rates.
Footnotes
References and Notes
- 1.Arnold ML. Natural Hybridization and Evolution. Oxford Univ. Press; Oxford: 1997. [Google Scholar]
- 2.Mishler BD, Donoghue MJ. Syst Zool. 1982;31:491. [Google Scholar]
- 3.Ehrlich PR, Raven PH. Science. 1969;165:1228. doi: 10.1126/science.165.3899.1228. [DOI] [PubMed] [Google Scholar]
- 4.Rieseberg LH, Wood TE, Baack EJ. Nature. 2006;440:524. doi: 10.1038/nature04402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morjan CL, Rieseberg LH. Mol Ecol. 2004;13:1341. doi: 10.1111/j.1365-294X.2004.02164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whitlock MC. Genetics. 2003;164:767. doi: 10.1093/genetics/164.2.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McDaniel SF, Shaw AJ. Mol Ecol. 2005;14:1121. doi: 10.1111/j.1365-294X.2005.02484.x. [DOI] [PubMed] [Google Scholar]
- 8.Bakker EG, et al. Mol Ecol. 2006;15:1405. doi: 10.1111/j.1365-294X.2006.02884.x. [DOI] [PubMed] [Google Scholar]
- 9.Ramsey J, Bradshaw HD, Schemske DW. Evolution Int J Org Evolution. 2003;57:1520. doi: 10.1111/j.0014-3820.2003.tb00360.x. [DOI] [PubMed] [Google Scholar]
- 10.Stebbins GL. Variation and Evolution in Plants. Columbia Univ. Press; New York: 1950. [Google Scholar]
- 11.Sargent RD, Otto SP. Am Nat. 2006;167:67. doi: 10.1086/498433. [DOI] [PubMed] [Google Scholar]
- 12.Gavrilets S, Vose A. Mol Ecol. 2007;16:2910. doi: 10.1111/j.1365-294X.2007.03304.x. [DOI] [PubMed] [Google Scholar]
- 13.Ungerer MC, Baird SJE, Pan J, Rieseberg LH. Proc Natl Acad Sci USA. 1998;95:11757. doi: 10.1073/pnas.95.20.11757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fishman L, Kelly AJ, Willis JH. Evolution Int J Org Evolution. 2002;56:2138. doi: 10.1111/j.0014-3820.2002.tb00139.x. [DOI] [PubMed] [Google Scholar]
- 15.Rieseberg LH, Church S, Morjan CL. New Phytol. 2004;161:59. doi: 10.1046/j.1469-8137.2003.00933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orr HA. Genetics. 1998;149:2099. doi: 10.1093/genetics/149.4.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Igic B, Kohn JR. Proc Natl Acad Sci USA. 2001;98:13167. doi: 10.1073/pnas.231386798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stebbins GL. Adv Genet. 1958;9:147. doi: 10.1016/s0065-2660(08)60162-5. [DOI] [PubMed] [Google Scholar]
- 19.Rieseberg LH. Trends Ecol Evol. 2001;16:351. doi: 10.1016/s0169-5347(01)02187-5. [DOI] [PubMed] [Google Scholar]
- 20.Werth CR, Windham MD. Am Nat. 1991;137:515. [Google Scholar]
- 21.Lai Z, et al. Genetics. 2005;171:291. doi: 10.1534/genetics.105.042242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Noor MAF, Grams KL, Bertucci LA, Reiland J. Proc Natl Acad Sci USA. 2001;98:12084. doi: 10.1073/pnas.221274498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kirkpatrick M, Barton N. Genetics. 2006;173:419. doi: 10.1534/genetics.105.047985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moyle LC, Graham EB. Genetics. 2005;169:355. doi: 10.1534/genetics.104.029546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fishman L, Willis JH. Evolution Int J Org Evolution. 2006;60:1372. doi: 10.1554/05-708.1. [DOI] [PubMed] [Google Scholar]
- 26.Gillman JD, Bentolila S, Hanson MR. Plant J. 2007;49:217. doi: 10.1111/j.1365-313X.2006.02953.x. [DOI] [PubMed] [Google Scholar]
- 27.Bomblies K, Weigel D. Nat Rev Genet. 2007;8:382. doi: 10.1038/nrg2082. [DOI] [PubMed] [Google Scholar]
- 28.Christie P, Macnair MR. Evolution Int J Org Evolution. 1987;41:571. doi: 10.1111/j.1558-5646.1987.tb05827.x. [DOI] [PubMed] [Google Scholar]
- 29.Sweigart AL, Mason AR, Willis JH. Evolution Int J Org Evolution. 2007;61:141. doi: 10.1111/j.1558-5646.2007.00011.x. [DOI] [PubMed] [Google Scholar]
- 30.Gross BL, Rieseberg LH. J Hered. 2005;96:241. doi: 10.1093/jhered/esi026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hegarty MJ, Hiscock SJ. New Phytol. 2005;165:411. doi: 10.1111/j.1469-8137.2004.01253.x. [DOI] [PubMed] [Google Scholar]
- 32.Buerkle CA, Morris RJ, Asmussen MA, Rieseberg LH. Heredity. 2000;84:441. doi: 10.1046/j.1365-2540.2000.00680.x. [DOI] [PubMed] [Google Scholar]
- 33.Otto SP, Whitton J. Annu Rev Genet. 2000;34:401. doi: 10.1146/annurev.genet.34.1.401. [DOI] [PubMed] [Google Scholar]
- 34.Stebbins GL. Chromosomal Evolution in Higher Plants. Edward Arnold; London: 1971. [Google Scholar]
- 35.Soltis DE, et al. Taxon. 2007;56:13. [PMC free article] [PubMed] [Google Scholar]
- 36.Cui L, et al. Genome Res. 2006;16:738. doi: 10.1101/gr.4825606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramsey J, Schemske DW. Annu Rev Ecol Syst. 1998;29:467. [Google Scholar]
- 38.Levin DA. Taxon. 1975;24:35. [Google Scholar]
- 39.Rodriguez DJ. Am Nat. 1996;147:33. [Google Scholar]
- 40.Baack EJ. Heredity. 2005;94:538. doi: 10.1038/sj.hdy.6800656. [DOI] [PubMed] [Google Scholar]
- 41.Rausch JH, Morgan MT. Evolution Int J Org Evolution. 2005;59:1867. [PubMed] [Google Scholar]
- 42.Mallet J. Nature. 2007;446:279. doi: 10.1038/nature05706. [DOI] [PubMed] [Google Scholar]
- 43.Levin DA. Am Nat. 1983;122:1. [Google Scholar]
- 44.Hodges SA, Arnold ML. Proc R Soc London B Biol Sci. 1995;262:343. [Google Scholar]
- 45.Coyne JA, Orr HA. Speciation. Sinauer Assoc; Sunderland, MA: 2004. [Google Scholar]
- 46.Bolmgren K, Eriksson O, Linder HP. Evolution Int J Org Evolution. 2003;57:2001. doi: 10.1111/j.0014-3820.2003.tb00380.x. [DOI] [PubMed] [Google Scholar]
- 47.Mittelbach GG, et al. Ecol Lett. 2007;10:315. doi: 10.1111/j.1461-0248.2007.01020.x. [DOI] [PubMed] [Google Scholar]
- 48.Nagy ES, Rice KJ. Evolution Int J Org Evolution. 1997;51:1079. doi: 10.1111/j.1558-5646.1997.tb03955.x. [DOI] [PubMed] [Google Scholar]
- 49.Grundt HH, et al. Proc Natl Acad Sci USA. 2006;103:972. doi: 10.1073/pnas.0510270103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schemske DW, Bradshaw HD. Proc Natl Acad Sci USA. 1999;96:11910. doi: 10.1073/pnas.96.21.11910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kruger J, et al. Science. 2002;296:744. doi: 10.1126/science.1069288. [DOI] [PubMed] [Google Scholar]
- 52.Rieseberg LH, et al. Science. 2003;301:1211. doi: 10.1126/science.1086949. [DOI] [PubMed] [Google Scholar]
- 53.Wang J, Tian L, Lee HS, Chen ZJ. Genetics. 2006;173:965. doi: 10.1534/genetics.106.056580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hughes C, Eastwood R. Proc Natl Acad Sci USA. 2006;103:10334. doi: 10.1073/pnas.0601928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.We thank the great botanical naturalists of the 20th century who provided the foundation for current studies of plant speciation. These include the founding fathers of ecological genetics (J. Clausen, D. Keck, and W. Hiesey), the grand synthesizers (G. L. Stebbins and Verne Grant), and the hybridization enthusiast, Edgar Anderson. We also thank members of the Rieseberg and Willis laboratories and three referees for useful comments on an earlier version of this paper. The authors’ research on speciation has been supported by NSF, NIH, USDA, and the National Sciences and Engineering Research Council of Canada.