FIP200, a key signaling node to coordinately regulate various cellular processes (original) (raw)
. Author manuscript; available in PMC: 2009 May 1.
Abstract
A central question in cell biology is how various cellular processes are coordinately regulated in normal cell and how dysregulation of the normal signaling pathways leads to diseases such as cancer. Recent studies have identified FIP200 as a crucial signaling component to coordinately regulate different cellular events by its interaction with multiple signaling pathways. This review will focus on the cellular functions of FIP200 and its interacting proteins, as well as the emerging roles of FIP200 in embryogenesis and cancer development. Further understanding of FIP200 function might provide novel therapeutic targets for human diseases such as cancer.
1. Introduction
During metazoan development, the increase of cell size and number and thus the growth of an organ or organism are coordinately regulated by several important biological processes, including cell growth, cell proliferation and cell survival/ death. Cell size is regulated by cell growth, whereas cell number is determined by the balance between cell proliferation and cell survival/death [1]. Cell growth is a prerequisite for cell proliferation during normal organ growth, and sustained cell proliferation must be coupled to appropriate cell growth. With appropriate cell growth, a net increase in cell number in a growing organ depends on the rate at which they are generated by cell proliferation and eliminated by cell death. Furthermore, cell spreading and migration also play important roles in multiple processes during metazoan development, including embryogenesis, wound healing, angiogenesis and inflammatory immune response [2]. Perturbation of these processes could lead to malformation or embryonic death, as well as a variety of diseases in adult life, such as cancer. In contrast to our understanding of the molecular mechanisms that regulate these cellular processes, relatively less is known about the mechanisms how these cellular processes are coordinately regulated during the development of an organism.
FIP200 (FAK-family Interacting Protein of 200 kDa) has emerged from recent studies as a critical signaling node to coordinately regulate cell growth, cell proliferation, cell survival and cell spreading/migration. There is also increasing evidence that dysregulation of FIP200 is linked to embryonic death and cancer development in adult life. Here, we focus on our current understanding of FIP200 signaling pathway, with an emphasis on the diverse cellular functions and interacting proteins regulated by FIP200. We will then explore the emerging roles of FIP200 in embryogenesis and cancer development. Finally, we will identify and discuss the important questions remaining to be addressed.
2. FIP200 gene expression, protein structure and subcellular localization
FIP200 was initially identified as a Pyk2 interacting protein through yeast two-hybrid screen in our laboratory in 2000 [3]. FIP200 was shown to inhibit Pyk2 kinase activity and negatively regulate Pyk2 functions [3]. In 2002, FIP200 has also been independently identified as a potential regulator of the RB1 gene (designated by this group as RB1CC1 for RB1-inducible coiled-coil 1) by Dr. Chano and his colleagues[4]. Prior to these studies, the full length cDNA for FIP200 was already isolated (named as KIAA0203), but without any further characterization or any functions ascribed to it, in projects of sequencing human cDNA clones which correspond to long transcripts in the mid-90's [5]. Fragments of FIP200 have also been identified as a stathmin interacting protein (named as CC1) [6] and as a binding partner of the listeria monocytogenes surface protein ActA (designated as LaXp180) [7] in a number of previous studies. For clarity and simplicity, the gene name “_FIP200_” will be used throughout the review.
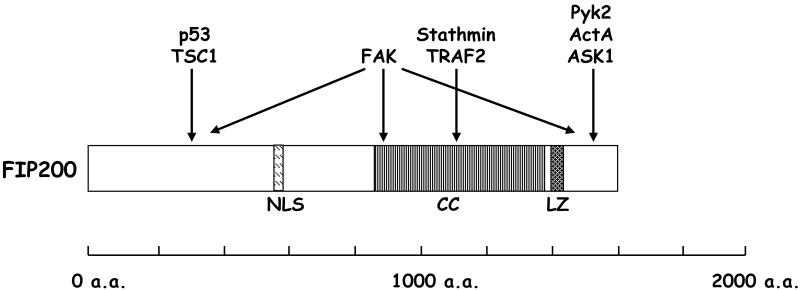
Human FIP200 gene localizes in chromosome 8q11, whereas mouse FIP200 gene localizes in chromosome 1A2–4, which is syntenic to human chromosome 8q11. Both human and mouse FIP200 genes contain 24 exons and share similar genomic structures. FIP200 encodes a 200kDa protein (1591 aa) characterized with a large coiled-coil region (residues 860-1391) containing a leucine zipper motif (residues 1371-1391) (Figure 1). FIP200 also contains a putative nuclear localization signal (NLS), suggesting that it might localize in nucleus (Figure 1). Human and mouse FIP200 proteins share around 90% identity and similar domain structure [8]. FIP200 is an evolutionarily conserved protein present in human, mouse, rat, Xenopus laevis, Drosophila melanogaster and Caenorhabditis elegans. However, the function of FIP200 orthologues in other species remains unknown currently. FIP200 is widely expressed in various human and mouse tissues. In particular, FIP200 is abundantly expressed in heart, testis and musculoskeletal systems [8, 9]. Furthermore, FIP200 is abundantly expressed throughout mouse embryonic development [10], suggesting that FIP200 might play an important role in embryogenesis.
Figure 1. Structural domains and binding proteins of FIP200.
FIP200 consists of a putative nuclear localization signal (NLS) at N-terminus, a large coiled-coil (CC) domain and a leucine zipper (LZ) motif located at C-terminus. The proteins known to interact with specific regions of FIP200 are also shown.
Most studies so far showed that FIP200 is a cytoplasmic protein [3, 6, 7, 11, 12]. However, FIP200 has also been shown to localize in focal adhesion [13] and nucleus [8, 14]. The nuclear localization of FIP200 is consistent with the putative NLS found in FIP200 sequence. However, whether this NLS indeed regulates FIP200 shuttling between cytoplasm and nucleus remains elusive. It is possible that FIP200 might show differential localization in different cell types or its subcellular localization is dynamically regulated by certain stimulus.
3. FIP200 function in various cellular processes
So far, totally 8 proteins have been identified as FIP200 interacting proteins, including stathmin [6], Pyk2 [3], FAK [13], ActA [7], p53 [14], TSC1 [11, 15], ASK1 and TRAF2 [12] (Table 1). It should be noted that not all FIP200 interacting proteins have been confirmed at the endogenous level and therefore the significance of some interactions should be more rigorously studied. Nevertheless, this short list of interacting proteins provides us valuable information about FIP200 function. It seems that FIP200 interacting partners mediate very diverse, largely non-overlapping cellular functions, including regulation of microtubule dynamics, cell adhesion and migration signaling, stress response, cell proliferation and cell growth, which is consistent with the idea that FIP200 might function to integrate different signaling pathways. Six out of 8 FIP200 interacting proteins mainly localize in the cytoplasm, consistent with the data from immunofluorescence and biochemical fractionation that FIP200 mainly localizes in cytoplasm. However, the fact that FIP200 can also interact with FAK in focal adhesions and p53 in nucleus suggests that a fraction of FIP200 can also localize in these subcellular compartments. Finally, it seems that FIP200 plays very diverse biochemical functions with different binding partners (Table 1). FIP200 functions to directly inhibit FAK and Pyk2 kinase activities [3, 13], disrupt TSC1-TSC2 complex formation [11], regulate p53 and TSC1 protein stability [14, 15], serve as a scaffolding to facilitate TRAF2-ASK1 signaling to JNK activation [12], and regulate the transcription of Rb1 when localizing in nucleus [4]. Since most of these binding partners associate with different regions of FIP200 (Figure 1), it is possible that FIP200 can exert multiple biochemical functions through different domains. With more binding proteins identified and more sophisticated structure information of FIP200 obtained in the near future, a much in-depth understanding of FIP200 biochemical function will be appreciated.
Table 1. Summary of FIP200 interacting proteins.
| Binding protein | Functional significance of the interaction | Confirmed by endogenous co-IP | Reference |
|---|---|---|---|
| FAK | Inhibit FAK activity and associated cellular functions, including cell proliferation, cell spreading and migration. | Yes | [13] |
| Pyk2 | Suppress Pyk2 activity and Pyk2-induced apoptosis. | Yes | [3] |
| TSC1 | Disrupt TSC1-TSC2 interaction and promote TSC1 degradation. Promote mTOR activation and cell growth. | Yes | [11, 15] |
| p53 | stabilize p53, upregulate p21 expression and inhibit cell proliferation. | Yes | [14] |
| ASK1 | Regulate TRAF2-ASK1 interaction and promote JNK activation-induced by TNFα. | Yes | [12] |
| TRAF2 | Regulate TRAF2-ASK1 interaction and promote JNK activation-induced by TNFα. | Yes | [12] |
| Stathmin | Unknown | No | [6] |
| ActA | Unknown | No | [7] |
3.1. FIP200 function in cell survival
The first cellular function linked to FIP200 signaling pathway is cell survival/ cell death. As mentioned earlier, FIP200 initially was identified as a Pyk2 interacting protein [3]. Pyk2 is a cytoplasmic tyrosine kinase implicated to play a role in multiple intracellular signaling pathways, including regulation of apoptosis [16]. Overexpression of Pyk2 has been shown to induce apoptosis in a number of cell lines [17]. FIP200 was shown to inhibit the kinase activity of Pyk2 and suppress Pyk2-induced apoptosis, which correlates with FIP200 binding to Pyk2. In addition, activation of Pyk2 by several biological stimuli correlates with the dissociation of endogenous FIP200–Pyk2 interaction, suggesting that FIP200 dissociation from Pyk2 may mediate activation of Pyk2 by some biological stimuli [3]. Together, this study suggests that FIP200 functions to promote cell survival through its inhibition of Pyk2 activity (Figure 2).
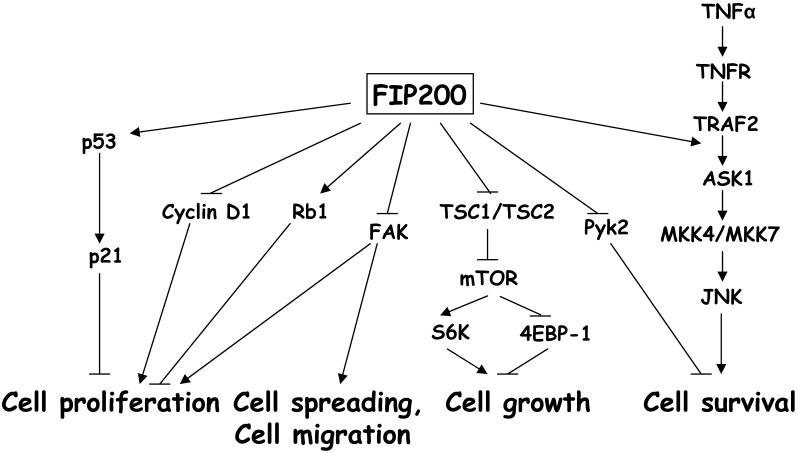
Figure 2. FIP200 signaling pathway.
FIP200 functions to regulate diverse cellular processes, including cell proliferation, cell spreading, cell migration, cell growth and cell survival. FIP200 suppresses cell proliferation by regulation of p53-p21, Cyclin D1, Rb1 and FAK pathways; FIP200 negatively regulates cell spreading and migration by its inhibition of FAK function; FIP200 promotes mTOR activation and cell growth through its interaction with TSC1-TSC2 complex; FIP200 positively regulates cell survival by its inhibition of Pyk2 activity and regulation of TNFα-JNK signaling cascade (see text for details).
The pro-survival function of FIP200 is further evidenced by recent FIP200 knockout (KO) mice study. Homozygous deletion of FIP200 in mouse leads to embryonic lethality associated with massive apoptosis in liver and heart [12]. FIP200 KO MEFs have been used to further understand the mechanism of regulation of apoptosis by FIP200. FIP200 KO MEFs do not show increased apoptosis under normal culture condition or various apoptosis induction conditions, such as energy deprivation, sorbitol treatment. In contrast, FIP200 KO MEFs and fetal liver cells show an elevated sensitivity to TNFα-induced apoptosis. Defective JNK signaling, but not NF-κB pathway, was subsequently identified to be responsible for increased TNFα-stimulated apoptosis in FIP200 KO MEFs as well as liver cells [12]. TNFα activates JNK through TNFR-TRAF2-ASK1-MKK4/MKK7-JNK signaling cascade [18-21]. Further mechanistic studies showed that FIP200 interacts with ASK1 and TRAF2, functions as a scaffold to orchestrate TRAF2-ASK1 signaling to JNK activation induced by TNFα [12]. In summary, FIP200 positively regulates JNK activation and cell survival in TNFα signaling pathway through the association of FIP200 with ASK1 and TRAF2. FIP200 deletion in mouse leads to reduced JNK activation and decreased cell survival (Figure 2). It appears that FIP200 inhibition of Pyk2 is independent of FIP200 regulation of TNFα-induced JNK activation and apoptosis, as TNFα stimulation does not affect Pyk2 activation and similar phosphorylation levels of Pyk2 were observed in FIP200 KO and WT MEFs under TNFα stimulation condition. It would be important to identify the physiological conditions under which Pyk2 activation is altered upon FIP200 deletion and to further study the exact role of Pyk2 in FIP200-mediated cell survival pathway.
3.2. FIP200 function in protein synthesis and cell growth
Cell growth (increase in cell mass and size) is a carefully orchestrated cellular process, which must be tightly regulated by both intracellular and extracellular stimuli. Cells promote macromolecular (such as protein and lipid) synthesis and thus increase cell mass and size in response to growth factor, nutrient stimuli or when energy and oxygen conditions are favorable. Conversely, cells inhibit macromolecular synthesis and restrain cell mass increase when nutrient and growth factors are limited or under energy stress and hypoxia conditions [1]. The studies from the last 10 years have identified the mammalian target of rapamycin (mTOR) as a critical regulator of cell growth by regulation of a variety of cellular functions, including initiation of mRNA translation, ribosome synthesis, expression of metabolism-related genes and autophagy [22]. Considerable bodies of evidence indicate that mTOR functions to positively regulate cell growth by phosphorylation of two key downstream targets: ribosomal S6 kinase (S6K) and eukaryotic initiation factor 4E binding protein-1 (4EBP-1) [23]. The discovery 5-6 years ago that tumor suppressor TSC1 and TSC2 function as negative regulator of mTOR signaling greatly expanded our understanding of how mTOR integrates with upstream signaling to regulate cell growth and how dysregulation of cell growth might lead to cancer development [24]. TSC1 and TSC2 (or hamartin and tuberin, respectively) are both tumor suppressor genes and mutation in either gene causes tuberous sclerosis (TSC) that occurs in ∼1 in 6,000 of the population and is defined by the formation of hamartomas in a wide range of tissues [24]. TSC1 and TSC2 form the complex and function as the GAP of the small GTPase Rheb (Ras homolog enriched in brain), a positive upstream regulator of mTOR. TSC1-TSC2 complex inhibits mTOR activity and restrain cell growth by stimulating Rheb GTP hydrolysis [24]. Mutation of either TSC1 or TSC2 results in hyperactivation of mTOR and unrestrained cell growth, which most likely leads to hamartomas development in TSC patients. This has stimulated great interest in clinical trials to use mTOR inhibitors to treat TSC patients [25].
We recently identified FIP200 as a TSC1 interacting protein through yeast two-hybrid screen [11]. Overexpression of FIP200 upregulates mTOR activation and increases cell size whereas RNAi knockdown of endogenous FIP200 suppresses mTOR activation and reduces cell size. FIP200 regulation of mTOR activation and cell growth correlates with FIP200 interaction with TSC1. Moreover, the regulation is abolished in TSC1 KO cells, strongly suggesting that the regulation of mTOR signaling and cell growth by FIP200 requires TSC1. FIP200 negatively regulates TSC function possibly through disruption of TSC1-TSC2 complex formation. In addition, FIP200 was shown to be important in nutrient stimulation-induced, but not energy- or serum-induced, mTOR activation [11]. Intriguingly, FIP200 was also independently identified as the regulator of TSC-mTOR signaling by a different approach [15]. It was noticed that the expression level of FIP200 is high in large cell types such as neuromuscular cells, but low in smaller cells such as leukocytes, suggesting the potential function of FIP200 in the regulation of cell size. Knockdown of FIP200 decreases mTOR activation and cell size whereas overexpression of FIP200 promotes mTOR activation and cell growth under nutrient deprived condition. Importantly, RNAi knockdown of FIP200 in mouse muscle cells reduces the cell size, suggesting that FIP200 might play a role in the regulation of muscle hypertrophy/ atrophy in vivo [15]. Overall, the consensus from these two independent studies is that FIP200 functions to positively regulate mTOR signaling and cell growth through its interaction with TSC1 and inhibition of TSC1-TSC2 complex function and that FIP200 might function in the nutrient signaling pathway to regulate cell growth. However, different mechanisms were proposed in these studies. Although our study suggests that FIP200 functions to regulate TSC1-TSC2 complex formation [11], the other study shows that FIP200 inhibits TSC by promoting TSC1 degradation through the ubiquitin-proteasomal pathway [15]. These suggest that FIP200 might regulate TSC function differentially under different contexts or in different cell types.
3.3. FIP200 function in cell proliferation
Our lab initially showed that overexpression of FIP200 potently inhibits cell proliferation in NIH3T3 cells. Furthermore, the inhibition of cell proliferation by FIP200 could be rescued by coexpression of FAK. Disruption of the functional interaction between endogenous FIP200 and FAK leads to increased FAK phosphorylation and partial restoration of cell cycle progression in cells plated on poly-l-lysine [13]. These results suggest that FIP200 interaction with FAK at least partially accounts for the inhibition of cell proliferation by FIP200. Numerous studies have suggested that FAK expression or phosphorylation correlates with the development of cancer [26-28]. The critical role of FAK in tumorigenesis is further evidenced by the data from the murine model that conditional deletion of FAK in mice suppresses chemically induced skin tumor formation and blocks malignant progression and thus provides the direct evidence implicating the role of FAK in tumorigenesis [29]. These studies raise the interesting possibility that the inhibition of FAK activity and cell proliferation by FIP200 might play a role in its potential tumor suppressor function as will be discussed below.
Many cell cycle regulators, such as p53, ARF, PTEN, can interact with a number of downstream components and regulate a variety of downstream signaling events, which mediate their tumor suppressor functions in different physiological settings. Consistent with this notion, FIP200 can regulate cell proliferation through FAK dependent and independent mechanisms. Overexpression of FIP200 has been shown to upregulate RB1 expression and inhibit cell proliferation in human leukemic cells, such as K562 and Jurkat cells [30]. In addition, RB1 expression correlates with FIP200 expression in the breast cancer cells with FIP200 truncation, suggesting that FIP200 regulation of RB1 expression might play a role in its tumor suppression [31]. Our recent study demonstrated that FIP200 could also inhibit cell proliferation in human breast cancer cells [14]. FIP200 was found to increase p21 and decrease cyclin D1 protein levels in breast cancer cells. In addition, FIP200 was shown to interact with p53, stabilize p53 and upregulate p21 expression. FIP200 can also decrease cyclin D1 protein half-life by promoting proteasome-dependent degradation of cyclin D1. These results suggest that FIP200 regulation of both p53 and cyclin D1 are critical for FIP200-induced G1 arrest in breast cancer cells [14]. Importantly, it was found that FIP200 regulation of p53 function is independent of FAK in the breast cancer cells studied [14]. In aggregate, these studies provide convincing evidence that FIP200 functions as a negative regulator of cell cycle progression and suggest that FIP200 regulation of cell proliferation involves different downstream signaling pathways in different cellular contexts.
3.4. FIP200 function in cell spreading and migration
Cell spreading and migration play essential roles in many biological processes such as embryogenesis, wound repair, angiogenesis, inflammatory immune response, and cancer metastasis [2, 32]. FAK, a major mediator of signal transduction by integrins, has been shown to play important roles in the regulation of cell spreading and migration[33, 34]. In contrast to our extensive knowledge of the FAK downstream signaling pathways to regulate cell spreading and migration, relatively little is known about the mechanisms of the regulation of FAK activity and its function in cell spreading and migration. FIP200 has been shown to colocalize with FAK at focal adhesions, interact with FAK kinase domain, inhibit FAK kinase activity and autophosphorylation as well as function in cell spreading and migration [13]. Coexpression of FAK could rescue the inhibition of cell spreading and migration by FIP200, suggesting that FIP200 regulation of cell spreading and migration is at least partially through its inhibition of FAK function. Moreover, endogenous interaction between FAK and FIP200 is detected in suspended cells and the interaction is decreased upon cell attachment on fibronectin (FN) [13]. Therefore, the association of endogenous FIP200 with FAK correlates with FAK inactivation upon cell detachment, suggesting that integrin signaling negatively regulates FIP200-FAK interaction and activates FAK activity and its downstream signaling by disrupting FAK-FIP200 interaction.
Other studies have identified some protein tyrosine phosphatases, such as PTEN, as FAK inhibitor by dephosphorylation of FAK [35, 36]. However, all these inhibitory events require the enzymatic activities of the phosphatases. In contrast, FIP200 inhibits FAK through the binding to FAK kinase domain and inhibiting its kinase activity, thus representing a novel regulatory mechanism of FAK function.
There are several interesting implications and questions regarding FIP200 function in cell spreading and migration, which await further investigation. Since activation of FAK has been implicated in diseases such as cancer metastasis [37], generation of small peptides or their derivatives from FIP200 as inhibitors for FAK could be of potential therapeutic significance. Although the data available so far suggest that FIP200 regulates cell spreading and migration through its interaction with FAK and inhibition of FAK activity, it is likely that FIP200 interaction with other proteins might also play a role in its regulation of cell spreading and migration. Identification of other FIP200 interacting proteins would create avenues to address this question.
3.5. FIP200 function in cell differentiation
The potential role of FIP200 in the regulation of cell differentiation was first implicated by its differential expression in human embryonic musculoskeletal cells [9]. FIP200 expresses at low level in immature embryonic musculoskeletal cells and its expression level increases concomitantly with the maturation of musculoskeletal cells [9]. Furthermore, FIP200 gene expression is induced in C2C12 cells upon myoblast differentiation, which also correlates with the upregulation of Rb1 and other myoblast differentiation marker proteins, such as Myhc [38]. Finally, RNAi knockdown of endogenous FIP200 suppresses the expression of Rb1 and Myhc during C2C12 myoblast differentiation [38]. Collectively, these data identified FIP200 as a potential regulator of myogenic differentiation, although the molecular mechanism by which FIP200 regulates cell differentiation and its in vivo significance remains to be illustrated.
4. The role of FIP200 in embryonic development
FIP200 is abundantly expressed throughout mouse embryonic development [10], suggesting that FIP200 might play an important role in embryogenesis. To study the potential function of FIP200 in embryogenesis, we recently generated FIP200 KO mouse. Targeted deletion of FIP200 in the mouse leads to embryonic death at mid/late gestation associated with heart failure and liver degeneration [12]. The heart ventricular wall in FIP200 KO embryos at E14.5 and E15.5 loses much of the normal trabecular and external compact myocytes, and the ventricular wall is significantly thinner and contains fewer cells when compared to the WT littermates. Majority of the FIP200 KO embryos also show severe liver degeneration characterized by loosely arranged hepatocytes mixed with numerous red blood cells. Hepatocytes are separated from each other with disrupted architecture due to dissecting hemorrhage in the liver. Furthermore, FIP200 KO embryos show massive apoptosis in liver and heart [12]. Thus defects in the formation and development of the myocardium and liver are the most likely cause of the embryonic lethality observed in FIP200 KO embryos.
As discussed above, FIP200 has been shown to regulate cell size through its interaction with TSC1-TSC2 complex. Interestingly, TSC1 and TSC2 KO embryos also exhibit severe heart defects with thickened ventricular walls [39, 40]. The opposite defective cardiac phenotypes suggest that FIP200 might function to antagonize TSC complex in the regulation of the thickness of ventricular wall during heart development. In support of this idea, FIP200 KO cardiomyocytes also show decreased mTOR activation and reduced cell size[12]. Therefore, a role of FIP200 in the regulation of cell size/cell growth may contribute to its function in heart development.
The liver degeneration phenotype observed in FIP200 KO embryos is similar to the KO embryo phenotypes by deletion of several components in TNFα survival signaling pathways including Rel A, IKK-β, IKK-γ, GSK-3, MKK4, MKK7, or c-Jun, which are characterized by mid/late gestational lethality associated with increased apoptosis in liver [41-47]. Furthermore, FIP200 KO MEFs exhibit increased apoptosis upon TNFα treatment, which might be explained by the loss of FIP200 interaction with ASK1 and TRAF2, regulation of TRAF2-ASK1 interaction and ASK1 phosphorylation [12]. These results raise the intriguing possibility that increased apoptosis and liver degeneration defects in FIP200 KO embryos may be caused by an increased susceptibility to TNFα- induced apoptosis.
5. The potential role of FIP200 in cancer development
Human FIP200 gene localizes in 8q11 chromosome, a region containing several loci of putative tumor suppressor genes, and loss of heterozygosity (LOH) of this region has been associated with various human cancers, including breast cancer [48, 49]. It was found that 20% (7 of 35) of primary breast cancers that were screened contain large deletion mutations in FIP200 that are predicted to generate markedly truncated proteins lacking the NLS, the leucine zipper motif and the coiled-coil region [31]. In all seven cases, both FIP200 alleles were inactivated with two primary breast cancers containing compound heterozygous deletions in both alleles and the other five showing loss of heterozygosity (LOH) of FIP200 [31]. Furthermore, LOH of FIP200 was also observed in several primary breast cancer samples in another study [50]. These genetic data thus provide evidence that FIP200 might function as a tumor suppressor gene.
The observation that FIP200 functions to negatively regulate cell proliferation, cell migration and spreading is consistent with its tumor suppressor function. On the other hand, the pro-survival function for FIP200 should not be in conflict with the potential role of FIP200 as a tumor suppressor, as deletion of several other tumor suppressors also results in increased apoptosis under certain contexts [51-54]. For example, TSC1, TSC2 and Lkb1 KO MEFs exhibit increased apoptosis under energy stress condition [55, 56]. Homozygous deletion of Rb in mouse leads to embryonic lethality associated with massive cell death in the nervous system [51-53]. Furthermore, Rb triple KO MEFs (lacking all three Rb family proteins pRb, p107, and p130) exhibit increased apoptosis under serum starvation condition [57].
Although the implication of FIP200 in the regulation of cancer development is of vital interest, there are several questions remaining to be answered to firmly establish FIP200 as a bona fide tumor suppressor. First, if FIP200 indeed functions as a tumor suppressor, the mutant version of FIP200 derived from cancer cells would function in a loss-of-function manner for its tumor suppressor function. It would be interesting to test whether any of the truncation mutants of FIP200 found in those breast cancer cells would function as a loss-of-function mutant in any of its cellular functions as discussed above, which would be of extreme importance in addressing the underlying mechanism of FIP200 function as a tumor suppressor. Furthermore, a major challenge in the field of human cancer genomics is to distinguish the mutations driving the neoplastic transformation process (drivers) from the incidental, irrelevant mutations (passengers) [58]. Although 8q11 deletion is frequently observed in various cancer types, there remains the daunting challenge of target cancer gene identification in this large regional loss. Much more studies are needed to firmly establsh FIP200 mutation is a true “driver” event in cancer development, including searching for point mutations of FIP200 in tumors and enlarging the tumor sample size and spectrum for mutation search. The subsequent copy number alterations (CNAs) and gene expression correlation study, both gain-of-function and loss-of-function studies in human cancer cells will help to validate whether FIP200 functions as target cancer gene. Finally, mouse cancer models will be very instructive to study FIP200 function in tumorigenesis. Heterozygous deletion of FIP200 has not led to development of mammary or any other tumors [12]. Future studies using FIP200 conditional KO mice combined with crossing with other mice tumor models will be necessary to overcome the embryonic lethality of homozygous deletion of FIP200 and clarify its potential role as a tumor suppressor in vivo.
6. Future prospects
Over the past several years, the important role of FIP200 in coordinating signaling pathways has emerged. However, much remains to be learned about the exact biochemical and cellular functions of FIP200 and its connection to human diseases. This review intends to summarize our current view of FIP200 signaling, identify important questions and provide guidance for future studies. Since many questions have been raised and discussed throughout the review, only a few will be highlighted here.
Although our knowledge of FIP200 downstream signaling pathways has increased considerably recently, virtually nothing is known about how FIP200 is regulated by upstream stimuli. Is the expression level of FIP200 or its phosphorylation status, protein stability, subcellular localization regulated by certain signaling, especially those in which FIP200 has been shown to play a role, such as cell adhesion, nutrient or growth factor signaling? If so, what is the underlying biochemical mechanism? Addressing these questions should provide critical insight into how FIP200 functions to coordinate various signaling pathways. In this regard, other genomics and proteomics studies might provide unbiased and important information of FIP200 regulation. A recent study applied the powerful mass spectrometric technology to study global in vivo phosphoproteome and its temporal dynamics upon growth-factor stimulation and has identified more than 6000 phosphorylation sites on 2000 or so proteins upon EGF stimulation [59]. Intriguingly, this study identified 6 phosphorylation sites in FIP200, at least one of which is strongly regulated by EGF stimulation. The phosphorylation sites can be further predicted to be phosphorylated by kinases such as GSK3 and PKA [59]. Although the identity and functional significance of these phosphorylation events need to be further confirmed, it provides strong evidence that FIP200 is tightly regulated by growth factor-mediated phosphorylation signaling events.
Second, our current knowledge of FIP200 is largely built upon in vitro studies and its in vivo relevance needs more rigorous investigation. The related questions for future studies include whether FIP200 indeed functions as a bona fide tumor suppressor, whether FIP200 play any role in the regulation of atrophy/hypertrophy based on its function in the regulation of mTOR signaling and cell growth, whether FIP200 regulates FAK signaling and cancer metastasis in vivo. Generation and analysis of various tissue specific KO mice models and compound mutant mice by crossing with other mice models will allow a critical determination of the in vivo functions of FIP200.
Finally, numerous studies from other model organisms, including yeast, worm and fly, have provided incredible insight into the protein functions in human and the molecular mechanisms of human diseases, including cancer development. Since FIP200 is an evolutionarily conserved protein present in worm and fly, utilization of these model organisms in the future study would be of vital importance in deciphering FIP200 function. We expect that there would be many exciting breakthroughs elucidating the mechanism of FIP200 signaling and its implications in human diseases in the foreseeable future.
Acknowledgments
This work is supported by NIH grant GM52890 to JL Guan. We thank our colleagues Huijun Wei, Huaping Fan, Fei Liu, Chun-Chi Liang and Heui Jin Ho for critical reading and helpful suggestions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Conlon I, Raff M. Cell. 1999;96(2):235–244. doi: 10.1016/s0092-8674(00)80563-2. [DOI] [PubMed] [Google Scholar]
- 2.Lauffenburger DA, Horwitz AF. Cell. 1996;84(3):359–369. doi: 10.1016/s0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]
- 3.Ueda H, Abbi S, Zheng C, Guan JL. J Cell Biol. 2000;149(2):423–430. doi: 10.1083/jcb.149.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chano T, Ikegawa S, Kontani K, Okabe H, Baldini N, Saeki Y. Oncogene. 2002;21(8):1295–1298. doi: 10.1038/sj.onc.1205178. [DOI] [PubMed] [Google Scholar]
- 5.Nagase T, Seki N, Ishikawa K, Ohira M, Kawarabayasi Y, Ohara O, Tanaka A, Kotani H, Miyajima N, Nomura N. DNA Res. 1996;3(5):321–329. 341–354. doi: 10.1093/dnares/3.5.321. [DOI] [PubMed] [Google Scholar]
- 6.Maucuer A, Camonis JH, Sobel A. Proc Natl Acad Sci U S A. 1995;92(8):3100–3104. doi: 10.1073/pnas.92.8.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pfeuffer T, Goebel W, Laubinger J, Bachmann M, Kuhn M. Cell Microbiol. 2000;2(2):101–114. doi: 10.1046/j.1462-5822.2000.00034.x. [DOI] [PubMed] [Google Scholar]
- 8.Chano T, Ikegawa S, Saito-Ohara F, Inazawa J, Mabuchi A, Saeki Y, Okabe H. Gene. 2002;291(12):29–34. doi: 10.1016/s0378-1119(02)00585-1. [DOI] [PubMed] [Google Scholar]
- 9.Chano T, Saeki Y, Serra M, Matsumoto K, Okabe H. Am J Pathol. 2002;161(2):359–364. doi: 10.1016/S0002-9440(10)64190-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bamba N, Chano T, Taga T, Ohta S, Takeuchi Y, Okabe H. Int J Mol Med. 2004;14(4):583–587. [PubMed] [Google Scholar]
- 11.Gan B, Melkoumian ZK, Wu X, Guan KL, Guan JL. J Cell Biol. 2005;170(3):379–389. doi: 10.1083/jcb.200411106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gan B, Peng X, Nagy T, Alcaraz A, Gu H, Guan JL. J Cell Biol. 2006;175(1):121–133. doi: 10.1083/jcb.200604129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abbi S, Ueda H, Zheng C, Cooper LA, Zhao J, Christopher R, Guan JL. Mol Biol Cell. 2002;13(9):3178–3191. doi: 10.1091/mbc.E02-05-0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Melkoumian ZK, Peng X, Gan B, Wu X, Guan JL. Cancer Res. 2005;65(15):6676–6684. doi: 10.1158/0008-5472.CAN-04-4142. [DOI] [PubMed] [Google Scholar]
- 15.Chano T, Saji M, Inoue H, Minami K, Kobayashi T, Hino O, Okabe H. Int J Mol Med. 2006;18(3):425–432. [PubMed] [Google Scholar]
- 16.Avraham H, Park SY, Schinkmann K, Avraham S. Cell Signal. 2000;12(3):123–133. doi: 10.1016/s0898-6568(99)00076-5. [DOI] [PubMed] [Google Scholar]
- 17.Xiong W, Parsons JT. J Cell Biol. 1997;139(2):529–539. doi: 10.1083/jcb.139.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis RJ. Cell. 2000;103(2):239–252. doi: 10.1016/s0092-8674(00)00116-1. [DOI] [PubMed] [Google Scholar]
- 19.Nishitoh H, Saitoh M, Mochida Y, Takeda K, Nakano H, Rothe M, Miyazono K, Ichijo H. Mol Cell. 1998;2(3):389–395. doi: 10.1016/s1097-2765(00)80283-x. [DOI] [PubMed] [Google Scholar]
- 20.Tobiume K, Matsuzawa A, Takahashi T, Nishitoh H, Morita K, Takeda K, Minowa O, Miyazono K, Noda T, Ichijo H. EMBO Rep. 2001;2(3):222–228. doi: 10.1093/embo-reports/kve046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee SY, Reichlin A, Santana A, Sokol KA, Nussenzweig MC, Choi Y. Immunity. 1997;7(5):703–713. doi: 10.1016/s1074-7613(00)80390-8. [DOI] [PubMed] [Google Scholar]
- 22.Schmelzle T, Hall MN. Cell. 2000;103(2):253–262. doi: 10.1016/s0092-8674(00)00117-3. [DOI] [PubMed] [Google Scholar]
- 23.Wullschleger S, Loewith R, Hall MN. Cell. 2006;124(3):471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 24.Kwiatkowski DJ, Manning BD. Hum Mol Genet. 2005;14(Spec No 2):R251–258. doi: 10.1093/hmg/ddi260. [DOI] [PubMed] [Google Scholar]
- 25.Guertin DA, Sabatini DM. Cancer Cell. 2007;12(1):9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 26.Owens LV, Xu L, Craven RJ, Dent GA, Weiner TM, Kornberg L, Liu ET, Cance WG. Cancer Res. 1995;55(13):2752–2755. [PubMed] [Google Scholar]
- 27.Agochiya M, Brunton VG, Owens DW, Parkinson EK, Paraskeva C, Keith WN, Frame MC. Oncogene. 1999;18(41):5646–5653. doi: 10.1038/sj.onc.1202957. [DOI] [PubMed] [Google Scholar]
- 28.Jones RJ, Brunton VG, Frame MC. Eur J Cancer. 2000;36(13 Spec No):1595–1606. doi: 10.1016/s0959-8049(00)00153-2. [DOI] [PubMed] [Google Scholar]
- 29.McLean GW, Komiyama NH, Serrels B, Asano H, Reynolds L, Conti F, Hodivala-Dilke K, Metzger D, Chambon P, Grant SG, Frame MC. Genes Dev. 2004;18(24):2998–3003. doi: 10.1101/gad.316304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kontani K, Chano T, Ozaki Y, Tezuka N, Sawai S, Fujino S, Saeki Y, Okabe H. Int J Mol Med. 2003;12(5):767–769. [PubMed] [Google Scholar]
- 31.Chano T, Kontani K, Teramoto K, Okabe H, Ikegawa S. Nat Genet. 2002;31(3):285–288. doi: 10.1038/ng911. [DOI] [PubMed] [Google Scholar]
- 32.Christopher RA, Guan JL. Int J Mol Med. 2000;5(6):575–581. doi: 10.3892/ijmm.5.6.575. [DOI] [PubMed] [Google Scholar]
- 33.Mitra SK, Hanson DA, Schlaepfer DD. Nat Rev Mol Cell Biol. 2005;6(1):56–68. doi: 10.1038/nrm1549. [DOI] [PubMed] [Google Scholar]
- 34.Abbi S, Guan JL. Histol Histopathol. 2002;17(4):1163–1171. doi: 10.14670/HH-17.1163. [DOI] [PubMed] [Google Scholar]
- 35.Tamura M, Gu J, Matsumoto K, Aota S, Parsons R, Yamada KM. Science. 1998;280(5369):1614–1617. doi: 10.1126/science.280.5369.1614. [DOI] [PubMed] [Google Scholar]
- 36.Yu DH, Qu CK, Henegariu O, Lu X, Feng GS. J Biol Chem. 1998;273(33):21125–21131. doi: 10.1074/jbc.273.33.21125. [DOI] [PubMed] [Google Scholar]
- 37.Mitra SK, Schlaepfer DD. Curr Opin Cell Biol. 2006;18(5):516–523. doi: 10.1016/j.ceb.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 38.Watanabe R, Chano T, Inoue H, Isono T, Koiwai O, Okabe H. Virchows Arch. 2005;447(3):643–648. doi: 10.1007/s00428-004-1183-1. [DOI] [PubMed] [Google Scholar]
- 39.Kobayashi T, Minowa O, Sugitani Y, Takai S, Mitani H, Kobayashi E, Noda T, Hino O. Proc Natl Acad Sci U S A. 2001;98(15):8762–8767. doi: 10.1073/pnas.151033798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kobayashi T, Minowa O, Kuno J, Mitani H, Hino O, Noda T. Cancer Res. 1999;59(6):1206–1211. [PubMed] [Google Scholar]
- 41.Beg AA, Sha WC, Bronson RT, Ghosh S, Baltimore D. Nature. 1995;376(6536):167–170. doi: 10.1038/376167a0. [DOI] [PubMed] [Google Scholar]
- 42.Hilberg F, Aguzzi A, Howells N, Wagner EF. Nature. 1993;365(6442):179–181. doi: 10.1038/365179a0. [DOI] [PubMed] [Google Scholar]
- 43.Hoeflich KP, Luo J, Rubie EA, Tsao MS, Jin O, Woodgett JR. Nature. 2000;406(6791):86–90. doi: 10.1038/35017574. [DOI] [PubMed] [Google Scholar]
- 44.Li Q, Van Antwerp D, Mercurio F, Lee KF, Verma IM. Science. 1999;284(5412):321–325. doi: 10.1126/science.284.5412.321. [DOI] [PubMed] [Google Scholar]
- 45.Nishina H, Vaz C, Billia P, Nghiem M, Sasaki T, De la Pompa JL, Furlonger K, Paige C, Hui C, Fischer KD, Kishimoto H, Iwatsubo T, Katada T, Woodgett JR, Penninger JM. Development. 1999;126(3):505–516. doi: 10.1242/dev.126.3.505. [DOI] [PubMed] [Google Scholar]
- 46.Rudolph D, Yeh WC, Wakeham A, Rudolph B, Nallainathan D, Potter J, Elia AJ, Mak TW. Genes Dev. 2000;14(7):854–862. [PMC free article] [PubMed] [Google Scholar]
- 47.Wada T, Joza N, Cheng HY, Sasaki T, Kozieradzki I, Bachmaier K, Katada T, Schreiber M, Wagner EF, Nishina H, Penninger JM. Nat Cell Biol. 2004;6(3):215–226. doi: 10.1038/ncb1098. [DOI] [PubMed] [Google Scholar]
- 48.Dahiya R, Perinchery G, Deng G, Lee C. Int J Oncol. 1998;12(4):811–816. doi: 10.3892/ijo.12.4.811. [DOI] [PubMed] [Google Scholar]
- 49.Perinchery G, Bukurov N, Nakajima K, Chang J, Hooda M, Oh BR, Dahiya R. Int J Oncol. 1999;14(3):495–500. doi: 10.3892/ijo.14.3.495. [DOI] [PubMed] [Google Scholar]
- 50.Schmidt-Kittler O, Ragg T, Daskalakis A, Granzow M, Ahr A, Blankenstein TJ, Kaufmann M, Diebold J, Arnholdt H, Muller P, Bischoff J, Harich D, Schlimok G, Riethmuller G, Eils R, Klein CA. Proc Natl Acad Sci U S A. 2003;100(13):7737–7742. doi: 10.1073/pnas.1331931100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Clarke AR, Maandag ER, van Roon M, van der Lugt NM, van der Valk M, Hooper ML, Berns A, te Riele H. Nature. 1992;359(6393):328–330. doi: 10.1038/359328a0. [DOI] [PubMed] [Google Scholar]
- 52.Jacks T, Fazeli A, Schmitt EM, Bronson RT, Goodell MA, Weinberg RA. Nature. 1992;359(6393):295–300. doi: 10.1038/359295a0. [DOI] [PubMed] [Google Scholar]
- 53.Lee EY, Chang CY, Hu N, Wang YC, Lai CC, Herrup K, Lee WH, Bradley A. Nature. 1992;359(6393):288–294. doi: 10.1038/359288a0. [DOI] [PubMed] [Google Scholar]
- 54.Grisendi S, Bernardi R, Rossi M, Cheng K, Khandker L, Manova K, Pandolfi PP. Nature. 2005;437(7055):147–153. doi: 10.1038/nature03915. [DOI] [PubMed] [Google Scholar]
- 55.Inoki K, Zhu T, Guan KL. Cell. 2003;115(5):577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 56.Shaw RJ, Kosmatka M, Bardeesy N, Hurley RL, Witters LA, DePinho RA, Cantley LC. Proc Natl Acad Sci U S A. 2004;101(10):3329–3335. doi: 10.1073/pnas.0308061100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sage J, Mulligan GJ, Attardi LD, Miller A, Chen S, Williams B, Theodorou E, Jacks T. Genes Dev. 2000;14(23):3037–3050. doi: 10.1101/gad.843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, Edkins S, O'Meara S, Vastrik I, Schmidt EE, Avis T, Barthorpe S, Bhamra G, Buck G, Choudhury B, Clements J, Cole J, Dicks E, Forbes S, Gray K, Halliday K, Harrison R, Hills K, Hinton J, Jenkinson A, Jones D, Menzies A, Mironenko T, Perry J, Raine K, Richardson D, Shepherd R, Small A, Tofts C, Varian J, Webb T, West S, Widaa S, Yates A, Cahill DP, Louis DN, Goldstraw P, Nicholson AG, Brasseur F, Looijenga L, Weber BL, Chiew YE, DeFazio A, Greaves MF, Green AR, Campbell P, Birney E, Easton DF, Chenevix-Trench G, Tan MH, Khoo SK, Teh BT, Yuen ST, Leung SY, Wooster R, Futreal PA, Stratton MR. Nature. 2007;446(7132):153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Cell. 2006;127(3):635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]