Yin-Yang 1 regulates effector cytokine gene expression and TH2 immune responses (original) (raw)
. Author manuscript; available in PMC: 2020 Mar 11.
Published in final edited form as: J Allergy Clin Immunol. 2008 Apr 18;122(1):195–201.e5. doi: 10.1016/j.jaci.2008.03.012
Abstract
Background:
The transcription factor Yin-Yang 1 (YY-1) binds to the promoter regions of several T-cell cytokine genes, but the expression and contribution of this factor to cytokine gene expression and T-cell activation in vivo is not clear. Objective: We sought to better define the role of YY-1 in T-cell gene regulation and allergic immune responses.
Methods:
We studied cytokine gene expression in T lymphocytes isolated from wild-type mice and heterozygous littermates bearing 1 targeted _yy_-1 allele (yy-1+/− mice). T cells were stimulated with anti-T-cell receptor (anti-TCR) plus CD28 antibodies or with peptide antigen plus antigen-presenting cells by using newly generated yy-1+/− TCR transgenic mice. We also studied ovalbumin-driven allergic immune responses in a mouse model of asthma and YY-1 expression in lung tissue from human asthmatic subjects.
Results:
CD4+ T cells from yy-1+/− mice secreted significantly less IL-4 and IFN-γ compared with wild-type littermates after TCR-dependent activation, whereas IL-2 production was not significantly affected. Both airway inflammation and recall splenocyte IL-4 production were inhibited in yy-1+/− mice, as was antigen-driven T-cell proliferation. YY-1 expression was higher in airway biopsy specimens from asthmatic compared with control subjects.
Conclusion:
These data indicate that YY-1 regulates T-cell cytokine gene expression and allergic immune responses in a gene dose-dependent manner. (J Allergy Clin Immunol 2008;122:195–201.)
Keywords: T lymphocyte, cytokine gene regulation, transcription factors, allergic inflammation, asthma
Yin-Yang 1 (YY-1) is an ubiquitously expressed zinc-finger DNA-binding transcription factor that influences the expression of a wide variety of cellular and viral genes.1 YY-1 is a versatile factor that can either initiate, activate, or repress transcription, dependent on the promoter context,1,2 and was recently implicated in genomic imprinting and in mitochondrial function.3,4 YY-1 interacts with a diverse array of other transcription factors and chromatin remodeling complexes that influence its transacting ability.1,2,5–7 The observation that YY-1 is a negative regulator of p53 helped explain why genetic deletion or knockdown of YY-1 in tumor cells resulted in growth arrest, apoptosis, or both.8 Recent studies using conditional deletion in mice discovered a critical role for YY-1 in the differentiation of oligodendrocytes and in B-cell development.9,10
Although YY-1 is widely expressed in cells of the immune system, the function of this factor in immune responses is only beginning to be understood. Conditional deletion of YY-1 early in B-cell development impairs VDJ recombination and immunoglobulin locus contraction, resulting in profoundly reduced numbers of mature B cells in the periphery.10 YY-1 might play a role in the pathogenesis of allergic diseases because single nucleotide polymorphisms in several asthma- and allergy-associated genes affect YY-1 binding.11–14 Several T-cell cytokine gene-promoter regions contain consensus YY-1 binding sites, including IL-4,15 IFN-γ,16,17 and IL-5,18,19 but most research to date has been performed by using cell lines, and it remains unclear whether or how YY-1 regulates the activation of primary T cells or immune responses in vivo. Cameron et al14 recently showed that YY-1 binds to a polymorphic site in the IL-13 promoter and positively regulates IL-13 expression. Using electrophoretic mobility shift assays, we showed that the IL-4 promoter contains 4 YY-1 binding sites and that overexpressed YY-1 enhanced promoter activity and IL-4 gene expression in both Jurkat and primary T cells independently of nuclear factor of activated T cells (NFAT), a key regulator of T-cell cytokines.15 The function of YY-1 in IFN-γ gene expression remains confusing. Ye et al17 discovered 2 YY-1 binding sites in the IFN_-γ_ promoter that appeared to act independently to repress basal promoter activity in Jurkat cells. Using stably integrated IFN-γ promoter reporter constructs in transgenic mice, Soutto et al20 confirmed that the −110 to −225 region of the IFN-γ promoter, which contains the binding sites for YY-1 and other factors, repressed IFN-γ promoter activity in vivo. In contrast, Sweetser et al21 used site-directed mutagenesis and concluded that YY-1 interacted with adjacently bound NFAT to enhance IFN-γ expression in mitogen-stimulated mouse splenocytes. Thus the precise role that YY-1 plays in regulating CD4+ cytokine expression requires further study.
Homozygous deletion of the murine yy-1 gene results in early embryonic lethality caused by peri-implantation defects.22 Heterozygous mice survive and are fertile, although they possess axial skeletal abnormalities consistent with homeotic transformation.23 Mutant embryos were recently shown to have impaired growth and viability in direct proportion to the genetic complement of YY-1.24 Thus YY-1 can act in a gene dose-dependent manner to regulate gene expression and differentiation.
We wanted to explore the expression and potential role of YY-1 in TH cytokine gene regulation and allergic immune responses in vivo. Using heterozygous yy-1+/− mice, we report that compared with that seen in wild-type (WT) littermates, secretion of CD4+ cytokines, including IL-4 and IFN-γ, is significantly impaired by partial YY-1 deficiency, especially at later stages of T-cell activation. We generated new strains of T-cell receptor (TCR) transgenic mice bearing 1 targeted YY-1 allele and observed significant inhibition of IL-4 and IFN-γ gene expression after antigen-specific activation. Targeted microarray arrays revealed that YY-1 regulates other genes in T cells in addition to cytokines. Using a well-established model of allergen-driven TH2-dependent airway inflammation, we confirmed that deficiency of yy-1 attenuated allergen-driven IL-4 gene expression and airway inflammation in vivo.
METHODS
Mouse lines
Mice genetically deficient in yy-1 were generated and previously characterized by Donohoe et al.22 Heterozygous mice were maintained on the C57BL6×129 background or backcrossed more than 10 generations to a BALB/c background. OTII.225 and DO11.10 mice, transgenic for a TCR that recognizes the ovalbumin (OVA) peptide OVA323–339, were obtained from Dr Shaun Huang (Johns Hopkins University) and Taconic Laboratories, respectively. Heterozygous yy-1+/− mice were bred with OT-II.2 and DO11.10 mice to generate offspring that were TCR transgenic but heterozygous for yy-1. Animals were maintained at the animal facilities of the Johns Hopkins Medical School and University of Rochester Medical Center in compliance with institutional guidelines and used at 6 to 12 weeks of age.
Cell isolation and analysis by means of flow cytometry
Form more information, see the Methods section in the Online Repository at www.jacionline.org.
CD4+ T-cell isolation and stimulation
CD4+ cells were isolated from mouse spleens with a CD4+ negative selection kit (Miltenyi Biotec, Bergisch Gladbach, Germany). Cells (1 × 106 per well in 24-well plates) were stimulated with coated anti-CD3 plus soluble anti-CD28 antibodies (2 μg/mL each; BD PharMingen, Franklin Lakes, NJ) or calcium ionophore A23187 (0.5 μmol/L) plus phorbol ester phorbol 12-myristate 13-acetate (PMA; 40 ng/mL; Calbiochem, San Diego, Calif). WT and yy-1+/− heterozygous TCR transgenic CD4+ T cells were incubated with antigen-presenting cells (APCs) plus OVA peptide for up to 4 days, and cytokine secretion was analyzed by means of ELISA. APCs were WT C57BL/ 6 bone marrow–derived dendritic cells, which were obtained by culturing bone marrow in recombinant mouse GM-CSF (25 ng/mL) plus mIL-4 (10 ng/mL; R&D Systems, Minneapolis, Minn) for 7 days. Loosely adherent cells were collected, washed, and loaded with different concentrations of OVA323–339 peptide (Peptides International, Louisville, Ky) and then incubated with OTII.2 CD4+ T cells at a dendritic/T cell ratio of 1:5 for 48 hours, followed by analysis of cytokine secretion by means of ELISA.
Cytokine ELISA or cytometric bead array
For more information, see the Methods section in the Online Repository.
Chromatin immunoprecipitation assays
For more information, see the Methods section in the Online Repository.
Quantitative RT-PCR–targeted array
For more information, see the Methods section in the Online Repository.
OVA sensitization and challenge protocol
Female WT and yy-1+/− heterozygous littermate mice 6 to 8 weeks of age on average were sensitized on days 0 and day 7 by means of intraperitoneal injection with 100 μL of 20 μg of OVA (Grade V; Sigma, St Louis, Mo) and 5 mg of aluminum hydroxide (Pierce, Cheshire, United Kingdom) in saline. Vehicle control mice were injected intraperitoneally with 100 μL of saline. On day 14, mice were challenged with intratracheal OVA solution (2.5 μg in 10 μL of saline) or saline alone as a control. Mice were killed 24 or 48 hours after OVA challenge for bronchoalveolar lavage (BAL) and isolation of splenocytes. Briefly, after anesthesia with ketamine (6 mg/kg) plus xylazine (0.25 mg/kg), BAL was performed by gently injecting 0.8 mL of PBS into the lung 3 times. BAL fluid cells were placed in pellets and washed, and differential cell counts were prepared by using a cytospin (Shandon, Pittsburgh, Pa) and fixed and stained with Diff-Quik (American Scientific Products, McGraw Park, Ill). Two counts of at least 250 cells were done for each sample in a blinded fashion.
Analysis of cell proliferation
Splenocytes (1 × 106 per well) from WT and yy-1+/− OTII.2 mice were labeled with 5, 6-carboxyfluorescein diacetate succinimidyl ester (CFSE) (1 mg/mL) for 10 minutes and incubated for 72 hours with peptide-loaded APCs and analyzed by means of flow cytometry for CFSE dilution as a marker of cell division.
Analysis of YY-1 in lung tissue by means of immunohistochemistry
Human asthmatic and control subjects were recruited from the Québec Respiratory Health Network consisting of the Montreal Chest Institute, Sacré Côeur Hospital, and Laval University. Atopic asthma (n = 6) was defined on the basis of clinical history, including intermittent chest tightness, cough, or wheezing according to national guidelines26; positive skin prick test results to 1 or more common aeroallergens; and total serum IgE concentrations of greater than 100 IU/mL. Nonatopic asthmatic subjects (n = 6) had a clinical history of perennial symptoms of asthma with no clear allergic trigger, negative skin prick test results with positive histamine reactions, and total serum IgE concentrations of less than 100 IU/mL. Control subjects (n = 6) were asymptomatic and nonatopic, defined as a negative skin prick test result with a positive histamine reaction, and also had normal spirometric results. Bronchial biopsy specimens were obtained at segmental divisions from all patients according to American Thoracic Society guidelines.27 Tissues were placed in acetone/methanol, blocked in optimal cutting temperature embedding medium (Sakura Finetechnical, Tokyo, Japan), and then fixed in 4% paraformaldehyde followed by paraffin embedding. Immunohistochemistry was performed on 5-mm sections by using a peroxidase protocol, as previously described,28 and antibodies directed against YY-1 (H-10; Santa Cruz Biotechnology, Santa Cruz, Calif), CD3 (NCL-CD3-PS1; Novocastra, Newcastle Upon Tyne, United Kingdom), or isotype controls. Slides were stained with diaminobenzidine or fast red chromogens (DAKO, Glostrup, Denmark) yielding brown and red colors and counterstained with hematoxylin. Positive signals were quantified with light microscopy at ×20 magnification by using a grid attachment of 0.2 mm2. Data were acquired by counting the number of immunoreactive cells and reported as the mean ± SEM of 6 to 8 grids.
RESULTS
Characterization of yy-1+/− mice
Because homozygous yy-1 deficiency is lethal during embryonic development,22 we used heterozygous yy-1+/− mice for these studies. To confirm that YY-1 expression was reduced in yy-1+/− mice, we analyzed spleen CD4+ T cells by means of RT-PCR and Western blotting and observed partial reduction in YY-1 mRNA and protein expression from yy-1+/− versus WT littermates (see Fig E1 in the Online Repository at www.jacionline.org). We next studied the size and cellular composition of the thymus, spleen, and lymph nodes in WT and yy-1+/− littermates and observed no significant differences in the composition of CD4+ and CD8+ single- or dual-positive subsets in the thymus (see Fig E1) or of the proportion of B lymphocytes, natural killer cells, and T cells in the spleens and peripheral lymph nodes from WT and yy-1+/− mice (data not shown). These data suggest that yy-1+/− mice are a valid model to study peripheral immune responses.
Reduced cytokine expression after polyclonal stimulation in yy-1+/− T cells
We next studied how partial YY-1 deficiency affected T-cell production of IL-4 and IFN-γ compared with production of IL-2, which is not known to be regulated by YY-1. Fig 1 shows that IL-4 production was significantly attenuated in anti-CD3/CD28–stimulated yy-1+/− CD4+ T cells compared with WT control cells, especially at later stages of T-cell activation (Fig 1, A; P < .05 for yy-1+/− vs WT; days 2–4), whereas IL-2 secretion was not significantly affected (Fig 1, B). Interestingly, IFN-γ secretion was also reduced, although not to the degree of IL-4 secretion (Fig 1, C). The delayed effects of yy-1 deficiency on IL-4 gene expression could reflect differences in the expression of other YY-1–dependent factors required for maximal IL-4 expression or delayed occupancy of the IL-4 promoter. To distinguish between these possibilities, we analyzed binding of YY-1 to the IL-4 promoter by using chromatin immunoprecipitation (ChIP) assays in T cells activated with anti-CD3 and anti-CD28 antibodies and found that YY-1 bound to the IL-4 promoter with delayed kinetics, peaking after approximately 48 hours of T-cell activation (Fig 1, D). Consistent with reduced expression of YY-1 and IL-4 in yy-1+/− mice, binding of YY-1 to the IL-4 promoter was barely detectable in yy-1+/− T cells. Thus these data are compatible with a model in which YY-1 directly activates IL-4 gene expression in acutely stimulated T cells.
FIG 1.
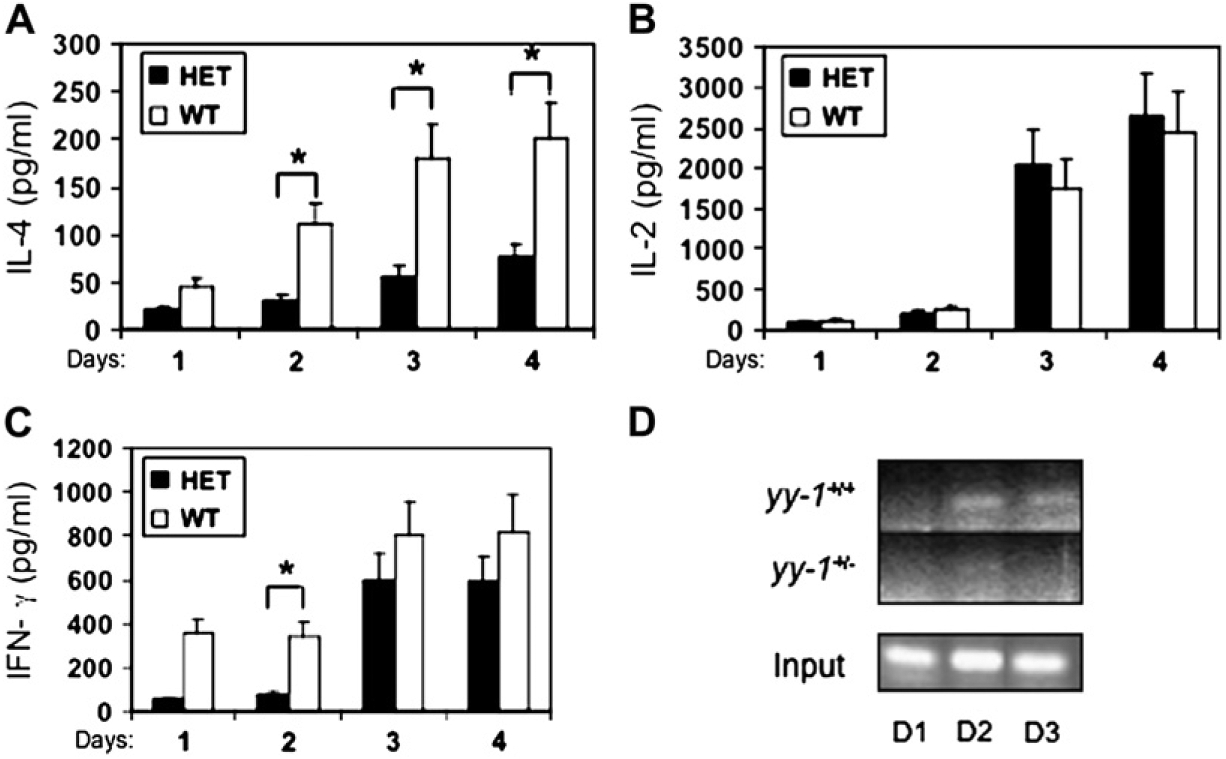
Effector cytokine secretion, YY-1 protein expression, and promoter occupancy are reduced in CD4+ T cells from yy-1+/− mice. CD4+ T cells were purified from WT and yy-1+/− heterozygous (HET) spleen cells and stimulated with anti-CD3/CD28 antibodies as indicated, followed by analysis of cytokine secretion by means of ELISA (A-C) or ChIP assay and PCR primers spanning the proximal IL-4 promoter (D). Results are the means ± SEMs of 4 to 5 mice per genotype analyzed in duplicate (Fig 1, A-C) or from 1 experiment representative of 3 (Fig 1, D).
We next analyzed the secretion of 5 cytokines by using a multiplex bead array (see the Methods section) and also analyzed CD4+ T cells isolated from WT and yy-1+/− mice on the BALB/c background. Similar to the case of C57BL/6×129 yy-1+/− mice (Fig 1), the secretion of IL-4 and IFN-γ, but not that of IL-2, was significantly attenuated by using BALB/c yy-1+/− CD4+ T cells (see Fig E2 in the Online Repository at www.jacionline.org). Interestingly, the secretion of other TH2 cytokines, including IL-5 and IL-13, was also reduced, indicating that YY-1 deficiency compromises TH2 cytokine secretion in general (see Fig E2).
Partial YY-1 deficiency affects the expression of other T-cell genes
We next isolated total RNA from WT and yy-1+/− CD4+ T cells and studied the expression of gene products implicated in TH differentiation by using a quantitative RT-PCR array. We considered gene products whose expressions differed by 2-fold or greater with a P value of .05 to be significantly different: 8 genes met these criteria, all of which were underexpressed in yy-1+/− versus WT T cells (see Fig E3 in the Online Repository at www.jacionline.org). Strikingly, the 2 most highly downregulated genes were IL-13 (0.19-fold) and IL-4 (0.25-fold; see Fig E3). Other significantly reduced genes included IFN-γ, the cytokine receptor tumor necrosis factor receptor superfamily, member 8, and the signaling molecules IFN regulatory factor 4, Junb, and suppressor of cytokine signaling 1 (see Fig E3). Thus YY-1 deficiency compromises T-cell activation in a selective manner, with a notable reduction in effector cytokine gene expression.
YY-1 is required for IL-4 and IFN-γ expression in antigen-activated CD4+ T cells
To rigorously test the idea that YY-1 is involved in antigen-specific CD4+ T-cell responses, we next generated 2 TCR transgenic mouse lines by crossing OTII.2 and DO11.10 TCR transgenic mice, which express a TCR specific for the OVA323–339 peptide, with yy-1+/− mice. The transgene in OTII.2 mice, which are on the C57BL/6 background, is integrated into the Y chromosome, and we confirmed that only male mice expressed an expanded population of CD4+V_α_2+ lymphocytes (data not shown). In coculture with peptide-loaded APCs, we found the secretion of IL-4 and IFN-γ from OTII.2 x yy-1+/− T cells was markedly attenuated when compared with that of WT control cells, whereas IL-2 secretion was not significantly affected (Fig 2). A similar pattern was observed with DO11.10 x yy-1+/− TCR transgenic T cells on the BALB/c background (see Fig E2), indicating that partial yy-1 deficiency attenuates TCR-dependent IL-4 and IFN-γ secretion, regardless of genetic background.
FIG 2.
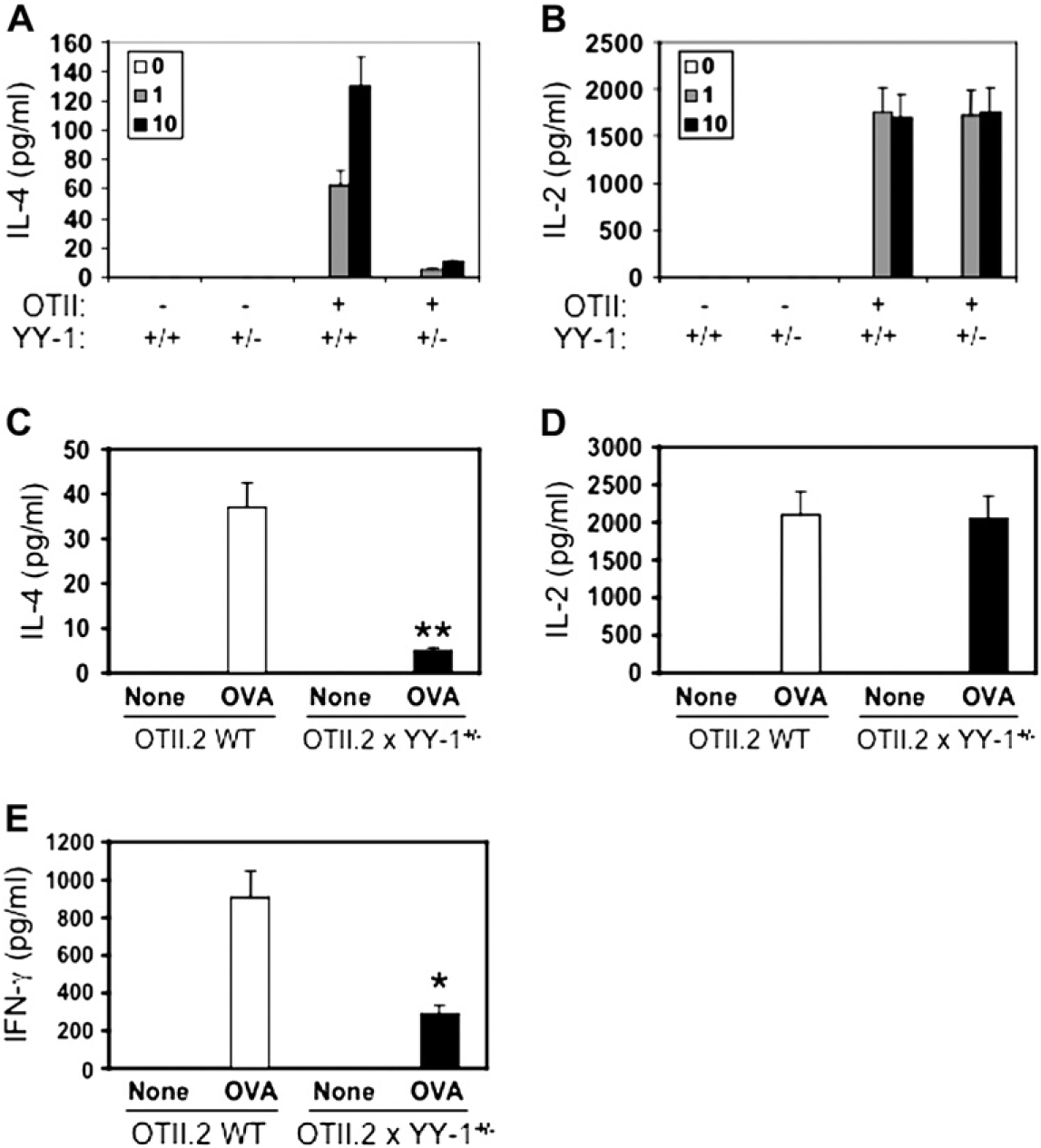
Antigen-driven CD4+ T-cell cytokine secretion is attenuated by partial yy-1 deficiency. Spleen CD4+ T cells were purified from WT or yy-1+/− (heterozygous) OTII.2 TCR transgenic mice and incubated with WT APCs and OVA323–339 peptide for 48 hours (see the Methods section). After 48 hours, supernatants were harvested, and cytokine secretion was analyzed by means of ELISA for IL-4 (A and C), IL-2 (B and D), and IFN-γ (E). Data in Fig 2, A and B, are from 1 experiment run in triplicate (representative of 3), whereas data in Fig 2, C through E, are the means ± SEMs of 4 mice per genotype. *P < .05 and **P < .01 for differences between genotypes.
Role of YY-1 in TH2 differentiation and allergic immune responses
We focused the remaining experiments on IL-4 production because this gene was one of the most consistently inhibited by deficiency of yy-1. To determine whether partial yy-1 deficiency affected IL-4 production during TH cell differentiation, we isolated naive CD62L+CD4+ cells from WT and yy-1 mice and polarized them in vitro under neutral (TH0), TH1, and TH2 conditions (see the Methods section). These experiments revealed that IL-4 secretion was reduced, especially under TH2-polarizing conditions (see Fig E4 in the Online Repository at www.jacionline.org), indicating that partial deficiency of yy-1 attenuates the differentiation of naive T cells toward the TH2 phenotype. To test the possibility that yy-1 deficiency attenuated TH2-driven immune responses in vivo, we used a well-established model of allergic airway inflammation characterized by antigen-driven TH2 immunity to inhaled OVA (see the Methods section). When compared with WT littermates, yy-1+/− mice demonstrated significantly lower influx of both eosinophils and lymphocytes into the lung after OVA sensitization and challenge (Fig 3, A and B), which is indicative of a reduced allergic immune response. Levels of the TH2 cytokines IL-4 and IL-13 were at or below the detection limit of ELISA in lung lavage fluids from both WT and yy-1+/− mice (data not shown), whereas recall IL-4 secretion and mRNA expression were both reduced in spleen cells from yy-1+/− mice compared with WT littermates (Fig 3, D). Expression of GATA-3, the master TH2 transcription factor, was unchanged. Taken together, these data indicate that deficiency of YY-1 attenuates allergen-driven IL-4 gene expression independently of the TH2 lineage marker GATA-3.
FIG 3.
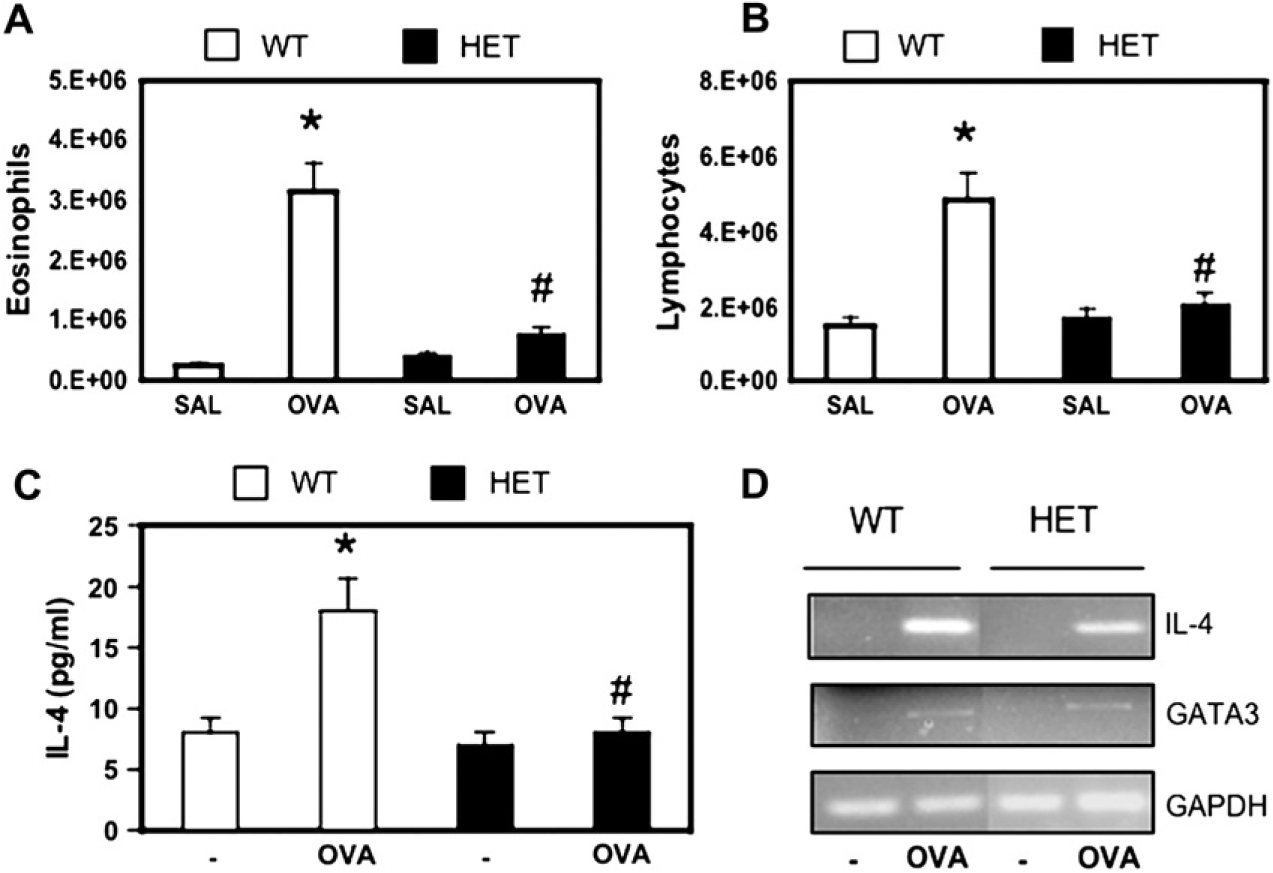
Airway inflammation and allergen-driven cytokine production are attenuated in yy-1+/− mice in a mouse model of asthma. WT and yy-1+/− (HET) mice were sensitized and challenged with OVA as indicated (see the Methods section), followed 48 hours later by analysis of lung lavage fluids by means of cytospin for eosinophils (A) and lymphocytes (B). Splenocytes were incubated with or without OVA (20 μg/mL) for 48 hours, and recall IL-4 expression was measured by means of ELISA (C) or RT-PCR compared with GATA-3 and GAPDH (D). Data are the means ± SEMs of 15 (Fig 3, A and B) or 20 (Fig 3, C) mice per genotype.
Because YY-1 deficiency attenuated allergic lung inflammation in mice, we wanted to study the expression of this factor in human subjects with and without asthma. We obtained endo-bronchial biopsy specimens from 6 subjects each with atopic and nonatopic asthma defined by using standard criteria and 6 nonasthmatic nonatopic control subjects (see the Methods section). By means of immunohistochemical analysis, we found that YY-1 expression was significantly increased in the airways in subjects with asthma compared with that seen in healthy control subjects (Fig 4). YY-1+ cells were equally increased in both atopic and nonatopic asthmatic subjects (8.8 vs 9.7 immunoreactive cells per square millimeter, respectively). The number of airway CD3+ T cells was not significantly different in asthmatic versus control subjects (data not shown), which is in keeping with numerous prior studies.29 In ongoing studies we found that YY-1 expression is also increased in lung tissues from subjects with sarcoidosis and idiopathic pulmonary fibrosis (data not shown). Taken together, these data indicate that YY-1 expression is increased in the airways in asthma and that this is not simply due to an increased number of T cells.
FIG 4.
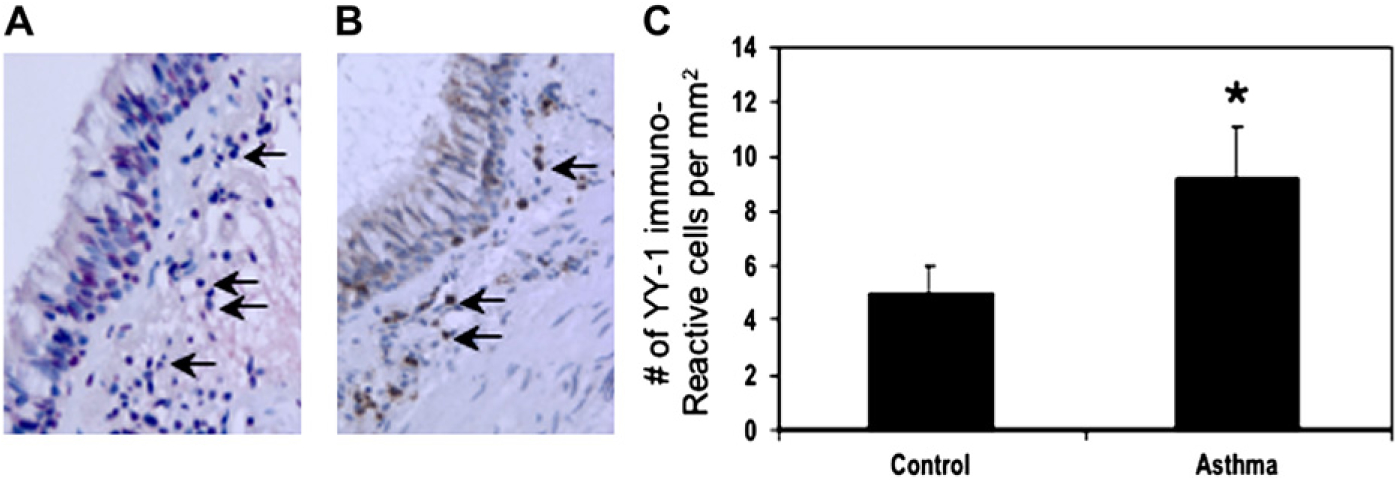
YY-1 expression is increased in the airways in asthmatic subjects. Endobronchial biopsy specimens were obtained from healthy and asthmatic subjects and analyzed by using immunohistochemistry for expression of YY-1+ and CD3+ cells (see the Methods section). A and B show representative staining of YY-1 and CD3, respectively, from a subject with asthma. C shows the mean ± SEMs of 6 healthy and 12 asthmatic subjects, respectively.
YY-1 deficiency inhibits T-cell proliferation
Because partial YY-1 deficiency was recently shown to inhibit cell proliferation,8 we wondered whether T cells from yy-1+/− deficient mice would proliferate normally. To address this, CFSE-labeled spleen cells from WT and heterozygous OTII mice were stimulated with antigen-loaded APCs, and CD4+ T cell proliferation was analyzed by means of immunofluorescence and flow cytometry. As shown in Fig 5, YY-1 deficiency inhibited antigen-driven T-cell proliferation.
FIG 5.
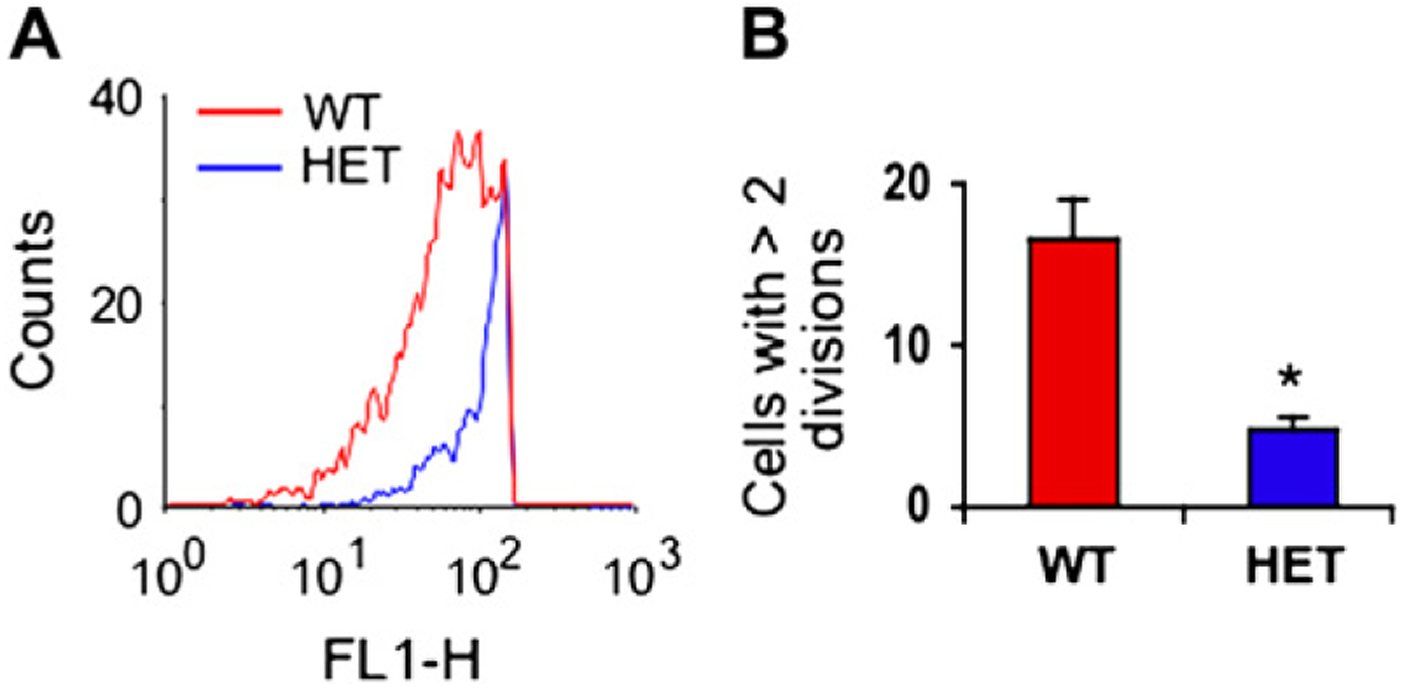
Partial YY-1 deficiency inhibits T-cell proliferation. Proliferation of spleen CD4+ T cells from WT and heterozygous (HET) mice was analyzed by monitoring CFSE dilution by means of immunofluorescence and flow cytometry. A shows representative data from 1 experiment, and B is the average ± SEMs of 3 experiments using antigen-stimulated TCR transgenic T cells from WT (red) and yy-1+/− (HET; blue) mice studied after 72 hours. *P < .05.
DISCUSSION
YY-1 is a pleiotropic transcription factor that regulates the expression of a diverse array of genes. Although YY-1 can bind to the promoter regions of several cytokine genes, the role of this factor as a transcriptional repressor or activator during immune responses has been difficult to determine with certainty. Here we provide evidence that YY-1, in a gene dose-dependent manner, regulates the expression of effector cytokines and allergen-driven TH2-dependent immune responses in vivo. In acutely stimulated CD4+ T cells, partial deficiency of yy-1 attenuates the expression of both IL-4 and IFN-γ. In the case of IL-4, this was especially apparent at later stages of T-cell activation due in part to delayed occupancy of the IL-4 promoter by YY-1. YY-1 deficiency also attenuated IL-5 and IL-13 gene expression, as well as airway inflammation in an allergen-driven TH2-dependent immune response.
Expression of the IL-4 gene in T lymphocytes is tightly regulated at the level of gene transcription by numerous transcription factors that bind to a proximal promoter region, as well as to other regulatory cis elements.30,31 For example, in the promoter region alone, the rate of IL-4 transcription is activated by binding of NFAT,32,33 activator protein 1,34 c-Maf,35 nuclear factor Y,33 and CCAAT enhancer binding protein-β36 to their cognate elements, whereas IL-4 transcription is repressed by binding of signal transducer and activator of transcription 6,37,38 Oct-1,39 and other still poorly defined factors40,41 (for recent review, see Li-Weber et al40). IL-4 gene expression is also controlled at the level of chromatin remodeling, especially during TH2 differentiation.42–45 An open chromatin structure is required for maximal IL-4 gene expression in TH2 cells and appears to be inherited as an epigenetic trait (for review, see Lee et al46). We previously reported that the IL-4 promoter contains 4 YY-1 binding sites that appear to act in an NFAT-independent manner to enhance IL-4 transcription.15 The data presented in the current report support the idea that YY-1 is a positive regulator of IL-4 gene expression, especially at later stages of T-cell activation. YY-1 is intimately involved in the regulation of chromatin structure at different genetic loci,2 and although we cannot rule out the possibility that partial deficiency of yy-1 attenuates IL-4 gene expression by affecting chromatin remodeling, we did not detect major differences in histone H3 or H4 lysine acetylation at the IL-4 promoter when comparing resting and activated yy-1+/+ versus yy-1+/− T cells (data not shown). We currently favor a model in which YY-1 directly regulates IL-4 gene expression by regulating transcriptional activation.
Three lines of evidence support the idea that YY-1 is involved in TCR-dependent cytokine gene expression. First, we noticed significantly reduced IL-4 and IFN-γ production after T-cell activation with anti-CD3 and anti-CD28 antibodies (Fig 1). Second, TCR transgenic T cells secreted significantly less IL-4 and IFN-γ after stimulation with antigen (Fig 2). Third, when we used calcium ionophore plus PMA to bypass receptor-mediated signaling pathways and acutely stimulate cells, differences in IL-4 secretion from WT versus yy-1+/− T cells were attenuated (data not shown). Therefore these results suggest that YY-1 integrates with physiologic signals emanating from the TCR or costimulatory receptors. YY-1 is generally considered to be a constitutively nuclear phosphoprotein, although it is subject to receptor-mediated posttranslational modifications indifferent cell types.47–50 It remains to be determined how T-cell signaling cascades affect YY-1 or its cofactors during T-cell activation. One potential mechanism for reduced IL-4 production in T cells from yy-1+/− mice is due to attenuated cell proliferation. Indeed, we found that yy-1+/− T cells stimulated with antigen or anti-CD3 plus anti-CD28 antibodies proliferated less efficiently compared with their WT counterparts (Fig 5). This was not due to defects in IL-2 secretion, which was equivalent between groups. The relationship between T-cell proliferation and cytokine gene expression is complex. Although T-cell proliferation can influence IL-4 expression probably by affecting chromatin structure at the TH2 locus,51,52 cycle progression is not absolutely required for full effector cytokine gene expression.52–54 Future experiments in which the level of YY-1 can be inducibly regulated in T cells should provide new insights into the role of this factor in T-cell proliferation and activation.
Asthma is a complex inflammatory disease of the airways involving the secretion of numerous gene products from cells of both the innate and adaptive immune systems.55 Interestingly, the promoter regions of several asthma-associated genes contain single nucleotide polymorphisms that generate YY-1 binding consensus sequences, including IL-13,14 IL-10,11 monocyte chemoattractant protein 4,12 and TGF-β1.13 Using a well-established mouse model of allergic airway inflammation, we also found that partial YY-1 deficiency attenuated airway inflammation and a TH2-dependent immune response in vivo. By means of immunohistochemistry, we also found that YY-1 expression was increased in the asthmatic airway. Thus in addition to signal transducer and activator of transcription 6,56 GATA-3,57 and T-bet,58 YY-1 provides another example of a transcription factor involved in allergic immune responses and asthma. Understanding how YY-1 expression is regulated and how it interacts with other transcription factors to reproduce different features of the asthmatic phenotype should prove worthwhile in future studies.
METHODS
Cell isolation and analysis by means of flow cytometry
Thymocytes and spleen and lymph node cells from yy-1+/− heterozygous mice at age 6 to 8 weeks and sex-matched littermate WT control animals were obtained by using standard protocols. Single-cell suspensions were analyzed by means of immunofluorescence and flow cytometry with saturating concentrations of isotype-matched controls and anti-CD4 (no. 553049), CD8 (no. 553031) and B220 (no. 553090, all from BD PharMingen) and NK1.1 (no. 11-5941-82; eBioscience, San Diego, Calf). To monitor TCR transgenic T cells from OTII mice, spleen cells were labeled with anti-CD4 antibodies together with anti-Vα2 (no. 553289) and anti-Vβ5 (no. 553190, BD Pharmingen).
Cytokine ELISA or cytometric bead array
ELISA kits were purchased from eBioscience (IFN-γ) and Biolegend (San Diego, Calif; IL-2 and IL-4) and used according to the manufacturer’s specification. A Bio-Plex mouse cytokine assay (Bio-Rad, Hercules, Calif) was performed by using a 96-well filtration plate and 50-μL aliquots of premixed beads (Fig 2). Samples and controls were read with a Bio-Plex 200 suspension array system, and the data were analyzed with Bio-Plex Manager software with 5PL curve fitting.
ChIP assays
For more information, see Fig 1. Solubilized chromatin was prepared from CD4+ splenic T cells (1 × 106 per antibody condition) isolated from WT and yy-1+/− heterozygous littermates by using an Acetyl H3 Histone ChIP assay kit (Upstate Biotechnologies, Lake Placid, NY), as previously described.E1 After reversion of cross-links at 65°C for 4 hours, the immunoprecipitated DNA was extracted, and PCR was performed with the following primers specific for the murine IL-4 promoter: mIL4p263F sense, tctgaaaggccgattatggtg; antisense, taacaatgcaatgctggcaga.
TH polarization
For more information, see Fig E4. Naive CD62L+CD4+ T cells were isolated from WT and yy-1+/− heterozygous splenocytes by means of positive (anti-CD62L) and negative (anti-CD4) immunomagnetic selection (Miltenyi Biotec). Cells (0.5 × 106 per well in 6-well plates precoated with 2 μg/mL anti-CD3 antibodies) were incubated with recombinant mIL-2 (50 ng/mL) under the following conditions: neutral (no other added cytokines or antibodies), TH1 (5 ng/mL mIL-12 plus 3 mg/mL anti-mIL-4), or TH2 (10 ng/mL mIL-4 plus 3 μg/mL anti-mIFN-γ). All recombinant murine cytokines were purchased from R&D Systems, and anti-cytokine antibodies were from Biolegend. After 7 days, cells were washed 3 times and restimulated with A23187 plus PMA for 18 hours, followed by cytokine secretion by means of ELISA.
PCR and RT-PCR
For moreinformation, see Fig E1. Genomic DNA and total cellular RNA from splenocytes was obtained by using TRIzol reagent (Life Technologies, Grand Island, NY), respectively, as per the manufacturer’s instructions. RNA samples were reverse transcribed, and gene-specific primers (Invitrogen, Carlsbad, Calif) were used to amplify selected regions of each gene. To verify that equal amounts of RNAwere added in each RT-PCR, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a control. Amplified PCR products were detected by using ethidium bromide gel electrophoresis. Primers used for PCR were as follows: neomycin forward, 5′ATGAACTGCAGGACGAGGCAGCG3′; neomycin reverse, 5′GGCGATAGAAGGCGATGCGCTG3′; YY-1 forward, 59TCGCGCTGCAGCCGCTGGTGAC39; YY-1 reverse, 5′CGCCACGGTGA CCAGCGTCTGC3′; IL-4 forward, 5′CACTTGAGAGAGATCATCGGC3′; IL-4 reverse, 5′GAGTCTCTGCAGCTCCATGAG3′; GATA-3 forward, 5′GCC GAGGACATGGAGGTGA3′; GATA-3 reverse, 5′CTAACCCATGGCGGTG ACCATG3′; GAPDH forward, 5′CAATTCAACGGCACAGTCAAG3′; GAP DH reverse, 5′CCTCACCCATTTGATGTTAGTG3′.
Western blotting
For more information, see Fig E1. Whole-cell lysates (30 μg per lane) were separated on 4% to 20% SDS-polyacrylamide gels, electroblotted onto nitro-cellulose membranes (Hybond-C extra; Amersham Pharmacia Biotech, Little Chalfont, Bucks, United Kingdom), blocked for 1 hour at room temperature in blocking buffer (1× PBS, 0.1% Tween 20, and 5% [wt/vol] Carnation non-fat dry milk), and washed 3 times for 10 minutes each in washing buffer PBST (PBS and 0.1% Tween 20). Blots were incubated with a 1:1000 dilution of primary antibody in PBST buffer overnight at 4°C with anti-YY-1 (H-10, Santa Cruz Biotechnology) or anti-GAPDH antibodies (AB8246, Abcam, Cambridge, Mass) followed by washing in PBST buffer at room temperature. The secondary antibody, (anti-mouse IgG-horseradish peroxidase, NA931V, Amersham Pharmacia Biotech, and anti-goat IgG horseradish peroxidase, R&D Systems) were diluted 1:10,000 in PBST buffer, incubated for 30 minutes at room temperature, and then washed 3 times in PBST buffer. Secondary antibody–horseradish peroxidase was detected by means of chemiluminescence (Western Blotting Luminol Reagent sc-2048, Santa Cruz Biotechnology).
Quantitative RT-PCR targeted array
For more information, see Fig E2. CD4+ T cells from 6-to 9-week-old WT and yy-1+/− mice (n = 3 each) were incubated with immobilized anti-CD3 on magnetic beads (2 μg/mL) and soluble anti-CD28 (2 μg/mL) for 48 hours, and total RNAwas isolated with RNA Easy kits (Qiagen, Valencia, Calif). RT-PCR was performed with a FirstStrand cDNA Synthesis Kit (SuperArray, Gaithersburg, Md), and quantitative gene expression was analyzed with a 96-well RT- PCR array (Mouse TH1/TH2/TH3 Pathway Array, SuperArray) and an IQ5 thermocycler (Bio-Rad). Relative gene expression was calculated by using the 2−ΔΔCt method, in which Ct indicates the fractional cycle number at which the fluorescent signal reaches the detection threshold. The ΔΔ method uses the normalized ΔCt value of each sample calculated by using a total of 5 endogenous control genes (18S rRNA, HPRT1, RPL13A, GAPDH, and ACTB). Values are presented as an average fold change of 2−(average ΔΔCt) for genes in CD4+ T cells from yy-1+/− versus WT mice.
Supplementary Material
1
Acknowledgments
We thank Dr Mark Boothby for helpful suggestions and advice, Dr Vincenzo Casolaro for guidance, and Jessica Roman and Krystal Matthews for expert technical assistance.
Supported by research grants from the National Institutes of Health/National Heart, Lung, and Blood Institute (R01HL073952) and the American Lung Association.
Abbreviations used
APC
Antigen-presenting cell
BAL
Bronchoalveolar lavage
CFSE
5, 6-Carboxyfluorescein diacetate succinimidyl ester
ChIP
Chromatin immunoprecipitation
GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
NFAT
Nuclear factor of activated T cells
OVA
Ovalbumin
PMA
Phorbol 12-myristate 13-acetate
TCR
T-cell receptor
WT
Wild-type
YY-1
Yin-Yang 1
Footnotes
Disclosure of potential conflict of interest: The authors have declared that they have no conflict of interest.
Clinical implications: YY-1 can contribute to allergic immune responses by regulating T-cell expansion and cytokine gene expression.
REFERENCES
- 1.Shi Y, Lee JS, Galvin KM. Everything you have ever wanted to know about Yin Yang 1. Biochim Biophys Acta 1997;1332:F49–66. [DOI] [PubMed] [Google Scholar]
- 2.Thomas MJ, Seto E. Unlocking the mechanisms of transcription factor YY1: are chromatin modifying enzymes the key? Gene 1999;236:197–208. [DOI] [PubMed] [Google Scholar]
- 3.Kim JD, Hinz AK, Bergmann A, Huang JM, Ovcharenko I, Stubbs L, et al. Identification of clustered YY1 binding sites in imprinting control regions. Genome Res 2006;16:901–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, Puigserver P. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature 2007;450:736–40. [DOI] [PubMed] [Google Scholar]
- 5.Gordon S, Akopyan G, Garban H, Bonavida B. Transcription factor YY1: structure, function, and therapeutic implications in cancer biology. Oncogene 2006;25:1125–42. [DOI] [PubMed] [Google Scholar]
- 6.Wilkinson FH, Park K, Atchison ML. Polycomb recruitment to DNA in vivo by the YY1 REPO domain. Proc Natl Acad Sci U S A 2006;103:19296–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai Y, Jin J, Yao T, Gottschalk AJ, Swanson SK, Wu S, et al. YY1 functions with INO80 to activate transcription. Nat Struct Mol Biol 2007;14:872–4. [DOI] [PubMed] [Google Scholar]
- 8.Sui G, Affar el B, Shi Y, Brignone C, Wall NR, Yin P, et al. Yin Yang 1 is a negative regulator of p53. Cell 2004;117:859–72. [DOI] [PubMed] [Google Scholar]
- 9.He Y, Dupree J, Wang J, Sandoval J, Li J, Liu H, et al. The transcription factor Yin Yang 1 is essential for oligodendrocyte progenitor differentiation. Neuron 2007;55:217–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu H, Schmidt-Supprian M, Shi Y, Hobeika E, Barteneva N, Jumaa H, et al. Yin Yang 1 is a critical regulator of B-cell development. Genes Dev 2007;21:1179–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hobbs K, Negri J, Klinnert M, Rosenwasser LJ, Borish L. Interleukin-10 and transforming growth factor-beta promoter polymorphisms in allergies and asthma. Am J Respir Crit Care Med 1998;158:1958–62. [DOI] [PubMed] [Google Scholar]
- 12.Kalayci O, Birben E, Wu L, Oguma T, Storm Van’s Gravesande K, Subramaniam V, et al. Monocyte chemoattractant protein-4 core promoter genetic variants: influence on YY-1 affinity and plasma levels. Am J Respir Cell Mol Biol 2003;29:750–6. [DOI] [PubMed] [Google Scholar]
- 13.Silverman ES, Palmer LJ, Subramaniam V, Hallock A, Mathew S, Vallone J, et al. Transforming growth factor-beta1 promoter polymorphism C-509T is associated with asthma. Am J Respir Crit Care Med 2004;169:214–9. [DOI] [PubMed] [Google Scholar]
- 14.Cameron L, Webster RB, Strempel JM, Kiesler P, Kabesch M, Ramachandran H, et al. Th2 cell-selective enhancement of human IL13 transcription by IL13–1112C>T, a polymorphism associated with allergic inflammation. J Immunol 2006;177:8633–42. [DOI] [PubMed] [Google Scholar]
- 15.Guo J, Casolaro V, Seto E, Yang WM, Chang C, Seminario MC, et al. Yin-Yang 1 activates interleukin-4 gene expression in T cells. J Biol Chem 2001;276:48871–8. [DOI] [PubMed] [Google Scholar]
- 16.Ye J, Ghosh P, Cippitelli M, Subleski J, Hardy KJ, Ortaldo JR, et al. Characterization of a silencer regulatory element in the human interferon-gamma promoter. J Biol Chem 1994;269:25728–34. [PubMed] [Google Scholar]
- 17.Ye J, Cippitelli M, Dorman L, Ortaldo J, Young H. The nuclear factor YY1 suppresses the human gamma interferon promoter through two mechanisms: inhibition of AP1 binding and activation of a silencer element. Mol Cell Biol 1996;16:4744–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mordvinov VA, Schwenger GT, Fournier R, De Boer ML, Peroni SE, Singh AD, et al. Binding of YY1 and Oct1 to a novel element that downregulates expression of IL-5 in human T cells. J Allergy Clin Immunol 1999;103:1125–35. [DOI] [PubMed] [Google Scholar]
- 19.Schwenger GT, Fournier R, Hall LM, Sanderson CJ, Mordvinov VA. Nuclear factor of activated T cells and YY1 combine to repress IL-5 expression in a human T-cell line. J Allergy Clin Immunol 1999;104:820–7. [DOI] [PubMed] [Google Scholar]
- 20.Soutto M, Zhang F, Enerson B, Tong Y, Boothby M, Aune TM. A minimal IFN-gamma promoter confers Th1 selective expression. J Immunol 2002;169:4205–12. [DOI] [PubMed] [Google Scholar]
- 21.Sweetser MT, Hoey T, Sun YL, Weaver WM, Price GA, Wilson CB. The roles of nuclear factor of activated T cells and yin-yang 1 in activation-induced expression of the interferon-gamma promoter in T cells. J Biol Chem 1998;273:34775–83. [DOI] [PubMed] [Google Scholar]
- 22.Donohoe ME, Zhang X, McGinnis L, Biggers J, Li E, Shi Y. Targeted disruption of mouse Yin Yang 1 transcription factor results in peri-implantation lethality. Mol Cell Biol 1999;19:7237–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lorente M, Perez C, Sanchez C, Donohoe M, Shi Y, Vidal M. Homeotic transformations of the axial skeleton of YY1 mutant mice and genetic interaction with the Polycomb group gene Ring1/Ring1A. Mech Dev 2006;123:312–20. [DOI] [PubMed] [Google Scholar]
- 24.Affar el B, Gay F, Shi Y, Liu H, Huarte M, Wu S, et al. Essential dosage-dependent functions of the transcription factor yin yang 1 in late embryonic development and cell cycle progression. Mol Cell Biol 2006;26:3565–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barnden MJ, Allison J, Heath WR, Carbone FR. Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunol Cell Biol 1998;76:34–40. [DOI] [PubMed] [Google Scholar]
- 26.National Asthma Education and Prevention Program. Expert Panel Report: guidelines for the diagnosis and management of asthma. Bethesda (MD): National Institutes of Health; 1997. [Google Scholar]
- 27.American Thoracic Society. Guidelines for fiberoptic bronchoscopy in adults. Medical Section of the American Lung Association. Am Rev Respir Dis 1987;136:1066. [DOI] [PubMed] [Google Scholar]
- 28.Bergeron C, Boulet LP, Page N, Laviolette M, Zimmermann N, Rothenberg ME, et al. Influence of cigarette smoke on the arginine pathway in asthmatic airways: increased expression of arginase I. J Allergy Clin Immunol 2007;119: 391–7. [DOI] [PubMed] [Google Scholar]
- 29.Djukanovic R, Roche W, Wilson J, Beasley C, Twentyman O, Howarth P, et al. Mucosal inflammation in asthma. Am Rev Respir Dis 1990;142:434–57. [DOI] [PubMed] [Google Scholar]
- 30.Li B, Tournier C, Davis RJ, Flavell RA. Regulation of IL-4 expression by the transcription factor JunB during T helper cell differentiation. EMBO J 1999;18:420–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ho IC, Glimcher LH. Transcription: tantalizing times for T cells. Cell 2002; 109(suppl):S109–20. [DOI] [PubMed] [Google Scholar]
- 32.Bruhn K, Nelms K, Boulay J, Paul W, Lenardo M. Molecular dissection of the murine interleukin-4 promoter. Proc Natl Acad Sci U S A 1993;90:9707–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szabo SJ, Jacobson NG, Dighe AS, Gubler U, Murphy KM. Developmental commitment to the Th2 lineage by extinction of IL-12 signaling. Immunity 1995;2:665–75. [DOI] [PubMed] [Google Scholar]
- 34.Rooney J, Hodge M, McCaffrey P, Rao A, Glimcher L. A common factor regulates both Th1- and Th2-specific cytokine gene expression. EMBO J 1994;13:625–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ho AM, Jain J, Rao A, Hogan PG. Expression of the transcription factor NFATp in a neuronal cell line and in the murine nervous system. J Biol Chem 1994;269:28181–6. [PubMed] [Google Scholar]
- 36.Davydov I, Krammer P, Li-Weber M. Nuclear factor-IL-6 activates the human IL-4 promoter in T cells. J Immunol 1995;155:5273–9. [PubMed] [Google Scholar]
- 37.Georas SN. Inhaled glucocorticoids, lymphocytes, and dendritic cells in asthma and obstructive lung diseases. Proc Am Thorac Soc 2004;1:215–21. [DOI] [PubMed] [Google Scholar]
- 38.Dorado B, Jerez MJ, Flores N, Martin-Saavedra FM, Duran C, Ballester S. Autocrine IL-4 gene regulation at late phases of TCR activation in differentiated Th2 cells. J Immunol 2002;169:3030–7. [DOI] [PubMed] [Google Scholar]
- 39.Pfeuffer I, Klein-Hebling S, Heinfling A, Chuvpilo S, Escher C, Brabletz T, et al. Octamer factors exert a dual effect on the IL-2 and IL-4 promoters. J Immunol 1994;153:5572–85. [PubMed] [Google Scholar]
- 40.Li-Weber M, Davydov I, Krafft H, Krammer P. The role of NF-Y and IRF-2 in the regulation of human IL-4 gene expression. J Immunol 1994;153:4122–33. [PubMed] [Google Scholar]
- 41.Georas S, Cumberland J, Burke T, Chen R, Park E, Ono S, et al. Characterization of a novel repressor element in the interleukin 4 promoter. Leukemia 2000;14:629–35. [DOI] [PubMed] [Google Scholar]
- 42.Agarwal S, Viola JP, Rao A. Chromatin-based regulatory mechanisms governing cytokine gene transcription. J Allergy Clin Immunol 1999;103:990–9. [DOI] [PubMed] [Google Scholar]
- 43.Valapour M, Guo J, Schroeder JT, Keen J, Cianferoni A, Casolaro V, et al. Histone deacetylation inhibits IL4 gene expression in T cells. J Allergy Clin Immunol 2002; 109:238–45. [DOI] [PubMed] [Google Scholar]
- 44.Fields PE, Lee GR, Kim ST, Bartsevich VV, Flavell RA. Th2-specific chromatin remodeling and enhancer activity in the Th2 cytokine locus control region. Immunity 2004;21:865–76. [DOI] [PubMed] [Google Scholar]
- 45.Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. Interchromosomal associations between alternatively expressed loci. Nature 2005;435:637–45. [DOI] [PubMed] [Google Scholar]
- 46.Lee GR, Kim ST, Spilianakis CG, Fields PE, Flavell RA. T helper cell differentiation: regulation by cis elements and epigenetics. Immunity 2006;24:369–79. [DOI] [PubMed] [Google Scholar]
- 47.Garban HJ, Bonavida B. Nitric oxide inhibits the transcription repressor Yin-Yang 1 binding activity at the silencer region of the Fas promoter: a pivotal role for nitric oxide in the up-regulation of Fas gene expression in human tumor cells. J Immunol 2001;167:75–81. [DOI] [PubMed] [Google Scholar]
- 48.Santiago FS, Lowe HC, Bobryshev YV, Khachigian LM. Induction of the transcriptional repressor Yin Yang-1 by vascular cell injury. Autocrine/paracrine role of endogenous fibroblast growth factor-2. J Biol Chem 2001;276:41143–9. [DOI] [PubMed] [Google Scholar]
- 49.Yao YL, Yang WM, Seto E. Regulation of transcription factor YY1 by acetylation and deacetylation. Mol Cell Biol 2001;21:5979–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hiromura M, Choi CH, Sabourin NA, Jones H, Bachvarov D, Usheva A. YY1 is regulated by O-linked N-acetylglucosaminylation (O-glcNAcylation). J Biol Chem 2003;278:14046–52. [DOI] [PubMed] [Google Scholar]
- 51.Bird J, Brown D, Mullen A, Moskowitz N, Mahowald M, Sider J, et al. Helper T cell differentiation is controlled by the cell cycle. Immunity 1998;9:229–37. [DOI] [PubMed] [Google Scholar]
- 52.Richter A, Lohning M, Radbruch A. Instruction for cytokine expression in T helper lymphocytes in relation to proliferation and cell cycle progression. J Exp Med 1999;190:1439–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Laouar Y, Crispe IN. Functional flexibility in T cells: independent regulation of CD4+ T cell proliferation and effector function in vivo. Immunity 2000;13:291–301. [DOI] [PubMed] [Google Scholar]
- 54.Ben-Sasson SZ, Gerstel R, Hu-Li J, Paul WE. Cell division is not a “clock” measuring acquisition of competence to produce IFN-gamma or IL-4. J Immunol 2001;166:112–20. [DOI] [PubMed] [Google Scholar]
- 55.Cohn L, Elias JA, Chupp GL. Asthma: mechanisms of disease persistence and progression. Annu Rev Immunol 2004;22:789–815. [DOI] [PubMed] [Google Scholar]
- 56.Kuperman D, Schofield B, Wills-Karp M, Grusby MJ. Signal transducer and activator of transcription factor 6 (Stat6)-deficient mice are protected from antigen-induced airway hyperresponsiveness and mucus production. J Exp Med 1998;187:939–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang DH, Yang L, Cohn L, Parkyn L, Homer R, Ray P, et al. Inhibition of allergic inflammation in a murine model of asthma by expression of a dominant-negative mutant of GATA-3. Immunity 1999;11:473–82. [DOI] [PubMed] [Google Scholar]
- 58.Finotto S, De Sanctis GT, Lehr HA, Herz U, Buerke M, Schipp M, et al. Treatment of allergic airway inflammation and hyperresponsiveness by antisense-induced local blockade of GATA-3 expression. J Exp Med 2001;193:1247–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E1.Valapour M, Guo J, Schroeder JT, Keen J, Cianferoni A, Casolaro V, et al. Histone deacetylation inhibits IL4 gene expression in T cells. J Allergy Clin Immunol 2002;109:238–45. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1