Role of Interferon in the Replication of Human Parainfluenza Virus Type 1 Wild Type and Mutant Viruses in Human Ciliated Airway Epithelium (original) (raw)
Abstract
Human parainfluenza virus type 1 (HPIV1) is a significant cause of pediatric respiratory disease in the upper and lower airways. An in vitro model of human ciliated airway epithelium (HAE), a useful tool for studying respiratory virus-host interactions, was used in this study to show that HPIV1 selectively infects ciliated cells within the HAE and that progeny virus is released from the apical surface with little apparent gross cytopathology. In HAE, type I interferon (IFN) is induced following infection with an HPIV1 mutant expressing defective C proteins with an F170S amino acid substitution, rHPIV1-CF170S, but not following infection with wild-type HPIV1. IFN induction coincided with a 100- to 1,000-fold reduction in virus titer, supporting the hypothesis that the HPIV1 C proteins are critical for the inhibition of the innate immune response. Two recently characterized live attenuated HPIV1 vaccine candidates expressing mutant C proteins were also evaluated in HAE. The vaccine candidates, rHPIV1-CR84G/Δ170HNT553ALY942A and rHPIV1-CR84G/Δ170HNT553ALΔ1710-11, which contain temperature-sensitive (ts) attenuating (att) and non-ts att mutations, were highly restricted in growth in HAE at permissive (32°C) and restrictive (37°C) temperatures. The viruses grew slightly better at 37°C than at 32°C, and rHPIV1-CR84G/Δ170HNT553ALY942A was less attenuated than rHPIV1-CR84G/Δ170HNT553ALΔ1710-11. The level of replication in HAE correlated with that previously observed for African green monkeys, suggesting that the HAE model has potential as a tool for the preclinical evaluation of HPIV1 vaccines, although how these in vitro data will correlate with vaccine virus replication in seronegative human subjects remains to be seen.
Human parainfluenza viruses (HPIVs) are enveloped, nonsegmented, single-stranded, negative-sense RNA viruses belonging to the family Paramyxoviridae. This group of viruses includes HPIV serotype 1 (HPIV1), HPIV2, and HPIV3, which collectively are the second leading cause of pediatric respiratory hospitalizations following respiratory syncytial virus (RSV) infections (25, 36). HPIV1 is responsible for approximately 6% of pediatric hospitalizations due to respiratory tract disease (25). Clinical manifestations range from mild disease, including rhinitis, pharyngitis, and otitis media, to more severe disease, including croup, bronchiolitis, and pneumonia (11, 22, 23, 25, 32, 46). Currently, licensed vaccines for the prevention of disease caused by any HPIV serotype are not available.
The HPIV1 genome is 15,600 nucleotides in length and contains six genes in the order 3′-N-P/C-M-F-HN-L-5′ (40). Each gene encodes a single protein, with the exception of the P/C gene, which encodes the phosphoprotein, P, in one open reading frame (ORF) and up to four accessory C proteins, C′, C, Y1, and Y2, in a second ORF. The C proteins initiate at four separate translational start codons in the C ORF in the order C′, C, Y1, and Y2 and are carboxy coterminal (25), although it is unclear whether the Y2 protein is actually expressed during HPIV1 infection (45). C proteins are expressed by viruses of the genera Respirovirus, Morbillivirus, and Henipavirus but not by viruses of the genera Rubulavirus and Avulavirus. The C proteins of Sendai virus (SeV), a member of the Respirovirus genus and the closest homolog of HPIV1, are perhaps the most extensively studied and have been shown to have multiple functions, including the inhibition of the host innate immune response by acting as interferon (IFN) antagonists (16, 20, 26).
To date, the HPIV1 C proteins have not been extensively studied, although recent studies provide evidence for a role for these proteins in the evasion of the host innate immune response. In A549 cells, a human lung adenocarcinoma epithelial cell line, it was previously shown that type I IFN production was not detected during infection with wild-type HPIV1 (HPIV1 wt) (59). Since type I IFN was induced during infection of A549 cells with a recombinant HPIV1 (rHPIV1) mutant with C proteins bearing an F170S amino acid substitution (rHPIV1-CF170S), a role for the C proteins as antagonists of the type I IFN response was suggested (59). This function was demonstrated to affect the innate immune response at the level of type I IFN induction and IFN signaling (59). Another study independently confirmed the role of the HPIV1 C proteins as antagonists of the type I IFN response, demonstrating that HPIV1 infection could inhibit STAT1 nuclear translocation and overcome an established IFN-induced antiviral state in MRC-5 human lung fibroblast cells and, furthermore, that HPIV1 C-protein expression was sufficient to inhibit STAT1 nuclear translocation in A549 cells (8). In contrast to the first study, the latter study demonstrated that type I IFN was induced during infection of MRC-5 cells with HPIV1 wt (8), which suggests that the inhibition of type I IFN induction is cell type specific. Therefore, further studies are required to better define the host IFN response in relevant cell types that are infected during HPIV1 infection in humans. This is particularly important because vaccine candidates with defective C proteins are being prepared for clinical trials, as described below.
In vitro models of the human airway epithelium (HAE) that closely mimic the morphological and physiological features of the HAE in vivo are now well characterized (43, 67). These models use freshly isolated human airway cells grown at an air-liquid interface to generate a differentiated, pseudostratified, mucociliary epithelium that bears close structural and functional similarity to HAE in vivo. Such models were previously used to demonstrate that paramyxoviruses such as RSV and HPIV3 preferentially infect ciliated cells, suggesting that these cells play a critical role in paramyxovirus replication and pathogenesis in the respiratory tract (66, 67). In addition, HAE models have been used to evaluate the attenuation of RSV vaccines (61).
In the present study, we attempted to validate the in vitro HAE model for HPIV1 infection. In order to do this, we first evaluated the levels of replication, cell tropism, and gross pathogenic effects of HPIV1 wt infection in HAE in an experimental setting (including infection with a low multiplicity of infection [MOI] and multicycle growth) that mimics virus infection of the lower conducting cartilaginous airways of humans. Second, we evaluated the abilities of HPIV1 wt and rHPIV1-CF170S mutant viruses to induce a type 1 IFN response and examined the role of the induced IFN in restricting the replication of HPIV1 in HAE cultures. In this way, the phenotypes previously associated with HPIV1 C mutants in cell culture and in vivo in African green monkeys (AGMs) were characterized in HAE cultures. These data suggested that HPIV1 replication in the HAE model is predictive of HPIV1 infection and growth in vivo. Therefore, we next compared the levels of attenuation of replication of two HPIV1 vaccine candidates in the airways of AGMs (2, 3, 34, 39) to that seen in the HAE model. This comparison revealed that the levels of replication of the two vaccine candidates are similar in HAE cells and in AGMs and, in addition, unexpectedly revealed the ability of the HAE system to detect an attenuating effect of mutations in genes such as L that are not revealed by other in vitro cell culture systems. The ability of the HAE model to predict the attenuation of vaccine viruses for humans will be further analyzed once clinical studies of humans are completed, and the levels of replication of the vaccine candidates in AGMs, in HAE cells, and in humans can be compared. Clinical studies for one of the vaccine candidates, rHPIV1-CR84G/Δ170 HNT553A LY942A, are currently in progress. The current findings suggest that HAE cultures are a useful model system for the preclinical evaluation of live attenuated HPIV vaccines.
MATERIALS AND METHODS
Cells and viruses.
Human airway tracheobronchial epithelial cells were isolated from airway specimens from patients without underlying lung disease, provided by the National Disease Research Interchange (Philadelphia, PA), or as excess tissue following lung transplantation under University of North Carolina at Chapel Hill (UNC) Institutional Review Board-approved protocols by the UNC Cystic Fibrosis Center Tissue Culture Core. Primary cells derived from single-patient sources were expanded on plastic to generate passage 1 cells and plated at a density of 3 × 105 cells per well on permeable Transwell-Col (12-mm-diameter) or 2 × 105 cells per well on permeable Millicell (12-mm-diameter) supports. HAE cultures were grown in custom medium with the provision of an air-liquid interface for 4 to 6 weeks to form differentiated, polarized cultures that resemble in vivo pseudostratified mucociliary epithelium, as previously described (43). LLC-MK2 cells (ATCC CCL 7.1) and HEp-2 cells (ATCC CCL 23) were maintained in Opti-MEM I (Gibco-Invitrogen, Inc., Grand Island, NY) supplemented with 5% fetal bovine serum and gentamicin sulfate (50 μg/ml). Vero cells (ATCC CCL-81) were maintained in Opti-PRO SFM (Gibco-Invitrogen, Inc.) supplemented with 50 μg/ml gentamicin sulfate and 4 mM l-glutamine. Media used for HPIV1 propagation and infection in LLC-MK2 cells contained 1.2% TrypLESelect, a recombinant trypsin (Gibco-Invitrogen, Inc.), without fetal bovine serum, in order to activate the HPIV1 F protein.
Biologically derived HPIV1 wt strain Washington/20993/1964, the parent for the recombinant virus, was isolated previously from a clinical sample in primary AGM kidney cells and passaged two additional times in primary AGM kidney cells (37) and once in LLC-MK2 cells (3). This preparation has a wild-type phenotype in AGMs, was previously designated HPIV1LLC1 (3), and will be referred to here as HPIV1 wt or its recombinant version, rHPIV1 wt. The construction of the rHPIV1 mutants rHPIV1-CF170S, rHPIV1-CR84G/Δ170HNT553ALY942A, and rHPIV1-CR84G/Δ170HNT553ALΔ1710-11 was described previously (2, 4). Each of the mutant viruses is named according to the mutations that it contains: CF170S refers to the indicated amino acid substitution in the C protein and confers a non-temperature-sensitive (ts) attenuation (att) phenotype; CΔ170 refers to a six-nucleotide deletion spanning codon 170 in C and confers a non-ts att phenotype; CR84G and HNT553A refer to amino acid substitutions in C and HN that, in combination, confer a non-ts att phenotype but individually have no attenuation phenotype; LY942A refers to the indicated amino substitution in L and confers a ts att phenotype; and LΔ1710-11 has the deletion of the indicated residues in L and confers a ts att phenotype. The CF170S mutation is silent in the overlapping P protein. The rHPIV1-CF170S mutant tested both here and in previous studies contains the nonattenuating HNT553A mutation (2, 59). Since previous studies referred to this virus simply as rHPIV1-CF170S (2, 59), we will employ the same nomenclature here for the purpose of comparison. Purified virus stocks were obtained by infecting LLC-MK2 cells and purifying the supernatant by centrifugation and banding in discontinuous 30% to 60% (wt/vol) sucrose gradients, steps designed to minimize contamination by cellular factors, especially IFN. Purification also removes exogenous trypsin from the virus preparation; however, since the viruses were prepared in trypsin medium, it is likely that the F proteins of the inoculum virus were cleaved. The vesicular stomatitis virus (VSV) used was a recombinant VSV-green fluorescent protein (GFP), originally obtained from John Hiscott (55). Stocks of VSV were propagated in Vero cells and sucrose purified, as indicated above.
Virus titers in samples were determined by 10-fold serial dilution of virus in 96-well LLC-MK2 monolayer cultures using 2 to 4 wells per dilution. After 7 days at 32°C, infected cultures were detected by hemadsorption with guinea pig erythrocytes, as described previously (52). Virus titers are expressed as log10 50% tissue culture infectious doses (TCID50) per ml. VSV stock titers were determined by plaque assay on Vero cells under 0.8% methyl cellulose overlay.
Viral inoculation of HAE.
HAE cultures were washed with phosphate-buffered saline (PBS) to remove apical surface secretions, and fresh medium was supplied to the basolateral compartments prior to infection. HPIV1s were applied to the apical surface of HAE for inoculation at a low-input MOI (0.01 TCID50/cell) or a high MOI (5.0 TCID50/cell), and VSV was applied to the basolateral surface at an MOI of 4.2 PFU/cell in a 100-μl inoculum. The inoculum was removed 2 h postinoculation at either 32°C or 37°C. The cells were then washed once for 5 min with PBS and incubated at 32°C or 37°C, as indicated. Samples were harvested from the apical or basolateral surfaces of HAE for the determination of virus titer or the amount of type I IFN produced. Apical samples were collected by incubating the apical surface with 300 μl of medium for 30 min at 32°C or 37°C, after which the remaining fluid was recovered. Basolateral samples were collected directly from the basolateral compartment, and the volume removed was replaced with fresh medium. Samples were stored at −80°C prior to analysis.
Histology and immunostaining of paraffin-embedded sections.
HAE cultures were fixed in 4% paraformaldehyde overnight and embedded in paraffin, and 5-μm histological sections were prepared. Sections were then either stained with hematoxylin and eosin for analysis by light microscopy or subjected to standard immunofluorescence protocols. Briefly, sections were blocked with 3% bovine serum albumin (BSA) in PBS++ (containing 1 mM CaCl2 and 1 mM MgCl2) and incubated with primary antibodies diluted in 1% BSA. Primary antibodies included a 1:4,000 dilution of rabbit anti-HPIV1 obtained from fluid present in subcutaneous chambers of rabbits immunized with purified HPIV1 (hemagglutination inhibition titer of 1:2,048), as described previously (9), and mouse anti-acetylated alpha-tubulin (1:2,000; Zymed, San Francisco, CA). Secondary antibodies used were fluorescein isothiocyanate-conjugated goat anti-rabbit immunoglobulin G (IgG) (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA) and Texas Red-conjugated goat anti-mouse IgG (Jackson ImmunoResearch Laboratories, Inc.). After a final wash, cells were overlaid with VectaShield mounting medium (Vector Laboratories, Inc., Burlingame, CA). Images were acquired using a Leica DMIRB inverted fluorescence microscope equipped with a cooled color charge-coupled-device digital camera (MicroPublisher; Q-Imaging, Burnaby, British Columbia, Canada).
En face staining and confocal microscopy.
HAE cultures were fixed overnight in 4% paraformaldehyde and permeabilized with 2.5% Triton X-100. The cultures were then blocked with 3% BSA-PBS++, and apical surfaces were incubated with primary antibodies diluted in 1% BSA. An additional primary antiserum used for en face staining was a rabbit anti-HPIV1 polyclonal antiserum obtained by vaccinating whiffle ball-implanted rabbits. The primary rabbit anti-HPIV1 serum was used at a 1:100 dilution, and the mouse anti-acetylated alpha-tubulin antibodies were used at a dilution of 1:500. Secondary antibodies were fluorescein isothiocyanate-conjugated goat anti-rabbit IgG and Alexa Fluor 594 goat anti-mouse IgG (Molecular Probes). Fluorescent confocal xy optical sections were obtained using a Zeiss 510 Meta laser scanning confocal microscope.
Type I IFN bioassay.
The amount of type I IFN produced by infected HAE was determined by an IFN bioassay according to previously published methods (42). Briefly, clarified cell culture medium supernatants were treated at pH 2.0 for 24 h at 4°C to inactivate virus and acid-labile type II IFN, and the pH was adjusted to 7.0 by the addition of 2 M NaOH. Type I IFN concentrations were determined by measuring the restriction of replication of VSV-GFP on HEp-2 cell monolayers in comparison to a known concentration of a human IFN-β standard (Avonex; Biogen, Inc., Cambridge, MA). IFN-β standard (5,000 pg/ml) and IFN-containing samples were serially diluted 10-fold in duplicate in 96-well plates of HEp-2 cells. After 24 h, the cells were washed and infected with VSV-GFP at 6.5 × 104 PFU/well. Control cultures (no VSV-GFP, no IFN; VSV-GFP, no IFN) were performed in quadruplicate on each plate. After an additional 24 h, plates were read for total GFP expression on a Typhoon phosphorimager using a Typhoon 8600 scanner (Molecular Dynamics Inc., Sunnyvale, CA) control program (fluorescence settings and 526-SP green fluorescein filter). The dilution at which the level of GFP expression was approximately 50% of that in untreated cultures was determined as the end point. The end point of the Avonex standard was compared to the end point of the unknown samples, and IFN concentrations were determined and expressed as means ± standard errors (pg/ml). According to the manufacturers, using the World Health Organization natural IFN-β standard, the Avonex IFN-β has a specific activity of approximately 1 IU of antiviral activity per 5 pg.
IFN mRNA qRT-PCR.
The levels of IFN-α and IFN-β mRNA in HAE cultures infected with HPIV1 wt or rHPIV1 mutants relative to those of mock-infected cultures were determined by quantitative reverse transcription (qRT)-PCR, as previously described (53). Briefly, total intracellular RNA was extracted from cell cultures using the RNeasy total RNA isolation kit (Qiagen, Valencia, CA). RNA was reverse transcribed using an oligo(dT) primer and reagents from the Brilliant qRT-PCR kit (Stratagene, La Jolla, CA). The PCR primers and Taqman probes used to detect human IFN-β, two specific sets of human IFN-α, and β-actin were described previously (53). IFN-α primer set 1 was specific for human IFN-α1, IFN-α6, and IFN-α13, and IFN-α set 2 was specific for human IFN-α4, IFN-α5, IFN-α8, IFN-α10, IFN-α14, IFN-α17, and IFN-α21. Duplex quantitative PCRs were performed to allow comparisons between IFN and the housekeeping gene β-actin. The probe for β-actin was labeled with the reporter dye 5-carboxyfluorescein at the 5′ end, and all IFN probes were labeled with the reporter dye 5′-hexachloro-6-carboxy-fluorescein at the 5′ end and Black Hole Quencher 1 at the 3′ end. Reaction mixtures contained a passive reference dye, which was not involved in the amplification of IFN or β-actin but was used to normalize the probe reporter signals. Positive control curves were generated using preparations known to contain high levels of IFN cDNA to ensure reaction efficiency. Each reaction signal was corrected individually for the β-actin signal. In addition, signals from all reaction mixtures from virus-infected samples were corrected against the signal generated in a mock-infected well, resulting in a β-actin-corrected measurement of expression over mock.
RESULTS
Characterization of rHPIV1 wt replication in HAE cultures.
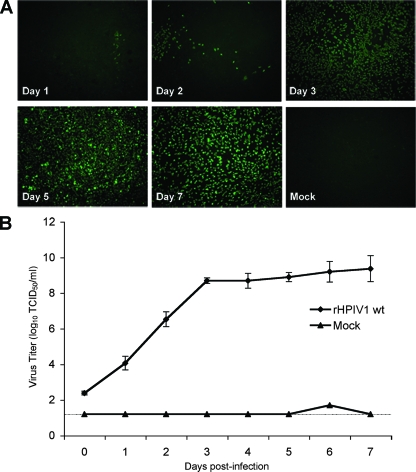
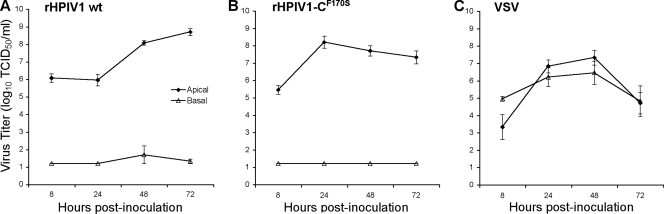
It was previously shown that infection of HAE with RSV and HPIV3 was specific to the ciliated cells of the surface epithelium and was not associated with overt cytotoxicity, whereas infection with influenza A virus resulted in the complete destruction of HAE cultures within 48 h (61, 67). In the present paper, we characterized both the ability of HPIV1 wt to infect HAE and the response to infection in these cultures. rHPIV1 wt efficiently infected HAE cells following apical inoculation with rHPIV1 wt at a low MOI (0.01 TCID50/cell) (Fig. 1). HPIV1 could be detected in HAE cells by en face immunostaining for HPIV1 antigen (Fig. 1A), and the increase in apical wash titers from day 0 to day 1 postinfection (p.i.) provided evidence of active replication and secretion of rHPIV1 wt (Fig. 1). By day 3 p.i., virus had efficiently spread throughout the culture, with significant numbers of cells staining positive for HPIV1 through day 7 p.i. (Fig. 1A). Virus titers correlated with the numbers of cells staining positive for viral antigen in the en face immunostaining (Fig. 1). rHPIV1 also replicated efficiently in a single-step growth curve following apical inoculation at a high-input MOI (5.0 TCID50/cell) (Fig. 2A). These growth curves were performed in the absence of added trypsin (Fig. 1 and 2). Since HPIV1 typically requires added trypsin for cleavage and infectivity when grown in cell lines such as Vero cells, this suggests that a trypsin-like enzyme capable of cleaving HPIV1 F is provided by HAE cultures. Influenza virus, another virus requiring serine protease activity at the apical surface for multicycle replication, also spread efficiently in HAE models in the absence of exogenous trypsin (33, 58). Attempts to isolate the required proteases that are responsible for cleaving viral proteins present in such models have identified some of these proteases (7), but there are most likely many proteases present in the HAE. Viral titers during infection were determined for both the apical and basolateral compartments, representing virus shed into the airway lumen and the serosal side of the epithelium, respectively. In general, viruses causing disease limited to the respiratory tract release virus via the apical surface only, whereas viruses released from both the apical and basolateral surfaces are typical of viruses that are able to spread systemically and cause disease in other tissues. This was demonstrated here by comparing growth curves for rHPIV1 wt to those for VSV (Fig. 2). As might be expected for a virus that is strongly pneumotropic, rHPIV1 wt was detected in apical washes but not in basolateral compartments. In contrast, VSV, which is capable of systemic infection, was released into both sites after basolateral inoculation (Fig. 2A and C).
FIG. 1.
rHPIV1 wt infects HAE cells, spreads throughout the culture, and replicates efficiently. HAE cells were mock infected or infected with rHPIV1 wt at a low MOI (0.01 TCID50/cell). At days 1 to 7 p.i., cells were fixed and stained en face for HPIV1 antigen (green) (A), and virus titers in the apical compartments were determined (B). Virus titers shown are the means of 3 to 11 cultures from a single donor ± standard errors (SE). The limit of detection is 1.2 log10 TCID50/ml, as indicated by the dashed line.
FIG. 2.
Comparison of single-cycle virus growth curves in HAE inoculated with rHPIV1 wt (A) or rHPIV1-CF170S (B) at an MOI of 5.0 TCID50/cell or with VSV (C) at an MOI of 4.2 PFU/cell at 37°C. Virus titers in the apical and basolateral compartments were determined at 8, 24, 48, and 72 h p.i. Virus titers shown are the means of cultures from two donors ± SE, and the limit of detection is 1.2 log10 TCID50/ml.
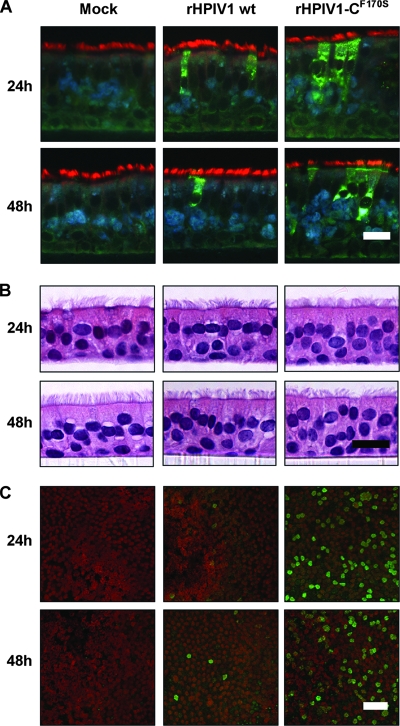
The ciliated cells of the HAE have been shown to be major targets for other respiratory viruses including influenza virus, severe acute respiratory syndrome coronavirus, and paramyxoviruses such as RSV and HPIV3 (33, 51, 58, 66, 67). Similarly, we show here that HPIV1 wt infects ciliated cells in HAE cultures, as observed by immunostaining of histological sections of HAE (Fig. 3A and C). HAE supports HPIV1 wt infection, and the pattern of infection seemed to mimic that observed for other paramyxoviruses such as RSV and HPIV3. Therefore, HAE are a good model for studying HPIV1 infection. This model is further used here to characterize innate host responses in human airway epithelial tissues to infection and to determine the attenuation phenotypes of potential HPIV1 vaccine candidates. In this study, infection with rHPIV1 did not induce any gross changes in morphology or the integrity of the epithelium or any other evidence of a cytopathic effect within 48 h of inoculation in comparison to mock-treated cells (Fig. 3B).
FIG. 3.
HPIV1 infection of ciliated cells without overt cytotoxicity. HAE were inoculated with HPIV1 wt or rHPIV1-CF170S at an MOI of 5.0 TCID50/cell or were mock infected, and cells were processed at 24 and 48 h p.i. for histological analysis in cross section by immunofluorescence (A) or hematoxylin and eosin staining (B) at ×40 magnification or stained en face (C). For histological immunofluorescence (A and C), antibodies to HPIV1 (green) and acetylated alpha-tubulin (red) were used to detect virus antigen and ciliated cells, respectively. Scale bars represent 20 μm (A and B) and 40 μm (C).
Induction of type I IFN during virus infection in HAE.
The HPIV1 wt C accessory proteins inhibit the type I IFN response during infection, and this function is eliminated by a point mutation, F170S, in the C ORF, which is present in all four species of C protein, C′, C, Y1, and Y2 (8, 59). In A549 cells, type I IFN was detected during infection with rHPIV1-CF170S but not rHPIV1 wt (59). We therefore compared levels of replication and IFN induction by rHPIV1-CF170S and rHPIV1 wt in HAE. As was observed for HPIV1 wt, rHPIV1-CF170S targeted ciliated cells in HAE (Fig. 3A). In addition, rHPIV1-CF170S grew at least as efficiently as HPIV1 wt in a high-MOI single-cycle growth curve (Fig. 2). However, although both viruses reached similar peak titers, the kinetics of replication were somewhat different. rHPIV1-CF170S reached a peak in titer by 24 h p.i., at which point its titer was 100-fold higher than that of rHPIV1 wt. In contrast, rHPIV1 wt titers rose steadily until 72 h p.i. (Fig. 2). The differences in the kinetics of virus replication between the two viruses in HAE correlated with en face staining, which demonstrated that a higher proportion of cells were positive for viral antigen following infection with rHPIV1-CF170S compared to that following infection with rHPIV1 wt at both 24 and 48 h p.i. (Fig. 3C). This finding was consistent for two independent donor sources of HAE (data not shown). To investigate the initial higher replication of rHPIV1-CF170S, we redetermined titers of mutant and wild-type virus stocks for three different cell lines, LLC-MK2, A549, and Vero cells. This showed that there were no cell line-specific differences in titers or infectivities between the two viruses. Furthermore, the ratios of infectious virus to hemagglutination titer were similar for the three viruses, indicating that they were comparable in infectivity (data not shown). Thus, the increased level of replication of the mutant virus did not appear to be due to a difference in the amount of input virus or its infectivity but may reflect a difference in the level of gene expression (see Discussion).
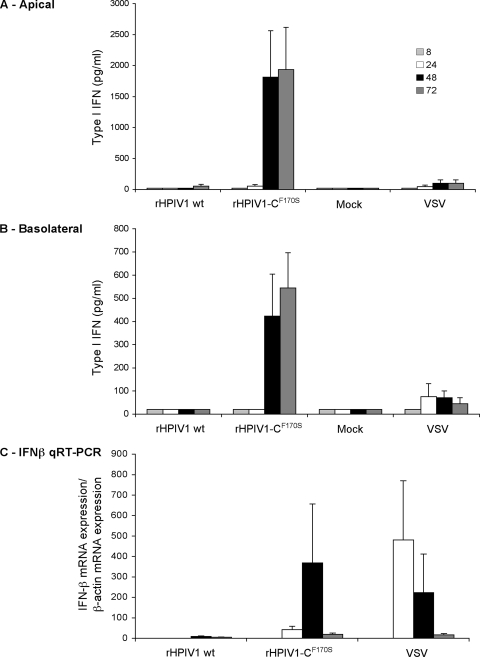
Type I IFN could be readily detected following high-MOI infection of HAE with rHPIV1-CF170S but not rHPIV1 wt (Fig. 4). Specifically, type I IFN secretion was detected in the apical and basolateral compartments of rHPIV1-CF170S-infected HAE by 48 h p.i., as determined by a type I IFN bioassay (Fig. 4A and B). In addition, significant IFN-β mRNA expression was detected as early as 24 h p.i. in this virus group, as determined by qRT-PCR (Fig. 4C), whereas IFN-α mRNA was not detected in any group (data not shown). VSV was used as a positive control for IFN induction, and low levels of type I IFN protein and IFN-β mRNA were detected by 24 h following VSV infection (Fig. 4). These results indicate that there is both lumenal and basolateral/systemic release of type I IFN after infection of HAE with rHPIV1-CF170S or VSV. The expression of IFN-β but not IFN-α following infection with rHPIV1-CF170S is consistent with our previous findings using A549 cells and implies a strong block of IFN-mediated signaling (8, 59). The lack of expression of IFN-β by HAE cells following infection with rHPIV1 wt also is consistent with results with A549 cells but differs from previous results reported for MRC-5 cells (8, 59). To investigate the role of the released type I IFN in the spread of HPIV1 in HAE, we next initiated infection at a lower MOI (0.01 TCID50/ml) in longer-term infections.
FIG. 4.
Comparison of the type I IFN response in HAE inoculated with rHPIV1 wt and rHPIV1-CF170S. HAE were inoculated with rHPIV1s (MOI = 5.0 TCID50/cell) or VSV (MOI = 4.2 PFU/cell) or were mock infected, and levels of type I IFN mRNA and secreted protein were quantitated at 8, 24, 48, and 72 h p.i. A type I IFN bioassay was used to quantitate levels of secreted type I IFN in the apical (A) and basolateral (B) compartments compared to those of an IFN-β standard. Type I IFN concentrations are expressed in pg/ml ± SE and are the means of data from duplicate cultures. The IFN-β standard has a specific activity of approximately 1 IU of antiviral activity per 5 pg. The limit of detection for type I IFN was 20.2 pg/ml. IFN-β mRNA expression was quantitated by qRT-PCR (C). Total RNA was extracted from HAE at 8, 24, 48, and 72 h p.i., and IFN-β mRNA was measured by qRT-PCR using specific primers and Taqman probes that were previously described (53). For each sample, the level of IFN-β mRNA was relative to that of β-actin and expressed as the increase compared to that for the mock-inoculated sample.
Evaluation of the role of type I IFN in multicycle replication of rHPIV1-CF170S in HAE.
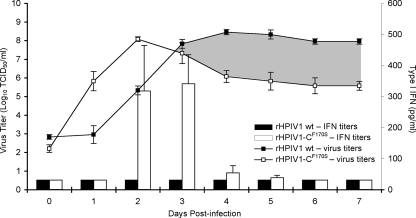
During natural HPIV1 infection, infection is likely initiated at a low MOI via luminal inoculation of the airways. Therefore, in order to mimic natural infection, replications of HPIV1 wt and mutant virus were evaluated using a multiple-cycle growth curve in HAE at 37°C at a low MOI of 0.01 (Fig. 5). Both rHPIV1 wt and rHPIV1-CF170S grew efficiently in HAE, reaching peak titers of 8.5 and 8.1 log10 TCID50/ml, respectively. The kinetics of replication and extent of infection mirrored those observed in the high-MOI growth curves (Fig. 2A and B and 3C). rHPIV1 wt reached a peak titer at day 4 p.i., which remained at a plateau of about 8.5 log10 TCID50/ml until day 7 p.i. (Fig. 5). In comparison, rHPIV1-CF170S reached a peak titer of 8.1 log10 TCID50/ml much earlier, at day 2 p.i., and virus replication then dropped dramatically by day 4 p.i. to plateau at 5.6 log10 TCID50/ml (Fig. 5). In addition, determinations of type I IFN concentrations in apical compartments by bioassay demonstrated no detectable type I IFN during HPIV1 wt infection, whereas type I IFN was detected from days 2 to 4 p.i. in cells infected with rHPIV1-CF170S. Interestingly, the decrease in virus titer during rHPIV1-CF170S infection followed the detection of type I IFN secretion. We have shown that cells infected with rHPIV1-CF170S are able to express type I IFN, and secreted IFN likely acts on neighboring cells to establish an antiviral state. Since HPIV1 is sensitive to an established type I IFN-induced antiviral state (59), these data suggest that type I IFN secretion from virus-infected cells protected neighboring cells from virus infection. This protection can be seen in the approximately 300-fold reduction of rHPIV1-CF170S replication in comparison to that of rHPIV1 wt (as indicated by the shaded area in Fig. 5).
FIG. 5.
Virus replication and type I IFN production during multicycle growth curves in HAE inoculated with rHPIV1 wt and rHPIV1-CF170S at an MOI of 0.01 TCID50/cell at 37°C. Virus titers (log10 TCID50/ml) (line graph) and type I IFN concentrations (pg/ml) (bar graph) in apical washes were determined on each day from day 0 to day 7 p.i. The titers shown are means of data from duplicate donor cultures ± SE. The limit of detection for virus titers was 1.2 log10 TCID50/ml, and that for type I IFN was 31.1 pg/ml. The area shaded in gray represents the overall difference in virus replication between rHPIV1 wt and rHPIV1-CF170S after day 2 p.i.
Replication of HPIV1 vaccine candidates in HAE.
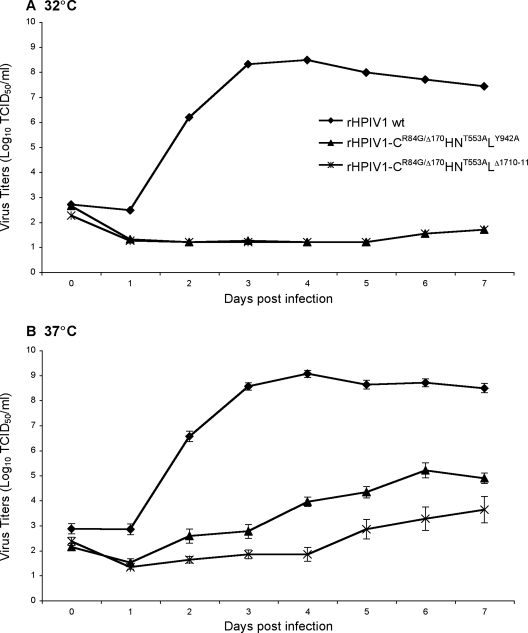
Since the HAE culture model is a useful in vitro tool for evaluating HPIV1 replication in a setting that closely resembles that of in vivo replication in seronegative humans, this model could be used for the preclinical evaluation of HPIV1 vaccine candidates. In order to determine if the level of virus replication in AGMs is reflected in the level of virus replication in HAE cells, two attenuated HPIV1 mutants, rHPIV1-CR84G/Δ170HNT553ALY942A and rHPIV1-CR84G/Δ170HNT553ALΔ1710-11 (named according to their attenuating mutations) (see Materials and Methods), were chosen for study in the HAE model. These viruses were chosen since they were previously characterized in vivo (Table 1) (4) and are currently being considered as live attenuated virus vaccine candidates for HPIV1. Clinical trials using rHPIV1-CR84G/Δ170HNT553ALY942A are currently in progress. These viruses were previously shown to possess a ts phenotype with in vitro shutoff temperatures (e.g., the lowest temperature at which there is a ≥100-fold reduction in replication compared to that of the wild-type virus) of 38°C and 35°C, respectively, and both mutants were attenuated for replication in AGMs (4). Here, we evaluated their replication in vitro in HAE. In order to simulate natural virus infection, HAE were inoculated with the vaccine candidates at a low MOI, and replication was compared to rHPIV1 wt replication at 32°C and 37°C, temperatures indicative of those in the upper and lower respiratory tracts, respectively (Fig. 6). Viral titers determined in the apical washes over a 7-day period showed that rHPIV1 wt replicated efficiently at both temperatures, reaching peak titers of 8.5 and 9.1 log10 TCID50/ml, respectively, by day 4 p.i. at 32°C and 37°C. However, both of the vaccine candidate viruses were severely restricted for replication in HAE and, unexpectedly, grew to higher titers at 37°C than at 32°C (Fig. 6A and B). At 32°C, both viruses demonstrated little to no replication, even though this temperature is fully permissive for both viruses in monolayer cell lines, while at 37°C, there was low-level replication, with mean peak titers of 5.1 and 3.2 log10 TCID50/ml for rHPIV1-CR84G/Δ170HNT553ALY942A and rHPIV1-CR84G/Δ170HNT553ALΔ1710-11, respectively (Fig. 6B). Thus, the rHPIV1-CR84G/Δ170HNT553ALΔ1710-11 virus demonstrated a higher degree of attenuation than did rHPIV1-CR84G/Δ170HNT553ALY942A. Since both vaccine candidates were more attenuated than the virus containing only the CF170S point mutation, which was previously shown to share the same phenotype as CΔ170 (2, 59), these data demonstrate that HAE cells are also sensitive to the attenuation phenotype specified by other mutations, specifically, mutations in the L gene. The attenuating effect of these mutations was not revealed by any other in vitro method. Interestingly, there was no type I IFN detected in the apical or basolateral compartments of cultures infected with these viruses (data not shown), which is likely due to the low levels of virus replication. Based on these data, it is possible that these viruses will be overattenuated in humans. However, the level of virus replication in the human upper respiratory tract in vivo remains to be determined. These data correlate well with previous data demonstrating the level of attenuation of these viruses in AGMs (Table 1) (4) and suggest that the HAE model might be a valuable preclinical tool. However, the ultimate validation of the HAE model for HPIV1 will require the screening of a larger sample of viruses with various levels of attenuation and comparison of this information with similar data on their levels of replication in seronegative humans.
TABLE 1.
Virus replication of HPIV1 wt and rHPIV1 mutants in AGMs and human airway epithelial cells
| Virus | Lower respiratory tract of AGMsa | Apical surface of HAE cells at 37°Cb | ||
|---|---|---|---|---|
| Mean peak virus titer (log10 TCID50/ml) | Reduction in replication vs HPIV1 wt (log10) | Virus titer on day 5 (log10 TCID50/ml) | Reduction in replication vs HPIV1 wt (log10) | |
| HPIV1 wt | 3.9 | 8.6 | ||
| rHPIV1-CF170S | 2.7 | 1.2 | 5.8 | 2.8 |
| rHPIV1-CR84G/Δ170HNT553ALY942A | 0.6 | 3.3 | 4.3 | 4.3 |
| rHPIV1-CR84G/Δ170HNT553ALΔ1710-11 | ≤0.5 | ≥3.4 | 2.5 | 6.1 |
FIG. 6.
The ability of HPIV1 vaccine candidates to replicate in HAE at 32°C and 37°C was determined by multicycle growth curves. HAE were inoculated with rHPIV1 wt, rHPIV1-CR84G/Δ170HNT553ALY942A, or rHPIV1-CR84G/Δ170HNT553ALΔ1710-11 at an MOI of 0.01 TCID50/cell at 32°C and 37°C. Virus titers (log10 TCID50/ml) in apical and basolateral compartments were determined each day from days 0 to 7 p.i. The virus titers shown are the means of data from triplicate donor cultures ± SE for apical washes (samples from the basolateral compartments were negative for virus), and the limit of detection is 1.2 log10 TCID50/ml.
DISCUSSION
HAE models provide useful tools for studying respiratory viruses in vitro. These cell culture systems mimic many morphological features of the human conducting airway epithelium, permitting the study of virus-host interactions in a relevant species as they occur at the site of virus infection. In this study, we have evaluated the ability of HPIV1 wt to infect HAE, and subsequently, we have determined a role for the HPIV1 C proteins in the regulation of the innate immune response in this system. HPIV1 wt readily infected HAE and replicated to high titers but failed to induce the production of type I IFN. The virus was released exclusively at the apical surface of HAE, like many other respiratory viruses that do not cause viremia (or spread systemically) (58, 66, 67). In contrast, VSV, which is not a common human pathogen but which is associated with systemic disease, albeit mild, in humans (30), was released at both the apical and basolateral surfaces. Ciliated cells of the HAE have been shown to be the target for many respiratory viruses including influenza virus, severe acute respiratory syndrome coronavirus, and paramyxoviruses such as RSV and HPIV3 (33, 51, 58, 66, 67). Furthermore, ciliated cells were previously identified as being initiators of cytokine secretion during RSV infection (35). We have shown here that HPIV1 can efficiently infect HAE cells and that it specifically targets ciliated cells. These studies indicate that the HAE model is well suited to study the pathogenesis of HPIV1 mutants. One such HPIV1 mutant, rHPIV1-CF170S, expresses defective C proteins and induces moderate to high levels of type I IFN, which in turn restricts the replication of rHPIV1-CF170S in HAE. Thus, the HPIV1 C proteins are critical regulators of the innate immune response in differentiated primary human epithelial cells in vitro.
Most viruses, including HPIV1, are susceptible to antiviral effects induced by IFN, and for this reason, many viruses have evolved strategies to evade the IFN response (10, 18, 19, 59, 60). In humans, IFN-β is the major type I IFN subtype produced, along with IFN-α1, as the first step in the IFN response (29, 63). Following virus infection, cytoplasmic viral RNA is recognized by host cell proteins such as the retinoic acid-inducible gene I (RIG-I) protein (24, 44, 65). RIG-I subsequently associates with and activates mitochondrial antiviral signaling (also known as IFN-β promoter stimulator 1 protein or virus-induced signaling adaptor protein) (48, 62). In epithelial cells, viral double-stranded RNA is also recognized by Toll-like receptor 3 (5) present in endosomes, resulting in the activation of TRIF-mediated pathways (13, 49). The activation of both the RIG-I and Toll-like receptor 3 pathways leads to the activation of kinases (IKK-ɛ and TBK-1) that phosphorylate IFN regulatory factor 3 (IRF-3), resulting in IRF-3 dimerization and nuclear translocation. IRF-3 along with nuclear factor κB (NF-κB) and ATF-2/c-Jun bind to the IFN-β gene promoter to activate IFN-β transcription (64). IFN-β is then released from the cell and binds to type I IFN receptors on the surface of virus-infected or neighboring cells to switch on the Janus kinase/signal transducer and activator of transcription (JAK/STAT) signaling pathway (18, 21, 28, 47, 57, 60). This pathway culminates in the transcriptional induction of type I IFN response genes, which comprise more than 300 genes that are able to exert a broad range of antiviral, antiproliferative, and immunomodulatory functions and include proteins such as double-stranded RNA-activated protein kinase, 2′,5′-oligoadenylate synthetase, and the Mx proteins (28). In this way, type I IFN can mediate the host immune response in virally infected cells and establish an antiviral state in neighboring uninfected cells in order to limit replication. The paramyxoviruses express nonstructural accessory proteins that are able to inhibit various stages of the type I IFN response. For example, the V proteins of several parainfluenza viruses (e.g., HPIV2 and simian virus 5), RSV NS1 and NS2 proteins, and SeV and HPIV1 C proteins have been shown to inhibit IRF-3 activation (1, 6, 26, 53, 54, 59). In addition, the C proteins of SeV, HPIV1, and HPIV3 and the V proteins of HPIV2 and simian virus 5 have been shown to block the IFN signaling cascade by various mechanisms, including STAT binding, the inhibition of STAT activation, and STAT destabilization/degradation (8, 12, 14, 17, 20, 27, 31, 41, 56, 59).
The role of the HPIV1 C accessory proteins in inhibiting the type I IFN response was recently characterized in vitro (8, 59); however, it was uncertain whether these in vitro observations would translate to similar functions in vivo. In HAE, both rHPIV1 wt and rHPIV1-CF170S replicated efficiently and reached similar mean peak titers. However, rHPIV1-CF170S reached its peak titer by day 2, compared to day 4 for rHPIV1 wt, which was true at both high and low MOIs. Furthermore, immunostaining of HAE infected at a high MOI also demonstrated a clear quantitative difference between the viruses at day 2 p.i., with many more cells staining positive in the rHPIV1-CF170S-infected cultures than in the rHPIV1 wt-infected cultures. Interestingly, a similar phenomenon was previously observed in monolayer cultures with an SeV mutant containing the same mutation in the SeV C proteins (15). The F170S mutation in SeV had the effect of increasing gene transcription fourfold compared to that of its parent virus, with corresponding increases in RNA replication and virus replication (15). This presumably accounts for the initial increased level of viral replication and viral antigen synthesis (detected by immunofluorescence) observed here for rHPIV1-CF170S in HAE cells.
The antiviral role of type I IFN became evident in HAE infected at a low MOI, and the quantitative impact of the type I IFN response was next assessed. HPIV1 wt virus replicated to high titers, and type I IFN was not induced. The HPIV1 wt virus titer persisted at a high level, presumably due to the absence of IFN production in the cultures throughout the duration of the study. In contrast, rHPIV1-CF170S replicated efficiently until IFN was detected, and titers then decreased by a factor of 100 to 1,000. While IFN is not the sole factor involved in viral clearance, the difference in replication between the two viruses in this model system is most likely due to the induction of IFN in HAE infected with rHPIV1-CF170S, as indicated by the shaded area in Fig. 5. It is likely that the type I IFN secreted by virus-infected HAE cells is acting on the neighboring uninfected cells to activate the JAK-STAT signaling pathway and ultimately establish an antiviral state in these cells by inducing the expression of type I IFN response genes. This would lead to some level of inhibition of virus replication and spread and would explain the decrease in virus titers that we observed. Recently, STAT1 activation in mouse airway epithelial cells was associated with the inhibition of cell-to-cell spread of SeV and the protection of uninfected cells from infection (50). Interestingly, rHPIV1-CF170S induced the expression of IFN-β mRNA, while the expression of IFN-α species was below the level of detection. This confirms findings observed for A549 cells and would be consistent with rHPIV1-CF170S retaining the ability to inhibit IFN-mediated signaling through the JAK/STAT pathway, which is necessary for the expression of most of the species of IFN-α and for the amplification of the IFN response.
The 100- to 1,000-fold difference in replication of rHPIV1-CF170S and HPIV1 wt in HAE was similar to that observed in the upper and lower respiratory tract of AGMs (Table 1), suggesting that HAE provide a reliable model for the role that type I IFN plays in restricting the replication of HPIV1 in vivo (2). It is evident that IFN production was associated with a reduction in virus replication; however, it was not sufficient to completely inhibit virus growth. A recent study using influenza A virus wild type and NS1-deficient viruses to infect murine epithelial cells yielded similar quantitative effects (38). The level of production of IFN-β was significantly elevated during infection with a human influenza A virus mutant expressing a defective NS1 protein, a known inhibitor of RIG-I-mediated type I IFN induction, compared to wild-type influenza A virus, and IFN-β production correlated with a reduction in the level of replication of this virus in murine airway epithelial cells (38). The role of the HPIV1 C proteins in inhibiting the type I IFN response makes these proteins good targets for attenuating viruses by mutagenesis, and A549 cells and HAE cultures appear to be suitable systems to study the effect of such mutations on replication. Mutations of the C proteins are included in current HPIV1 vaccine candidates (4).
The levels of replication of two HPIV1 vaccine candidates in HAE were evaluated to determine whether the replication of HPIV1 vaccine viruses in these cells might be predictive of their level of replication in humans. Two live attenuated vaccine candidates for HPIV1, namely, rHPIV1-CR84G/Δ170HNT553ALY942A and rHPIV1-CR84G/Δ170HNT553ALΔ1710-11 (4), are being considered for evaluation in humans, and each mutant contains mutations in C that specify the same IFN phenotype and attenuation phenotype as the CF170S point mutation (2, 39, 59). In addition, both vaccine viruses contain a ts attenuating mutation in the L polymerase gene that restricts replication at 37°C. Both vaccine candidates replicate efficiently at 32°C in Vero cells, the substrate for vaccine manufacture. We had anticipated that each vaccine candidate would replicate efficiently at 32°C in HAE but would be restricted in replication at 37°C due to the presence of the ts mutation. Surprisingly, the viruses were completely attenuated for replication in HAE at 32°C following inoculation at a low MOI and grew to very low levels at 37°C. The C and L gene mutations may collaborate to restrict replication at 32°C by a mechanism that is undefined but that can be addressed in HAE using mutants in which the various attenuating mutations are segregated. However, it is important that there was an additive effect in the level of attenuation specified by the combination of attenuating mutations in the P/C and L genes. This demonstrates that HAE cells are sensitive to attenuation specified by different mechanisms and underscores its validity as a tool for the preclinical testing of viral vaccine candidates. How these findings in HAE relate to replication in humans will be determined in clinical trials. Both vaccine candidates appear to be safe for evaluation in humans based on their highly restricted levels of replication in AGMs and HAE, but one or both might be overattenuated for humans. rHPIV1-CR84G/Δ170HNT553ALY942A replicated to slightly higher titers at 37°C than rHPIV1-CR84G/Δ170HNT553ALΔ1710-11 but was still significantly attenuated compared to rHPIV1 wt. IFN was not detected in cells infected with these viruses, which is most likely due to their highly restricted growth resulting in a poor induction of the innate immune response. Comparing data from this in vitro study with those from previous in vivo studies, we have shown that attenuation, i.e., restriction in replication in HAE, correlates with the reduction in mean peak titers in AGMs (Table 1) (2, 4). Although it is possible that these viruses will be overattenuated in humans, we have selected the rHPIV1-CR84G/Δ170HNT553ALY942A candidate for further clinical evaluation since it is attenuated in AGMs, it replicates to some extent in HAE, and it protects AGMs against challenge with HPIV1 wt. In addition to evaluating the potential of this virus as a vaccine, clinical trials will provide information on whether virus replication in the HAE model correlates with the level of attenuation of the virus in humans. A strong correlation would highlight the usefulness of the HAE model as a preclinical research tool and enable more efficient screening of HPIV1 and potentially other paramyxovirus vaccine candidates.
Acknowledgments
We are grateful to the directors and teams of the UNC Cystic Fibrosis Center Tissue Culture Core, the Morphology and Morphometry Core, and the Michael Hooker Microscopy Facility for supplying reagents and technical expertise and to Susan Burkett for technical assistance.
This project was funded as a part of the NIAID Intramural Program and National Institutes of Health grant NIH R01 HL77844-1.
Footnotes
▿
Published ahead of print on 4 June 2008.
REFERENCES
- 1.Andrejeva, J., K. S. Childs, D. F. Young, T. S. Carlos, N. Stock, S. Goodbourn, and R. E. Randall. 2004. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc. Natl. Acad. Sci. USA 10117264-17269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartlett, E. J., E. Amaro-Carambot, S. R. Surman, P. L. Collins, B. R. Murphy, and M. H. Skiadopoulos. 2006. Introducing point and deletion mutations into the P/C gene of human parainfluenza virus type 1 (HPIV1) by reverse genetics generates attenuated and efficacious vaccine candidates. Vaccine 242674-2684. [DOI] [PubMed] [Google Scholar]
- 3.Bartlett, E. J., E. Amaro-Carambot, S. R. Surman, J. T. Newman, P. L. Collins, B. R. Murphy, and M. H. Skiadopoulos. 2005. Human parainfluenza virus type I (HPIV1) vaccine candidates designed by reverse genetics are attenuated and efficacious in African green monkeys. Vaccine 234631-4646. [DOI] [PubMed] [Google Scholar]
- 4.Bartlett, E. J., A. Castano, S. R. Surman, P. L. Collins, M. H. Skiadopoulos, and B. R. Murphy. 2007. Attenuation and efficacy of human parainfluenza virus type 1 (HPIV1) vaccine candidates containing stabilized mutations in the P/C and L genes. Virol. J. 467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beutler, B. 2004. Inferences, questions and possibilities in Toll-like receptor signalling. Nature 430257-263. [DOI] [PubMed] [Google Scholar]
- 6.Bossert, B., S. Marozin, and K. K. Conzelmann. 2003. Nonstructural proteins NS1 and NS2 of bovine respiratory syncytial virus block activation of interferon regulatory factor 3. J. Virol. 778661-8668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bottcher, E., T. Matrosovich, M. Beyerle, H. D. Klenk, W. Garten, and M. Matrosovich. 2006. Proteolytic activation of influenza viruses by serine proteases TMPRSS2 and HAT from human airway epithelium. J. Virol. 809896-9898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bousse, T., R. L. Chambers, R. A. Scroggs, A. Portner, and T. Takimoto. 2006. Human parainfluenza virus type 1 but not Sendai virus replicates in human respiratory cells despite IFN treatment. Virus Res. 12123-32. [DOI] [PubMed] [Google Scholar]
- 9.Clemons, D. J., C. Besch-Williford, E. K. Steffen, L. K. Riley, and D. H. Moore. 1992. Evaluation of a subcutaneously implanted chamber for antibody production in rabbits. Lab. Anim. Sci. 42307-311. [PubMed] [Google Scholar]
- 10.Conzelmann, K. K. 2005. Transcriptional activation of alpha/beta interferon genes: interference by nonsegmented negative-strand RNA viruses. J. Virol. 795241-5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Counihan, M. E., D. K. Shay, R. C. Holman, S. A. Lowther, and L. J. Anderson. 2001. Human parainfluenza virus-associated hospitalizations among children less than five years of age in the United States. Pediatr. Infect. Dis. J. 20646-653. [DOI] [PubMed] [Google Scholar]
- 12.Didcock, L., D. F. Young, S. Goodbourn, and R. E. Randall. 1999. The V protein of simian virus 5 inhibits interferon signalling by targeting STAT1 for proteasome-mediated degradation. J. Virol. 739928-9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fitzgerald, K. A., S. M. McWhirter, K. L. Faia, D. C. Rowe, E. Latz, D. T. Golenbock, A. J. Coyle, S. M. Liao, and T. Maniatis. 2003. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4491-496. [DOI] [PubMed] [Google Scholar]
- 14.Garcin, D., J. Curran, and D. Kolakofsky. 2000. Sendai virus C proteins must interact directly with cellular components to interfere with interferon action. J. Virol. 748823-8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcin, D., M. Itoh, and D. Kolakofsky. 1997. A point mutation in the Sendai virus accessory C proteins attenuates virulence for mice, but not virus growth in cell culture. Virology 238424-431. [DOI] [PubMed] [Google Scholar]
- 16.Garcin, D., J. B. Marq, S. Goodbourn, and D. Kolakofsky. 2003. The amino-terminal extensions of the longer Sendai virus C proteins modulate pY701-Stat1 and bulk Stat1 levels independently of interferon signaling. J. Virol. 772321-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcin, D., J. B. Marq, L. Strahle, P. le Mercier, and D. Kolakofsky. 2002. All four Sendai virus C proteins bind Stat1, but only the larger forms also induce its mono-ubiquitination and degradation. Virology 295256-265. [DOI] [PubMed] [Google Scholar]
- 18.Goodbourn, S., L. Didcock, and R. E. Randall. 2000. Interferons: cell signalling, immune modulation, antiviral response and virus countermeasures. J. Gen. Virol. 812341-2364. [DOI] [PubMed] [Google Scholar]
- 19.Gotoh, B., T. Komatsu, K. Takeuchi, and J. Yokoo. 2002. Paramyxovirus strategies for evading the interferon response. Rev. Med. Virol. 12337-357. [DOI] [PubMed] [Google Scholar]
- 20.Gotoh, B., K. Takeuchi, T. Komatsu, and J. Yokoo. 2003. The STAT2 activation process is a crucial target of Sendai virus C protein for the blockade of alpha interferon signaling. J. Virol. 773360-3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grandvaux, N., B. R. tenOever, M. J. Servant, and J. Hiscott. 2002. The interferon antiviral response: from viral invasion to evasion. Curr. Opin. Infect. Dis. 15259-267.12015460 [Google Scholar]
- 22.Heikkinen, T., M. Thint, and T. Chonmaitree. 1999. Prevalence of various respiratory viruses in the middle ear during acute otitis media. N. Engl. J. Med. 340260-264. [DOI] [PubMed] [Google Scholar]
- 23.Henderson, F. W., A. M. Collier, M. A. Sanyal, J. M. Watkins, D. L. Fairclough, W. A. Clyde, Jr., and F. W. Denny. 1982. A longitudinal study of respiratory viruses and bacteria in the etiology of acute otitis media with effusion. N. Engl. J. Med. 3061377-1383. [DOI] [PubMed] [Google Scholar]
- 24.Hornung, V., J. Ellegast, S. Kim, K. Brzozka, A. Jung, H. Kato, H. Poeck, S. Akira, K. K. Conzelmann, M. Schlee, S. Endres, and G. Hartmann. 2006. 5′-Triphosphate RNA is the ligand for RIG-I. Science 314994-997. [DOI] [PubMed] [Google Scholar]
- 25.Karron, R. A., and P. L. Collins. 2007. Parainfluenza viruses, p. 1497-1526. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Strauss (ed.), Fields virology, 5th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 26.Komatsu, T., K. Takeuchi, J. Yokoo, and B. Gotoh. 2004. C and V proteins of Sendai virus target signaling pathways leading to IRF-3 activation for the negative regulation of interferon-beta production. Virology 325137-148. [DOI] [PubMed] [Google Scholar]
- 27.Komatsu, T., K. Takeuchi, J. Yokoo, and B. Gotoh. 2002. Sendai virus C protein impairs both phosphorylation and dephosphorylation processes of Stat1. FEBS Lett. 511139-144. [DOI] [PubMed] [Google Scholar]
- 28.Levy, D. E., and A. Garcia-Sastre. 2001. The virus battles: IFN induction of the antiviral state and mechanisms of viral evasion. Cytok. Growth Factor Rev. 12143-156. [DOI] [PubMed] [Google Scholar]
- 29.Lin, R., P. Genin, Y. Mamane, and J. Hiscott. 2000. Selective DNA binding and association with the CREB binding protein coactivator contribute to differential activation of alpha/beta interferon genes by interferon regulatory factors 3 and 7. Mol. Cell. Biol. 206342-6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lyles, D. S., and C. E. Rupprecht. 2007. Rhabdoviridae, p. 1363-1408. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Strauss (ed.), Fields virology, 5th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 31.Malur, A. G., S. Chattopadhyay, R. K. Maitra, and A. K. Banerjee. 2005. Inhibition of STAT1 phosphorylation by human parainfluenza virus type 3 C protein. J. Virol. 797877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marx, A., T. J. Torok, R. C. Holman, M. J. Clarke, and L. J. Anderson. 1997. Pediatric hospitalizations for croup (laryngotracheobronchitis): biennial increases associated with human parainfluenza virus 1 epidemics. J. Infect. Dis. 1761423-1427. [DOI] [PubMed] [Google Scholar]
- 33.Matrosovich, M. N., T. Y. Matrosovich, T. Gray, N. A. Roberts, and H. D. Klenk. 2004. Human and avian influenza viruses target different cell types in cultures of human airway epithelium. Proc. Natl. Acad. Sci. USA 1014620-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McAuliffe, J. M., S. R. Surman, J. T. Newman, J. M. Riggs, P. L. Collins, B. R. Murphy, and M. H. Skiadopoulos. 2004. Codon substitution mutations at two positions in the L polymerase protein of human parainfluenza virus type 1 yield viruses with a spectrum of attenuation in vivo and increased phenotypic stability in vitro. J. Virol. 782029-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mellow, T. E., P. C. Murphy, J. L. Carson, T. L. Noah, L. Zhang, and R. J. Pickles. 2004. The effect of respiratory syncytial virus on chemokine release by differentiated airway epithelium. Exp. Lung Res. 3043-57. [DOI] [PubMed] [Google Scholar]
- 36.Murphy, B. R., G. A. Prince, P. L. Collins, K. V. W. Coelingh, R. A. Olmsted, M. K. Spriggs, R. H. Parrott, H. W. Kim, C. D. Brandt, and R. M. Chanock. 1988. Current approaches to the development of vaccines effective against parainfluenza and respiratory syncytial viruses. Virus Res. 111-15. [DOI] [PubMed] [Google Scholar]
- 37.Murphy, B. R., D. D. Richman, E. G. Chalhub, C. P. Uhlendorf, S. Baron, and R. M. Chanock. 1975. Failure of attenuated temperature-sensitive influenza A (H3N2) virus to induce heterologous interference in humans to parainfluenza type 1 virus. Infect. Immun. 1262-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Newby, C. M., L. Sabin, and A. Pekosz. 2007. The RNA binding domain of influenza A virus NS1 protein affects secretion of tumor necrosis factor alpha, interleukin-6, and interferon in primary murine tracheal epithelial cells. J. Virol. 819469-9480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Newman, J. T., J. M. Riggs, S. R. Surman, J. M. McAuliffe, T. A. Mulaikal, P. L. Collins, B. R. Murphy, and M. H. Skiadopoulos. 2004. Generation of recombinant human parainfluenza virus type 1 vaccine candidates by importation of temperature-sensitive and attenuating mutations from heterologous paramyxoviruses. J. Virol. 782017-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Newman, J. T., S. R. Surman, J. M. Riggs, C. T. Hansen, P. L. Collins, B. R. Murphy, and M. H. Skiadopoulos. 2002. Sequence analysis of the Washington/1964 strain of human parainfluenza virus type 1 (HPIV1) and recovery and characterization of wild-type recombinant HPIV1 produced by reverse genetics. Virus Genes 2477-92. [DOI] [PubMed] [Google Scholar]
- 41.Parisien, J. P., J. F. Lau, J. J. Rodriguez, B. M. Sullivan, A. Moscona, G. D. Parks, R. A. Lamb, and C. M. Horvath. 2001. The V protein of human parainfluenza virus 2 antagonizes type I interferon responses by destabilizing signal transducer and activator of transcription 2. Virology 283230-239. [DOI] [PubMed] [Google Scholar]
- 42.Park, M. S., A. Garcia-Sastre, J. F. Cros, C. F. Basler, and P. Palese. 2003. Newcastle disease virus V protein is a determinant of host range restriction. J. Virol. 779522-9532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pickles, R. J., D. McCarty, H. Matsui, P. J. Hart, S. H. Randell, and R. C. Boucher. 1998. Limited entry of adenovirus vectors into well-differentiated airway epithelium is responsible for inefficient gene transfer. J. Virol. 726014-6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Plumet, S., F. Herschke, J. M. Bourhis, H. Valentin, S. Longhi, and D. Gerlier. 2007. Cytosolic 5′-triphosphate ended viral leader transcript of measles virus as activator of the RIG I-mediated interferon response. PLoS ONE 2e279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Power, U. F., K. W. Ryan, and A. Portner. 1992. The P genes of human parainfluenza virus type 1 clinical isolates are polycistronic and microheterogeneous. Virology 189340-343. [DOI] [PubMed] [Google Scholar]
- 46.Reed, G., P. H. Jewett, J. Thompson, S. Tollefson, and P. F. Wright. 1997. Epidemiology and clinical impact of parainfluenza virus infections in otherwise healthy infants and young children < 5 years old. J. Infect. Dis. 175807-813. [DOI] [PubMed] [Google Scholar]
- 47.Samuel, C. E. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14778-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seth, R. B., L. Sun, C. K. Ea, and Z. J. Chen. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122669-682. [DOI] [PubMed] [Google Scholar]
- 49.Sharma, S., B. R. tenOever, N. Grandvaux, G. P. Zhou, R. Lin, and J. Hiscott. 2003. Triggering the interferon antiviral response through an IKK-related pathway. Science 3001148-1151. [DOI] [PubMed] [Google Scholar]
- 50.Shornick, L. P., A. G. Wells, Y. Zhang, A. C. Patel, G. Huang, K. Takami, M. Sosa, N. A. Shukla, E. Agapov, and M. J. Holtzman. 2008. Airway epithelial versus immune cell Stat1 function for innate defense against respiratory viral infection. J. Immunol. 1803319-3328. [DOI] [PubMed] [Google Scholar]
- 51.Sims, A. C., R. S. Baric, B. Yount, S. E. Burkett, P. L. Collins, and R. J. Pickles. 2005. Severe acute respiratory syndrome coronavirus infection of human ciliated airway epithelia: role of ciliated cells in viral spread in the conducting airways of the lungs. J. Virol. 7915511-15524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Skiadopoulos, M. H., T. Tao, S. R. Surman, P. L. Collins, and B. R. Murphy. 1999. Generation of a parainfluenza virus type 1 vaccine candidate by replacing the HN and F glycoproteins of the live-attenuated PIV3 cp45 vaccine virus with their PIV1 counterparts. Vaccine 18503-510. [DOI] [PubMed] [Google Scholar]
- 53.Spann, K. M., K. C. Tran, B. Chi, R. L. Rabin, and P. L. Collins. 2004. Suppression of the induction of alpha, beta, and lambda interferons by the NS1 and NS2 proteins of human respiratory syncytial virus in human epithelial cells and macrophages. J. Virol. 784363-4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spann, K. M., K. C. Tran, and P. L. Collins. 2005. Effects of nonstructural proteins NS1 and NS2 of human respiratory syncytial virus on interferon regulatory factor 3, NF-κB, and proinflammatory cytokines. J. Virol. 795353-5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stojdl, D. F., B. D. Lichty, B. R. tenOever, J. M. Paterson, A. T. Power, S. Knowles, R. Marius, J. Reynard, L. Poliquin, H. Atkins, E. G. Brown, R. K. Durbin, J. E. Durbin, J. Hiscott, and J. C. Bell. 2003. VSV strains with defects in their ability to shutdown innate immunity are potent systemic anti-cancer agents. Cancer Cell 4263-275. [DOI] [PubMed] [Google Scholar]
- 56.Takeuchi, K., T. Komatsu, J. Yokoo, A. Kato, T. Shioda, Y. Nagai, and B. Gotoh. 2001. Sendai virus C protein physically associates with Stat1. Genes Cells 6545-557. [DOI] [PubMed] [Google Scholar]
- 57.Taniguchi, T., and A. Takaoka. 2002. The interferon-alpha/beta system in antiviral responses: a multimodal machinery of gene regulation by the IRF family of transcription factors. Curr. Opin. Immunol. 14111-116. [DOI] [PubMed] [Google Scholar]
- 58.Thompson, C. I., W. S. Barclay, M. C. Zambon, and R. J. Pickles. 2006. Infection of human airway epithelium by human and avian strains of influenza A virus. J. Virol. 808060-8068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Van Cleve, W., E. Amaro-Carambot, S. R. Surman, J. Bekisz, P. L. Collins, K. C. Zoon, B. R. Murphy, M. H. Skiadopoulos, and E. J. Bartlett. 2006. Attenuating mutations in the P/C gene of human parainfluenza virus type 1 (HPIV1) vaccine candidates abrogate the inhibition of both induction and signaling of type I interferon (IFN) by wild-type HPIV1. Virology 35261-73. [DOI] [PubMed] [Google Scholar]
- 60.Weber, F., G. Kochs, and O. Haller. 2004. Inverse interference: how viruses fight the interferon system. Viral Immunol. 17498-515. [DOI] [PubMed] [Google Scholar]
- 61.Wright, P. F., M. R. Ikizler, R. A. Gonzales, K. N. Carroll, J. E. Johnson, and J. A. Werkhaven. 2005. Growth of respiratory syncytial virus in primary epithelial cells from the human respiratory tract. J. Virol. 798651-8654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu, L. G., Y. Y. Wang, K. J. Han, L. Y. Li, Z. Zhai, and H. B. Shu. 2005. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell 19727-740. [DOI] [PubMed] [Google Scholar]
- 63.Yeow, W. S., W. C. Au, W. J. Lowther, and P. M. Pitha. 2001. Downregulation of IRF-3 levels by ribozyme modulates the profile of IFNA subtypes expressed in infected human cells. J. Virol. 753021-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yie, J., K. Senger, and D. Thanos. 1999. Mechanism by which the IFN-beta enhanceosome activates transcription. Proc. Natl. Acad. Sci. USA 9613108-13113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yoneyama, M., M. Kikuchi, T. Natsukawa, N. Shinobu, T. Imaizumi, M. Miyagishi, K. Taira, S. Akira, and T. Fujita. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5730-737. [DOI] [PubMed] [Google Scholar]
- 66.Zhang, L., A. Bukreyev, C. I. Thompson, B. Watson, M. E. Peeples, P. L. Collins, and R. J. Pickles. 2005. Infection of ciliated cells by human parainfluenza virus type 3 in an in vitro model of human airway epithelium. J. Virol. 791113-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang, L., M. E. Peeples, R. C. Boucher, P. L. Collins, and R. J. Pickles. 2002. Respiratory syncytial virus infection of human airway epithelial cells is polarized, specific to ciliated cells, and without obvious cytopathology. J. Virol. 765654-5666. [DOI] [PMC free article] [PubMed] [Google Scholar]