Cyclin-Specific Control of Ribosomal DNA Segregation (original) (raw)
Abstract
Following chromosome duplication in S phase of the cell cycle, the sister chromatids are linked by cohesin. At the onset of anaphase, separase cleaves cohesin and thereby initiates sister chromatid separation. Separase activation results from the destruction of its inhibitor, securin, which is triggered by a ubiquitin ligase called the anaphase-promoting complex (APC). Here, we show in budding yeast that securin destruction and, thus, separase activation are not sufficient for the efficient segregation of the repetitive ribosomal DNA (rDNA). We find that rDNA segregation also requires the APC-mediated destruction of the S-phase cyclin Clb5, an activator of the protein kinase Cdk1. Mutations that prevent Clb5 destruction are lethal and cause defects in rDNA segregation and DNA synthesis. These defects are distinct from the mitotic-exit defects caused by stabilization of the mitotic cyclin Clb2, emphasizing the importance of cyclin specificity in the regulation of late-mitotic events. Efficient rDNA segregation, both in mitosis and meiosis, also requires APC-dependent destruction of Dbf4, an activator of the protein kinase Cdc7. We speculate that the dephosphorylation of Clb5-specific Cdk1 substrates and Dbf4-Cdc7 substrates drives the resolution of rDNA in early anaphase. The coincident destruction of securin, Clb5, and Dbf4 coordinates bulk chromosome segregation with segregation of rDNA.
Cyclin-dependent kinases (Cdks) determine the order of cell cycle events by phosphorylating different substrates at different points in the cell cycle. The temporal control of substrate phosphorylation is made possible by the periodic synthesis and destruction of multiple cyclin cofactors that bind and activate the Cdk (17). Different cyclins have different functional capacities, in part because they target the associated Cdk to specific substrates (2, 4). In the budding yeast Saccharomyces cerevisiae, for example, a complex of the S-phase cyclin Clb5 and Cdk1 displays high specificity for a small subset of Cdk1 substrates, due to a docking site on Clb5 that greatly increases its affinity for those substrates (16). Many Clb5-specific substrates are involved in DNA synthesis, helping to explain why Clb5-Cdk1 drives early cell cycle events.
The final events of M phase require dephosphorylation of Cdk1 targets, which results from a combination of cyclin destruction, induction of Cdk1 inhibitors, and activation of phosphatases, such as Cdc14 of budding yeast, that remove phosphates from Cdk1 sites (30). Clb5 and the major mitotic cyclin Clb2 are both targeted for destruction by the anaphase-promoting complex (APC), a ubiquitin ligase that is activated in metaphase by association with the cofactor Cdc20 (APCCdc20) (22, 32). APCCdc20 triggers the complete destruction of Clb5 before anaphase but promotes only partial destruction of Clb2, which is completely destroyed later in mitosis when the APC is activated by Cdh1 (24, 39). These differences in the timing of cyclin destruction might provide a mechanism to allow the dephosphorylation of Clb5-specific substrates in early anaphase, while the general substrates of Clb2-Cdk1 remain phosphorylated until later (30). The Clb5-specific substrates Fin1 and Ase1, for example, are dephosphorylated in early anaphase, allowing them to help initiate important changes in mitotic spindle dynamics (12, 40).
If the order of cyclin destruction determines the timing of late-mitotic events, then the stabilization of different cyclins should have distinct effects on mitotic processes. Indeed, the stabilization of metazoan cyclin A (by mutation of sequences required for APC recognition) causes phenotypes very different than those caused by stabilized cyclin B (21, 25). In budding yeast, stabilization or overexpression of Clb2 causes defects in spindle breakdown and mitotic exit, whereas partially stabilized Clb5 has little apparent effect on late-mitotic events (39). Overexpression of Clb5 is highly toxic (11), but the cellular defects that result from excess Clb5-Cdk1 activity are not clear.
The initiating event of anaphase is sister chromatid separation, which results primarily from the proteolytic cleavage of cohesin by separase (19). In budding yeast, however, separase-mediated cohesin destruction is not sufficient for resolution of the highly repetitive ribosomal DNA (rDNA) locus on chromosome XII. Segregation of the rDNA also requires separase-dependent activation of the phosphatase Cdc14 (5, 28), suggesting that dephosphorylation of Cdk1 substrates is required.
It remains unclear if destruction of APCCdc20 substrates other than securin is required for resolution of the rDNA in anaphase. Here, we investigate this question and find that the degradation of two additional APCCdc20 substrates, Clb5 and Dbf4, contributes to rDNA segregation. Our results also provide evidence for cyclin specificity in late-mitotic control, as rDNA resolution depends on loss of Clb5 activity but not on loss of activity of the major mitotic cyclin Clb2. We also show that degradation of Clb5 is critical for origin relicensing and, hence, DNA replication. Finally, we demonstrate that the molecular mechanisms that govern loss of rDNA cohesion in mitosis are similar to those in anaphase I of the meiotic program.
MATERIALS AND METHODS
All mitotic experiments were performed in strain W303, and all meiotic experiments were performed in strain SK1. Epitope tagging of endogenous genes and gene deletions was performed by gene targeting using PCR products. The CLB5 gene, along with three C-terminal hemagglutinin epitopes and 451 bp of the CLB5 promoter, was amplified from budding yeast and cloned to allow integration of an extra copy of CLB5-HA3 at the URA3 locus. This plasmid was modified to generate the N-terminal deletion, CLB5_-Δ_N, by removal of the coding sequence for amino acids 2 to 95. For expression in meiosis, the coding sequence of CLB5_-Δ_N-TAP was placed in front of 341 bp of the DMC1 promoter to allow integration of pDMC1-CLB5_-Δ_N-TAP at the HIS3 locus. To express high levels of cyclins in mitosis, CLB5_-Δ_N-TAP or CLB2_-Δ_N-HA3 (encoding Clb2 lacking amino acids 2 to 176) was cloned in front of the GAL1-10 promoter. To make stabilized Dbf4, a plasmid that contained 523 bp of the DBF4 promoter and a portion of the DBF4 gene with the coding sequence for amino acids 2 to 65 removed was created. This plasmid was linearized to integrate at the DBF4 locus and to switch the endogenous gene to DBF4_-Δ_N and was confirmed by PCR.
Conditions for growth and release of synchronous cultures from arrest in G1 by α-factor were performed as described previously (16). In the GAL-CDC20 shutoff experiment (Fig. 1), cells were grown in raffinose/galactose media for α-factor arrest and released into glucose media. In the GAL-CLB experiment (Fig. 2), cells were grown in raffinose media for α-factor arrest and released into raffinose/galactose media. In experiments involving GAL-SIC1, cells were grown in raffinose/galactose media for α-factor arrest (Fig. 3, 4, and 5). Once arrested, glucose was added for 30 min, and cells were released from the arrest into fresh glucose media. Synchronous meiotic experiments were performed as described previously (29). Western blotting, budding counts, and immunofluorescence analysis were performed as described previously (28). The antibodies used were α-Nop1 clone MCA28F2 (Encor), α-tubulin YOL1/34 (Serotec), α-hemagglutinin clone 16B12 (Babco), and α-myc clone 9E10 (Babco).
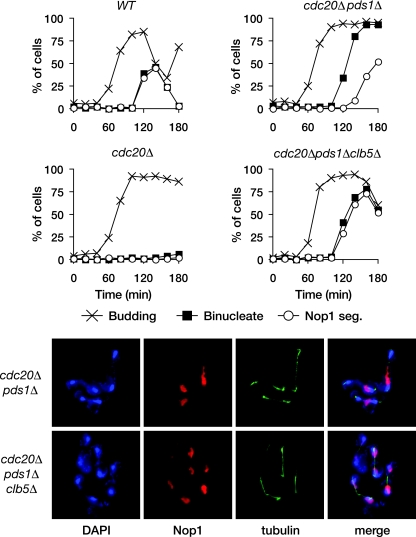
FIG. 1.
APCCdc20-dependent degradation of securin and Clb5 allows rDNA segregation. Wild-type (WT), _cdc20_Δ, _cdc20_Δ _pds1_Δ, and _cdc20_Δ _pds1_Δ _clb5_Δ yeast expressing GAL-CDC20 were arrested in G1 with α-factor and then released into fresh dextrose media to repress CDC20 transcription. Samples were collected every 20 min for analysis of budding, binucleate formation (bulk chromosome segregation, assessed by DAPI [4′,6-diamidino-2-phenylindole] staining), and Nop1 segregation (a marker of nucleolar rDNA segregation) (top panel). The bottom panel shows representative images of _cdc20_Δ _pds1_Δ and _cdc20_Δ _pds1_Δ _clb5_Δ yeast 140 min after release from G1.
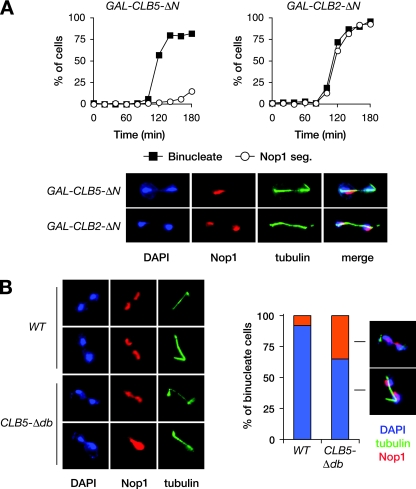
FIG. 2.
Clb5 inhibits rDNA segregation. (A) Cells carrying either GAL-CLB5_-Δ_N or GAL-CLB2_-Δ_N were arrested in G1 with α-factor and then released into fresh galactose media to induce cyclin expression. Samples were collected every 15 min for analysis of binucleate formation and Nop1 segregation. Representative images are shown of GAL-CLB5_-Δ_N yeast and GAL-CLB2_-Δ_N yeast 120 min after release from G1. (B) Asynchronous CLB5 (WT) and CLB5_-Δ_db cells were collected and binucleates analyzed for the number of segregated and unsegregated Nop1 masses. Representative images are shown (left).
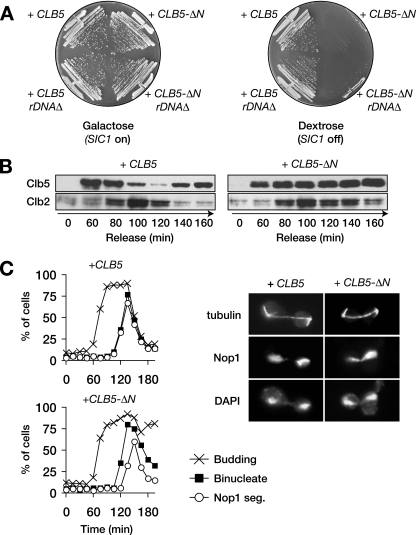
FIG. 3.
CLB5-ΔN is lethal and causes an rDNA segregation delay. (A) Wild-type or _rDNA_Δ cells, carrying GAL-SIC1 and an extra copy of the CLB5 or CLB5_-Δ_N gene (expressed from the CLB5 promoter), were plated on either galactose or dextrose media. (B) GAL-SIC1 cells carrying an extra copy of CLB5 or CLB5_-Δ_N were arrested in G1 by α-factor treatment and released into fresh dextrose media to repress SIC1 transcription. Samples were taken every 20 min, and the levels of Clb5 and Clb2 were analyzed by Western blotting. (C) Procedure was as in panel B, but α-factor was added 90 min after release to arrest cells in the following G1. Samples were collected every 15 min for analysis of budding, binucleate formation, and Nop1 segregation. Images on the right show representative cells 140 min after release from G1.
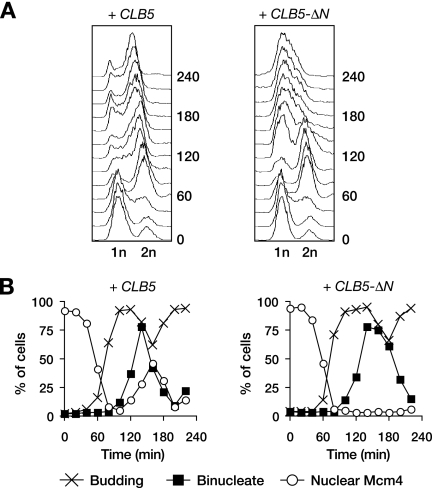
FIG. 4.
CLB5_-Δ_N causes DNA replication defects. (A) CLB5 and CLB5_-Δ_N cells were released from G1 arrest in the absence of SIC1 as in Fig. 3B, and flow cytometry was used to analyze cellular DNA content. (B) Procedure was as in panel A but with MCM4-GFP cells, which were analyzed for budding, binucleates, and the localization of the Mcm4-GFP protein.
FIG. 5.
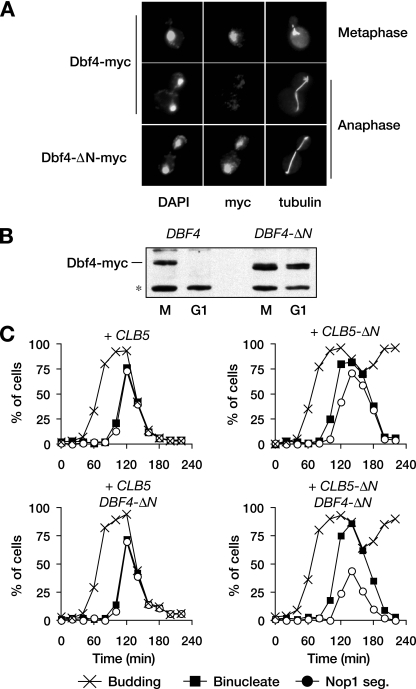
Stabilization of Dbf4 contributes to the rDNA segregation delay. (A) Representative images of Dbf4-myc in metaphase or anaphase from wild-type or DBF4_-Δ_N cells. (B) DBF4 and DBF4_-Δ_N cells were arrested in M phase by nocodazole treatment or G1 by α-factor treatment, and the levels of Dbf4 were analyzed. The asterisk indicates a nonspecific background band recognized by the anti-myc antibody. (C) DBF4 and DBF4-ΔN cells, carrying an extra copy of either CLB5 or CLB5_-Δ_N, were released from a G1 arrest as in Fig. 3C, and α-factor was re-added 80 min after release to arrest cells in the following G1. Samples were collected every 20 min for analysis of budding, binucleate formation, and Nop1 segregation.
RESULTS
Resolution of the rDNA requires APCCdc20-mediated degradation of Clb5 and securin.
To test whether the destruction of APCCdc20 targets other than securin governs rDNA resolution, we examined chromosome segregation in budding-yeast cells lacking Cdc20 and securin (Pds1 in yeast). Wild-type, _cdc20_Δ, and _cdc20_Δ _pds1_Δ strains, expressing Cdc20 from the repressible GAL1-10 promoter, were arrested in G1 by α-factor treatment and then released into dextrose media to repress transcription of CDC20. Budding and binucleate formation were scored. To examine rDNA segregation, we used immunofluorescence analysis of Nop1, a nucleolar protein that is involved in rDNA processing. Nop1 associates with the rDNA throughout the budding-yeast cell cycle, and its division into two distinct foci at anaphase is a useful marker of rDNA segregation. Wild-type cells segregated bulk DNA and rDNA with roughly equivalent timing, while _cdc20_Δ cells failed to segregate either (Fig. 1). In contrast, bulk chromosome segregation in _cdc20_Δ _pds1_Δ cells occurred with normal timing, but rDNA segregation was delayed. Destruction of APCCdc20 targets other than securin might therefore be necessary for efficient rDNA segregation.
Deletion of CLB5 is known to restore viability to _cdc20_Δ _pds1_Δ mutants (24). As Clb5 is an APCCdc20 substrate, this suggests that the failure of _cdc20_Δ _pds1_Δ yeast to efficiently segregate their rDNA might be due to the continued presence of Clb5. Consistent with this possibility, we found that deletion of CLB5 almost completely abolished the rDNA segregation delay in the _cdc20_Δ _pds1_Δ strain (Fig. 1). Thus, APCCdc20 promotes rDNA segregation at anaphase onset by targeting both securin and Clb5 for destruction.
rDNA cohesion is controlled specifically by Clb5-Cdk1 activity.
The rDNA segregation defect caused by Clb5 could be due to either a specific role for Clb5-Cdk1 or a general increase in Cdk1 activity. To distinguish between these possibilities, we compared the effects of different cyclins on rDNA segregation. Cells were released from G1 in the presence of galactose-inducible Clb5 or Clb2 protein (each stabilized by removal of amino-terminal APC-targeting domains, as discussed below). Cells overexpressing either cyclin arrested as large-budded cells with segregated DNA masses (Fig. 2A), and examination of chromosome III segregation revealed that bulk chromatin segregated accurately (data not shown). Strikingly, however, rDNA segregation was almost completely blocked in cells overexpressing Clb5 but was unperturbed in cells overexpressing Clb2. This result indicates that rDNA cohesion is mediated specifically by Clb5-Cdk1 activity.
Stabilized Clb5 mutants cause rDNA segregation defects.
If efficient rDNA resolution requires Clb5 destruction, then segregation defects should result when stabilized mutant forms of Clb5 are expressed at physiological levels. APCCdc20 recognizes its substrates through interactions with specific sequence motifs such as the D-box (22). The amino-terminal region of Clb5 contains a D-box, the removal of which partly stabilizes it in anaphase (the Clb5-Δdb mutant) (39). Clb5-Δdb is not lethal and has no clear effect on mitotic progression. We examined rDNA segregation in asynchronous populations of wild-type and CLB5_-Δ_db cells. 92% of wild-type cells that had segregated their chromosomes had also segregated their rDNA, whereas this was the case for only 65% of CLB5_-Δ_db cells (Fig. 2B). Thus, partly stabilized Clb5 slows rDNA segregation.
Many APC substrates contain multiple destruction determinants in addition to the D-box. Full stabilization of cyclins, for example, often requires the deletion of their entire N-terminal regions. To produce fully stabilized Clb5, we removed its 95 amino-terminal residues, leaving its Cdk1-binding domain intact (the Clb5-ΔN mutant). Transformation of wild-type cells with a plasmid carrying CLB5_-Δ_N under the control of its own promoter was lethal. This lethality was suppressed by overexpression of SIC1 from the GAL1-10 promoter (Fig. 3A). When expression of GAL-SIC1 was repressed by plating on glucose media, cells carrying the CLB5_-Δ_N formed inviable microcolonies. To confirm that Clb5 was stabilized by the removal of its N terminus, cells containing an extra copy of either CLB5 or CLB5_-Δ_N were released from G1 arrest in the absence of SIC1 expression. Levels of wild-type Clb5 fell at anaphase onset, while the amount of Clb5-ΔN was unchanged (Fig. 3B). Thus, removal of the N terminus of Clb5 stabilizes it in anaphase and causes cell lethality.
We analyzed anaphase progression in CLB5 and CLB5_-Δ_N cells, again shutting off SIC1 expression after release from G1. α-Factor was added back to the cultures after the initiation of budding. In CLB5-ΔN cells, budding and chromosome segregation occurred with normal timing (Fig. 3C). However, there was a significant delay in rDNA segregation in these cells, demonstrating that Clb5 degradation is necessary for timely resolution of the rDNA. Unlike wild-type cells, CLB5_-Δ_N cells failed to arrest in the following G1, suggesting that Clb5-ΔN prevented the G1 arrest mediated by mating pheromone.
We considered the possibility that Clb5-ΔN prevents rDNA segregation by blocking activation of the phosphatase Cdc14, which is required for rDNA segregation (5, 28). However, the phenotypes of the CLB5_-Δ_N mutant are clearly distinct from those of temperature-sensitive cdc14 mutants. cdc14-1 cells, in addition to delaying rDNA segregation, fail to break down the anaphase spindle, fail to undergo cytokinesis and do not enter the next cell cycle (26). In contrast, CLB5_-Δ_N cells, like CLB5_-Δ_db cells (39), showed no significant defects in mitotic exit, as judged by spindle disassembly, cytokinesis, or new cycle entry, although Clb2 destruction was slightly delayed (Fig. 3B). Furthermore, previous studies of _cdc20_Δ _pds1_Δ cells showed that Cdc14 is released efficiently from the nucleolus at anaphase onset even though Clb5 levels remain high (24). Together, these lines of evidence suggest that the delayed rDNA segregation of CLB5_-Δ_N cells is not due to problems with Cdc14 activation but instead that Cdk1-Clb5 promotes rDNA cohesion through another mechanism.
One possibility is that Clb5 opposes the effects of Cdc14 on the phosphorylation state of Cdk1 targets involved in rDNA cohesion. Consistent with this possibility, we found that deletion of CLB5 partly suppressed rDNA segregation defects in cdc14-1 cells. When arrested for 2.5 h at 37°C, rDNA segregation occurred in 44% of cdc14-1 cells and 68% of cdc14-1 Δ_clb5_ cells in three separate experiments.
We next tested if the lethality of the CLB5_-Δ_N mutation results from a failure to efficiently segregate chromosome XII, which contains the rDNA. We used a yeast strain from which the entire rDNA locus is deleted and viability is supported by multiple copies of a plasmid carrying an rDNA repeat (36). A GAL-SIC1 plasmid was introduced into this strain, along with a plasmid carrying either CLB5 or CLB5_-Δ_N. Deletion of the rDNA locus did not restore the viability of CLB5_-Δ_N cells when SIC1 expression was repressed on dextrose media (Fig. 3A). We conclude that rDNA segregation defects are not the sole cause of CLB5_-Δ_N lethality.
DNA replication fails in CLB5_-Δ_N cells.
Clb5-Cdk1 is a critical regulator of DNA replication. To investigate whether DNA replication is aberrant in cells expressing stabilized Clb5, we measured the DNA content of cells expressing CLB5 or CLB5_-Δ_N after release from G1 in the absence of Sic1. Both strains completed the first round of DNA replication and progressed into mitosis with similar timing, revealing no significant defects in the first cycle of DNA replication (Fig. 4A). The CLB5 strain then entered the next cell cycle and completed another S phase and progressed through the second and subsequent cell cycles. In contrast, CLB5_-Δ_N cells failed to undergo a second round of DNA replication and arrested as large-budded cells with a 1n DNA content (Fig. 4A). Thus, the continued presence of Clb5 after anaphase prevents DNA replication and thereby causes cell death.
We suspected that Clb5-ΔN blocked DNA replication by inhibiting the formation of prereplicative complexes (pre-RCs) at replication origins. From early S to late M phases, Cdk1 blocks pre-RC assembly by phosphorylating and thereby inhibiting several pre-RC subunits; Cdk1 inactivation in late mitosis then triggers pre-RC assembly (1, 20). A key pre-RC component is the Mcm complex, the phosphorylation of which prevents its nuclear import (14, 15). We examined the localization of a GFP-tagged Mcm complex in cells released from a G1 arrest. In cells expressing wild-type CLB5, the Mcm complex moved from the nucleus to the cytoplasm during S phase and reentered the nucleus in late mitosis (Fig. 4B). In contrast, late-mitotic nuclear reentry of the Mcm complex did not occur in CLB5_-Δ_N cells, and it remained cytoplasmic. Thus, the continued presence of Clb5 after anaphase blocks the nuclear import of the Mcm complex, presumably preventing efficient pre-RC formation. It seems likely that this problem leads to defects in the initiation of DNA replication, and the resulting replication stress triggers a DNA damage response that blocks cell cycle progression.
APCCdc20-mediated destruction of Dbf4 contributes to timely rDNA disjunction.
We noted earlier that _cdc20_Δ _pds1_Δ cells exhibited a 30-min delay in rDNA segregation (Fig. 1). However, expression of Clb5-ΔN in otherwise wild-type cells caused only a 15-min delay in rDNA segregation (Fig. 3C). One explanation for this difference is that Clb5-ΔN is not fully stabilized, but we could find no evidence for that (Fig. 3B). An alternate possibility is that degradation of an additional APCCdc20 substrate contributes to rDNA segregation. One appealing candidate was Dbf4, the activating subunit for the protein kinase Cdc7. Dbf4 is known to be an APC substrate whose levels drop in mitosis, although the precise timing of its destruction and the APC cofactor involved are not known (7). We found that APCCdc20 ubiquitinates Dbf4 in vitro and that Dbf4 is destroyed at anaphase onset, independently of CDH1 (Fig. 5A and data not shown), suggesting that Dbf4 is an APCCdc20 substrate. Removal of its amino-terminal destruction sequences stabilized Dbf4 in mitosis and G1 (Fig. 5A and B).
To assess the contribution of Dbf4 to rDNA segregation, we constructed a strain in which the endogenous DBF4 gene was replaced with DBF4_-Δ_N. This strain displayed no apparent defects in cell viability, mitotic progression, or rDNA segregation (Fig. 5C). We then introduced either CLB5 or CLB5_-Δ_N in combination with GAL-SIC1 and examined cell cycle progression. Notably, cells carrying both CLB5_-Δ_N and DBF4_-Δ_N exhibited an rDNA segregation defect that was more severe than that seen in CLB5_-Δ_N cells. The average delay in rDNA segregation relative to bulk chromatin in the CLB5_-Δ_N DBF4_-Δ_N cells was similar to that in _cdc20_Δ _pds1_Δ cells, and many cells failed to separate their nucleolus before cytokinesis. Thus, Clb5 and Dbf4 are APCCdc20 targets that must both be destroyed for efficient rDNA segregation.
Meiotic rDNA segregation fails in the continued presence of Clb5 and Dbf4.
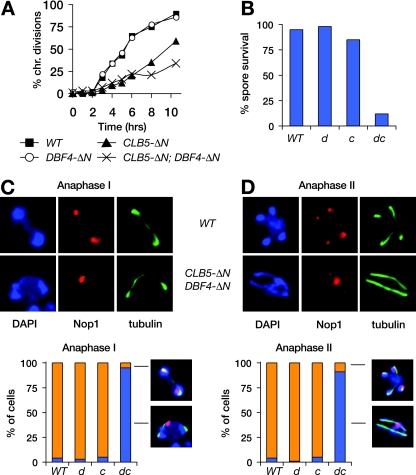
In anaphase I of meiosis, homologs are resolved by loss of cohesin on the chromosome arms. However, rDNA cohesion must also be resolved to allow segregation of the chromosome XII homologs. We investigated whether meiotic rDNA segregation involves the same mechanisms as those we identified in mitosis. Immunofluorescence analysis revealed that both Clb5 and Dbf4 were present at high levels in metaphase I and absent in anaphase I, consistent with degradation via APCCdc20 (data not shown). We then expressed stabilized Clb5 and/or Dbf4 in the SK1 budding-yeast strain, which allows analysis of synchronous meiosis. Replacement of endogenous DBF4 with DBF4_-Δ_N had no apparent impact on meiosis (Fig. 6A and B). Expression of CLB5_-Δ_N from the meiosis-specific DMC1 promoter caused a delay in meiotic progression and a small decrease in spore viability (Fig. 6A and B) but no apparent defect in rDNA segregation in anaphase I. However, cells expressing both Clb5-ΔN and Dbf4-ΔN exhibited defects in meiotic progression and particularly in spore viability. Strikingly, rDNA segregation failed completely in the double mutant cells, as can be seen clearly in both anaphase I and II cells, where only a single, unsegregated nucleolar mass was visible (Fig. 6C and D). Thus, APCCdc20-mediated destruction of both Clb5 and Dbf4 contributes to rDNA segregation in meiosis, and a failure to destroy both in anaphase I completely prevents rDNA disjunction.
FIG. 6.
Meiotic rDNA segregation fails in the presence of stabilized Dbf4 and Clb5. Diploid DBF4 and DBF4_-Δ_N SK1 cells, with or without pDMC1-CLB5_-Δ_N, were followed through synchronous meiosis. Chromosome divisions were scored (A), as were the survival rates of resulting spores (B) and the segregation patterns of Nop1 through both anaphase I (C) and anaphase II (D). Histograms are labeled WT (wild type), d (DBF4_-Δ_N), c (pDMC1-CLB5_-Δ_N), and dc (DBF4-ΔN with pDMC1-CLB5_-Δ_N).
DISCUSSION
Sister chromatid cohesion depends primarily on cohesin linkages (19). In budding yeast, an additional mechanism operates specifically at the rDNA locus of chromosome XII. Removal of both cohesive mechanisms requires APCCdc20-dependent degradation of securin and the liberation of separase, which cleaves a subunit of cohesin and also promotes dissolution of rDNA cohesion by activating the phosphatase Cdc14 (5, 27, 28, 31). Although it is well established that the only critical role of APCCdc20 in cohesin removal is the promotion of securin degradation, it was not known if the destruction of additional targets is necessary for the resolution of rDNA. Our results demonstrate that efficient rDNA resolution also depends on APCCdc20-dependent destruction of Clb5 and Dbf4.
Although delayed, rDNA segregation does eventually occur in cells expressing stabilized forms of Clb5 and Dbf4, indicating that the destruction of these proteins is not essential for rDNA segregation in mitosis. In meiosis, however, rDNA segregation fails completely in the presence of stabilized Clb5 and Dbf4, revealing a critical function for the inactivation of these kinases in meiotic anaphase.
Previous evidence that the phosphatase Cdc14 promotes rDNA segregation (5, 28) suggested that rDNA cohesion depends on the phosphorylation of Cdk1 substrates. Our results are consistent with this possibility and also suggest that rDNA resolution depends specifically on the dephosphorylation of Clb5-Cdk1 targets, as rDNA segregation was not significantly affected by Clb2 overexpression. Given the known timing of Clb5 and Clb2 destruction, this cyclin specificity supports a model (Fig. 7) in which Clb5 destruction and Cdc14 activation trigger the dephosphorylation in early anaphase of specific Cdk1 targets involved in rDNA segregation. The significant levels of Clb2 that remain in the anaphase cell are apparently insufficient to maintain the phosphorylation of these targets in the face of separase-dependent Cdc14 activation. However, we speculate that Clb2-Cdk1 activity in anaphase does maintain the phosphorylation state of other Cdk1 targets involved in spindle disassembly and mitotic exit; the dephosphorylation of these targets occurs after anaphase as a result of Clb2 destruction, perhaps coupled with the increased Cdc14 activity that results from activation of the mitotic-exit network (30). Stabilization of Clb2 therefore blocks mitotic exit but not rDNA segregation or other anaphase processes.
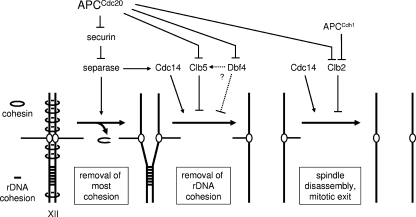
FIG. 7.
Model of regulatory mechanisms governing chromosome segregation in budding yeast. Bulk chromosome separation occurs when cohesin is cleaved by separase, the activation of which results from the APCCdc20-dependent destruction of securin. Efficient removal of rDNA cohesion also requires the dephosphorylation of Clb5-specific Cdk1 targets, which results from the activation of Cdc14 by separase and the destruction of Clb5 via APCCdc20. By uncertain mechanisms, APCCdc20-dependent Dbf4 destruction also contributes. Destruction of Clb2 allows dephosphorylation of its targets after anaphase, leading to spindle disassembly and mitotic exit.
What are the key targets of Clb5-Cdk1 in rDNA cohesion? The molecular basis of rDNA cohesion is not well understood but seems to depend on complex mechanisms involving topoisomerase II, sumoylation, condensin, and RNA polymerase I (5, 10, 18, 28, 33, 34, 38). Clb5-Cdk1 may act through one or multiple substrates to govern these mechanisms. We identified several Clb5-specific Cdk1 substrates in our previous work (16), but none of these appear to be a clear candidate for the control of rDNA cohesion. A significant future challenge will be the systematic identification of all Clb5-specific Cdk1 substrates, which should allow discovery of the critical Clb5 substrates involved in rDNA resolution and other anaphase processes.
We also identified the Cdc7-binding partner Dbf4 as a potential regulator of rDNA cohesion in budding yeast. Dbf4-Cdc7 is known primarily as a regulator of the initiation of DNA synthesis, and no mitotic roles have been described for this kinase. How might Dbf4-Cdc7 regulate rDNA cohesion? The fact that the DBF4_-Δ_N mutant has no apparent cell cycle defect alone but enhances the segregation delay from the CLB5_-Δ_N mutant suggests that one function of Dbf4-Cdc7 might be to enhance Clb5-Cdk1 function, either by increasing its intrinsic kinase activity or by enhancing its activity toward some Clb5-Cdk1 target. Alternatively, Dbf4-Cdc7 could promote cohesion by directly phosphorylating some component of the rDNA machinery. Notably, recent work suggests that Dbf4-Cdc7 helps promote DNA recombination in meiotic prophase and monopolar chromosome attachment in meiosis I (23, 35, 37). Dbf4-Cdc7 might therefore be involved in the control of multiple processes other than DNA replication.
In fission yeast and higher eukaryotes, it is not clear if there are cohesin-independent chromosome linkages like those operating at budding-yeast rDNA. The fission yeast ortholog of Cdc14, Clp1, is not required for rDNA segregation (3), but it remains possible that other phosphatases contribute to the dephosphorylation of Cdk1 substrates involved in rDNA cohesion. In mammalian cells, noncohesin rDNA linkages, if they exist, may be lost early in mitosis as a result of the “prophase” pathway that drives chromosome arm resolution and separation before metaphase (8, 13). Alternatively, rDNA resolution in animal cells might depend on the degradation of cyclin A in prometaphase. In some cell types, expression of APC-resistant cyclin A leads to a metaphase-like arrest with unseparated sister chromatids (6, 9, 21, 25), and cohesin-independent linkages may contribute to this separation defect.
Future models of mitosis should take into account the qualitative contributions of individual cyclin-Cdk1 complexes, in terms of both phosphorylation events early in the cell cycle and dephosphorylation events as cyclins are sequentially destroyed in late mitosis. Early destruction of cyclins that display strong substrate specificity, such as Clb5 and cyclin A, may be required for the correct timing of anaphase events. The substrate specificity of phosphatases such as Cdc14 provides another layer of complexity in the control of Cdk substrate dephosphorylation. These mechanisms collaborate to ensure that key Cdk substrates involved in processes such as chromosome segregation and spindle elongation are dephosphorylated at anaphase onset, while dephosphorylation of other Cdk substrates is delayed until later in mitosis.
Acknowledgments
We are grateful to A. Amon, F. Cross, J. Diffley, K. Nasmyth, M. Nomura, and F. Uhlmann for reagents and to members of the Morgan laboratory for discussions and comments on the manuscript.
This work was supported by funding from the National Institute of General Medical Sciences.
Footnotes
▿
Published ahead of print on 30 June 2008.
REFERENCES
- 1.Arias, E. E., and J. C. Walter. 2007. Strength in numbers: preventing rereplication via multiple mechanisms in eukaryotic cells. Genes Dev. 21497-518. [DOI] [PubMed] [Google Scholar]
- 2.Bloom, J., and F. R. Cross. 2007. Multiple levels of cyclin specificity in cell-cycle control. Nat. Rev. Mol. Cell Biol. 8149-160. [DOI] [PubMed] [Google Scholar]
- 3.Chen, C. T., M. P. Peli-Gulli, V. Simanis, and D. McCollum. 2006. S. pombe FEAR protein orthologs are not required for release of Clp1/Flp1 phosphatase from the nucleolus during mitosis. J. Cell Sci. 1194462-4466. [DOI] [PubMed] [Google Scholar]
- 4.Cross, F. R., M. Yuste-Rojas, S. Gray, and M. D. Jacobson. 1999. Specialization and targeting of B-type cyclins. Mol. Cell 411-19. [DOI] [PubMed] [Google Scholar]
- 5.D'Amours, D., F. Stegmeier, and A. Amon. 2004. Cdc14 and condensin control the dissolution of cohesin-independent chromosome linkages at repeated DNA. Cell 117455-469. [DOI] [PubMed] [Google Scholar]
- 6.den Elzen, N., and J. Pines. 2001. Cyclin A is destroyed in prometaphase and can delay chromosome alignment and anaphase. J. Cell Biol. 153121-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferreira, M. F., C. Santocanale, L. S. Drury, and J. F. Diffley. 2000. Dbf4p, an essential S phase-promoting factor, is targeted for degradation by the anaphase-promoting complex. Mol. Cell. Biol. 20242-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gandhi, R., P. J. Gillespie, and T. Hirano. 2006. Human Wapl is a cohesin-binding protein that promotes sister-chromatid resolution in mitotic prophase. Curr. Biol. 162406-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geley, S., E. Kramer, C. Gieffers, J. Gannon, J. M. Peters, and T. Hunt. 2001. Anaphase-promoting complex/cyclosome-dependent proteolysis of human cyclin A starts at the beginning of mitosis and is not subject to the spindle assembly checkpoint. J. Cell Biol. 153137-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirano, T., G. Konoha, T. Toda, and M. Yanagida. 1989. Essential roles of the RNA polymerase I largest subunit and DNA topoisomerases in the formation of fission yeast nucleolus. J. Cell Biol. 108243-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacobson, M. D., S. Gray, M. Yuste-Rojas, and F. R. Cross. 2000. Testing cyclin specificity in the exit from mitosis. Mol. Cell. Biol. 204483-4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khmelinskii, A., C. Lawrence, J. Roostalu, and E. Schiebel. 2007. Cdc14-regulated midzone assembly controls anaphase B. J. Cell Biol. 177981-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kueng, S., B. Hegemann, B. H. Peters, J. J. Lipp, A. Schleiffer, K. Mechtler, and J. M. Peters. 2006. Wapl controls the dynamic association of cohesin with chromatin. Cell 127955-967. [DOI] [PubMed] [Google Scholar]
- 14.Labib, K., J. F. Diffley, and S. E. Kearsey. 1999. G1-phase and B-type cyclins exclude the DNA-replication factor Mcm4 from the nucleus. Nat. Cell Biol. 1415-422. [DOI] [PubMed] [Google Scholar]
- 15.Liku, M. E., V. Q. Nguyen, A. W. Rosales, K. Irie, and J. J. Li. 2005. CDK phosphorylation of a novel NLS-NES module distributed between two subunits of the Mcm2-7 complex prevents chromosomal rereplication. Mol. Biol. Cell 165026-5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loog, M., and D. O. Morgan. 2005. Cyclin specificity in the phosphorylation of cyclin-dependent kinase substrates. Nature 434104-108. [DOI] [PubMed] [Google Scholar]
- 17.Morgan, D. O. 2007. The cell cycle: principles of control. New Science Press, London, United Kingdom.
- 18.Nakazawa, N., T. Nakamura, A. Kokubu, M. Ebe, K. Nagao, and M. Yanagida. 2008. Dissection of the essential steps for condensin accumulation at kinetochores and rDNAs during fission yeast mitosis. J. Cell Biol. 1801115-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nasmyth, K. 2002. Segregating sister genomes: the molecular biology of chromosome separation. Science 297559-565. [DOI] [PubMed] [Google Scholar]
- 20.Nguyen, V. Q., C. Co, and J. J. Li. 2001. Cyclin-dependent kinases prevent DNA re-replication through multiple mechanisms. Nature 4111068-1073. [DOI] [PubMed] [Google Scholar]
- 21.Parry, D. H., and P. H. O'Farrell. 2001. The schedule of destruction of three mitotic cyclins can dictate the timing of events during exit from mitosis. Curr. Biol. 11671-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peters, J. M. 2006. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat. Rev. Mol. Cell Biol. 7644-656. [DOI] [PubMed] [Google Scholar]
- 23.Sasanuma, H., K. Hirota, T. Fukuda, N. Kakusho, K. Kugou, Y. Kawasaki, T. Shibata, H. Masai, and K. Ohta. 2008. Cdc7-dependent phosphorylation of Mer2 facilitates initiation of yeast meiotic recombination. Genes Dev. 22398-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shirayama, M., A. Toth, M. Galova, and K. Nasmyth. 1999. APC-Cdc20 promotes exit from mitosis by destroying the anaphase inhibitor Pds1 and cyclin Clb5. Nature 402203-207. [DOI] [PubMed] [Google Scholar]
- 25.Sigrist, S., H. Jacobs, R. Stratmann, and C. F. Lehner. 1995. Exit from mitosis is regulated by Drosophila fizzy and the sequential destruction of cyclins A, B and B3. EMBO J. 144827-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stegmeier, F., and A. Amon. 2004. Closing mitosis: the functions of the Cdc14 phosphatase and its regulation. Annu. Rev. Genet. 38203-232. [DOI] [PubMed] [Google Scholar]
- 27.Stegmeier, F., R. Visintin, and A. Amon. 2002. Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell 108207-220. [DOI] [PubMed] [Google Scholar]
- 28.Sullivan, M., T. Higuchi, V. L. Katis, and F. Uhlmann. 2004. Cdc14 phosphatase induces rDNA condensation and resolves cohesin-independent cohesion during budding yeast anaphase. Cell 117471-482. [DOI] [PubMed] [Google Scholar]
- 29.Sullivan, M., and D. O. Morgan. 2007. A novel destruction sequence targets the meiotic regulator Spo13 for anaphase-promoting complex-dependent degradation in anaphase I. J. Biol. Chem. 28219710-19715. [DOI] [PubMed] [Google Scholar]
- 30.Sullivan, M., and D. O. Morgan. 2007. Finishing mitosis, one step at a time. Nat. Rev. Mol. Cell Biol. 8894-903. [DOI] [PubMed] [Google Scholar]
- 31.Sullivan, M., and F. Uhlmann. 2003. A non-proteolytic function of separase links the onset of anaphase to mitotic exit. Nat. Cell Biol. 5249-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thornton, B. R., and D. P. Toczyski. 2006. Precise destruction: an emerging picture of the APC. Genes Dev. 203069-3078. [DOI] [PubMed] [Google Scholar]
- 33.Tomson, B. N., D. D'Amours, B. S. Adamson, L. Aragon, and A. Amon. 2006. Ribosomal DNA transcription-dependent processes interfere with chromosome segregation. Mol. Cell. Biol. 266239-6247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Torres-Rosell, J., F. Machin, A. Jarmuz, and L. Aragon. 2004. Nucleolar segregation lags behind the rest of the genome and requires Cdc14p activation by the FEAR network. Cell Cycle 3496-502. [PubMed] [Google Scholar]
- 35.Valentin, G., E. Schwob, and F. Della Seta. 2006. Dual role of the Cdc7-regulatory protein Dbf4 during yeast meiosis. J. Biol. Chem. 2812828-2834. [DOI] [PubMed] [Google Scholar]
- 36.Wai, H. H., L. Vu, M. Oakes, and M. Nomura. 2000. Complete deletion of yeast chromosomal rDNA repeats and integration of a new rDNA repeat: use of rDNA deletion strains for functional analysis of rDNA promoter elements in vivo. Nucleic Acids Res. 283524-3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wan, L., H. Niu, B. Futcher, C. Zhang, K. M. Shokat, S. J. Boulton, and N. M. Hollingsworth. 2008. Cdc28-Clb5 (CDK-S) and Cdc7-Dbf4 (DDK) collaborate to initiate meiotic recombination in yeast. Genes Dev. 22386-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang, B. D., V. Yong-Gonzalez, and A. V. Strunnikov. 2004. Cdc14p/FEAR pathway controls segregation of nucleolus in S. cerevisiae by facilitating condensin targeting to rDNA chromatin in anaphase. Cell Cycle 3960-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wäsch, R., and F. Cross. 2002. APC-dependent proteolysis of the mitotic cyclin Clb2 is essential for mitotic exit. Nature 418556-562. [DOI] [PubMed] [Google Scholar]
- 40.Woodbury, E. L., and D. O. Morgan. 2007. Cdk and APC activities limit the spindle-stabilizing function of Fin1 to anaphase. Nat. Cell Biol. 9106-112. [DOI] [PubMed] [Google Scholar]