Expression and Adhesion Profiles of SynCAM Adhesion Molecules Indicate Distinct Neuronal Functions (original) (raw)
. Author manuscript; available in PMC: 2009 Sep 1.
Published in final edited form as: J Comp Neurol. 2008 Sep 1;510(1):47–67. doi: 10.1002/cne.21773
Abstract
Cell-cell interactions through adhesion molecules play key roles in the development of the nervous system. SynCAMs (Synaptic Cell Adhesion Molecules) comprise a group of four immunoglobulin (Ig) superfamily members that mediate adhesion and are prominently expressed in the brain. Although SynCAMs have been implicated in the differentiation of neurons, there has been no comprehensive analysis of their expression patterns. We here examine the spatiotemporal expression patterns of SynCAMs using RT-PCR, in situ hybridization, and immunohistological techniques. SynCAMs 1–4 are widely expressed throughout the developing and adult central nervous system. They are prominently expressed in neurons throughout the brain and present in both excitatory and inhibitory neurons. Investigation of different brain regions in the developing and mature mouse brain indicates that each SynCAM exhibits a distinct spatiotemporal expression pattern. This is observed in all regions analyzed and particularly notable in the cerebellum, where SynCAMs display highly distinct expression in cerebellar granule and Purkinje cells. These unique expression profiles are complemented by specific heterophilic adhesion patterns of SynCAM family members as shown by cell overlay experiments. Three prominent interactions are observed, mediated by the extracellular domains of SynCAMs 1/2, 2/4, and 3/4. These expression and adhesion profiles of SynCAMs together with their previously reported functions in synapse organization indicate that SynCAM proteins contribute importantly to the synaptic circuitry of the central nervous system.
Keywords: SynCAM, CADM, TSLC, nectin-like molecule, adhesion, synapse
INTRODUCTION
SynCAMs (Synaptic Cell Adhesion Molecules) are four membrane proteins belonging to the immunoglobulin (Ig) superfamily that are encoded by the CADM1–4 (Cell Adhesion Molecule) genes (Biederer, 2006). Due to their identification in different contexts, these proteins have also been named TSLC (tumor suppressor in lung carcinoma) proteins (Kuramochi et al., 2001) and Necls (nectin-like molecules) (Shingai et al., 2003). SynCAM proteins consist of an extracellular sequence with three Ig-like domains that mediate adhesive interactions followed by a single transmembrane region and a short cytosolic sequence containing binding motifs for FERM and PDZ domains (Biederer, 2006). SynCAM 1 protein is predominantly expressed in brain, but also found in lung and testis (Fogel et al., 2007) as well as in cancer cell lines (Kuramochi et al., 2001) and mast cells (Furuno et al., 2005). SynCAM 2, 3, and 4 proteins are detected solely in the nervous system, consistent with brain-specific functions of these adhesion molecules (Fogel et al., 2007). In the developing hippocampus, all SynCAMs appear to be expressed predominantly in neurons and localize to synapses in vitro, consistent with their fractionation as synaptic plasma membrane proteins in brain (Fogel et al., 2007). These observations are supported by immunolocalization studies using an antibody that simultaneously recognizes SynCAM 1–3, showing that these family members are expressed at both pre- and postsynaptic membrane specializations in vivo (Biederer et al., 2002). In synaptic membranes, SynCAM 1 and 2 form a heterophilic adhesion complex (Fogel et al., 2007). Here, both proteins organize synapses by driving formation of functional presynaptic terminals and promote excitatory synaptic transmission (Biederer et al., 2002; Sara et al., 2005; Fogel et al., 2007).
Through their extracellular Ig-like domains, the four SynCAM proteins engage in multiple, calcium-independent adhesive interactions both within and outside the family. Specifically, SynCAMs 1, 2, and 3 are homophilic adhesion molecules, while two heterophilic adhesion complexes comprised of SynCAM 1/2 and 3/4 have been demonstrated more recently (Biederer et al., 2002; Shingai et al., 2003; Kakunaga et al., 2005; Fogel et al., 2007; Maurel et al., 2007; Spiegel et al., 2007). Outside the SynCAM family, extracellular interaction partners of SynCAM 1 are the T-cell receptor CRTAM, which mediates activation of natural killer and T cells in the immune system (Arase et al., 2005; Boles et al., 2005), and the protein nectin 3 (Shingai et al., 2003). Other identified partners of SynCAM 3 are the proteins nectin 1 and 3 (Kakunaga et al., 2005). While the CRTAM and nectin interaction partners are non-homologous to SynCAMs, they also belong to the Ig superfamily of proteins. The biological significance of their interactions in brain remains to be elucidated.
SynCAMs are enriched in synaptic plasma membranes, can engage each other as trans-synaptic adhesion molecules, and share synapse-organizing activities. Based on these properties, synaptic SynCAM adhesion complexes have been proposed to subserve development of the central nervous system (Fogel et al., 2007). Additional evidence for a role of SynCAM proteins in interactions of neurons with glia supports broad functions of these proteins in the development of the nervous system. In the central nervous system, SynCAM 3 is present not only at contact sites between neurons and axons, but also at adhesion sites between neurons and astrocyte processes at cerebellar synapses (Kakunaga et al., 2005). In the peripheral nervous system, SynCAM 3 is present along myelinated axons while SynCAM 4 is expressed in Schwann cells, and both proteins mediate adhesion at internodes between axons and Schwann cells that is required for proper myelination (Maurel et al., 2007; Spiegel et al., 2007). The SynCAM family therefore plays important roles in the development of both the central and peripheral nervous systems.
Though there are several reports of the activities of these proteins in the nervous system, there is no comprehensive overview of SynCAM expression in the brain. We therefore examined the distribution of these adhesion molecules in the developing and mature mouse central nervous system using a combination of approaches. We find that all SynCAMs are expressed throughout the brain and are prominently neuronal, found in both excitatory as well as inhibitory neurons. However, these molecules are divergently expressed between neuronal populations. These differential SynCAM expression patterns are largely maintained from the developing to the mature brain. We combine these expression studies with an exhaustive analysis of SynCAM heterophilic interactions and report three major heteromeric complexes. These distinct spatial patterns of expression and their differential adhesion implicate the SynCAM family of adhesion molecules as important players in establishing local circuits throughout the central nervous system.
MATERIALS AND METHODS
Tissue
All animal procedures undertaken in this study were approved by the Yale University Institutional Animal Care and Use Committee and were in compliance with NIH guidelines.
Probes and antibodies
Real-time reverse transcription (RT) PCR oligonucleotide primer sets specific for SynCAM 1–4 and the control β-actin were obtained from Applied Biosystems (Foster City, CA) and are described in Supplemental Table 1. The in situ hybridization riboprobes used in this study are described in Supplemental Table 2 and were previously published (Fogel et al., 2007). They were generated as full-length riboprobes to match sequences present in all splice forms of SynCAM 1–4 (Biederer, 2006). SynCAM-encoding sequences were obtained by PCR amplification from a mouse cDNA library as previously described (Fogel et al., 2007) and were subcloned into the vectors indicated in Supplemental Table 2 following standard molecular biological procedures. All PCR products were confirmed by sequencing. Specific riboprobes for detection of VGLUT1 and 2 as well as GAD67 transcripts were previously published (Katarova et al., 2000; Landry et al., 2004). For detection of neuronal nuclei in sections processed for in situ hybridization, a monoclonal anti-NeuN antibody (Millipore MAB377, Billerica, MA) was employed that was raised in mice against brain cell nuclei and that specifically recognizes a neuronal 46–48 kDa nuclear protein in immunoblotting and immunostaining (Mullen et al., 1992; Lind et al., 2005). This anti-NeuN antibody stained all neuronal nuclei with exception of mitral cells in the olfactory bulb and Purkinje cells in the cerebellum as described previously (Mullen et al., 1992). For detection of Purkinje cell bodies in sections processed for in situ hybridization, an anti-inositol 1,4,5-trisphosphate (Ins(1,4,5)P3) receptor antibody was employed that was raised in rabbits against a peptide with the sequence RIGLLGHPPHMNVNPQQPA, corresponding to the 19 carboxyl-terminal amino acids of the protein, recognizing a 260 kDa membrane protein in immunoblotting of cerebellar lysates (Mignery et al., 1989). Strong staining of Purkinje cells in cerebellum with low levels of immunoreactivity in other brain regions was observed with this antibody as described previously (Mignery et al., 1989). This antibody was a gift from Dr. Pietro De Camilli (Yale University, Department of Cell Biology). For detection of Bergmann glia cell bodies in sections processed for in situ hybridization, a specific anti-Brain Lipid-Binding Protein (BLBP) antibody (Millipore AB9558, Billerica, MA) was employed that was raised in rabbits against full-length mouse BLBP heterologously expressed in E.coli and recognizes a 15 kDa brain protein in immunoblotting of cerebellar lysates (Feng et al., 1994). In cerebellar sections, strong cellular staining in the Purkinje cell layer was observed that morphologically corresponded to Bergmann glia cells as described previously (Feng et al., 1994). This antibody was a gift from Dr. Angelique Bordey (Yale University, Department of Neurosurgery).
Adhesion analysis by cell overlay
To detect protein interactions on cell surfaces using an overlay approach, HEK 293 cells were transfected with full-length CFP-tagged SynCAM proteins or soluble CFP as negative control, and then overlaid with soluble proteins corresponding to the complete SynCAM extracellular sequences. Full-length SynCAM proteins were tagged with CFP within their intracellular sequence to minimally interfere with extracellular adhesion. These pCMV5 based expression vectors for SynCAM1(420)CFP, SynCAM2(391)CFP, SynCAM3(377)CFP, and SynCAM4(369)CFP were previously described (Fogel et al., 2007). Bracketed numbers indicate the amino acid preceding the inserted CFP sequence. For heterologous expression of full-length SynCAM proteins, HEK 293 cells were transfected with FuGENE 6 (Roche Applied Science, Indianapolis, IN). To obtain proteins corresponding to the soluble SynCAM extracellular sequences, COS7 cells were transfected by the DEAE-dextran method (Gorman, 1985) with pCMVIG9 expression vectors encoding fusions of SynCAM 1, 2, 3, or 4 extracellular sequences with human IgG1-Fc that were previously described (Fogel et al., 2007). Heterologously expressed extracellular SynCAM IgG1-Fc fusion proteins were purified from cell supernatants as described (Sugita et al., 2001). For adhesion analysis, HEK 293 cells were split on day 1, transfected on day 2 at 50% confluency, seeded into 24 well plates on day 3 and overlay was performed on day 4. HEK 293 cells were overlaid with extracellular SynCAM IgG1-Fc fusion proteins purified from COS7 cell supernatants or control human IgG (Sigma, St. Louis, MO) at 10 µg/mL in culture medium supplemented with 25 mM HEPES. The IgG-Fc fusion tag was directly detected by including Protein A-Alexa 594 (6 µg/mL; Invitrogen, Carlsbad, CA) in this step. Incubation was performed for 20 min at 37° to allow for binding without endocytosis (Elior Peles, Weizmann Institute of Science, Israel, personal communication). The medium was replaced with DMEM and the cells were imaged immediately with a Hamamatsu Orca camera attached to a Nikon Eclipse TE2000-U microscope. Camera settings were kept constant within experiments. For each image, the total amount of red fluorescence was quantified and divided by the total amount of cyan fluorescence. CFP and Protein-A signals were quantified using a custom ImageJ macro. Specifically, the CFP signal was used to generate a region of interest (ROI). Background was subtracted from all images and the remaining signal was integrated over the ROI for both channels. The Protein A signal was then divided by the CFP signal. This macro is available upon request.
Quantitative RT-PCR
RT-PCR analysis was performed on pooled tissue from three animals, then run in duplicate. RNA was isolated from various brain tissues using Trizol (Invitrogen), and genomic DNA was removed using the Turbo DNA-free kit (Ambion). RNA was further purified using the RNeasy MinElute Cleanup Kit (Qiagen, Valencia, CA). cDNA was prepared by the W.M. Keck Facility at Yale University using the High-Capacity cDNA Archive Kit (Applied Biosystems). We performed quantitative probe-based real-time RT PCR in duplicate using an ABI 7900 Sequence Detection System (Applied Biosystems) at the W.M. Keck Facility with primer sets specific for each of the four SynCAMs and the control β-actin (see Supplemental Table 1). The threshold cycle value Ct for each condition was selected using the Primer Express software package (Applied Biosystems) and ΔΔCt’s were calculated relative to β-actin for each sample and to P0 olfactory bulb. All steps were performed according to the manufacturers’ instructions.
In situ hybridization
In situ hybridizations were performed at least three times from at least three animals. P15 (postnatal day 15, where P1 is the day of birth) or adult Balb/C mice (Charles River, Wilmington, MA) were anesthetized with isoflurane and perfusion fixed with 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS; 0.1 M phosphate buffer, 0.9% NaCl, pH 7.4), followed by immersion fixing in the same solution at 4°C overnight. For collection of spinal cord from newborn pups at P2, hypothermic anesthesia was induced and was followed by rapid decapitation. All tissue was rinsed for a minimum of 2 hrs in PBS after fixation before processing for microscopy. Tissue was cryoprotected by immersion in 30% sucrose in PBS at 4°C until tissue sank. Tissue was embedded in OCT compound (Sakura Finetek, Torrance, CA) and frozen and stored at −80° until sectioning. The tissue was serially sectioned in the sagittal or coronal plane (for spinal cord sections only) using a Reichert-Jung 2800 Frigocut B cryostat. 40 µm free-floating sections were cut on the cryostat, rinsed in PBS, and stored at −20°C in antifreeze (20% glycerol and 30% ethylene glycol in 0.05 M phosphate buffer, pH 7.4) until needed.
For detection, riboprobes labeled with digoxigenin-11-UTP or fluorescein-12-UTP (both Roche Applied Science) were generated using a MAXIscript kit (Ambion) as per the directions of the kit. In situ hybridization employing chromogenic or fluorescence detection was performed as follows. For the chromogenic detection, sections were rinsed three times in PBS. Tissue was then treated with a 10 minute immersion in 0.2 M HCl. Tissue was immersed in 1% Triton X-100 in PBS for 2 minutes, followed by two 1 minute washes in PBS. Tissue was next treated with 50% formamide/5X SSC (1X SSC = 15 mM sodium citrate, 150 mM sodium chloride, pH 7.0) for 10 minutes. Finally, sections were incubated in the appropriate digoxigenin-labeled riboprobe at 1 ng/µL in hybridization solution (50% formamide, 10mM Tris (pH 8.0), 200 µg/mL yeast tRNA, 10% dextran sulfate, 1x Denhardt’s Solution, 600 mM sodium chloride, 0.25% SDS, 1 mM EDTA) overnight at 65° C. Tissue was washed twice in 0.1x SSC at 65° C for 10 minutes each time. Tissue was blocked for 30 minutes in TNB (100 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.5% Blocking Reagent (Perkin Elmer, Fremont, CA)). Tissue was then incubated with a polyclonal antibody against digoxigenin conjugated to alkaline phosphatase (1:1000; Roche Applied Science) in TNB for 60 minutes and subsequently washed in TNT buffer (100 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.05% Tween-20). Sections were then immersed in AP buffer (100 mM Tris (pH 9.5), 100 mM sodium chloride, 5 mM magnesium chloride) for 10 minutes and incubated in NBT/BCIP (Roche Applied Science) diluted 1:50 in AP buffer containing 5% polyvinyl alcohol (Sigma, P1763). Tissue was rinsed in PBS to stop the development reaction and mounted in PBS without the use of mounting media.
For the fluorescence detection, sections were rinsed three times in PBS to remove the antifreeze. Tissue was then treated and washed as described above for the chromogenic detection. Sections were then blocked in TNB (100 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.5% Blocking Reagent (Perkin Elmer)) for 30 minutes, followed by 60 minute incubation in TNB with the appropriate primary antibodies. We employed either a mouse monoclonal anti-NeuN antibody (1:300; Millipore, MAB377), a rabbit polyclonal anti-Ins(1,4,5)P3 receptor antibody (1:250; a gift from Dr. Pietro De Camilli, Yale University), a rabbit polyclonal anti-BLBP antibody (1:250; Millipore, AB9558) antibody, or a sheep polyclonal antibody against digoxigenin conjugated to alkaline phosphatase (1:1000; Roche Applied Science, 11093274910). Sections were rinsed three times in TNT. Tissue was then incubated for one hour in TNB containing goat anti-mouse IgG1 conjugated to Alexa 488 (1:1000; Invitrogen) to detect NeuN staining, or goat anti-rabbit antibodies conjugated to Alexa 488 (1:1000; Invitrogen) to detect anti-Ins(1,4,5)P3 receptor and anti-BLBP staining. Sections were then rinsed three times in TNT followed by 10 minute incubation in AP buffer. Tissue was then incubated in HNPP/Fast Red (Roche Applied Science) diluted in AP buffer containing 5% polyvinyl alcohol (Sigma, P1763) for 5–30 minutes to detect the digoxigenin-labeled riboprobes. The reaction was stopped in PBS and sections were mounted in PBS, coverslipped, and immediately imaged.
For double fluorescent in situ hybridization, sections were treated as described above. Tissue was blocked in TNB for 30 minutes, followed by a 60 minute incubation in TNB containing the sheep polyclonal antibody against digoxigenin conjugated to alkaline phosphatase (1:1000; Roche Applied Science) and a sheep polyclonal antibody against fluorescein conjugated to horseradish peroxidase (1:100; Roche Applied Science, 11426346910). Tissue was rinsed three times in TNT buffer followed by a 10 minute incubation in Tyramide Signal Amplification Biotin Tyramide reagent (Perkin Elmer) diluted 1:50 in amplification diluent (Perkin Elmer). Sections were rinsed three times in TNT followed by detection of the fluorescein-labeled riboprobes using streptavidin conjugated to Alexa 488 in TNB for 30 minutes (1:300; Invitrogen). Tissue was rinsed three times in TNT buffer followed by submersion in AP buffer for 10 minutes. Sections were then incubated in HNPP/Fast Red (Roche Applied Science) diluted in AP buffer containing 5% polyvinyl alcohol for 5–30 minutes to detect the digoxigenin-labeled riboprobes. The reaction was stopped in PBS and sections were mounted in PBS, coverslipped, and immediately imaged.
Image acquisition
Fluorescent in situ hybridization images co-labeled with anti-NeuN, anti-Ins(1,4,5)P3 receptor or anti-BLBP antibodies were acquired with an Olympus Magnafire camera attached to an Olympus BX51 epifluorescence microscope. Double fluorescent in situ hybridization images were acquired with a Hamamatsu Orca ER camera attached to a Nikon Eclipse TE2000-U epifluorescence microscope. Sagittal overview images after chromogenic detection of in situ hybridization signals were obtained on a Leica MZ 16F microscope with a PLANOP 1.0X objective attached to a Canon PowerShot S70 camera. All digital images were color-balanced using Adobe Photoshop CS3 (San Jose, CA). The composition of the images was not altered. Plates were constructed using Adobe Illustrator CS3.
RESULTS
SynCAM proteins engage in three prominent heterophilic interactions
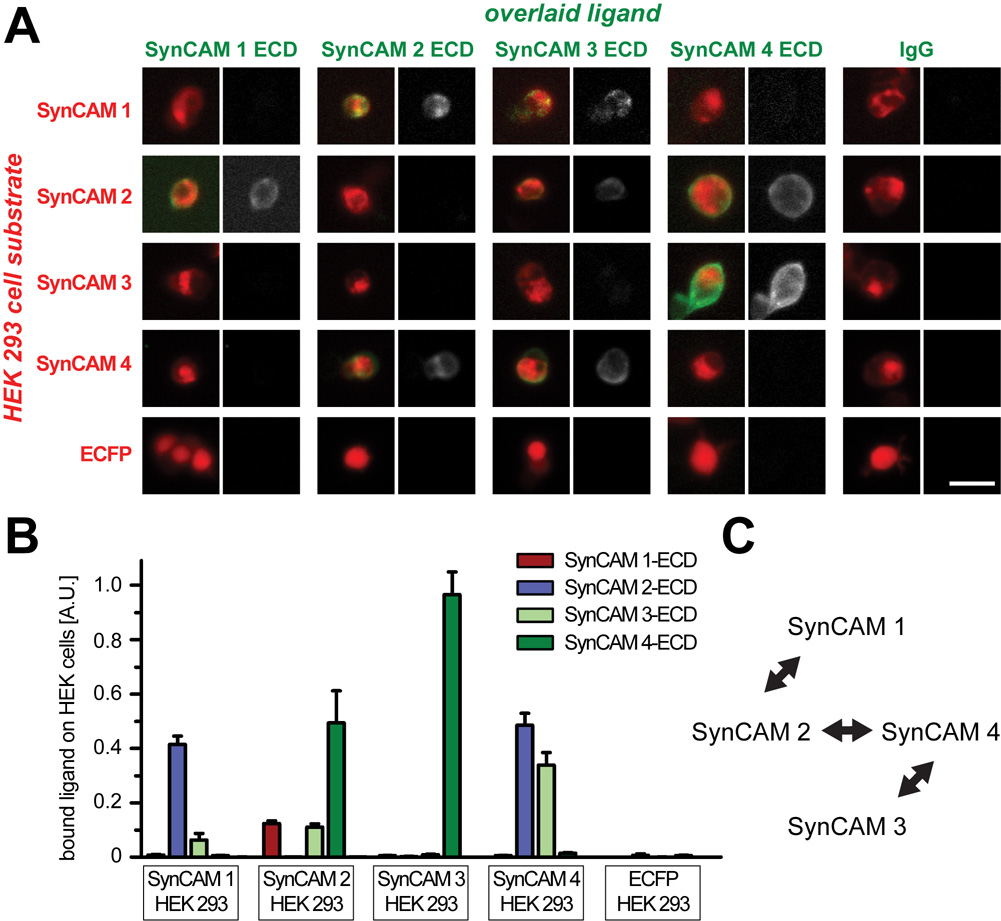
SynCAMs are encoded by the genes CADM1–4, and Table 1 provides the nomenclatures of these gene products. All four SynCAM proteins are broadly expressed in the developing brain, and function in neurons as synaptic cell adhesion molecules, consistent with the SynCAM acronym (Fogel et al., 2007). Their gene products will therefore be referred to in this study following the SynCAM 1–4 nomenclature. Evaluating their potential functions in brain regions (Fig. 2, 3) and cells (Fig. 4–11) requires insight into the distinguishing molecular properties of each SynCAM family member. Importantly, these proteins have earlier been identified as adhesion molecules through biochemical and cell-based assays, in agreement with their extracellular sequence consisting of three Ig-like domains. To define and compare the adhesive interactions of SynCAMs, we performed a comprehensive adhesion study employing a cell overlay approach that included all four SynCAM proteins (Fig. 1A). Adhesion analyses were conducted with HEK 293 cells heterologously expressing each full-length SynCAM protein tagged with CFP within the cytosolic sequence. The CFP tag was inserted within the cytosolic tail to minimally interfere with extracellular SynCAM interactions. This array of SynCAM 1–4 expressing HEK 293 cells was then individually incubated with the purified, soluble SynCAM extracellular domains fused to IgG. After the overlay, the retention of SynCAM extracellular domains on the HEK 293 cell surface was quantitated by fluorescence microscopy (Fig. 1B). Signals of retained proteins were normalized to the expression level of full-length SynCAM in the HEK 293 cells to account for transfection differences.
Table 1. Nomenclatures of genes and gene products analyzed in this study.
Human and mouse SynCAM proteins are encoded by the genes CADM 1–4 and Cadm 1–4, respectively. Different nomenclatures used for individual gene products are listed.
| Human gene symbol | Mouse gene symbol | Gene product (this study) | Alternate names | References |
|---|---|---|---|---|
| CADM1 | Cadm1 | SynCAM 1 | Necl-2, Tslc-1, SgIGSF, RA175 | Shingai et al., 2003; Kuramochi et al., 2001; Wakayama et al., 2001; Urase et al., 2001; Biederer, 2006 |
| CADM2 | Cadm2 | SynCAM 2 | Necl-3 | Ikeda et al., 2003; Biederer, 2006 |
| CADM3 | Cadm3 | SynCAM 3 | Necl-1, Tsll-1 | Kakunaga et al., 2005; Fukami et al., 2003; Biederer, 2006 |
| CADM4 | Cadm4 | SynCAM 4 | Tsll-2 | Fukami et al., 2003; Biederer, 2006 |
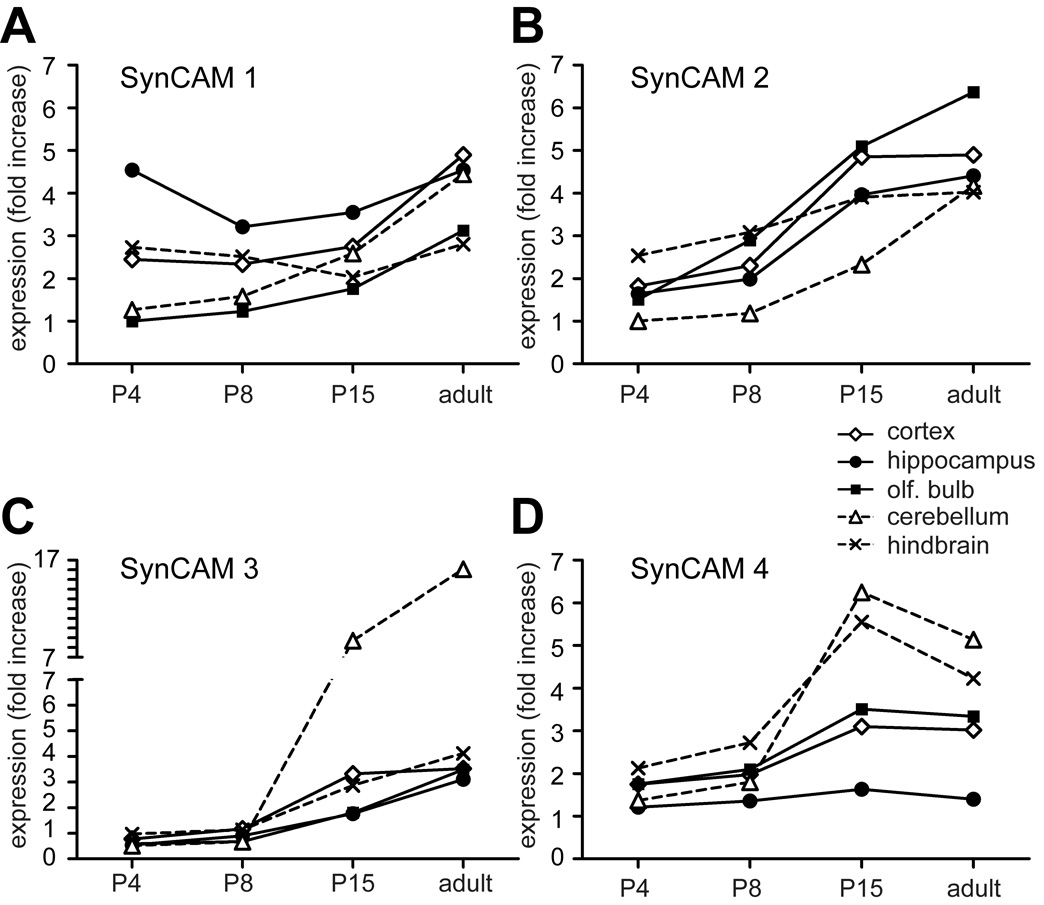
Fig. 2. Quantitative real-time RT-PCR of SynCAMs 1–4 in the central nervous system.
The four SynCAMs transcripts were detected in all brain regions examined, though each had a distinct expression profile during development. Results were normalized to actin and are presented as n-fold increase in mRNA expression of each SynCAM over four developmental stages (P4, P8, P15, and adult) in five different brain regions (cortex, hippocampus, olfactory bulb, cerebellum, and hindbrain). Results are expressed relative to SynCAM 1 levels in olfactory bulb at P4. The legend is shown on the top right, with solid lines indicating expression in forebrain regions. A. mRNA encoding SynCAM 1 was the most prominent of the four SynCAM transcripts in hippocampus at P4. B. mRNA encoding SynCAM 2 was increasingly expressed during postnatal development in all brain regions. C. mRNA encoding SynCAM 3 was increasingly expressed during postnatal development in all brain regions and was most prominent in cerebellum after P8. D. mRNA encoding SynCAM 4 was most prominent in cerebellum and hindbrain after P8 but displayed a low relative expression in hippocampus.
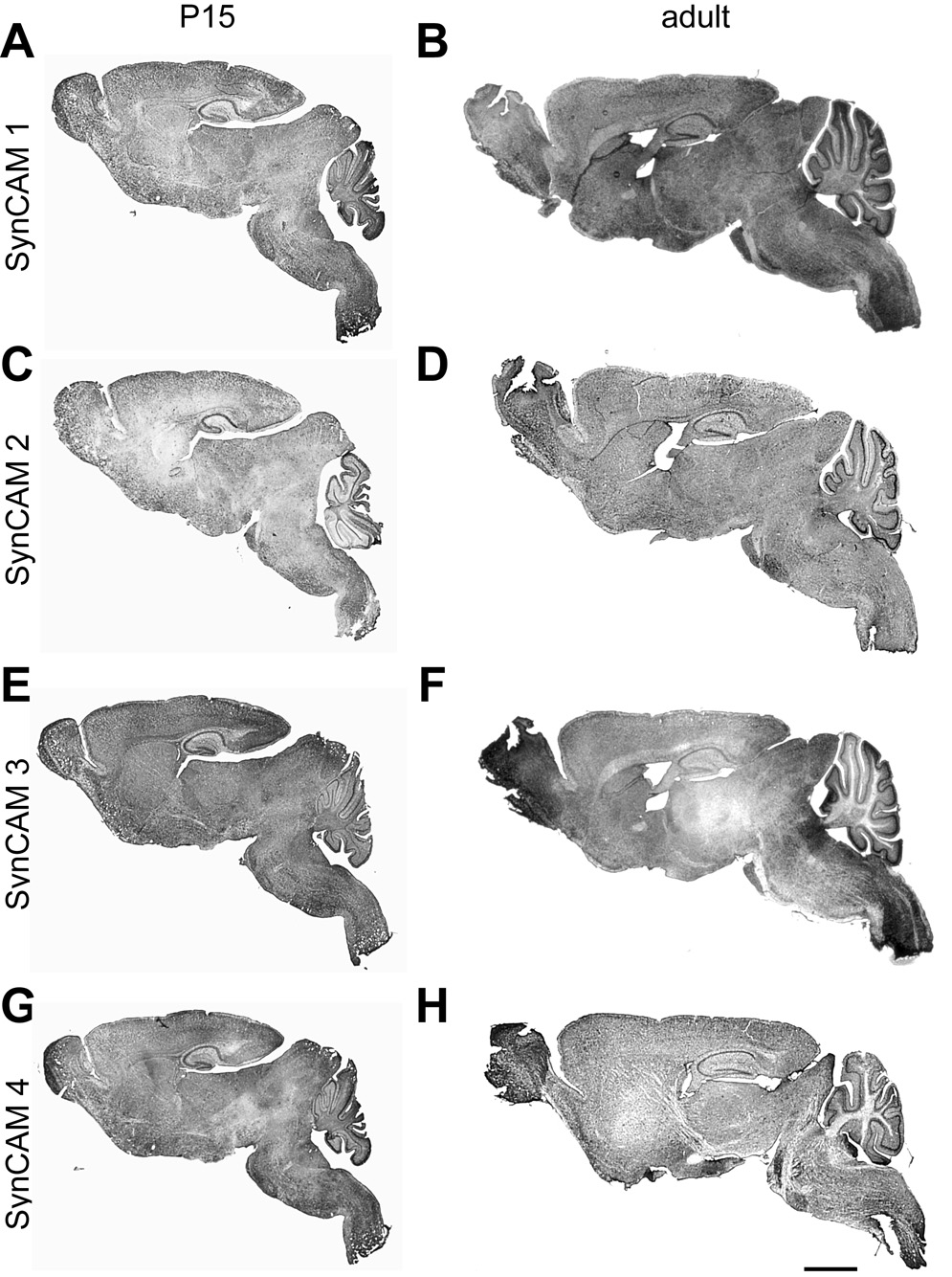
Fig. 3. SynCAM 1–4 in situ hybridization in sagittal sections at P15 and adult stages.
All SynCAMs are widely expressed throughout the central nervous system at both developmental time points. A, C, E, G. Expression overview at P15. SynCAM 1 and 2 are expressed in more caudal regions of the central nervous system, with relatively weaker staining observed for SynCAM 2 in all brain regions. High expression of SynCAM 3 was observed in cerebellum. B, D, F, H. Expression overview in adult. Expression patterns for all SynCAM proteins are similar to those at P15. Scale bar = 5 mm.
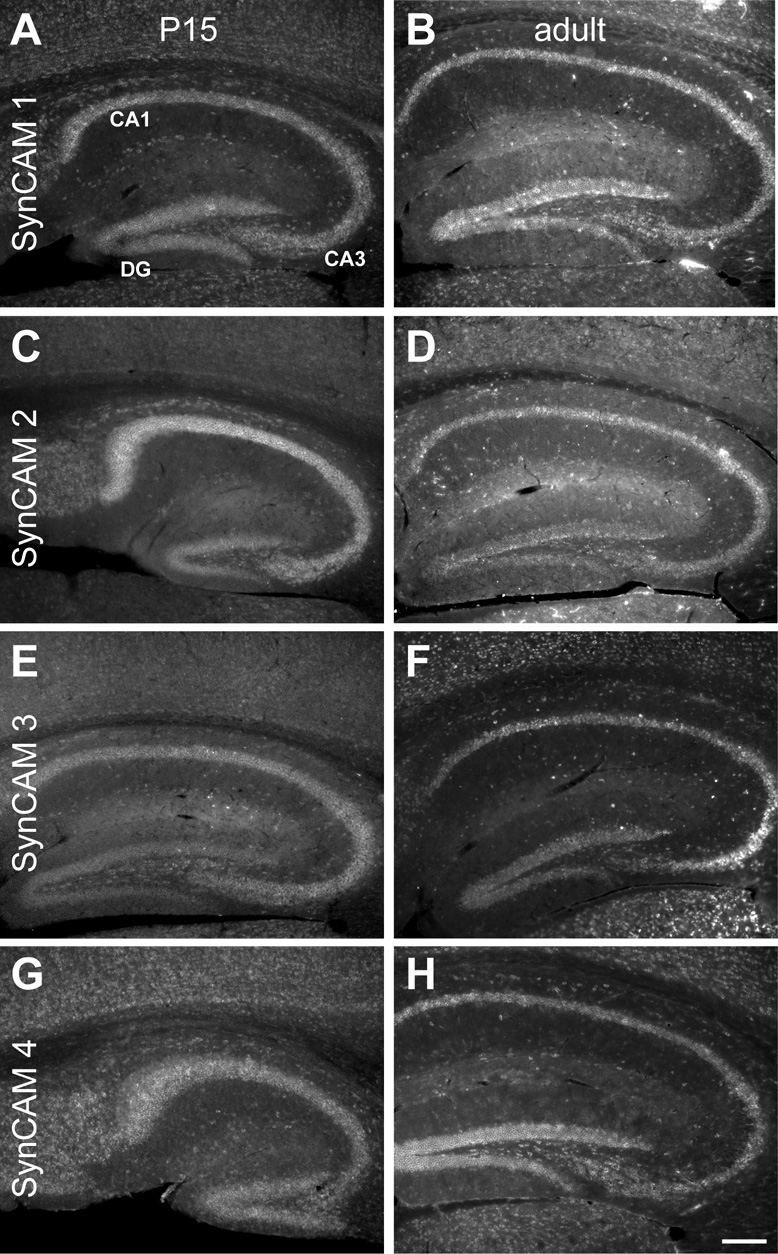
Fig. 4. SynCAM 1–4 in situ hybridization in hippocampus at P15 (A, C, E, G) and adult (B, D, F, H).
A. At P15, SynCAM 1 is expressed throughout the hippocampus, with strongest expression seen in the pyramidal cells of the CA fields and the granule cells of the dentate gyrus (DG). Expression was also seen in some cells of the corpus callosum. B. In the adult, SynCAM 1 expression was identical to that seen at P15. C. At P15, SynCAM 2 is most strongly expressed in the pyramidal cells of the CA fields. Reduced expression is seen in the granule cells of the dentate gyrus. Expression is also seen in many interneurons in the hippocampal formation. D. In adult, expression of SynCAM 2 is still high in the pyramidal cells of the CA fields and relatively low in the granule cells of the dentate gyrus. E. At P15, SynCAM 3 is strongly expressed in the pyramidal cells of the CA fields, with slightly elevated expression observed in the cells of CA3. Granule cells of the dentate gyrus show somewhat weaker expression. Interneurons of the hippocampal formation also express SynCAM 3. F. In the adult, expression of SynCAM 3 appears identical to P15. G. SynCAM 4 appears uniformly expressed in the pyramidal cells of the CA fields and the granule cells of the dentate gyrus at P15. Expression of SynCAM 4 is also seen in the interneurons throughout the hippocampal formation. Additionally, SynCAM 4 is expressed in some cells of the corpus callosum. H. In the adult, SynCAM 4 expression is identical to that seen at P15. Sections were obtained in a sagittal plane that was relatively medial. DG, dentate gyrus. Scale bar = 100 µm.
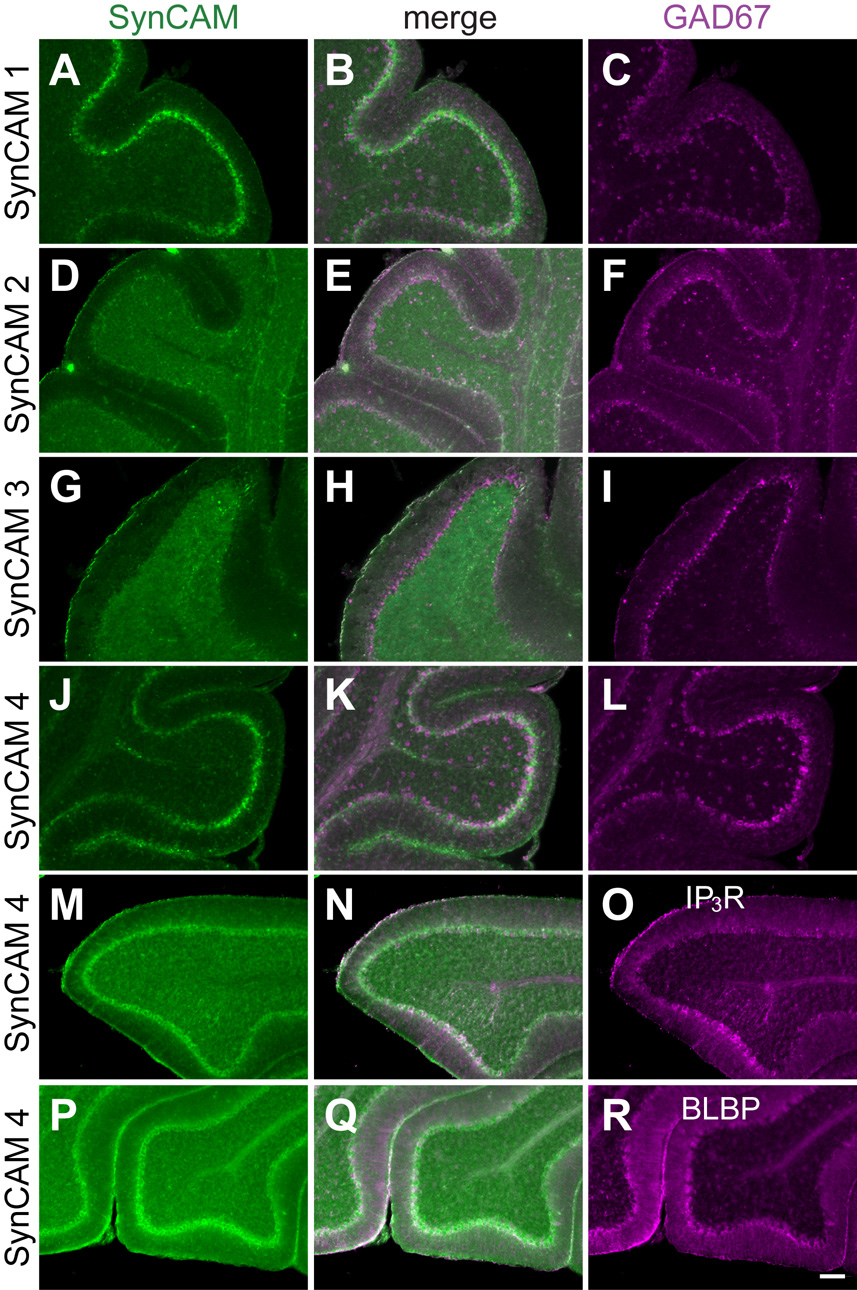
Fig. 11. Co-expression analysis of SynCAM 1–4 in P15 cerebellum.
A–C. Expression analysis of SynCAM 1 and GAD67 by double in situ hybridization in P15 cerebellum. SynCAM 1 expression (A) colocalizes with GAD67 (C) in the Purkinje cells of the cerebellum. GAD67-positive cells throughout the rest of the cerebellum do not express SynCAM 1. The center panel shows merged images (B). D–F. SynCAM 2 (D) is detected by double in situ hybridization in GAD67-positive Purkinje cells (F). Similarly to SynCAM 1, SynCAM 2 does not label GAD67-positive inhibitory neurons in the cerebellum, indicating that its expression in the granule cell layer is restricted to excitatory neurons. G–I. SynCAM 3 (G) is expressed exclusively in the granule cells of the cerebellum at P15 as shown by double in situ hybridization, with no coexpression with inhibitory Purkinje cells or interneurons that express GAD67 (I). J–L. SynCAM 4 (J) appears coexpressed by double in situ hybridization with GAD67-positive Purkinje cells (L). Other inhibitory interneurons positive for GAD67 do not coexpress SynCAM 4 throughout the cerebellum. M–O. SynCAM 4 is expressed in Purkinje cells of P15 cerebellum. SynCAM 4 in situ hybridization signal (M) colocalizes with Purkinje cells immunopositive for the Ins(1,4,5)P3 receptor (O). The center panel shows merged images (N). P–R. SynCAM 4 is not prominently expressed in Bergmann glia cells of the Purkinje cell layer in P15 cerebellum. SynCAM 4 in situ hybridization signal (P) does not colocalize with Bergmann glia cells immunopositive for the marker BLBP (R). IP3R, Ins(1,4,5)P3 receptor. Scale bar = 100 µm.
Fig. 1. Detection of specific heterophilic interactions between SynCAMs in a cell overlay assay.
A. Detection of soluble SynCAM extracellular domain interactions with full-length SynCAM proteins by cell overlay. HEK 293 cells expressing the indicated full-length SynCAM-CFP fusion proteins on their surface (red) serve as substrate for the indicated soluble fusion proteins of SynCAM extracellular domains with IgG (green). Identical protein amounts of each purified SynCAM extracellular domain were overlaid on HEK 293 cells, and bound protein was detected with Protein A-Alexa 594. Fluorescence microscopy images are shown, with the merged image on the left of each panel and the signal from the overlaid ligand in grayscale on the right. Images are representative of the quantification results below. Scale bar = 30 µm. B. Quantification of SynCAM cell overlay. Fluorescence images of SynCAM-CFP expressing HEK 293 cells overlaid with individual, soluble SynCAM extracellular domains were analyzed for the signal intensities of expressed SynCAM-CFP and of retained extracellular domains (n = 6 images per condition, with 3 images each obtained in two independent experiments; each analyzed image contained up to several hundred cells). Expression of each full-length SynCAM in the overlaid HEK293 cells was comparable (data not shown). Strong and reciprocal heterophilic interactions of SynCAM 1 and 2, of SynCAM 2 and 4, and of SynCAM 3 and 4 were detected. C. Diagram depicting the three SynCAM heterophilic complexes identified in this study.
We confirmed the heterophilic interaction of SynCAMs 3 and 4 as well as the binding of SynCAM 1 and 2 (Fogel et al., 2007; Maurel et al., 2007; Spiegel et al., 2007). SynCAM 2 had not been included in previous cell overlay studies. We observed that it assembles not only in a complex with SynCAM 1 as identified previously in affinity chromatography studies (Fogel et al., 2007), but also engages in a separate interaction with SynCAM 4. This novel SynCAM 2/4 complex therefore constitutes the third prominent heterophilic interaction between the members of this family of cell adhesion molecules alongside with SynCAM 3/4 and SynCAM 1/2 complexes (Fig. 1C). Reciprocal binding of full-length SynCAM and overlaid extracellular domains was detected for all three interactions, validating the described binding specificities. Additional interactions may occur between the SynCAM 3 extracellular domain and either full-length SynCAM 1 (Fig.1B) (Kakunaga et al., 2005; Fogel et al., 2007) or full-length SynCAM 2 (Fig. 1B). However, these two SynCAM 3 interactions were not confirmed in reciprocal experiments using full-length SynCAM 3 as substrate, presumably due to the high stringency of this adhesion analysis. They could therefore not be confidently assessed and will not be considered in our discussion of SynCAM expression patterns (see below). The homophilic interactions of SynCAM proteins were not detected, likely due to their relatively low strength (Fogel et al., 2007). Our cellular overlay results determine the differential adhesive properties of SynCAMs and identify the three distinct SynCAM 1/2, 2/4, and 3/4 complexes as the defining matrix of heterophilic SynCAM interactions.
Distinct developmental and regional SynCAM 1–4 expression
The existence of distinct SynCAM adhesion complexes motivated the question to which extent these molecules are differentially distributed across brain regions in development. Differential expression patterns could indicate that SynCAM-mediated adhesion may help to spatially define cellular interactions in the brain. To investigate the spatiotemporal expression levels of each SynCAM in the central nervous system, we conducted a quantitative real-time RT-PCR analysis in five brain regions at multiple developmental time points. Tissue was isolated from mice at postnatal day (P) 4, P8, P15, and adult to span critical stages throughout postnatal development. Notably, P15 was included as synaptogenesis and myelination occur at a rapid pace at this age. Tissue examined included the forebrain regions of the olfactory bulb, hippocampus, and cortex. Additionally, cerebella were dissected as were the pons and medulla, here collectively termed hindbrain.
SynCAM transcripts were detected in all brain regions examined, and relative SynCAM transcript levels were quantitated and normalized to actin (Fig. 2). Each brain region displayed a distinct SynCAM mRNA expression profile. SynCAM 1 was expressed strongly relatively early in hippocampus where it is maintained approximately at the same expression level at all examined ages (Fig. 2A). Similarly, in the hindbrain, SynCAM 1 expression levels did not change during development, with overall expression levels lower than in the hippocampus. In cerebellum, cortex, and olfactory bulb, however, its expression levels rose with time to a similar extent. SynCAM 2 expression increased with age in all examined brain regions to reach approximately equal levels in all adult regions (Fig. 2B). As with SynCAM 2, SynCAM 3 expression rose equally with age in most regions analyzed (Fig. 2C). The notable exception was the cerebellum, where its expression levels grew rapidly after P8 until the level there was roughly five times of those seen elsewhere in the nervous system. This high level of SynCAM 3 transcripts in cerebellum is consistent with its expression by the abundant granule cells (see Fig. 8) and with the large amount of SynCAM 3 protein in cerebellum (Gruber-Olipitz et al., 2006). SynCAM 4 expression underwent an increase in most brain regions after P8 (Fig. 2D). The sole exception was the hippocampus, where levels remained low at all examined ages. The sharpest increase of SynCAM 4 mRNA expression occurred in cerebellum and hindbrain. In the adult, its expression levels either remained the same (olfactory bulb, cortex, and hippocampus) or showed a slight decline (cerebellum and hindbrain) relative to the levels seen at P15. The spatiotemporal expression profiles of SynCAM family members in the developing brain are therefore divergent and characteristic for each of the SynCAM family members.
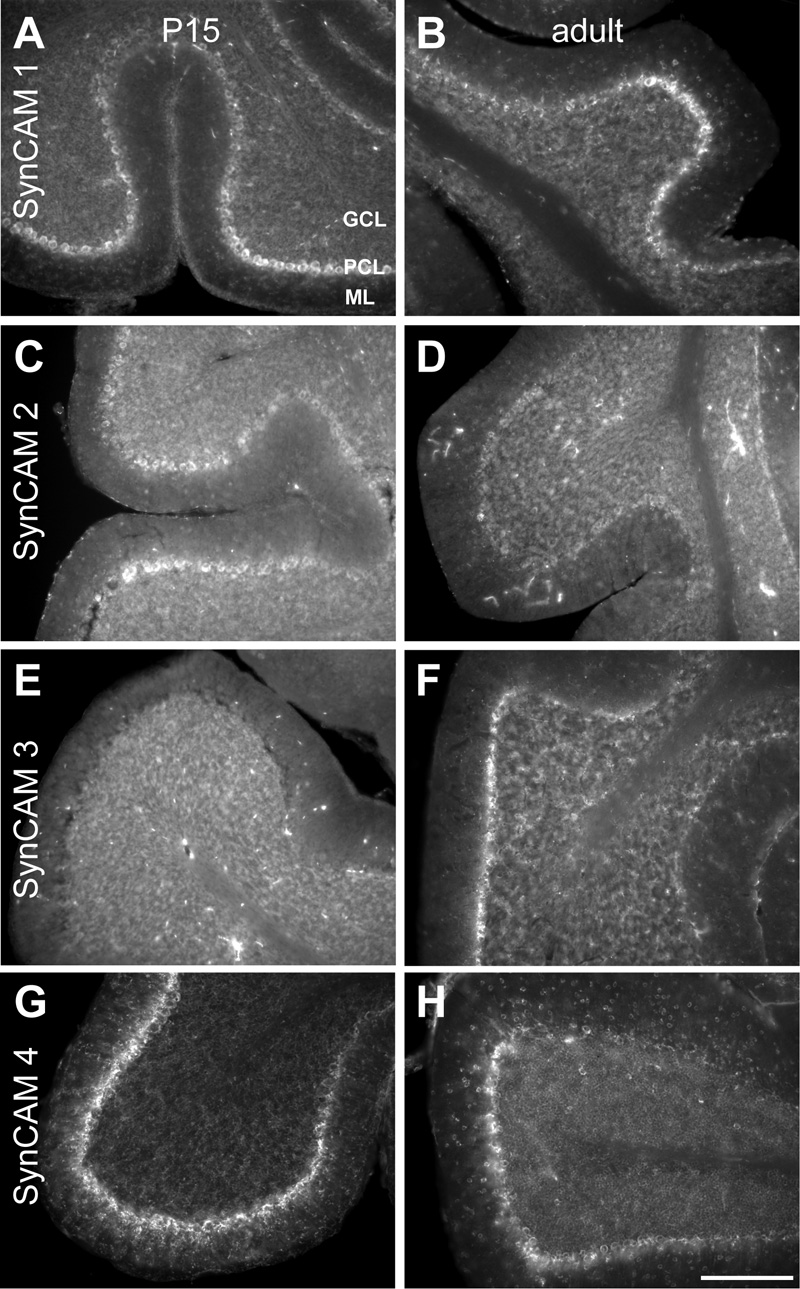
Fig. 8. SynCAM 1–4 in situ hybridization in cerebellum at P15 (A, C, E, G) and adult (B, D, F, H).
A. At P15, SynCAM 1 is most strongly expressed in Purkinje cells. Weak expression is seen in the granule cells. SynCAM 1 expression is also observed in the molecular layer (ml), indicating expression in interneurons or glia. B. In the adult, SynCAM 1 expression is similar to that seen at P15. C. At P15, SynCAM 2 is weakly expressed in Purkinje cells. D. In the adult, SynCAM 2 expression is uniformly weak throughout the cerebellum. E. At P15, SynCAM 3 is strongly expressed in the granule cells of the cerebellum and not detected in Purkinje cells. F. In the adult, SynCAM 3 expression remains high in the granule cells and is at this developmental time point also observed in Purkinje cells. SynCAM 3 labeling is additionally detectable in cells in the molecular layer, potentially interneurons or glia. G. At P15, SynCAM 4 expression is strong in the Purkinje cells, with no apparent expression in the granule cell layer H. In the adult, expression of SynCAM 4 remains strongest in the Purkinje cells and relatively weak in the granule cells. SynCAM 4 expression the molecular layer is increased in comparison to that at P15, to a level nearly as high as in the granule cells. Sections were obtained in a sagittal plane that was relatively medial. GCL, granule cell layer; PCL, Purkinje cell layer; ML, molecular layer. Scale bar = 100 µm.
Spatial profiles of SynCAM transcript expression in the postnatal and adult mouse brain
After defining the heterophilic interaction profiles of SynCAM proteins (Fig. 1), and having determined that SynCAMs are broadly expressed throughout the central nervous system at both the mRNA (Fig. 2) and protein levels (Fogel et al., 2007), we sought finer resolution of cellular SynCAM expression to elucidate potential sites and roles for these intercellular interactions in the brain. In a previous study (Biederer et al., 2002), the synaptic localization of SynCAMs was demonstrated in hippocampus, cerebellum, and spinal cord by immunohistochemistry and was confirmed by immuno-electron microscopy of hippocampal neurons using antibodies that recognize SynCAM 1, 2, and 3 equally well (Fogel et al., 2007). However, no specific antibodies are available for individual SynCAM family members that can be utilized for immunohistochemical studies of protein expression and localization. We therefore performed a comprehensive expression analysis by in situ hybridization of sections of hippocampus, cortex, olfactory bulb, cerebellum, and spinal cord.
Considering the potential roles of SynCAMs in early postnatal development of the central nervous system, specifically synapse formation, we conducted this study at different developmental time points. Synaptogenesis occurs at the highest rate in the developing brain, albeit at different rates in the distinct brain regions studied here. P15 is the peak period of synaptogenesis in the rodent hippocampus (Harris et al., 1992; Fiala et al., 1998), the brain region in which the synaptic functions of SynCAMs have been most extensively studied so far. In the rodent cortex, rapid excitatory synapse formation also occurs at this stage, with synapse densities increasing continuously during the first three weeks and doubling from P15 to reach a maximum at P20 (Dyson and Jones, 1980; Blue and Parnavelas, 1983). P15 – P20 correspond to the peak of glomerular synapse density in the rodent olfactory bulb, with synapse densities in the external plexiform layer also almost at maximum (Hinds and Hinds, 1976). In the rodent cerebellum, dendritic spines in the molecular layer rapidly increase in density from P6 to reach their maximum at P21 (Takacs and Hamori, 1994). P15 therefore corresponds to either the peak of synapse density or a period of maximal synapse formation in these differentiating brain regions, motivating us to conduct a study of SynCAM cellular expression at this time point. Due to the high degree of specificity of synapse formation in spinal cord (Chen et al., 2003), we included an analysis of SynCAM transcript expression at P2, the onset of maximal synapse density in this region (Weber and Stelzner, 1980). Together, the expression analyses at P15 and P2 aim to gain insights into SynCAM roles in cellular differentiation in different developing regions of the central nervous system. Adult ages were included to assess SynCAM expression in the mature central nervous system, when most cellular contacts have been stably established.
Our RT-PCR results indicated that all family members are broadly and highly expressed at these ages (Fig. 2), consistent with the expression profiles of SynCAM protein in the developing brain (Fogel et al., 2007). We confirmed by in situ hybridization of sagittal brain sections that each SynCAM family member was expressed throughout the central nervous system at both P15 and adult stages (Fig. 3). The SynCAM signals detected by in situ hybridization were specific as determined with sense probes (data not shown). SynCAM 1 and 2 were expressed in more rostral regions of the central nervous system, while SynCAM 3 and 4 were observed more caudal. These data are consistent with the results from the RT-PCR analysis of different brain regions (Fig. 2), especially highlighting the prominent expression of SynCAM 3 and 4 in the cerebellum. Multiple local variations, evident between the family members and across development, motivated further detailed investigation.
SynCAM transcript expression in hippocampus
A number of functional studies of SynCAM proteins have been conducted in dissociated hippocampal neurons, necessitating a detailed analysis of their expression in this region. It was previously shown at P15 that the expression of the four family members in dentate gyrus and CA fields is divergent (Fogel et al., 2007). To elucidate their expression in the hippocampus in more detail, we performed a comparative analysis at P15 and adult. These time points were chosen as they represent the peak of synaptogenesis and a stage with less intense synapse formation in this brain region, respectively.
At P15, SynCAM 1 expression was readily observed in the principal cells of the hippocampus, the granule cells of the dentate gyrus and the pyramidal cells of the CA fields (Fig. 4A). Expression was also observed in some cells of the hilus of the dentate gyrus. Cells in the stratum oriens, stratum radiatum, and stratum lacunosum-moleculare also expressed SynCAM 1. Expression of SynCAM 1 in the adult hippocampus was identical to that at P15 (Fig. 4B).
As described (Fogel et al., 2007), SynCAM 2 was most strongly expressed at P15 in the pyramidal cells of the CA1 field and also in pyramidal cells of the CA3 field, with reduced expression in the granule cells of the dentate gyrus (Fig. 4C). As with SynCAM 1, expression of SynCAM 2 was seen in the hilus of the dentate gyrus as well as in cells in the stratum oriens, stratum radiatum, and stratum lacunosum-moleculare. In the adult, SynCAM 2 expression remained high in the pyramidal cells in the CA fields, and low in the granule cells of the dentate gyrus (Fig. 4D). Fewer cells in hilus, stratum oriens, stratum radiatum, and stratum lacunosum-moleculare expressed SynCAM 2 as compared to P15.
At P15, SynCAM 3 was most strongly observed in pyramidal cells of the CA fields (Fig. 4E), with granule cells of the dentate gyrus showing relatively weaker levels of expression. SynCAM 3 signal was slightly elevated in the CA3 region. Expression of SynCAM 3 was also seen in the hilus, stratum oriens, stratum radiatum, and statum lacunosum-moleculare at P15. The apparently elevated expression in CA3 and relatively weaker expression in the dentate gyrus compared to CA fields that was seen at P15 was also observed in the adult (Fig. 4F). In the adult, SynCAM 3 expression was also seen in the hilus, stratum oriens, stratum radiatum, and stratum lacunosum-moleculare.
SynCAM 4, the least abundant SynCAM family member in hippocampus (Fig. 2), exhibited at both P15 and adult stages apparently uniform expression in the pyramidal cells of the CA fields and the granule cells of the dentate gyrus (Fig. 4G,H). As with SynCAMs 1, 2, and 3, expression was also seen in the hilus, stratum oriens, stratum radiatum, and stratum lacunosum-moleculare.
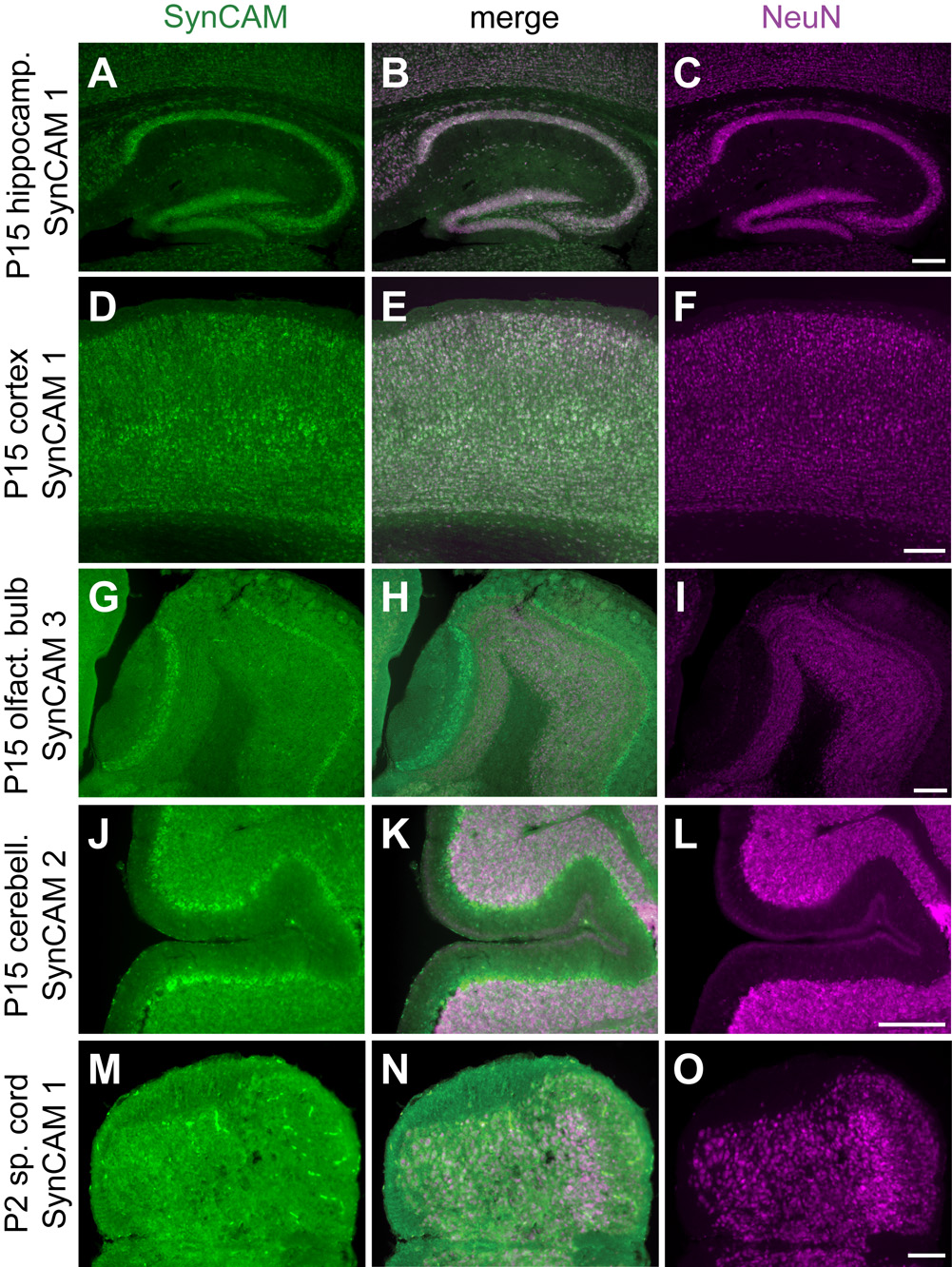
Notably, the vast majority of SynCAM 1–4 expressing cells in hippocampus at both P15 and adult were neurons as demonstrated by immunostaining of sections after in situ hybridization with antibodies against the neuronal nuclei marker NeuN (Fig. 5A–C, and data not shown). We were intrigued to find that almost all NeuN positive cells also expressed SynCAMs, showing that they are present in the majority of hippocampal neurons. Occasional expression of SynCAM 1 at P15 and of SynCAM 4 at both P15 and adult stages was also observed in some cells of the corpus callosum, indicating their potential additional expression in a subset of glial cells (Fig. 4A,G,H).
Fig. 5. SynCAM expression is predominantly neuronal and observed in the vast majority of neurons.
A–C. Coexpression (B) of SynCAM 1 observed by in situ hybridization (A) and NeuN by immunohistochemistry (C) in the hippocampus at P15. D–F. Coexpression (E) of SynCAM 1 (D) and NeuN (F) in P15 cortex. G–I. Coexpression (H) of SynCAM 3 (G) and NeuN (I) in main and accessory olfactory bulbs at P15. NeuN does not label the mitral cell bodies of the olfactory bulb. J–L. Coexpression (K) of SynCAM 2 (J) and NeuN (L) in P15 cerebellum. NeuN does not label the Purkinje cell bodies of the cerebellum. M–O. Coexpression (N) of SynCAM 1 (M) and NeuN (O) in P2 spinal cord. Scale bars = 100 µm.
SynCAM mRNA expression in cortex
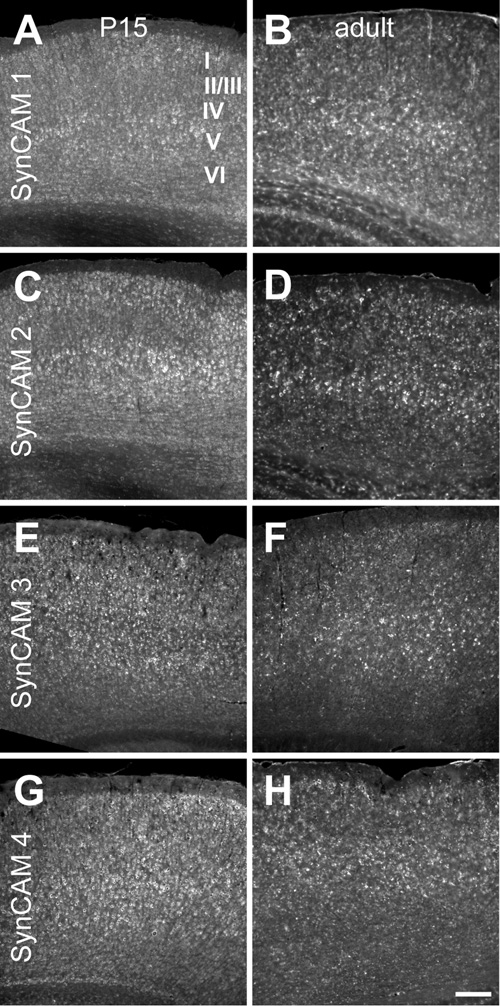
In cortex, SynCAMs were broadly expressed in both P15 and adult mice (Fig. 6). There were, however, some subtle differences. At P15 and adult, cells in layer V appeared to express SynCAM 1 at a slightly elevated level (Fig. 6A,B). At P15, cells in layers II/III and V showed an apparent increase in SynCAM 2 expression as compared to the other layers (Fig. 6C). In the adult, SynCAM 2 signal remained elevated in layer V, with uniformly lower expression seen in the other cortical layers (Fig. 6D). SynCAM 3 expression was relatively uniform at both time points (Fig. 6E,F). Likewise, SynCAM 4 was uniform in its expression (Fig. 6G,H).
Fig. 6. SynCAM 1–4 in situ hybridization in mouse cortex at P15 (A, C, E, G) and adult (B, D, F, H).
A. At P15, SynCAM 1 is expressed throughout cortex, but is expressed at a slightly elevated level in layer V. B. Expression of SynCAM 1 in the adult cortex shows a similar enrichment in layer V. C. At P15, SynCAM 2 expression was slightly elevated in layers II/III and V as compared to the other layers. D. In adult cortex, SynCAM 2 remained highly expressed in layer V with uniformly lower expression in other cortical layers. E. SynCAM 3 was uniformly expressed across all cell layers at P15. F. In the adult, SynCAM 3 was also expressed evenly in all layers of cortex. G. SynCAM 4 was broadly expressed across all layers at P15. H. SynCAM 4 expression was also uniformly expressed in all layers in the adult. Sections were obtained approximately in the middle of the rostral-caudal axis. Scale bar = 100 µm.
As observed in hippocampus, almost all SynCAM 1–4 expressing cells in cortex at both P15 and adult stages were positive for the neuronal marker NeuN, and the vast majority of NeuN positive cells also were positive for SynCAMs (Fig. 5D–F, and data not shown).
SynCAM transcript expression in olfactory bulb
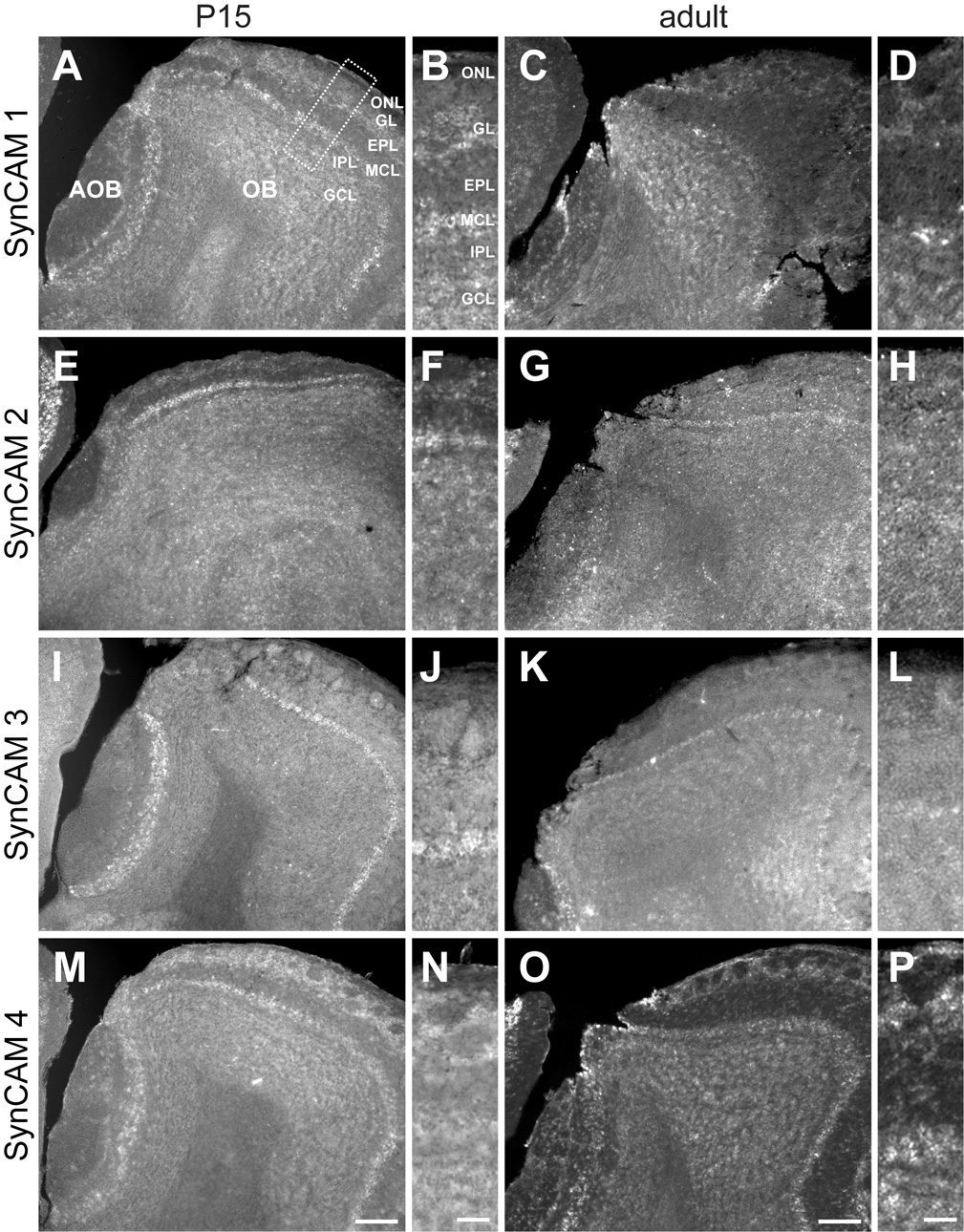
In the olfactory bulb, SynCAMs also displayed divergent expression profiles (Fig. 7). Here, SynCAM 1 was prominently expressed in mitral and tufted cells of both the main and accessory olfactory bulbs (Fig. 7A). In the main olfactory bulb, SynCAM 1 was expressed at P15 by juxtaglomerular cells on the deep surface of glomeruli, likely external tufted and short axon cells (Fig. 7B). By the adult timepoint, SynCAM 1 expression surrounded glomeruli, indicating expression by periglomerular cells as well (Fig. 7C,D). No other change in SynCAM 1 expression pattern in the olfactory bulb was seen.
Fig. 7. SynCAM 1–4 in situ hybridization in the main and accessory olfactory bulbs at P15 (A, B, E, F, I, J, M, N) and adult (C, D, G, H, K, L, O, P).
A. At P15, SynCAM 1 is expressed in the mitral and tufted cells of both the main (OB) and accessory (AOB) olfactory bulbs. Expression is also seen in the granule and deep juxtaglomerular cells. B. Enlarged panel (as indicated by the box in A) showing expression of SynCAM 1 in the different layers of the OB at P15. C. In the adult, SynCAM 1 expression was similar to that at P15, with expression also seen in periglomerular cells. D. Enlarged image of SynCAM 1 expression in adult OB. E. At P15, SynCAM 2 is predominantly expressed in mitral cells of the main olfactory bulb, with significantly weaker expression observed in the accessory olfactory bulb. F. Magnified view of SynCAM 2 expression at P15. G. In the adult olfactory bulb, nearly all cell populations appear to express SynCAM 2 at relatively uniform levels. H. Enlarged image of SynCAM 2 expression in adult OB. I. At P15, SynCAM 3 expression is strongest in the mitral cells of both the main and accessory olfactory bulbs. Faint expression is seen in other cell types. J. Magnified view of SynCAM 3 expression in the OB at P15. K. In the adult, SynCAM 3 expression remains strong and relatively uniform in the mitral and tufted cells of the main olfactory bulb. Granule cells demonstrate a relative increase in signal as compared to P15. L. Magnified panel of adult SynCAM 3 expression in the OB. M. At P15, strongest expression of SynCAM 4 was seen superficially in the periglomerular cells and olfactory ensheathing cells, with other cell types also showing expression. N. Enlarged image of SynCAM 4 expression in the OB at P15. O. In the adult, the pattern of SynCAM 4 expression was similar to that at P15. P. Magnified view of adult SynCAM 4 OB expression. Sections were obtained in a sagittal plane that was relatively medial. OB, main olfactory bulb; AOB, accessory olfactory bulb; ONL, olfactory nerve layer; GL, glomerular layer; EPL, external plexiform layer; MCL, mitral cell body layer; IPL, internal plexiform layer; GCL, granule cell layer. Scale bars = 100 µm (in M, O), 30 µm (in N, P) apply to the respective columns of panels.
SynCAM 2 was expressed predominantly by mitral cells of the main olfactory bulb at P15 (Fig. 7E,F). Little or no expression was seen in the mitral and tufted cells of the accessory olfactory bulb. In the adult, all cell populations were seen to express SynCAM 2 (Fig. 7G,H).
SynCAM 3 was mostly strongly expressed by mitral and tufted cells of both the main and accessory olfactory bulbs at both ages (Fig. 7I–L). Faint expression in most other cells was observed at P15. In the adult, only the granule cells expressed SynCAM 3 at a level comparable to that seen in the mitral and tufted cells; the remaining cell populations expressed SynCAM 3 only faintly (Fig. 7L).
SynCAM 4 expression was seen in all cell populations at both examined ages (Fig. 7M–P). Strongest expression was seen in juxtaglomerular cells and in cells in the olfactory nerve layer that are presumably olfactory ensheathing glia based on their distribution (Fig. 7N,P).
As observed in all brain regions analyzed, the vast majority of all SynCAM 1–4 expressing cells in olfactory bulb at both P15 and adult stages also expressed the neuronal marker NeuN, and almost all NeuN positive cells were positive for SynCAMs (Fig. 5G–I, and data not shown). In olfactory bulb, mitral cell bodies are not labeled by antibodies directed against NeuN (Mullen et al., 1992) but were identified by their layer-specific localization and characteristic size and morphology.
Cerebellar expression of SynCAMs
SynCAM 1, which was not highly expressed in cerebellum at either P15 or adult stages (Fig. 2), was expressed most notably in the Purkinje cell layer (Fig. 8A,B). Expression appeared not uniform among Purkinje cells, with patches showing stronger and weaker signal. Strong expression was generally, though not exclusively, observed in the Purkinje cells at the tips of the folia, with somewhat weaker expression seen in the deeper folds of the folia. Weak expression of SynCAM 1 was also detected in the granule cell layer at both P15 and adult stages. Additional expression was observed in the molecular layer, indicating expression by interneurons or glia.
At P15, expression of SynCAM 2 was pronounced in the Purkinje cell layer, but also detected in cerebellar granule cells (Fig. 8C). In adults, staining appeared uniformly low across all cell populations of the cerebellum (Fig. 8D).
SynCAM 3 is the most prominent family member in cerebellum (Fig. 2). At P15, its expression was limited to granule cells, which showed noticeably higher expression levels than other cell types in the brain (Fig. 8E). No Purkinje cell layer expression was detected at this developmental time point. In the adult, the granule cell expression of SynCAM 3 appeared even stronger (Fig. 8F). At the adult time point, significant expression of SynCAM 3 was now additionally observed in Purkinje cells. SynCAM 3 labeling was also detectable in the adult molecular layer, indicating expression by interneurons or glia.
In cerebellum, SynCAM 4 is the second most prominently expressed family member after SynCAM 3 (Fig. 2). Strikingly, SynCAM 4 showed at P15 an expression pattern inverse to SynCAM 3. Here, very high SynCAM 4 expression was evident in the Purkinje cell layer but not any other cell layers (Fig. 8G). This pattern persisted into adulthood, with SynCAM 4 expression very strong in the Purkinje cell layer and some weak expression by the granule cells (Fig. 8H). SynCAM 4 expression in cells of the molecular layer had increased by this age, with those cells expressing it at a low level comparable to that seen in cerebellar granule cells. Double-labeling with antibodies detecting Purkinje cells or Bergmann glia demonstrated that SynCAM 4 expression at P15 appeared specific for Purkinje cells (see below, Fig. 11M–R).
As observed in forebrain regions, almost all cells in cerebellum were positive for both SynCAM 1–4 transcripts and the neuronal marker NeuN (Fig. 5J–L, and data not shown). Purkinje cell bodies are not labeled by antibodies directed against NeuN (Mullen et al., 1992) but were identified by their layer-specific localization.
Expression of SynCAM transcripts in spinal cord
As synaptogenesis occurs earlier in the spinal cord than the rest of the central nervous system, we addressed SynCAM expression at P2, the onset of maximum synapse formation in this tissue (Weber and Stelzner, 1980). We also examined SynCAM expression in adult tissue. All SynCAM family members were observed at both developmental stages in lumbar spinal cord sections, with variations observed in cell type expression.
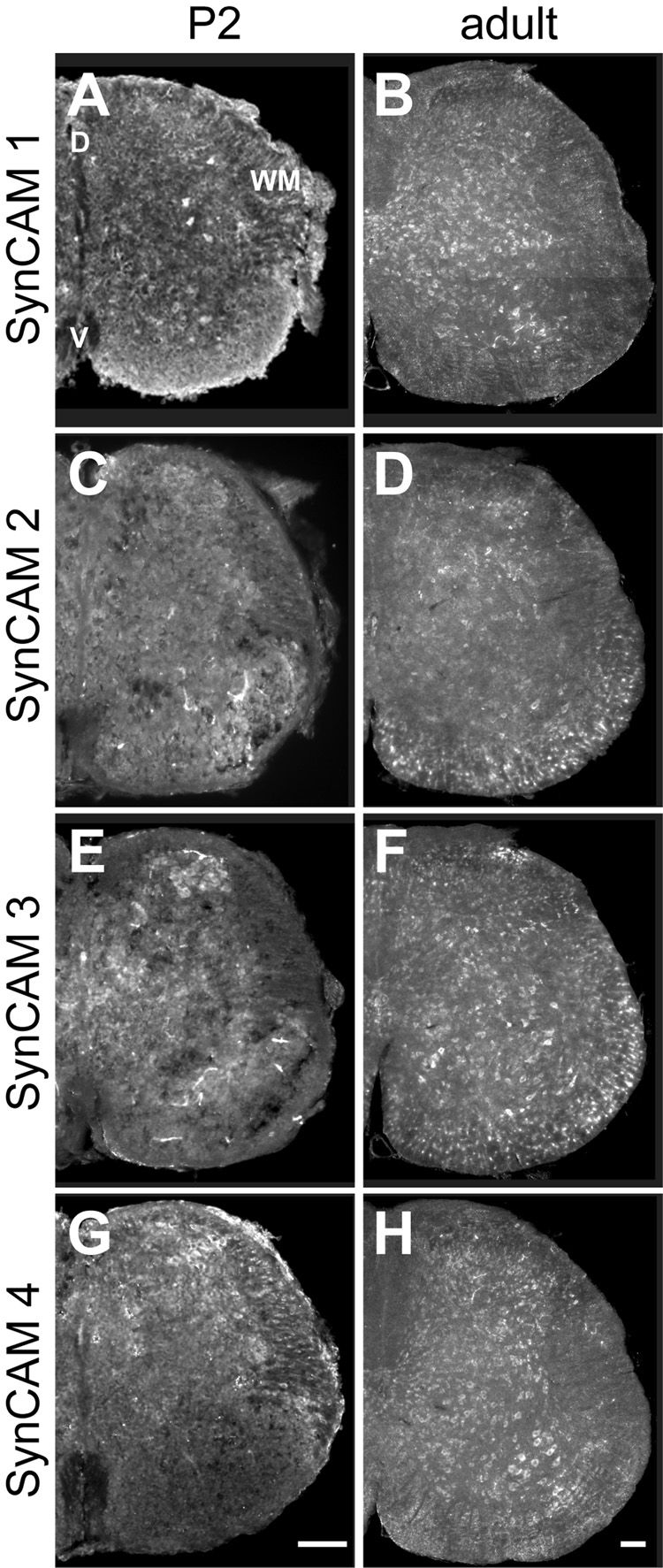
At P2, SynCAM 1 appears expressed in neurons in both dorsal and ventral spinal cord (Fig. 9A). Neuronal expression was confirmed using immunostaining against the neuronal nuclei marker NeuN (Fig. 5M–O). SynCAM 1 expression was also detected in small, NeuN-negative cells of white matter located ventrally in the spinal cord that may correspond to oligodendrocytes. In adult spinal cord, SynCAM 1 expression appeared limited to neurons, with uniform expression seen in dorsal and ventral areas (Fig. 9B). Expression of SynCAM 1 was apparent in the motor neuron pool in the ventral horn.
Fig. 9. SynCAM 1–4 in situ hybridization in spinal cord at P2 (A, C, E, G) and adult (B, D, F, H).
A. At P2, SynCAM 1 is expressed in neurons as well as presumptive oligodendrocytes of the white matter (WM) in both dorsal (D) and ventral (V) spinal cord. B. In the adult, expression of SynCAM 1 expression is restricted to neurons in the dorsal and ventral areas of the spinal cord. Expression is evident in the motor nuclei of the ventral horn. C. At P2, SynCAM 2 is expressed in neurons throughout the dorsal and ventral areas. D. In the adult, expression of SynCAM 2 is seen uniformly throughout the dorsal and ventral areas. Expression is now also observed in presumptive oligodendrocytes of the white matter. E. At P2, SynCAM 3 is expressed in neurons throughout the spinal cord. F. In the adult, SynCAM 3 is uniformly expressed in both dorsal and ventral spinal cord. Expression is also seen in presumptive oligodendrocytes. G. At P2, SynCAM 4 is expressed in neurons with higher levels of expression in dorsal spinal cord. H. In adult, SynCAM 4 expression is limited to neurons, with uniform staining in dorsal and ventral areas. Expression is apparent in the motor nuclei of the ventral horn. All sections are from the lumbar region of the spinal cord. D, dorsal; V, ventral; WM, white matter. Scale bars = 100 µm (in G, H) apply to the respective columns of panels.
In contrast, SynCAM 2 was detected solely in neurons at P2 (Fig. 9C). In the adult, expression was uniform throughout the dorsal and ventral areas. In addition to neuronal expression, SynCAM 2 was seen in NeuN-negative, presumptive oligodendrocytes of the white matter (Fig. 9D, and data not shown).
At P2, SynCAM 3 was also uniformly expressed in neurons throughout the spinal cord as determined by NeuN co-labeling of cells (Fig. 9E, and data not shown). In particular, SynCAM 3 signal was observed in small neurons of the dorsal horn. In adult, SynCAM 3 remained uniformly expressed in both dorsal and ventral spinal cord, with additional expression seen in NeuN-negative, presumptive oligodendrocytes of the white matter (Fig. 9F, and data not shown).
SynCAM 4 was expressed at P2 similar to SynCAM 1 in both neurons and presumptive oligodendrocytes, with higher level of expression seen in dorsal spinal cord (Fig. 9G). By adulthood, however, SynCAM 4 was expressed only in neurons, with uniform staining seen in both dorsal and ventral areas (Fig. 9H, and data not shown). As with SynCAM 1, expression of SynCAM 4 was evident in the motor neuron pool in the ventral horn.
Supplemental Figure 1 shows enlarged images from the ventral horn region of the spinal cord for further assessment of the overview images in Figure 9. As in the other regions of the central nervous system, the majority of NeuN-positive cells in spinal cord were also positive for each SynCAM transcript (Fig. 5M–O, and data not shown). However, apparent glial expression of SynCAMs such as in NeuN-negative, presumptive oligodendrocytes appeared more pronounced in spinal cord than elsewhere in the central nervous system.
SynCAMs are distinctly present in both excitatory and inhibitory neurons
For even finer resolution of the cell types expressing each SynCAM throughout the central nervous system, we performed double-fluorescent in situ hybridization of each SynCAM with either excitatory (data not shown) or inhibitory (Fig. 10, Fig. 11) neuronal markers. To define SynCAM expression at the peak of synaptogenesis, when they are prominent in the central nervous system, we performed our analysis in P15 mouse brains. We examined the hippocampus for excitatory neuronal expression and both hippocampus and cerebellum for inhibitory neurons to assess SynCAM expression patterns in these stereotypically organized brain regions.
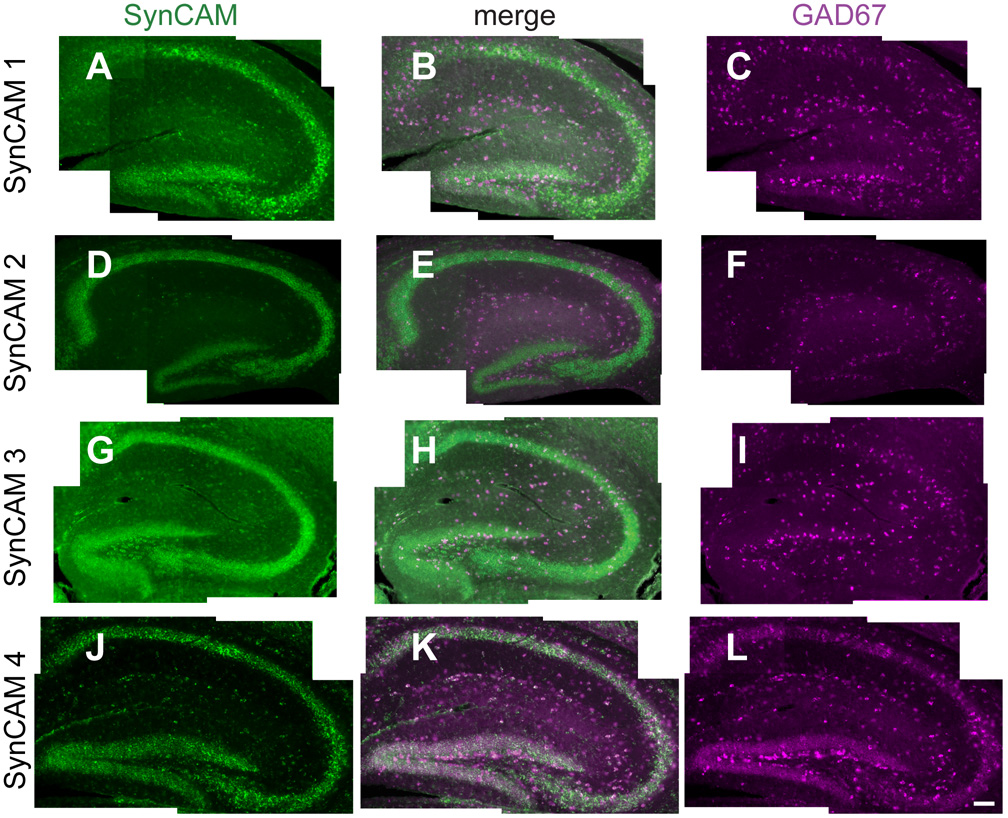
Fig. 10. Co-expression analysis of SynCAM 1–4 and GAD67 by double in situ hybridization in P15 hippocampus.
A–C. Expression of SynCAM 1 and GAD67 in P15 hippocampus. SynCAM 1-expressing cells (A) and a small number of GAD67-positive inhibitory interneurons (C) colocalize in the hippocampal formation. The center panel shows merged images (B). D–F. SynCAM 2 expression (D) is observed in a small number of GAD67-expressing cells (F). G–I. Expression of SynCAM 3 (G) labels some GAD67-positive inhibitory interneurons (I). J–L. SynCAM 4 (J) shows partial coexpression with GAD67 (L), with several GAD67-positive inhibitory interneurons also expressing SynCAM 4. Scale bar = 100 µm.
To detect excitatory neurons in hippocampus, we employed riboprobes for VGLUT1 and 2 transcripts, which encode vesicular glutamate transporters (Herzog et al., 2001; Sakata-Haga et al., 2001). SynCAM 1, 2, and 4 expression strongly colocalized with VGLUT 1 and 2 in CA fields and dentate gyrus. Strong colocalization with SynCAM 3 was detected only in CA fields due to the weaker expression of SynCAM 3 in the dentate gyrus (data not shown). In hippocampus, SynCAM expression was additionally detected in inhibitory neurons positive for transcripts of glutamic acid decarboxylase-67 (GAD67), the most prominently and ubiquitously expressed biosynthetic enzyme for the inhibitory neurotransmitter gamma-aminobutyric acid (Esclapez et al., 1993; Katarova et al., 2000). At P15, GAD67 expression was seen in the inhibitory interneurons of the hippocampal formation (Fig. 10C,F,I,L). As described above (Fig. 4), all SynCAMs showed some level of expression in the hilus of the dentate gyrus, stratum oriens, stratum radiatum and stratum lacunosum-moleculare. Some of this expression co-localized with the GAD67 expression in the hippocampal formation (Fig. 10B,E,H,K) but this was least apparent for SynCAMs 2 and 3.
The cerebellum represents another brain region with exemplary spatial separation of inhibitory neurons from excitatory ones, and was therefore also analyzed for SynCAM expression. Here, the strongest GAD67 expression is detectable in inhibitory Purkinje cells, with additional GAD67 expression seen in inhibitory interneurons throughout the rest of the cerebellum (Fig. 11C,F,I,L). Inhibitory interneurons in the granule cell layer of the cerebellum, marked by GAD67 expression, do not express any detectable SynCAM transcripts. SynCAM 3 shows no co-localization with GAD67, consistent with its apparently exclusive expression in the excitatory granule cells. SynCAM 1 and 4-positive neurons show co-labeling with GAD67 in the Purkinje cell layer (Fig. 11B,K).
The apparent expression of SynCAM 4 in GAD67-positive cells of the Purkinje cell layer (Fig. 11J, see also Fig. 8G) prompted us to investigate the extent of SynCAM 4 expression in Purkinje cells versus Bergmann glia. The majority of SynCAM 4 expression at P15 appeared to occur in Purkinje cells as demonstrated after SynCAM 4 in situ hybridization by its colocalization in immunostained sections with antibody staining against the Ins(1,4,5)P3 receptor, a Purkinje cell marker (Mignery et al., 1989) (Fig, 11M–O). In contrast, no colocalization of SynCAM 4 expression was observed with Bergmann glia cells immunopositive for the marker BLBP (Feng et al., 1994) (Fig. 11P–R). Results obtained for SynCAM 4 expression in Purkinje cells and Bergmann glia in adult tissue were identical to P15 (data not shown).
DISCUSSION
This comparative study was motivated by the recent observations that distinct heterophilic interactions distinguish SynCAM proteins from each other, indicating specific functions of these molecules in the brain (Kakunaga et al., 2005; Fogel et al., 2007; Maurel et al., 2007; Spiegel et al., 2007). We determined both the adhesion properties of SynCAM proteins and their mRNA expression profiles to study to which extent their adhesive interaction patterns complement their cellular expression profiles. Our five key findings are first, mRNA transcripts of all four SynCAMs are increasingly expressed throughout the regions of the developing brain. Second, SynCAM transcripts are prominently observed in neurons in the central nervous system. Third, the cellular SynCAM mRNA expression profiles are non-identical, which holds for all brain regions analyzed. Fourth, these distinct expression profiles are largely maintained throughout brain development from the peak period of synaptogenesis around P15 to later adult stages. Lastly, SynCAM proteins engage each other in distinct heterophilic interactions, consistent with specific adhesive properties conferred upon neurons that differentially express these molecules.
This adhesion analysis draws a comprehensive map of SynCAM family interactions. We here report the first analysis of the extracellular interactions of all four SynCAM family members using a cell overlay approach. Consistent with recent studies using this method (Maurel et al., 2007; Spiegel et al., 2007) or affinity chromatographical approaches (Fogel et al., 2007), heterophilic binding of SynCAM 1/2 and SynCAM 3/4 proteins is detected. In addition, we identify a novel, third heterophilic complex formed by SynCAMs 2 and 4. Prior biochemical studies had not detected the binding of SynCAM 2 and 4 (Fogel et al., 2007), which could be due to the fact that the relative orientation of extracellular domains is less favorable under affinity chromatographical conditions than in cell overlay studies. The weaker homophilic interactions of SynCAM 1, 2, and 3 (Fogel et al., 2007) were not observed in this assay under our conditions. This is consistent with this cell overlay assay serving as comparatively stringent method to evaluate extracellular protein interactions.
Due to the lack of specific antibodies applicable for immunohistochemical analysis of the localization of individual SynCAM family members, we conducted an analysis of their mRNA expression profiles. Our RT-PCR results demonstrate expression of the four SynCAMs throughout all brain regions analyzed. Interestingly, in most cases their transcript levels reach almost maximal amounts already at P15. This indicates that SynCAMs function in the early postnatal differentiation of neurons, including synaptogenesis. While their transcript amounts are often comparable, some regions display unusual expression levels, indicating that distinct expression profiles can characterize each SynCAM protein. Here, the cerebellum with its very high expression of SynCAM 3 in granule cells stands out. Another example for differing SynCAM amounts is the hippocampus with its overall low amounts of SynCAM 4 transcripts.
To resolve SynCAM expression in multiple regions of the central nervous system, we performed in situ hybridization studies in three forebrain regions, namely hippocampus, cortex, and olfactory bulb. Further, we analyzed the cerebellum and spinal cord. We conducted this analysis in the developing postnatal and in the adult brain to assess potential developmental changes. Sagittal sections confirmed the general expression of SynCAMs throughout the developing and adult brain. We additionally performed double labeling with antibodies against the neuronal marker NeuN to identify the cells expressing SynCAMs. Notably, the vast majority of central neurons in all brain regions express SynCAMs, indicating general functions of these adhesion molecules in neurons. Further, most SynCAM-expressing cells in the brain are NeuN-positive, in agreement with a predominantly neuronal expression and function of these molecules in the central nervous system. Future high-resolution studies employing double-labeling of SynCAMs and glial markers will be required to achieve a more quantitative assessment of their relative expression in glia. With respect to SynCAM 4, its prominent neuronal expression in the central nervous system differs from the peripheral nervous system, where it is expressed in myelinating Schwann cells (Maurel et al., 2007; Spiegel et al., 2007). It will be important to investigate to which extent SynCAM 4 also functions in subsets of glia cells of the central nervous system analogous to this role in the peripheral nervous system.
The cellular resolution of these in situ hybridization studies provides insights into differential expression of the four SynCAMs among neuronal populations within the same brain region. We here provide an analysis of SynCAM 1–4 expression at postnatal and adult stages that complements and extends previous and more general immunohistochemical studies (Biederer et al., 2002; Ohta et al., 2005) that utilized pleio-SynCAM antibodies recognizing SynCAM 1, 2, and 3 equally well (Fogel et al., 2007). Divergent SynCAM transcript expression is detected in all analyzed forebrain regions. In hippocampus, a potential role of SynCAM 1/2 interactions between neurons in different hippocampal subfields is indicated by the pronounced SynCAM 1 expression in the cell bodies of dentate granule cells, which appears equal to its expression in the CA fields. This contrasts with the relatively low SynCAM 2 expression in the dentate gyrus compared to neurons in the CA fields, and points toward a possible role of SynCAM 1/2 adhesion in the specification of mossy fiber inputs from SynCAM 1-expressing granule cells in the dentate gyrus to SynCAM 2-expressing neurons in the CA3 subregion. Weaker interactions of SynCAM 1 in dentate granule cells with SynCAM 3 in neurons of the CA3 subregion may subserve their developmental cellular interactions during input specification. In cortex and olfactory bulb, SynCAMs also display multiple layer-and region-specific expression differences.
Divergent expression of SynCAMs, however, is most apparent in cerebellum, where SynCAM 4 is distinctly expressed by Purkinje cells. Double labeling with antibodies against Purkinje cell and Bergmann glia markers supports a Purkinje cell expression of SynCAM 4 in this layer. This contrasts with the prominent expression of SynCAM 3 mRNA by cerebellar granule cells, a finding that complements the previous report of SynCAM 3 protein expression within the cerebellum’s molecular layer in parallel fiber axons and parallel fiber presynaptic terminals as well as in glial cells (Kakunaga et al., 2005). These observations suggest roles for SynCAM 3/4 adhesive interactions between cerebellar granule and Purkinje cells during cerebellar differentiation, including the specification of parallel fiber synapses. Future studies will address whether this differential SynCAM 3/4 expression indeed guides cerebellar synapse formation and organization. Next to SynCAM 3, SynCAM 2 is the second protein forming an adhesive complex with SynCAM 4. As SynCAM 2 is also expressed by cerebellar granule cells, its potential interaction with SynCAM 4 in Purkinje cells will also have to be assessed. It is notable that the peak of SynCAM 4 expression observed by RT-PCR in the third postnatal week coincides with the intense synapse formation by parallel fibers on Purkinje cells at that time in development (Takacs and Hamori, 1994). As SynCAM 4 decreases thereafter in the cerebellum, this expression profile may indicate a role of SynCAM 4 in Purkinje cells in the initiation but not maintenance of their parallel fiber inputs.
In spinal cord, all SynCAMs are found in neurons, albeit SynCAM 1 and 4 appear to be present at P2 also in presumptive oligodendrocytes, as are SynCAM 2 and 3 in adult spinal cord. Their expression patterns change more in the developing spinal cord than in other brain regions. This plastic expression is of interest in light of a recent report that neuronal SynCAM transcripts are downregulated after sciatic nerve transection of spinal motoneurons, indicating that they could contribute to the connectivity of spinal cord neurons (Zelano et al., 2007).
These results show that cognate SynCAMs are expressed in distinct neuronal populations in the brain. When suitable antibodies become available, co-localization studies will address whether the hypothesized SynCAM heterophilic interactions occur between these distinct neurons, and their potential roles in neuronal differentiation will be determined. Future studies will extend these results by analyzing the expression of other adhesion molecules reported as extracellular interaction partners of SynCAM proteins.
In summary, we here define three distinct heterophilic SynCAM adhesion complexes and combine their adhesive characterization with an in situ hybridization study of SynCAM expression in the developing and mature central nervous system. Our studies demonstrate that the four SynCAM adhesion molecules are expressed in characteristic and divergent profiles across the neuronal populations of multiple brain regions. The observation that these expression differences are mostly maintained from developing to adult stages indicates that SynCAMs could mediate comparable functions both in differentiating neurons and the mature brain, such as establishing and maintaining neuronal circuitry. Additional diversity may arise from the glycosylation of SynCAM extracellular domains, a posttranslational modification that can promote their adhesion (Fogel et al., 2007). Future studies will address to which extent the divergent expression profiles of SynCAMs and their distinct adhesive properties are complementary and provide for cell-type specific interactions in the brain. This direction has already been pursued in the central nervous system for the synaptic adhesion complex assembled by SynCAM 1 and 2 (Fogel et al., 2007).
Notably, the differential expression of adhesion molecules has been proposed to contribute to defining topographic maps and neuronal circuitry (Benson et al., 2001). This concept was initially motivated by the differential expression of cadherins in neuronal subpopulations throughout development (Redies and Takeichi, 1996; Benson et al., 2001) and was further strengthened by the region-dependent expression of protocadherin subfamily members in the developing cerebral cortex (Frank et al., 2005; Hirayama and Yagi, 2006). It should be noted that differential expression patterns of adhesion molecules may be further refined by dynamic changes in their posttranslational modification, which can modify their adhesive properties as established for the conjugation of polysialic acid to NCAM in the plastic nervous system (Chung et al., 1991; Rutishauser and Landmesser, 1996). The glycosylation of SynCAMs, which can be developmentally regulated, may analogously serve in regulating their adhesion in brain (Fogel et al., 2007). Together, the divergent expression of SynCAMs fits into this framework of differential expression of adhesion molecules in brain. Their comparative analysis is therefore required to gain insight into the roles of SynCAMs in neuronal development. In addition, these studies provide benchmarks to assess functional roles of these adhesion molecules in vitro and in vivo. Together, the expression of SynCAMs throughout the developing brain in unique spatiotemporal patterns indicates a potential key role in organizing the central nervous system and warrants a detailed analysis of multiple SynCAM-mediated adhesive interactions in the different brain regions.
Supplementary Material
Supp Fig s1
Supp Tab s1
Supp Tab s2
OTHER ACKNOWLEDGMENTS
We are grateful to Yuling Lei for excellent technical assistance. Riboprobes for detection of VGLUT1 and 2 were gifts from Dr. Marc Landry (Université Bordeaux, France), and for GAD67 from Dr. Zoya Katarova (Institute of Experimental Medicine, Budapest). We thank Drs. Charles Greer (Yale University, Department of Neurosurgery), Stephen Strittmatter (Yale University, Department of Neurology), Eric Frank (Tufts University, Department of Physiology), and Sreeganga Chandra (Yale University, Department of Neurology and Program in Cellular Neuroscience, Neurodegeneration, and Repair) for critical comments on this manuscript, and Drs. Greer and Strittmatter for access to microscopes. We thank Drs. Pietro De Camilli and Angelique Bordey for gifts of antibodies.
Acknowledgement for Support: National Institute of Health Grant; Grant number: R01 DA018928 (to T.B.); Grant sponsor: March of Dimes Foundation Award; Grant number 5-FY05-138 (to T.B.); Grant sponsor: The Brain Tumor Society (to T.B.); Grant sponsor: Anna Fuller Predoctoral Fellowship in Cancer Research (to L.A.T.)
LITERATURE CITED
- Arase N, Takeuchi A, Unno M, Hirano S, Yokosuka T, Arase H, Saito T. Heterotypic interaction of CRTAM with Necl2 induces cell adhesion on activated NK cells and CD8+ T cells. Int Immunol. 2005;17:1227–1237. doi: 10.1093/intimm/dxh299. [DOI] [PubMed] [Google Scholar]
- Benson DL, Colman DR, Huntley GW. Molecules, maps and synapse specificity. Nat Rev Neurosci. 2001;2:899–909. doi: 10.1038/35104078. [DOI] [PubMed] [Google Scholar]
- Biederer T. Bioinformatic characterization of the SynCAM family of immunoglobulin-like domain-containing adhesion molecules. Genomics. 2006;87:139–150. doi: 10.1016/j.ygeno.2005.08.017. [DOI] [PubMed] [Google Scholar]
- Biederer T, Sara Y, Mozhayeva M, Atasoy D, Liu X, Kavalali ET, Südhof TC. SynCAM, a synaptic adhesion molecule that drives synapse assembly. Science. 2002;297:1525–1531. doi: 10.1126/science.1072356. [DOI] [PubMed] [Google Scholar]
- Blue ME, Parnavelas JG. The formation and maturation of synapses in the visual cortex of the rat. II. Quantitative analysis. J Neurocytol. 1983;12:697–712. doi: 10.1007/BF01181531. [DOI] [PubMed] [Google Scholar]
- Boles KS, Barchet W, Diacovo T, Cella M, Colonna M. The tumor suppressor TSLC1/NECL-2 triggers NK cell and CD8+ T cell responses through the cell surface receptor CRTAM. Blood. 2005 doi: 10.1182/blood-2005-02-0817. 2005-2002-0817. [DOI] [PubMed] [Google Scholar]
- Chen HH, Hippenmeyer S, Arber S, Frank E. Development of the monosynaptic stretch reflex circuit. Curr Opin Neurobiol. 2003;13:96–102. doi: 10.1016/s0959-4388(03)00006-0. [DOI] [PubMed] [Google Scholar]
- Chung WW, Lagenaur CF, Yan YM, Lund JS. Developmental expression of neural cell adhesion molecules in the mouse neocortex and olfactory bulb. J Comp Neurol. 1991;314:290–305. doi: 10.1002/cne.903140207. [DOI] [PubMed] [Google Scholar]
- Dyson SE, Jones DG. Quantitation of terminal parameters and their inter-relationships in maturing central synapses: a perspective for experimental studies. Brain Res. 1980;183:43–59. doi: 10.1016/0006-8993(80)90118-3. [DOI] [PubMed] [Google Scholar]
- Esclapez M, Tillakaratne NJ, Tobin AJ, Houser CR. Comparative localization of mRNAs encoding two forms of glutamic acid decarboxylase with nonradioactive in situ hybridization methods. J Comp Neurol. 1993;331:339–362. doi: 10.1002/cne.903310305. [DOI] [PubMed] [Google Scholar]
- Feng L, Hatten ME, Heintz N. Brain lipid-binding protein (BLBP): a novel signaling system in the developing mammalian CNS. Neuron. 1994;12:895–908. doi: 10.1016/0896-6273(94)90341-7. [DOI] [PubMed] [Google Scholar]
- Fiala JC, Feinberg M, Popov V, Harris KM. Synaptogenesis via dendritic filopodia in developing hippocampal area CA1. J Neurosci. 1998;18:8900–8911. doi: 10.1523/JNEUROSCI.18-21-08900.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogel AI, Akins MR, Krupp AJ, Stagi M, Stein V, Biederer T. SynCAMs organize synapses through heterophilic adhesion. J Neurosci. 2007;27:12516–12530. doi: 10.1523/JNEUROSCI.2739-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank M, Ebert M, Shan WS, Phllips GR, Arndt K, Colman DR, Kemler R. Differential expression of individual gamma-protocadherins during mouse brain development. Mol Cell Neurosci. 2005;29:603–616. doi: 10.1016/j.mcn.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Furuno T, Ito A, Koma Y-i, Watabe K, Yokozaki H, Bienenstock J, Nakanishi M, Kitamura Y. The spermatogenic Ig superfamily/Synaptic Cell Adhesion Molecule mast-cell adhesion molecule promotes interaction with nerves. J Immunol. 2005;174:6934–6942. doi: 10.4049/jimmunol.174.11.6934. [DOI] [PubMed] [Google Scholar]
- Gorman C. DNA cloning. A practical approach. Oxford: IRL Press; 1985. [Google Scholar]
- Gruber-Olipitz M, Yang JW, Slavc I, Lubec G. Nectin-like molecule 1 is a high abundance protein in cerebellar neurons. Amino Acids. 2006;30:409–415. doi: 10.1007/s00726-006-0323-0. [DOI] [PubMed] [Google Scholar]
- Harris KM, Jensen FE, Tsao B. Three-dimensional structure of dendritic spines and synapses in rat hippocampus (CA1) at postnatal day 15 and adult ages: implications for the maturation of synaptic physiology and long-term potentiation. J Neurosci. 1992;12:2685–2705. doi: 10.1523/JNEUROSCI.12-07-02685.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog E, Bellenchi GC, Gras C, Bernard V, Ravassard P, Bedet C, Gasnier B, Giros B, El Mestikawy S. The existence of a second vesicular glutamate transporter specifies subpopulations of glutamatergic neurons. J Neurosci. 2001;21:RC181. doi: 10.1523/JNEUROSCI.21-22-j0001.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinds JW, Hinds PL. Synapse formation in the mouse olfactory bulb. I. Quantitative studies. J Comp Neurol. 1976;169:15–40. doi: 10.1002/cne.901690103. [DOI] [PubMed] [Google Scholar]
- Hirayama T, Yagi T. The role and expression of the protocadherin-alpha clusters in the CNS. Curr Opin Neurobiol. 2006;16:336–342. doi: 10.1016/j.conb.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Kakunaga S, Ikeda W, Itoh S, Deguchi-Tawarada M, Ohtsuka T, Mizoguchi A, Takai Y. Nectin-like molecule-1/TSLL1/SynCAM3: a neural tissue-specific immunoglobulin-like cell-cell adhesion molecule localizing at non-junctional contact sites of presynaptic nerve terminals, axons and glia cell processes. J Cell Sci. 2005;118:1267–1277. doi: 10.1242/jcs.01656. [DOI] [PubMed] [Google Scholar]
- Katarova Z, Sekerkova G, Prodan S, Mugnaini E, Szabo G. Domain-restricted expression of two glutamic acid decarboxylase genes in midgestation mouse embryos. J Comp Neurol. 2000;424:607–627. doi: 10.1002/1096-9861(20000904)424:4<607::aid-cne4>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- Kuramochi M, Fukuhara H, Nobukuni T, Kanbe T, Maruyama T, Ghosh HP, Pletcher M, Isomura M, Onizuka M, Kitamura T, Sekiya T, Reeves RH, Murakami Y. TSLC1 is a tumor-suppressor gene in human non-small-cell lung cancer. Nat Genet. 2001;27:427–430. doi: 10.1038/86934. [DOI] [PubMed] [Google Scholar]
- Landry M, Bouali-Benazzouz R, El Mestikawy S, Ravassard P, Nagy F. Expression of vesicular glutamate transporters in rat lumbar spinal cord, with a note on dorsal root ganglia. J Comp Neurol. 2004;468:380–394. doi: 10.1002/cne.10988. [DOI] [PubMed] [Google Scholar]
- Lind D, Franken S, Kappler J, Jankowski J, Schilling K. Characterization of the neuronal marker NeuN as a multiply phosphorylated antigen with discrete subcellular localization. J Neurosci Res. 2005;79:295–302. doi: 10.1002/jnr.20354. [DOI] [PubMed] [Google Scholar]
- Maurel P, Einheber S, Galinska J, Thaker P, Lam I, Rubin MB, Scherer SS, Murakami Y, Gutmann DH, Salzer JL. Nectin-like proteins mediate axon Schwann cell interactions along the internode and are essential for myelination. J Cell Biol. 2007;178:861–874. doi: 10.1083/jcb.200705132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignery GA, Sudhof TC, Takei K, De Camilli P. Putative receptor for inositol 1,4,5-trisphosphate similar to ryanodine receptor. Nature. 1989;342:192–195. doi: 10.1038/342192a0. [DOI] [PubMed] [Google Scholar]
- Mullen RJ, Buck CR, Smith AM. NeuN, a neuronal specific nuclear protein in vertebrates. Development. 1992;116:201–211. doi: 10.1242/dev.116.1.201. [DOI] [PubMed] [Google Scholar]
- Ohta Y, Itoh K, Yaoi T, Tando S, Fukui K, Fushiki S. Spatiotemporal patterns of expression of IGSF4 in developing mouse nervous system. Dev Brain Res. 2005;156:23–31. doi: 10.1016/j.devbrainres.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Redies C, Takeichi M. Cadherins in the developing central nervous system: an adhesive code for segmental and functional subdivisions. Dev Biol. 1996;180:413–423. doi: 10.1006/dbio.1996.0315. [DOI] [PubMed] [Google Scholar]
- Rutishauser U, Landmesser L. Polysialic acid in the vertebrate nervous system: a promoter of plasticity in cell-cell interactions. Trends Neurosci. 1996;19:422–427. doi: 10.1016/0166-2236(96)10041-2. [DOI] [PubMed] [Google Scholar]
- Sakata-Haga H, Kanemoto M, Maruyama D, Hoshi K, Mogi K, Narita M, Okado N, Ikeda Y, Nogami H, Fukui Y, Kojima I, Takeda J, Hisano S. Differential localization and colocalization of two neuron-types of sodium-dependent inorganic phosphate cotransporters in rat forebrain. Brain Res. 2001;902:143–155. doi: 10.1016/s0006-8993(01)02290-9. [DOI] [PubMed] [Google Scholar]
- Sara Y, Biederer T, Atasoy D, Chubykin A, Mozhayeva MG, Südhof TC, Kavalali ET. Selective capability of SynCAM and neuroligin for function al synapse assembly. J Neurosci. 2005;25:260–270. doi: 10.1523/JNEUROSCI.3165-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shingai T, Ikeda W, Kakunaga S, Morimoto K, Takekuni K, Itoh S, Satoh K, Takeuchi M, Imai T, Monden M, Takai Y. Implications of nectin-like molecule-2/IGSF4/RA175/SgIGSF/TSLC1/SynCAM1 in cell-cell adhesion and transmembrane protein localization in epithelial cells. J Biol Chem. 2003;278:35421–35427. doi: 10.1074/jbc.M305387200. [DOI] [PubMed] [Google Scholar]
- Spiegel I, Adamsky K, Eshed Y, Milo R, Sabanay H, Sarig-Nadir O, Horresh I, Scherer SS, Rasband MN, Peles E. A central role for Necl4 (SynCAM4) in Schwann cell-axon interaction and myelination. Nat Neurosci. 2007;10:861–869. doi: 10.1038/nn1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita S, Saito F, Tang J, Satz J, Campbell K, Sudhof TC. A stoichiometric complex of neurexins and dystroglycan in brain. J Cell Biol. 2001;154:435–445. doi: 10.1083/jcb.200105003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takacs J, Hamori J. Developmental dynamics of Purkinje cells and dendritic spines in rat cerebellar cortex. J Neurosci Res. 1994;38:515–530. doi: 10.1002/jnr.490380505. [DOI] [PubMed] [Google Scholar]
- Weber ED, Stelzner DJ. Synaptogenesis in the intermediate gray region of the lumbar spinal cord in the postnatal rat. Brain Res. 1980;185:17–37. doi: 10.1016/0006-8993(80)90667-8. [DOI] [PubMed] [Google Scholar]
- Zelano J, Wallquist W, Hailer NP, Cullheim S. Down-regulation of mRNAs for synaptic adhesion molecules neuroligin-2 and -3 and SynCAM1 in spinal motoneurons after axotomy. J Comp Neurol. 2007;503:308–318. doi: 10.1002/cne.21382. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supp Fig s1
Supp Tab s1
Supp Tab s2