Surveillance for Neuraminidase Inhibitor Resistance among Human Influenza A and B Viruses Circulating Worldwide from 2004 to 2008 (original) (raw)
Abstract
The surveillance of seasonal influenza virus susceptibility to neuraminidase (NA) inhibitors was conducted using an NA inhibition assay. The 50% inhibitory concentration values (IC50s) of 4,570 viruses collected globally from October 2004 to March 2008 were determined. Based on mean IC50s, A(H3N2) viruses (0.44 nM) were more sensitive to oseltamivir than A(H1N1) viruses (0.91 nM). The opposite trend was observed with zanamivir: 1.06 nM for A(H1N1) and 2.54 nM for A(H3N2). Influenza B viruses exhibited the least susceptibility to oseltamivir (3.42 nM) and to zanamivir (3.87 nM). To identify potentially resistant viruses (outliers), a threshold of a mean IC50 value + 3 standard deviations was defined for type/subtype and drug. Sequence analysis of outliers was performed to identify NA changes that might be associated with reduced susceptibility. Molecular markers of oseltamivir resistance were found in six A(H1N1) viruses (H274Y) and one A(H3N2) virus (E119V) collected between 2004 and 2007. Some outliers contained previously reported mutations (e.g., I222T in the B viruses), while other mutations [e.g., R371K and H274Y in B viruses and H274N in A(H3N2) viruses) were novel. The R371K B virus outlier exhibited high levels of resistance to both inhibitors (>100 nM). A substantial variance at residue D151 was observed among A(H3N2) zanamivir-resistant outliers. The clinical relevance of newly identified NA mutations is unknown. A rise in the incidence of oseltamivir resistance in A(H1N1) viruses carrying the H274Y mutation was detected in the United States and in other countries in the ongoing 2007 to 2008 season. As of March 2008, the frequency of resistance among A(H1N1) viruses in the United States was 8.6% (50/579 isolates). The recent increase in oseltamivir resistance among A(H1N1) viruses isolated from untreated patients raises public health concerns and necessitates close monitoring of resistance to NA inhibitors.
Influenza A and B viruses are respiratory pathogens that affect humans and that are responsible for substantial morbidity, mortality, and decreased productivity in the United States (29). Vaccination provides the primary means for protection from influenza virus infections. Due to the continuous evolution of major viral antigens, hemagglutinin (HA) and neuraminidase (NA), vaccine strains must be selected annually. This selection is based on global surveillance of A(H1N1), A(H3N2), and B influenza viruses circulating in humans. Antivirals provide a valuable addition to the available options used to control influenza infections. Two classes of these antiviral drugs, adamantanes and NA inhibitors, are currently licensed by the U. S. Food and Drug Administration (FDA) for the prophylaxis and treatment of influenza infections. The first and oldest class, adamantanes (amantadine and rimantadine), target the proton channel formed by the viral M2 protein. Due to the absence of this protein in influenza B viruses, adamantanes have no antiviral effect on this virus type (20). Surveillance of adamantane resistance among influenza A viruses is based on the detection of well-characterized molecular markers in the M2 protein transmembrane domain (3). The rapid spread of resistance to adamantanes in recent years (5, 10) diminished the usefulness of this class of drugs and prompted changes to the recommendations made by the Centers for Disease Control and Prevention (CDC) (4).
The two NA inhibitors, orally bioavailable oseltamivir and inhaled zanamivir, are the only drugs currently recommended for the treatment of both influenza A and B virus infections in the United States (31). Molecular markers of resistance to this newer class of drugs are not well defined, and the clinical relevance of some identified mutations remains uncertain (27). Mutations identified in the NAs of viruses selected in the presence of NA inhibitors vary depending on the NA antigenic type/subtype and on the drug (13). For these reasons, monitoring resistance to the NA inhibitors is based primarily on testing viruses by using an NA activity inhibition (NAI) assay in conjunction with an NA sequence analysis (11, 30, 32, 33, 38). Two synthetic substrates, the chemiluminogenic NA-Star (6) and the fluorogenic 2′-(4-methylumbelliferyl)-α-d-_N_-acetylneuraminic acid (MUNANA) (11, 21, 34), are the substrates most commonly utilized in various NAI assays (38, 42). The chemiluminescent assay was chosen in this study of drug resistance surveillance of seasonal influenza because of the linearity of its signal and its high sensitivity in measuring NA activity compared to that of other substrates (6, 33, 38).
The NA is a surface antigen containing an enzymatic active site that is targeted by NA inhibitors. Therefore, viruses with reduced drug susceptibility can emerge as a result of drug use and/or natural genetic variation in the NA. The emergence of drug resistance is accompanied by an increase in the drug concentration needed to inhibit the enzyme activity of the mutant compared to that needed for the wild-type virus (27). NA mutants emerging during antiviral treatment commonly acquire changes at conserved residues in the NA active site (15, 16, 23, 36). One of the major challenges in determining drug resistance is the substantial variation in susceptibility in the NAI assay result, even for viruses carrying an enzyme of the same antigenic subtype (26, 33). Moreover, there are no widely accepted criteria for resistance to NA inhibitors because of the variety of NAI assays and assay conditions utilized in different laboratories. In surveillance studies, the criteria are based on statistical analyses of data obtained with the NAI assays for a large number of viruses grouped by NA type (A or B) and subtype (N1 or N2) (11, 26, 30, 33). This approach is used to determine the range of 50% inhibitory concentration values (IC50s) for the population of nonresistant viruses. Sequence analysis of the NA of those viruses that are determined to be outliers in the NAI assay serves to confirm drug resistance if the known genetic markers associated with drug resistance are identified (26, 30). In this study, box plot analysis was done retrospectively for comparison with an elected threshold cutoff IC50 of greater than the mean IC50 + 3 standard deviations (SD), which was used to select extreme and mild outliers for retesting and sequencing.
The large-scale screening study (26) conducted by the Global Neuraminidase Inhibitor Susceptibility Network (42) of viruses isolated between 1996 and 1999, before the introduction of the NA inhibitors into markets, showed the general lack of primary resistance to these antivirals. Drug susceptibility surveillance studies (11, 30, 33) conducted in the subsequent seasons 1999 to 2002, 2000 to 2002, and 2005 to 2006 supported the low frequency of virus variants exhibiting reduced drug susceptibility in the NAI assays. Among the drug-resistant variants studied, only one contained the marker for oseltamivir resistance (i.e., H274Y), previously identified in viruses isolated from drug-treated patients (15).
The continuous evolution of influenza viruses and widespread resistance to adamantanes necessitated the close monitoring of resistance to NA inhibitors among seasonal influenza viruses. In the present study, we report the results of susceptibility testing of two licensed NA inhibitors against influenza A and B viruses, circulating worldwide during the three influenza seasons, from 1 October 2004 to 30 September 2007, as well as during the early months of the 2007 to 2008 season. The study was conducted with the use of a recently developed NA-Star kit that utilizes a chemiluminescent substrate (6). We report here the detection of drug-resistant virus variants carrying established genetic markers for oseltamivir resistance (i.e., H274Y in N1 and E119V in N2), as well as highly resistant viruses (e.g., R371K in influenza B) or moderate outliers with novel mutations in the NA active site. We also report here a noticeable rise in the frequency of the H274Y mutations among influenza A(H1N1) viruses circulating in the United States, starting from the end of October of 2007. These findings are in accord with reports of the emergence of oseltamivir resistance in A(H1N1) viruses in Europe and other countries (http://www.who.int/csr/disease/influenza/h1n1_table/en/index.html).
MATERIALS AND METHODS
Compounds.
Zanamivir was provided by GlaxoSmithKline. Oseltamivir carboxylate (an active metabolite of the prodrug oseltamivir) was provided by Hoffman-La Roche (an abbreviated name, oseltamivir, is used throughout the text for simplicity instead of its full name, oseltamivir carboxylate).
Viruses.
The influenza A and B viruses collected during three consecutive seasons (1 October 2004 to 30 September 2005; 1 October 2005 to 30 September 2006; and 1 October 2006 to 30 September 2007) and during the current ongoing influenza season (1 October 2007 to 21 March 2008) and submitted to the World Health Organization Collaborating Center for Surveillance, Epidemiology, and Control of Influenza at the CDC (Atlanta, GA) were tested. Surveillance for antiviral resistance is deemed to be a public health practice and is exempt from review by the CDC internal review board.
There was no intentional or systematic bias in the sample selection process, except during the 2007 to 2008 season, when a request for additional A(H1N1) viruses was issued to public health laboratories in the United States. No testing for susceptibility to NA inhibitors was done prior to the submission of isolates to the CDC.
The influenza A and B viruses used in the study were from 47 individual U.S. states and 62 foreign countries, including those in Africa (7), Asia (23), Europe (11), North America (2), Oceania (2), and South America (17). Antiviral testing was performed with viruses propagated in Madin-Darby canine kidney (MDCK) cells or embryonated chicken eggs, as part of ongoing influenza surveillance. Virus types and subtypes of HA were determined by the laboratories providing the viruses and confirmed at the CDC.
A panel of previously characterized drug-resistant viruses and their drug-sensitive counterparts were grown in MDCK cells and used as controls for a corresponding type and subtype of the NA. The panel included B/Memphis/20/96 and its R152K mutant; B/Rochester/2/2001 and its D198N mutant; A/Texas/36/91 (H1N1) and its H274Y mutant; A/turkey/Minnesota/833/80 (H4N2) and its R292K, E119G, E119A, and E119D mutants; and the A/Bethesda/956/2006 (H3N2) R292K mutant (28).
NAI assay.
The chemiluminescent NAI assay was conducted using a commercially available kit, NA-Star (Applied Biosystems, Foster City, CA), which includes NA-Star buffer (26 mM morpholineethanesulfonic acid, 4 mM CaCl2 [pH 6.0]), NA-Star substrate, NA-Star accelerator, and 96-well solid white plates. The working NA inhibitor dilutions of 10 half-log (0.03 to 1,000 nM) were prepared in NA-Star buffer and stored at 4°C for a maximum of 2 weeks. The protocol was performed essentially as recommended by the manufacturer, with minor modifications. NA activity was determined before the NAI assay was performed by testing serial twofold dilutions of the virus. Viruses were diluted to a final concentration in NA-Star assay buffer so that their signal-to-noise ratio fell between 10:1 and 40:1. All viruses were diluted at least fivefold to avoid signal quenching (38). Twenty-five microliters of each of the dilutions of NA inhibitor was added to each well of a plate, to which 25 μl of each virus dilution was then added. Plates were preincubated with the drug at room temperature for 10 to 30 min. Ten microliters of 10 mM NA-Star substrate, diluted 1:1,000 in NA-Star assay buffer (final concentration of 0.01 mM), was added to each well, and the plate was then incubated at room temperature for 10 to 30 min without shaking. Shaking was abolished to reduce manual manipulations, essential for high-throughput screening. Sixty microliters of NA-Star accelerator was injected into each well of the 96-well plate, and the luminescence was read immediately for 0.5 s at a single point. Luminescence was measured using a multiplate Victor3V (Perkin-Elmer, Shelton, CT) reader equipped with automatic injectors for the accelerator.
IC50 analysis.
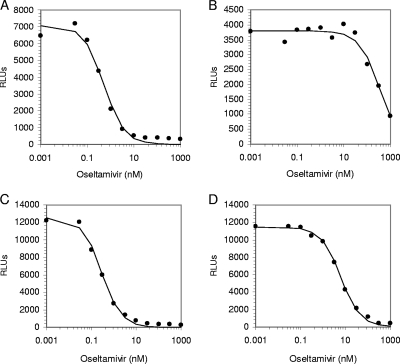
Curve fitting and IC50s (a concentration of drug needed to inhibit enzyme activity by 50%) were determined by the equation y = _V_max× {1 − [x/(K + x)]}, where _V_max is the maximum rate of metabolism, using Robosage software (38) (GlaxoSmithKline in-house program, kindly provided by Michael Lutz and Margaret Tisdale), x is the inhibitor concentration, y is the response being inhibited, and K is the IC50 for the inhibition curve (that is, y = 50% _V_max when x = K). As shown in Fig. 1, certain resistant viruses (e.g., the H274Y strain) can be identified easily based on IC50 data only; however, detection of other resistant viruses (e.g., the E119V strain) would require additional analysis (see below).
FIG. 1.
Assessment of the IC50s for oseltamivir, using the chemiluminescent NAI assay. (A) Oseltamivir-sensitive A/Massachusetts/6/2006 (H1N1) virus; (B) oseltamivir-resistant A/Massachusetts/5/2007 (H1N1) virus with the mutation H274Y; (C) oseltamivir-sensitive A/Washington/1/2007 (H3N2) virus; (D) oseltamivir-resistant A/Texas/12/2007(H3N2) virus with the E119V mutation. x axis, NA activity in relative light units (RLUs). y axis, oseltamivir concentrations (nM) on a logarithmic scale. Data points indicate actual activity measured at a single time point using a plate reader, Victor 3V. The data line represents the best-fit curve generated using Robosage software.
Statistical analysis.
The IC50s of enzyme susceptibility to either drug were analyzed using Microsoft Excel 2003 (Microsoft Corporation, Redmond, WA). Mean and SD values of IC50s were determined separately for each virus type/subtype for each influenza season and were analyzed cumulatively for three seasons, from 2004 through 2007. In previous surveillance studies (26, 30), the threshold value was set at 95% confidence (mean IC50 ± 2 SD) and between 1.5 and 3.0 times the interquartile range (IQR). The use of either criterion in our study resulted in a substantial number of isolates without genetic changes detected as outliers. Thus, the mean IC50 value + 3 SD criterion was selected as a cutoff value and was calculated for each NA type/subtype for each NA inhibitor. In this study, extreme outliers (isolates with IC50s more than 10-fold higher than the mean IC50 for each respective type/subtype and drug) were excluded from statistical analysis of the overall population. All viruses with IC50s outside the cutoff criterion but within 10-fold of the mean IC50 value were considered mild outliers. All outliers were retested with the NAI assay, followed by full NA sequence analysis. Retested viruses that were consistent outliers containing previously characterized NA mutations known to be associated with resistance to NA inhibitors that emerged in treated patients were considered drug resistant for this study.
A one-way analysis of variance was done to compare the IC50s among the seasons for each type/subtype and drug, using SAS 9.1 (SAS Institute, Inc., Cary, NC) software. Statistical significance was set at an α value of 0.05. A box plot analysis of all isolates by type/subtype and drug, excluding extreme outliers, was used to determine the IQR and to establish the statistical cutoff. Statistical cutoff was determined to be isolates with IC50s which were greater than three times the IQR to the right of the third quartile (_X_0.75) (IC50 > _X_0.75 + 3 IQR).
Reverse transcription-PCR and sequence analysis.
Viral RNA was extracted using either a QIAamp viral RNA mini-kit (Qiagen, Valencia, CA) or a MagNA Pure (Roche Diagnostics, Indianapolis, IN) kit. RT-PCR amplification of the NA gene was performed using a SuperScript III One-Step RT-PCR kit (Invitrogen, Carlsbad, CA). Sequences of primers are available upon request.
Nucleotide sequence accession numbers.
NA sequences of 50 viruses tested in this study were deposited into the GenBank database and/or Los Alamos National Laboratory Influenza Sequence Database (LANL). Sequence accession numbers are shown in Tables 4 and 5. All mutations of NA are given in N2 subtype numbering (7).
TABLE 4.
Characterization of extreme and mild outliers among A(H3N2) viruses from three previous seasons
| Season | Strain | Mean IC50 ± SD (nM)b | Outlier(s)/drug(s)c | NA mutation(s)d | Accession no. | |
|---|---|---|---|---|---|---|
| Oseltamivir | Zanamivir | |||||
| 2006-2007 | A/Texas/12/2007a | 9.18 ± 0.78e | 0.96 ± 0.46 | E/O | E119V | EU516111 |
| 2006-2007 | A/Montana/8/2007 | 3.50 ± 0.06 | 191.94 ± 11.83f | M/O, E/Z | D151V/D | EU516195 |
| 2004-2005 | A/Argentina/135/2005 | 1.52 ± 0.28 | 62.49 ± 32.89 | M/O, E/Z | D151V | EU879077 |
| 2006-2007 | A/Canada/270/2007 | 1.18 ± 0.39 | 47.60 ± 19.58 | E/Z | D151A/D | EU879074 |
| 2004-2005 | A/Oman/6943/2005 | 0.74 ± 0.28 | 33.79 ± 2.39 | E/Z | D151A | EU879069 |
| 2006-2007 | A/Canada/83/2006 | 0.90 ± 0.41 | 15.12 ± 5.76 | M/Z | V165I, H274N | EU879070 |
| 2006-2007 | A/Canada/309/2007 | 1.64 ± 0.51 | 14.31 ± 4.78 | M/O, M/Z | G248R, K249E | EU879076 |
| 2004-2005 | A/Singapore/8/2005 | 1.15 ± 0.08 | 13.98 ± 1.37 | M/Z | D151N | EU879072 |
| 2006-2007 | A/Canada/281/2007 | 1.39 ± 0.36 | 12.80 ± 4.24 | M/Z | D151N, K249E | EU879067 |
| 2004-2005 | A/Hong Kong/4653/2005 | 1.00 ± 0.18 | 12.28 ± 1.19 | M/Z | D151N, S315G, L338S | EU879075 |
| 2004-2005 | A/Singapore/2/2005 | 1.07 ± 0.21 | 10.76 ± 5.56 | M/Z | A27S, D151N/D, K249E, V313A | EU879068 |
| 2006-2007 | A/Canada/267/2007 | 1.40 ± 0.18 | 10.66 ± 3.31 | M/Z | D251G | EU879065 |
| 2005-2006 | A/Santiago/9491/2006 | 2.78 ± 2.77 | 9.54 ± 2.08 | M/O, M/Z | D151N | EU879071 |
| 2006-2007 | A/Canada/9/2006 | 0.46 ± 0.26 | 8.92 ± 2.99g | M/Z | D151G/D | EU879066 |
| 2006-2007 | A/Canada/1210/2006 | 0.61 ± 0.34 | 8.02 ± 2.88g | M/Z | D151G/D | EU879073 |
TABLE 5.
Characterization of extreme and mild outliers detected among influenza B viruses from three previous seasons
| Season | Strain | Mean IC50 ± SD (nM)a | Outlier(s)/drug(s)b | NA mutationc | Accession no. | |
|---|---|---|---|---|---|---|
| Oseltamivir | Zanamivir | |||||
| 2004-2005 | B/Hong Kong/36/2005 | 1,778.25 ± 729.78d | 154.85 ± 47.47d | E/O, E/Z | R371K | EU879085 |
| 2004-2005 | B/Michigan/20/2005 | 10.80 ± 2.90e | 1.54 ± 0.27 | M/O | H274Y | CY015375 |
| 2004-2005 | B/California/5/2005 | 10.48 ± 1.62 | 8.65 ± 1.46 | M/O | I222T | EU879086 |
| 2004-2005 | B/California/4/2005 | 8.60 ± 1.93 | 3.73 ± 1.72 | M/O | T325I | EU879084 |
RESULTS
The goals of this surveillance study were to determine the baseline susceptibility of influenza viruses circulating among humans in recent years to the two licensed NA inhibitors, to identify outliers and, thus, to detect likely drug-resistant viruses, and to track the emergence of drug-resistant viruses over time.
Testing previously characterized drug-resistant viruses with an NA-Star kit.
To carry out high-throughput screening for resistance to NA inhibitors, we utilized an NA-Star kit (described in Materials and Methods). Since conditions (buffer, substrate concentration, etc.) of the NAI assay are known to affect the IC50s (17, 38), it was essential to confirm our ability to detect the drug-resistant mutants that had previously established markers of resistance to oseltamivir and/or zanamivir (14, 17), using the NA-Star kit. Specifically, the IC50s were determined with zanamivir and oseltamivir for a set of eight influenza A and B mutants that were either recovered from drug-treated patients or were selected in cell culture in the presence of NA inhibitors. The mutants exhibited a noticeable increase in IC50s (at least fourfold) and were distinguishable from their corresponding drug-sensitive parents, with one exception: the zanamivir-selected R292K (N2) mutant, when tested with zanamivir, showed little or no difference in IC50 from its parent virus (Table 1). The mutant and the parent viruses were resequenced, and the presence of the lysine at residue 292 in the mutant was confirmed. Thus, using the current assay and protocol, the resistance to zanamivir conferred by the R292K mutation would not have been detected. Nevertheless, this mutant was readily detected using oseltamivir (970-fold). As both NA inhibitors are routinely used for testing, detection of variants carrying R292K does not present a challenge for surveillance. This limitation of the assay should not be interpreted as a demonstration of zanamivir sensitivity of the R292K mutants.
TABLE 1.
Inhibition of the enzyme activity of the reference viruses in the NAI assay with NA-Star substrate
| Virus strain | Virus NA type/subtype | NA mutation | Mean IC50 ± SD (nM) (fold increase)c | |
|---|---|---|---|---|
| Oseltamivir | Zanamivir | |||
| B/Memphis/20/96 | B | 2.01 ± 0.69 | 4.10 ± 1.79 | |
| Zanamivir-selected mutanta | B | R152K | 77.24 ± 23.94 (38) | 40.99 ± 27.88 (10) |
| B/Rochester/2/2001 | B | 3.82 ± 1.96 | 4.39 ± 1.82 | |
| Oseltamivir-selected mutanta | B | D198N | 28.92 ± 9.37 (8) | 42.08 ± 22.14 (10) |
| A/Texas/36/91 (H1N1) | A/N1 | 1.06 ± 0.36 | 1.55 ± 0.22 | |
| Oseltamivir-selected mutant | A/N1 | H274Y | 260.25 ± 55.82 (246) | 2.41 ± 0.81 (2) |
| A/turkey/Minnesota/833/80 (H4N2) | A/N2 | 0.82 ± 0.34 | 4.26 ± 2.17 | |
| Zanamivir-selected mutant | A/N2 | R292K | 795.50 ± 196.24 (970) | 6.36 ± 1.37 (1.5) |
| Zanamivir-selected mutant | A/N2 | E119G | 0.93 ± 0.71 (1) | 33.48 ± 6.41 (8) |
| Zanamivir-selected mutant | A/N2 | E119A | 1.44 ± 1.04 (2) | 17.82 ± 4.87 (4) |
| Zanamivir-selected mutant | A/N2 | E119D | 1.21 ± 0.72 (1) | 71.52 ± 16.79 (17) |
| A/Bethesda/956/2006 (H3N2) oseltamivir-selected mutanta,b | A/N2 | R292K | 480.64 ± 217.299 | 5.24 ± 2.31 |
Screening for drug resistance among seasonal influenza virus isolates.
During the three seasons (2004 to 2005, 2005 to 2006, and 2006 to 2007), a total of 3,261 influenza A and B virus isolates collected worldwide were screened for susceptibility to oseltamivir and zanamivir. The number of viruses tested per season during this period rose from 371 to 648 to 2,242, respectively. For the ongoing 2007 to 2008 season, 1,309 influenza A and B virus isolates have been tested to date. The mean IC50 for A(H1N1) viruses was low to both oseltamivir (0.91 nM) and zanamivir (1.06 nM) (Table 2). The mean IC50 for influenza A(H3N2) was similarly low to oseltamivir (0.44 nM) but was higher against zanamivir (2.54 nM). Influenza B virus isolates had greater mean IC50s to both oseltamivir and zanamivir (3.42 nM and 3.87 nM, respectively).
TABLE 2.
NAI data analysis of IC50s grouped according to season and virus type/subtype
| Drug | Virus | IC50 (nM) per season | Total IC50 (nM)a | P valueb | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2004-2005 | 2005-2006 | 2006-2007 | ||||||||||
| Range (n) | Mean ± SD | Range (n) | Mean ± SD | Range (n) | Mean ± SD | Range (n) | Mean ± SD | Cutoff | Statistical cutoff | |||
| Oseltamivir | A(H1N1) | 0.35-6.05 (53) | 0.96 ± 0.97 | 0.19-10.00 (258) | 1.15 ± 1.01 | 0.16-4.51 (817) | 0.84 ± 0.45 | 0.16-10.00 (1,128) | 0.91 ± 0.66 | 2.89 | 2.43 | <0.0001 |
| A(H3N2) | 0.11-3.45 (142) | 0.52 ± 0.34 | 0.08-3.05 (186) | 0.59 ± 0.46 | 0.04-5.16 (908) | 0.40 ± 0.32 | 0.04-5.16 (1,236) | 0.44 ± 0.35 | 1.50 | 1.51 | <0.0001 | |
| B | 1.24-11.63 (175) | 3.23 ± 1.63 | 0.70-7.95 (203) | 3.45 ± 1.36 | 0.19-10.48 (511) | 3.47 ± 1.24 | 0.19-11.63 (889) | 3.42 ± 1.35 | 7.47 | 8.64 | 0.122 | |
| Zanamivir | A(H1N1) | 0.50-6.48 (53) | 1.12 ± 0.88 | 0.20-13.50 (259) | 1.30 ± 1.34 | 0.06-11.60 (822) | 0.98 ± 0.74 | 0.06-13.50 (1,134) | 1.06 ± 0.93 | 3.85 | 3.32 | <0.0001 |
| A(H3N2) | 0.49-13.98 (140) | 3.66 ± 2.51 | 0.14-11.77 (186) | 2.62 ± 2.04 | 0.15-20.00 (907) | 2.35 ± 2.16 | 0.14-20.00 (1,233) | 2.54 ± 2.22 | 9.20 | 9.93 | <0.0001 | |
| B | 1.12-16.51 (175) | 4.21 ± 2.65 | 0.85-17.86 (203) | 4.84 ± 2.77 | 0.35-12.00 (511) | 3.36 ± 2.14 | 0.35-17.86 (889) | 3.87 ± 2.47 | 11.29 | 14.21 | <0.0001 |
Identification of outliers during the 2004 to 2005, 2005 to 2006, and 2006 to 2007 influenza seasons.
During the 2004 to 2005, 2005 to 2006, and 2006 to 2007 seasons, we identified 12 extreme outliers (Table 3 to 5) with IC50s more than 10-fold higher than the mean IC50 for that NA type/subtype. While there were statistically significant differences (P < 0.0001), except for B viruses tested against oseltamivir, among the data from the three seasons, there was no trend toward increased IC50s with time (Table 2). All IC50s were pooled to determine a mean IC50 and SD for each type and subtype. Overall, influenza A and B viruses were more sensitive to oseltamivir than to zanamivir. Influenza A viruses were more sensitive to both NA inhibitors than the influenza B viruses. These findings are in accord with those of reports of influenza drug susceptibility in previous years (26, 30, 33). Isolates that met the mild-outlier criterion for one or both drugs were retested with the NAI assay. The average IC50 was calculated for each virus after it was retested and compared again with the cutoff value. After viruses were retested, 16 mild outliers were identified.
TABLE 3.
Characterization of extreme and mild outliers among A(H1N1) viruses from three previous seasonsa
| Season | Strain | Mean IC50 ± SDc (nM) | Outlier(s)/drug(s)d | NA mutation(s)e | Accession no. | |
|---|---|---|---|---|---|---|
| Oseltamivir | Zanamivir | |||||
| 2005-2006 | A/Zhejiang/Xiangshan522/2006b | 131.85 ± 31.55 | 2.56 ± 0.01 | E/O | H274Y | EU879082 |
| 2006-2007 | A/Georgia/20/2006b | 383.01 ± 143.69f | 2.55 ± 1.02 | E/O | H274Y | EU516197i |
| 2006-2007 | A/Massachusetts/5/2007b | 258.48 ± 41.56g | 1.71 ± 0.28 | E/O | H274Y | EU516028 |
| 2006-2007 | A/Texas/31/2007b | 285.56 ± 18.05 | 1.14 ± 0.33 | E/O | H274Y | EU516027 |
| 2006-2007 | A/Minnesota/23/2007b | 225.61 ± 13.85 | 0.99 ± 0.05 | E/O | H274Y | EU516141 |
| 2006-2007 | A/Gansu/Chenguan/1129/2007b | 126.89 ± 1.96 | 0.49 ± 0.25 | E/O | H274Y | EU879064 |
| 2005-2006 | A/Guangdongluohu/1232/2006 | 8.76 ± 4.68 | 2.33 ± 1.47 | M/O | D79G, S247G | EU879081 |
| 2006-2007 | A/Hubei/Jiangan/1251/2006 | 6.75 ± 5.24 | 13.50 ± 12.24 | M/Z, M/O | H126N | EU879078 |
| 2006-2007 | A/Thailand/708/2006 | 4.51 ± 1.36 | 2.76 ± 0.01h | M/O | None | EU879083 |
| 2005-2006 | A/Zhejiang/Xiaocheng/1345/2006 | 4.21 ± 0.68 | 0.54 ± 0.12 | M/O | S247G | EU879079 |
| 2005-2006 | A/Thailand/674/2006 | 1.95 ± 0.69 | 7.12 ± 3.64 | M/Z | G248R | EU879080 |
A statistical cutoff for outliers was calculated based on the box plot analysis of the pooled IC50s from which the extreme outliers were excluded (Table 2). The cutoff of an IC50 of >_X_0.75 + 3 IQR for a statistical outlier was more restrictive than the elected criteria of an IC50 value greater than the mean IC50 + 3 SD for influenza A(H3N2) (zanamivir, 9.93 nM versus 9.20 nM) and B virus isolates (oseltamivir, 8.64 nM versus 7.47 nM; and zanamivir, 14.21 nM versus 11.29 nM) but less restrictive for A(H1N1) isolates (oseltamivir, 2.43 nM versus 2.89 nM; and zanamivir, 3.32 nM versus 3.85 nM). All outliers determined by the elected criteria of an IC50 value greater than the mean IC50 + 3 SD would still be outliers based on the statistical cutoff, with the exception of two strains, A/Santiago/9491/2006 (H3N2) and B/California/4/2005. The significance, if any, of the mutations in the NA of these two viruses has not been demonstrated.
Influenza A(H1N1) viruses.
Six of the 12 extreme outliers defined as resistant viruses were A(H1N1) strains from the 2004 to 2007 influenza seasons. They all exhibited elevated IC50s to oseltamivir, ranging from 126.89 nM to 383.00 nM (Table 3), but demonstrated low IC50s toward zanamivir. Of the six A(H1N1) viruses, four were isolated in the United States and two were isolated in China (Table 3). Sequence analysis revealed the H274Y mutation at the active site, which is known to confer oseltamivir resistance (15, 36, 37). These six extreme outliers were reported as resistant viruses because they met both criteria: a high IC50 and a previously described mutation in the NA, arising in drug-treated patients. For two of the viruses, counterpart isolates with matching NA sequences (identical except for H274Y) were identified, and their IC50s were determined for comparative analysis. The IC50s to oseltamivir were reduced 638- and 1,616-fold in counterpart viruses without this mutation (Table 3). In addition, two Chinese oseltamivir-resistant viruses were also resistant to adamantanes (amantadine and rimantadine), as evidenced by the presence of the S31N mutation in the M2 protein (results not shown).
The IC50s of the remaining A(H1N1) viruses ranged from 0.06 nM to 13.50 nM when tested against zanamivir and from 0.16 nM to 10.00 nM, tested against oseltamivir (Table 2). Five A(H1N1) viruses were identified as mild outliers to one or both NA inhibitors (Table 3). Among those five outliers, A/Hubei/Jiangan/1251/2006 had elevated IC50s for both zanamivir and oseltamivir and contained a single mutation, H126N, in the NA. Three of the oseltamivir outliers with IC50s ranging from 4.21 nM to 8.76 nM contained the change S247N or S274G, alone or in combination with at least one other amino acid change. The A/Thailand/674/2006 virus with elevated IC50 for zanamivir (7.12 nM) contained the previously described mutation G248R (30). Though another isolate from Thailand, A/Thailand/708/2006, repeatedly demonstrated elevated IC50s to oseltamivir (4.51 nM), no unique amino acid changes in the NA were detected by conventional sequence analysis.
Influenza A(H3N2) viruses.
One extreme oseltamivir outlier, A/Texas/12/2007 (IC50 = 9.18 nM) (Table 4), was detected; the NA of this virus contained E119V, a well-known mutation associated with oseltamivir resistance (23, 36). The IC50 of A/Texas/12/2007 was approximately 20-fold greater than the mean IC50 (0.44 nM) and more than 50-fold greater than the IC50 of the drug-sensitive counterpart A/Washington/1/2007 virus (Table 4). A/Texas/12/2007 was also resistant to adamantanes, as were the majority of A(H3N2) viruses circulating in the United States in 2007 (data not shown).
In addition, three extreme zanamivir outliers were detected that carried a mutation at position 151 in the NA. Two other extreme outliers, A/Oman/6943/2005 and A/Canada/270/2007, had the D151A or D151A/D (a mixture of D and A) change in the NA, respectively, which has not previously been described. A comparison of the IC50 of A/Montana/8/2007 that had the change D151V/D with that of its sensitive counterpart, A/Brazil/80/2007, revealed a 150-fold increase in IC50 (Table 4).
A total of eight mild outliers with either single mutations or a combination of mutations was detected. Five mild zanamivir outliers demonstrated the D151N change (Table 4) reported in a previous surveillance study (26), two of which had D151N as the only mutation. A mixture of G/D at position 151 was also observed; however, upon retesting, the IC50s of these mutants no longer qualified as outliers. The A/Canada/83/2006 virus showed a slightly elevated IC50 to zanamivir (15.12 nM) and, in addition to the V165I mutation, contained a novel mutation, H274N, in the NA active site. The two additional mild zanamivir outliers had the mutation D251G, described previously (18), and three other mild zanamivir outliers contained K249E in combination with either D151N or G248R (Table 4).
Influenza B viruses.
The IC50s for the 2004 to 2007 season influenza B viruses ranged from 0.35 nM to 17.86 nM for zanamivir and from 0.19 nM to 11.63 nM for oseltamivir (Table 5). One extreme outlier was detected, B/Hong Kong/36/2005, that exhibited high IC50s for both zanamivir (154.85 nM) and oseltamivir (1778.25 nM) (Table 5). This virus had a single novel amino acid change, R→K, at the highly conserved position 371 and had not been selected in the presence of any NA inhibitor, either in vitro or in vivo. Compared to a virus lacking this mutation (B/Hong Kong/45/2005), the R371K mutant displayed a 29-fold and a 407-fold reduction in sensitivity to zanamivir and oseltamivir, respectively.
Three mild outliers were detected among influenza B viruses (Table 5). B/Michigan/20/2005 contained a H→Y change at position 274. This mutation in influenza B virus NA has not been detected in earlier surveillance studies but has been detected after growth in cell culture in the presence of the NA inhibitor peramivir (1). The mild oseltamivir outlier B/California/5/2005 contained the previously reported change I222T at a conserved position of the NA active site (30). The B/California/4/2005 virus had the reverse change (T→I) at position 326 and exhibited the lowest IC50 among mild outliers for oseltamivir (Table 5).
Frequency of resistance in influenza viruses during the 2004 to 2005, 2005 to 2006, and 2006 to 2007 seasons.
Of the 3,261 viruses tested, we identified 16 (∼0.5%) viruses that exhibited mildly elevated IC50s and 12 viruses with extremely elevated IC50s with the chemiluminescent NAI assay (the extreme outliers were excluded from statistical analysis). Based on sequence analysis of the NA, 7 of 12 extreme outliers revealed mutations in the NA that had been described previously in isolates from treated patients and that can be reported as drug-resistant mutants. Among those seven resistant viruses, six A(H1N1) viruses carried a H274Y mutation, and one A(H3N2) virus had an E119V mutation. There was no resistance detected in the viruses from the 2004 to 2005 season, although the number of isolates tested was relatively small. Among A(H1N1) viruses isolated globally, the frequency of resistance was 0.4% (1/259) and 0.6% (5/822) in the 2005 to 2006 and 2006 to 2007 seasons, respectively. In the United States only, oseltamivir resistance was not detected during the 2004 to 2005 season or the 2005 to 2006 season and was 0.9% (5/584) during the 2006 to 2007 season. These data served as a baseline during our surveillance of influenza viruses isolated during the 2007 to 2008 season.
Rise of oseltamivir resistance in influenza A(H1N1) viruses during the 2007 to 2008 season.
Starting from late October 2007, a rise in the resistance of influenza A(H1N1) viruses to oseltamivir was observed. Fifty-seven of the 896 influenza A(H1N1) viruses collected from 1 October 2007 to 21 March 2008 had IC50s against oseltamivir ranging from 85.08 nM to 255.83 nM and were classified as extreme outliers. Sequence analysis showed the H274Y mutation in the NA of each isolate. Fifty of the oseltamivir-resistant viruses were collected from 15 U.S. states (n = 706), and seven oseltamivir-resistant viruses were from foreign isolates (n = 190). The frequency of apparent drug resistance has risen to 7.1% (50/706) among U.S. influenza A(H1N1) viruses isolated since the beginning of the 2007 to 2008 season; however, analysis is ongoing as isolates continue to be received and tested. All 2007 to 2008 oseltamivir-resistant viruses remained sensitive to zanamivir and were also sensitive to adamantanes (results not shown). Of the 232 influenza A(H3N2) and 181 influenza B viruses tested with the NAI assay, no outliers or viruses resistant to either NA inhibitor have been detected.
DISCUSSION
We report here the results of a large-scale surveillance for drug susceptibility among influenza A and B viruses isolated worldwide during the three previous influenza seasons and during the ongoing 2007 to 2008 season (1 October 2007 to 21 March 2008). The beginning of the 2007 to 2008 season was notable for an unprecedented increase in oseltamivir resistance in A(H1N1) viruses circulating in several European countries (i.e., 67% in Norway). Our study demonstrates that the approach to NA inhibitor susceptibility monitoring established at the CDC in previous seasons was instrumental in the timely detection of the oseltamivir resistance in the United States that began in October 2007.
Viruses were tested utilizing the chemiluminescent NAI assay. Based on the mean IC50s, the viruses of type A influenza were more drug sensitive than type B viruses, and viruses of the A(H3N2) subtype were more sensitive to oseltamivir than viruses of the A(H1N1) subtype. In this respect, the patterns observed for drug sensitivities were similar to those previously published (26, 30, 33). Assessment of drug resistance to NA inhibitors has been hindered by the lack of a clear definition of resistance. Moreover, a wide range of IC50s seen within the same subtype requires the determination of a threshold IC50 as an indicator of potential resistance. In this study, a virus isolate had to meet two criteria to be reported as drug resistant: (i) its IC50 should be above the threshold value determined for each subtype and drug; and (ii) its NA should contain the mutation(s) previously recognized in viruses isolated from patients treated with NA inhibitor. Seven influenza A viruses from the three previous seasons met both criteria. Moreover, the viruses reported as resistant in this study were extreme outliers (30), because their IC50s were more than 10-fold above the mean IC50s for oseltamivir.
Among those resistant viruses, one A(H3N2) virus carrying the E119V mutation was detected. There are several reports describing the emergence of E119V mutants in oseltamivir-treated patients (2, 22, 36). The other six resistant viruses belonged to the A(H1N1) subtype and shared the H274Y mutation. In early clinical studies conducted in the United States and Japan, the H274Y mutants were recovered from oseltamivir-treated children and adults (15, 36, 39). Therefore, the H274Y mutation was considered to be a previously established marker of oseltamivir resistance in A(H1N1) viruses. Based on the available information, two of the seven oseltamivir-resistant viruses from the 2006 to 2007 influenza season analyzed in the present study were recovered from patients following oseltamivir treatment, namely A/Texas/12/2007 (H3N2), with the E119V mutation, and A/Massachusetts/5/2007 (H1N1), with the H274Y mutation. The overall frequency of resistance for influenza viruses isolated globally was low (0.2%; 7/3,261) from 2004 to 2007. During the early part of the 2007 to 2008 season, the frequency of oseltamivir resistance in A(H1N1) influenza viruses isolated in the United States only was estimated at 7.1% (50 of 706), which is a 10-fold increase compared to 0.7% (4 of 588) for the previous season.
In addition, five viruses from the 2004 to 2007 season were determined to be extreme outliers and contained mutations at conserved residues in the NA active site. However, the identified variants (with a change at D151 or R371) have not been previously described among viruses isolated from NA inhibitor-treated patients, and, consequently, these extreme outliers were not defined as resistant viruses in this study. B/Hong Kong/36/2005 had the mutation R371K at a key residue in the NA active site and exhibited high IC50s to both oseltamivir and zanamivir. Residue 371 is a part of the arginine triad (R118, R292, and R371) shared by all influenza virus neuraminidases and other sialidases and is involved in catalysis of substrate (8, 25). When the R→K mutation was introduced into the equivalent position of the NA of the influenza A(H3N2) virus, it reduced NA enzyme activity (25). Nevertheless, an influenza A(H3N2) virus generated using reverse genetics with the R371K mutation was viable and exhibited resistance to both NA inhibitors in the NAI assay (40). In contrast to the N2 subtype enzyme, which requires two simultaneous nucleotide substitutions to replace R with K (40), a single nucleotide substitution is sufficient to achieve an equivalent change in the NA of type B viruses. Studies are under way to evaluate the impact of the R371K mutation on drug susceptibility and fitness of influenza B viruses.
Four influenza A(H3N2) viruses were extreme zanamivir outliers and contained either V or A at residue 151. Replacement of D151 with N, G, E, or V, but not A, was previously reported for type A and B outliers. Mutations at position 151 were common in the A(H3N2) viruses that circulated from 1996 to 1999 (26) but not in those circulating from 1999 to 2002 (30). According to a mutagenesis study, the mutation D151E in the N2 NA background led to ∼10-fold reduced sensitivity to oseltamivir (41). It is evident that more studies are needed to assess the possible effect of changes at position 151 on virus susceptibility in vivo. Without evidence for the emergence of position 151 mutants in treated patients, outliers with substitutions at position 151 cannot be qualified as NA inhibitor-resistant viruses at this time.
The A/Canada/83/2007 (H3N2) virus had a mutation at residue 274 (H→N) and was identified as a mild zanamivir outlier (IC50 = 15.12 nM). This finding is consistent with the reduced sensitivity to zanamivir, but not to oseltamivir, demonstrated in vitro with a recombinant N1 NA carrying the same mutation (35).
The definition of mild outlier depends on the mean IC50, which varies by type/subtype. Although the oseltamivir IC50 for B/Michigan/20/2005 is close to the IC50 of oseltamivir-resistant A/Texas/12/2007 (H3N2) virus (10.80 nM versus 9.18 nM; Table 5), this influenza B virus does not qualify as oseltamivir resistant. The mutation detected in the NA active site of B/Michigan/20/2005 was H274Y, which has not been reported previously in treated patients but was selected in the presence of an investigational NA inhibitor, peramivir (1). Another mild oseltamivir outlier, B/California/5/2005, contained an I222T mutation in the NA active site. This mutation was previously described in viruses recovered from untreated patients (18, 30).
Although not all outliers detected in the present study met the criteria for drug resistance, their detection is essential to improving our knowledge about changes in the NA, which have the potential to alter the virus' susceptibility to NA inhibitors in humans. This information also provides further insights into the mechanism of resistance and cross-resistance to this newer class of drugs. Criteria elected for outliers in the present study allow rigorous detection of virus variants with even slightly reduced drug susceptibility with the NAI assay. After mild outliers were retested, the IC50s of certain viruses were no longer above the cutoff value (e.g., A/Canada/9/2006). Nevertheless, we performed a thorough analysis of the NA sequences and found either no unique mutations or a mixed sequence in the NA (Table 3). According to our method of surveillance, outliers containing no established markers of drug resistance were recorded and investigated further. However, they were not reported as drug-resistant viruses without supporting experimental evidence.
In this study, the elected threshold criterion was able to promptly detect isolates with potentially reduced drug susceptibility (extreme and mild outliers) for further analysis and was applicable to the ongoing 2007 to 2008 season. The elected cutoff values were similar to statistical outlier cutoffs, based on box plot analyses of all three seasons. However, the box plot analysis was variable by season (data not shown) and can be done only retrospectively.
In conclusion, the rise in frequency of oseltamivir resistance during the 2007 to 2008 season is worrisome because oseltamivir is the antiviral agent most frequently prescribed in the United States for the control of seasonal influenza infections (12). Resistance was conferred by the H274Y change in the NA of the A(H1N1) viruses. This phenomenon strengthens previous concerns over the potential emergence of H274Y oseltamivir-resistant mutants among the highly virulent A(H5N1) viruses (9, 19, 24).
With the increased use of NA inhibitors to treat influenza infections, there is a need to establish vigorous drug resistance monitoring programs at both the national and international levels. Collaboration among laboratories conducting such drug resistance surveillance will promote harmonization of the drug resistance detection assays, refinement of criteria, and improvement in the overall interpretation of drug susceptibility data. Epidemiological and virological studies are also needed to elucidate mechanisms underlying the recent emergence and spread of oseltamivir-resistant viruses in humans.
Acknowledgments
We thank Wilina Lim, Centre for Health Protection (Hong Kong, SAR, China), and all collaborators in the WHO Global Influenza Surveillance Network, including the National Influenza Centers, for the submission of isolates. We thank Amanda Balish, Angela Foust, Henrietta Hall, Jan Mabry, Gregory Kocher, Zachary Reed, John Barnes, Allison Myrick, Catherine Smith, Michael Shaw, and other members of the Influenza Division for contributions to this project.
T.G.S. and M.O.-A. received financial support for this work from the Oak Ridge Institute for Science and Education, Oak Ridge, TN (ORISE).
We declare that we have no conflict of interest.
The findings and conclusions in the report are those of the authors and do not necessarily represent the views of the funding agency.
Footnotes
▿
Published ahead of print on 14 July 2008.
REFERENCES
- 1.Baum, E. Z., P. C. Wagaman, L. Ly, I. Turchi, J. Le, D. Bucher, and K. Bush. 2003. A point mutation in influenza B neuraminidase confers resistance to peramivir and loss of slow binding. Antivir. Res. 59**:**13-22. [DOI] [PubMed] [Google Scholar]
- 2.Baz, M., Y. Abed, J. McDonald, and G. Boivin. 2006. Characterization of multidrug-resistant influenza A/H3N2 viruses shed during 1 year by an immunocompromised child. Clin. Infect. Dis. 43**:**1555-1561. [DOI] [PubMed] [Google Scholar]
- 3.Bright, R. A., M. J. Medina, X. Xu, G. Perez-Oronoz, T. R. Wallis, X. M. Davis, L. Povinelli, L. Cox, and A. I. Klimov. 2005. Incidence of adamantane resistance among influenza A (H3N2) viruses isolated worldwide from 1994 to 2005: a cause for concern. Lancet 366**:**1175-1181. [DOI] [PubMed] [Google Scholar]
- 4.Bright, R. A., D. Shay, J. Bresee, A. Klimov, N. Cox, and J. Ortiz. 2006. High levels of adamantane resistance among influenza A(H3N2) viruses and interim guidelines for use of antiviral agents: United States, 2005-06 influenza season. MMWR Morb. Mortal. Wkly. Rep. 55**:**44-46. [PubMed] [Google Scholar]
- 5.Bright, R. A., D. K. Shay, B. Shu, N. J. Cox, and A. I. Klimov. 2006. Adamantane resistance among influenza A viruses isolated early during the 2005-2006 influenza season in the United States. JAMA 295**:**891-894. [DOI] [PubMed] [Google Scholar]
- 6.Buxton, R. C., B. Edwards, R. R. Juo, J. C. Voyta, M. Tisdale, and R. C. Bethell. 2000. Development of a sensitive chemiluminescent neuraminidase assay for the determination of influenza virus susceptibility to zanamivir. Anal. Biochem. 280**:**291-300. [DOI] [PubMed] [Google Scholar]
- 7.Colman, P. M. 1989. Neuraminidase: enzyme and antigen, p. 175-210. In R. M. Krug (ed.) The influenza viruses. Plenum Press, New York, NY.
- 8.Colman, P. M., P. A. Hoyne, and M. C. Lawrence. 1993. Sequence and structure alignment of paramyxovirus hemagglutinin-neuraminidase with influenza virus neuraminidase. J. Virol. 67**:**2972-2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Jong, M. D., T. T. Tran, H. K. Truong, M. H. Vo, G. J. Smith, V. C. Nguyen, V. C. Bach, T. Q. Phan, Q. H. Do, Y. Guan, J. S. Peiris, T. H. Tran, and J. Farrar. 2005. Oseltamivir resistance during treatment of influenza A (H5N1) infection. N. Engl. J. Med. 353**:**2667-2672. [DOI] [PubMed] [Google Scholar]
- 10.Deyde, V. M., X. Xu, R. A. Bright, M. Shaw, C. B. Smith, Y. Zhang, Y. Shu, L. V. Gubareva, N. J. Cox, and A. I. Klimov. 2007. Surveillance of resistance to adamantanes among influenza A(H3N2) and A(H1N1) viruses isolated worldwide. J. Infect. Dis. 196**:**249-257. [DOI] [PubMed] [Google Scholar]
- 11.Escuret, V., E. Frobert, M. Bouscambert-Duchamp, M. Sabatier, I. Grog, M. Valette, B. Lina, F. Morfin, and O. Ferraris. 2008. Detection of human influenza A (H1N1) and B strains with reduced sensitivity to neuraminidase inhibitors. J. Clin. Virol. 41**:**25-28. [DOI] [PubMed] [Google Scholar]
- 12.Fazio, D., A. Laufer, J. Meek, J. Palumbo, R. Lynfield, C. Morin, K. Vick, J. Baumbach, M. Mueller, R. Belflower, C. Long, and L. Kamimoto. 2008. Influenza-testing and antiviral-agent prescribing practices—Connecticut, Minnesota, New Mexico, and New York, 2006-07 influenza season. MMWR Morb. Mortal. Wkly. Rep. 57**:**61-65. [PubMed] [Google Scholar]
- 13.Ferraris, O., and B. Lina. 2008. Mutations of neuraminidase implicated in neuraminidase inhibitors resistance. J. Clin. Virol. 41**:**13-19. [DOI] [PubMed] [Google Scholar]
- 14.Gubareva, L. V., R. Bethell, G. J. Hart, K. G. Murti, C. R. Penn, and R. G. Webster. 1996. Characterization of mutants of influenza A virus selected with the neuraminidase inhibitor 4-guanidino-Neu5Ac2en. J. Virol. 70**:**1818-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gubareva, L. V., L. Kaiser, M. N. Matrosovich, Y. Soo-Hoo, and F. G. Hayden. 2001. Selection of influenza virus mutants in experimentally infected volunteers treated with oseltamivir. J. Infect. Dis. 183**:**523-531. [DOI] [PubMed] [Google Scholar]
- 16.Gubareva, L. V., M. N. Matrosovich, M. K. Brenner, R. C. Bethell, and R. G. Webster. 1998. Evidence for zanamivir resistance in an immunocompromised child infected with influenza B virus. J. Infect. Dis. 178**:**1257-1262. [DOI] [PubMed] [Google Scholar]
- 17.Gubareva, L. V., R. G. Webster, and F. G. Hayden. 2002. Detection of influenza virus resistance to neuraminidase inhibitors by an enzyme inhibition assay. Antivir. Res. 53**:**47-61. [DOI] [PubMed] [Google Scholar]
- 18.Hatakeyama, S., N. Sugaya, M. Ito, M. Yamazaki, M. Ichikawa, K. Kimura, M. Kiso, H. Shimizu, C. Kawakami, K. Koike, K. Mitamura, and Y. Kawaoka. 2007. Emergence of influenza B viruses with reduced sensitivity to neuraminidase inhibitors. JAMA 297**:**1435-1442. [DOI] [PubMed] [Google Scholar]
- 19.Hayden, F., A. Klimov, M. Tashiro, A. Hay, A. Monto, J. Kimm-Breschkin, C. Macken, A. Hampson, R. G. Webster, M. Amyard, and M. Zambon. 2005. Neuraminidase inhibitor susceptibility network position statement: antiviral resistance in influenza A/H5N1 viruses. Antivir. Ther. 10**:**873-877. [PubMed] [Google Scholar]
- 20.Hayden, F. G. 1996. Amantadine and rimantadine-clinical aspects, p. 59-77. In D. D. Richman (ed.), Antiviral drug resistance. John Wiley and Sons Ltd., San Francisco, CA.
- 21.Hurt, A. C., I. G. Barr, G. Hartel, and A. W. Hampson. 2004. Susceptibility of human influenza viruses from Australasia and South East Asia to the neuraminidase inhibitors zanamivir and oseltamivir. Antivir. Res. 62**:**37-45. [DOI] [PubMed] [Google Scholar]
- 22.Ison, M. G., L. V. Gubareva, R. L. Atmar, J. Treanor, and F. G. Hayden. 2006. Recovery of drug-resistant influenza virus from immunocompromised patients: a case series. J. Infect. Dis. 193**:**760-764. [DOI] [PubMed] [Google Scholar]
- 23.Kiso, M., K. Mitamura, Y. Sakai-Tagawa, K. Shiraishi, C. Kawakami, K. Kimura, F. G. Hayden, N. Sugaya, and Y. Kawaoka. 2004. Resistant influenza A viruses in children treated with oseltamivir: descriptive study. Lancet 364**:**759-765. [DOI] [PubMed] [Google Scholar]
- 24.Le, Q. M., M. Kiso, K. Someya, Y. T. Sakai, T. H. Nguyen, K. H. Nguyen, N. D. Pham, H. H. Ngyen, S. Yamada, Y. Muramoto, T. Horimoto, A. Takada, H. Goto, T. Suzuki, Y. Suzuki, and Y. Kawaoka. 2005. Avian flu: isolation of drug-resistant H5N1 virus. Nature 437**:**1108. [DOI] [PubMed] [Google Scholar]
- 25.Lentz, M. R., R. G. Webster, and G. M. Air. 1987. Site-directed mutation of the active site of influenza neuraminidase and implications for the catalytic mechanism. Biochemistry 26**:**5351-5358. [DOI] [PubMed] [Google Scholar]
- 26.McKimm-Breschkin, J., T. Trivedi, A. Hampson, A. Hay, A. Klimov, M. Tashiro, F. Hayden, and M. Zambon. 2003. Neuraminidase sequence analysis and susceptibilities of influenza virus clinical isolates to zanamivir and oseltamivir. Antimicrob. Agents Chemother. 47**:**2264-2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McKimm-Breschkin, J. L. 2005. Management of influenza virus infections with neuraminidase inhibitors: detection, incidence, and implications of drug resistance. Treat. Respir. Med. 4**:**107-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mishin, V. P., F. G. Hayden, and L. V. Gubareva. 2005. Susceptibilities of antiviral-resistant influenza viruses to novel neuraminidase inhibitors. Antimicrob. Agents Chemother. 49**:**4515-4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Molinari, N. A., I. R. Ortega-Sanchez, M. L. Messonnier, W. W. Thompson, P. M. Wortley, E. Weintraub, and C. B. Bridges. 2007. The annual impact of seasonal influenza in the US: measuring disease burden and costs. Vaccine 25**:**5086-5096. [DOI] [PubMed] [Google Scholar]
- 30.Monto, A. S., J. L. McKimm-Breschkin, C. Macken, A. W. Hampson, A. Hay, A. Klimov, M. Tashiro, R. G. Webster, M. Aymard, F. G. Hayden, and M. Zambon. 2006. Detection of influenza viruses resistant to neuraminidase inhibitors in global surveillance during the first 3 years of their use. Antimicrob. Agents Chemother. 50**:**2395-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moscona, A. 2005. Neuraminidase inhibitors for influenza. N. Engl. J. Med. 353**:**1363-1373. [DOI] [PubMed] [Google Scholar]
- 32.Mungall, B. A., X. Xu, and A. Klimov. 2003. Assaying susceptibility of avian and other influenza A viruses to zanamivir: comparison of fluorescent and chemiluminescent neuraminidase assays. Avian Dis. 47(Suppl. 3)**:**1141-1144. [DOI] [PubMed] [Google Scholar]
- 33.Mungall, B. A., X. Xu, and A. Klimov. 2004. Surveillance of influenza isolates for susceptibility to neuraminidase inhibitors during the 2000-2002 influenza seasons. Virus Res. 103**:**195-197. [DOI] [PubMed] [Google Scholar]
- 34.Potier, M., L. Mameli, M. Belisle, L. Dallaire, and S. B. Melancon. 1979. Fluorometric assay of neuraminidase with a sodium (4-methylumbelliferyl-alpha-D-N-acetylneuraminate) substrate. Anal. Biochem. 94**:**287-296. [DOI] [PubMed] [Google Scholar]
- 35.Wang, M. Z., C. Y. Tai, and D. B. Mendel. 2002. Mechanism by which mutations at His274 alter sensitivity of influenza a virus N1 neuraminidase to oseltamivir carboxylate and zanamivir. Antimircob. Agents Chemother. 46**:**3809-3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ward, P., I. Small, J. Smith, P. Suter, and R. Dutkowski. 2005. Oseltamivir (Tamiflu) and its potential for use in the event of an influenza pandemic. J. Antimicrob. Chemother. 55(Suppl. 1)**:**i5-i21. [DOI] [PubMed] [Google Scholar]
- 37.Weinstock, D. M., L. V. Gubareva, and G. Zuccotti. 2003. Prolonged shedding of multidrug-resistant influenza A virus in an immunocompromised patient. N. Engl. J. Med. 348**:**867-868. [DOI] [PubMed] [Google Scholar]
- 38.Wetherall, N. T., T. Trivedi, J. Zeller, C. Hodges-Savola, J. L. McKimm-Breschkin, M. Zambon, and F. G. Hayden. 2003. Evaluation of neuraminidase enzyme assays using different substrates to measure susceptibility of influenza virus clinical isolates to neuraminidase inhibitors: report of the neuraminidase inhibitor susceptibility network. J. Clin. Microbiol. 41**:**742-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whitley, R. J., F. G. Hayden, K. S. Reisinger, N. Young, R. Dutkowski, D. Ipe, R. G. Mills, and P. Ward. 2001. Oral oseltamivir treatment of influenza in children. Pediatr. Infect. Dis. J. 20**:**127-133. [DOI] [PubMed] [Google Scholar]
- 40.Yen, H. L., E. Hoffmann, G. Taylor, C. Scholtissek, A. S. Monto, R. G. Webster, and E. A. Govorkova. 2006. Importance of neuraminidase active-site residues to the neuraminidase inhibitor resistance of influenza viruses. J. Virol. 80**:**8787-8795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yen, H. L., N. A. Ilyushina, R. Salomon, E. Hoffmann, R. G. Webster, and E. A. Govorkova. 2007. Neuraminidase inhibitor-resistant recombinant A/Vietnam/1203/04 (H5N1) influenza viruses retain their replication efficiency and pathogenicity in vitro and in vivo. J. Virol. 81**:**12418-12426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zambon, M., F. G. Hayden, and the Global Neuraminidase Inhibitor Susceptibility Network. 2001. Position statement: global neuraminidase inhibitor susceptibility network. Antiviral Res. 49**:**147-156. [DOI] [PubMed] [Google Scholar]