Delta-like 4 is the essential, nonredundant ligand for Notch1 during thymic T cell lineage commitment (original) (raw)
Abstract
Thymic T cell lineage commitment is dependent on Notch1 (N1) receptor–mediated signaling. Although the physiological ligands that interact with N1 expressed on thymic precursors are currently unknown, in vitro culture systems point to Delta-like 1 (DL1) and DL4 as prime candidates. Using _DL1_- and DL4-lacZ reporter knock-in mice and novel monoclonal antibodies to DL1 and DL4, we show that DL4 is expressed on thymic epithelial cells (TECs), whereas DL1 is not detected. The function of DL4 was further explored in vivo by generating mice in which DL4 could be specifically inactivated in TECs or in hematopoietic progenitors. Although loss of DL4 in hematopoietic progenitors did not perturb thymus development, inactivation of DL4 in TECs led to a complete block in T cell development coupled with the ectopic appearance of immature B cells in the thymus. These immature B cells were phenotypically indistinguishable from those developing in the thymus of conditional N1 mutant mice. Collectively, our results demonstrate that DL4 is the essential and nonredundant N1 ligand responsible for T cell lineage commitment. Moreover, they strongly suggest that N1-expressing thymic progenitors interact with DL4-expressing TECs to suppress B lineage potential and to induce the first steps of intrathymic T cell development.
The thymus is continuously seeded by progenitors derived from hematopoietic stem cells, which reside in the BM. These progenitors migrate via the blood stream into the thymus, where they adopt a T cell fate, proliferate, and differentiate into mature functional T cells. This differentiation process is characterized by multiple developmental stages. The earliest thymic progenitors lack surface expression of CD4 and CD8 and are therefore referred to as double-negative (DN) thymocytes. They subsequently up-regulate both CD4 and CD8 coreceptors (double positive [DP]) before undergoing positive and negative selection, and maturing to CD4 and CD8 single-positive (SP) thymocytes that emigrate to the periphery. Immature DN thymocytes can be subdivided into four subpopulations according to the surface expression of CD117, CD44, and CD25. The most immature thymocyte progenitors (DN1) express CD117 and CD44 and are negative for CD25, followed by the DN2 population, which up-regulates CD25, and the DN3 cells, which down-regulate CD117 and CD44 before generating DN4 thymocytes lacking expression of all three markers (1, 2).
Over the last decade, many reports highlighted the importance of the evolutionarily conserved Notch cascade for the lymphoid system (3). Mammals possess 4 Notch receptors (N1–4), which are activated by two classes of transmembrane bound ligands named Jagged 1 and 2, and Delta-like (DL) 1, 3, and 4. Notch signaling is initiated upon ligand receptor interaction, which results in the proteolytic release of the Notch intracellular cytoplasmic domain (NICD) of Notch receptors. The liberated NICD translocates to the nucleus and binds to the transcription factor RBP-J (also known as CSL), thereby converting it from a transcriptional repressor into an activator. Mastermind-like proteins are required for Notch signaling, as they bind to the NICD–RBP-J complex and recruit additional coactivators.
Multiple genetic loss- and gain-of-function experiments show that signaling mediated through the N1 receptor plays an important role for T cell lineage commitment and maturation within the thymus. Inducible inactivation of N1 in BM progenitors results in a block in T cell development and ectopic B cell development in the thymus, suggesting that N1 instructs an early thymic progenitor to adopt a T cell fate (4, 5). An identical phenotype is observed in mice in which the RBP-J gene was inactivated in BM progenitors (6), strongly indicating that T cell specification is mediated by N1–RBP-J–dependent signaling. Interference with Notch signaling by transgenic expression of modulators (such as Fringe, Deltex, or Nrarp) or dominant-negative forms of transcriptional coactivators (mastermind-like 1) also blocks T cell development concomitant with B lymphopoiesis in the thymus (for review see reference 3). Reciprocal gain-of-function studies overexpressing NICD in BM progenitors result in ectopic T cell development at the expense of B cell development (3). Thus, both loss- and gain-of-function studies demonstrate that N1 is essential for T lineage commitment. In addition, N1–RBP-J signaling promotes the differentiation of pro–T cells into pre–T cells within the thymus by controlling rearrangement of the TCR β locus (7) through regulating chromatin accessibility (8), thereby assuring the successful generation of a pre-TCR complex, which is essential for thymocyte development.
Although N1 is unequivocally the key Notch receptor involved in T lineage commitment and thymic T cell maturation in vivo, the ligands that could be the physiological partners of N1 in these processes are still a matter of debate. The thymic epithelial microenvironment expresses all Notch ligands (mainly assessed by RT-PCR), except DL3, which is hardly found in this organ (9–12). However, both Jagged ligands can be excluded from playing an essential role, as Jagged2−/− and, more recently, Jagged1−/− mice have been shown to have normal T cell development (13, 14), thus implicating DL1 and/or DL4 ligands. Historically, DL1 has been favored as the potential N1 ligand in T cell fate specification, because DL1-expressing stromal cells can support the complete development of mature functional T cells from BM precursors in vitro (10). Moreover, when BM progenitors are co-cultured on stromal cells overexpressing DL1, B cell development is blocked (15). Surprisingly, in contrast with these promising data obtained in vitro, conditional inactivation of DL1 in thymocytes and/or thymic epithelial cells (TECs) does not inhibit T cell development (16). This discrepancy may be caused by the in vivo presence of DL4, which shares a high degree of homology with DL1. Data obtained in vitro support this hypothesis, because overexpression of DL4 in stromal cells can also direct T lymphopoiesis (16, 17). In addition, overexpression of DL4 in BM precursors transferred into γ-irradiated mice leads to ectopic T cell development in the BM as well as a severe defect in B cell development (17–19). Moreover, we recently described a hierarchy of Notch–Delta interactions showing that the avidity of DL4 to bind to immature thymocytes is much higher than that of DL1. DL4 binding is high from the DN1 to DN3 stages and declines in DN4 to become undetectable in subsequent DP and SP thymocytes. This binding pattern parallels the functional requirements of Notch signaling during thymocyte development (20, 21), suggesting that DL4 might play an important role during T cell development within the thymus.
In this report, we explore the function of DL4 within the thymus using a conditional loss-of-function approach. Our data show that _Foxn1-Cre_–mediated inactivation of DL4 within the thymic epithelium leads to a complete block in T cell development accompanied by ectopic B cell development within the thymus, which phenocopies mice with inducible inactivation of N1 in BM progenitors. Thus, DL4 is essential for T cell fate specification and thereby represents the physiological partner of N1 during this process.
RESULTS AND DISCUSSION
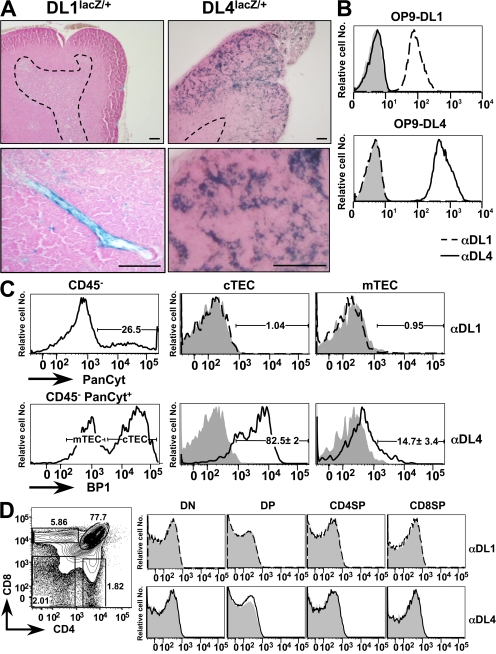
Because DL1- and DL4-expressing OP-9 stromal cells (10, 16, 17) or normal thymic stroma (22) can both support the complete development of mature functional T cells from BM precursors in vitro, it is conceivable that either or both of these proteins function as physiological ligands for the N1 receptor during thymic T cell lineage commitment. To gain further insight into the expression pattern of DL1 and DL4 in the thymus, we analyzed thymi of mice in which the lacZ gene was knocked into either the DL1 or DL4 locus (23, 24). LacZ staining on thymic sections derived from DL1lacZ/+ mice was only positive for rare endothelial cells within the thymus, whereas DL4LacZ/+ mice exhibited a strong reticular expression pattern within the outer cortex (characteristic of epithelial cells) and a weaker more punctuated staining within the medulla of the thymus (Fig. 1 A). This expression pattern was confirmed at the protein level using novel mAbs generated against the extracellular domains of DL1 and DL4. The specificity of the anti-DL1 and -DL4 mAbs was tested on DL1- and DL4-expressing OP-9 cells. Anti-DL1 mAbs bound specifically to DL1- but not to DL4-expressing OP-9 cells, whereas the anti-DL4 mAbs showed a reciprocal binding pattern (Fig. 1 B). These results indicate that both mAbs specifically bind to the corresponding Notch ligands and that they do not exhibit any cross-reactivity. These mAbs were then used to stain cell preparations enriched for TECs and analyzed by flow cytometry. TECs were identified as CD45−PanCytokeratin (PanCyt)+ (25) and were further subdivided into cortical TECs (cTECs) and medullary TECs (mTECs) based on BP1 expression (26). Anti-DL1 mAbs did not stain cTECs or mTECs above background. In contrast anti-DL4 mAbs showed significant staining of TECs, with brighter staining for cortical- (BP1+) versus medullary- (BP1−) derived populations (Fig. 1 C). Analysis of thymocyte subpopulations for the expression of DL1 and DL4 with these mAbs was negative (Fig. 1 D). These expression data are in general agreement with previous studies (18, 27, 28), with the notable exception of one report in which TECs were broadly stained by a commercial polyclonal antibody against DL1 (12). The specificity of this polyclonal reagent has already been challenged by others (29). In conclusion, the thymic expression pattern of DL1 and DL4 revealed by our novel mAbs confirms the results obtained with the lacZ knock-in mice, strongly suggesting that DL4 might be the physiological ligand expressed by TECs that interacts with N1-expressing T cell precursors to specify the T cell lineage.
Figure 1.
DL4 but not DL1 expression is detected on TECs. (A) LacZ staining on thymic sections derived from DL1 and DL4 lacZ knock-in mice. mRNA expression of the _DL1_-driven lacZ gene is confined to blood vessels within the thymus, whereas DL4 drives expression preferentially within the cortical epithelium. The dashed lines indicate cortical–medullary boundaries. Bars, 100 μm. (B) Anti-DL1 and -DL4 antibodies specifically bind their corresponding ligands without exhibiting cross-reactivity; OP-9–DL1–EGFP (top) and OP-9–DL4–EGFP (bottom) were stained with isotype Ctrl (shaded), anti-DL1 (dashed line), or anti-DL4 antibodies (continuous line; dilution, 1:100). The analysis was performed on gated EGFP-positive cells, and representative histograms are shown. (C) Anti-DL1 and -DL4 antibody staining on TECs. Enriched TECs extracted from wild-type thymi were stained for BP1 (3C6), CD45, and isotype Ctrl, anti-DL1, or anti-DL4 antibodies, followed by intracellular staining for PanCyt (C11) and flow cytometric analysis. Total TECs were gated as PanCyt+CD45− (top left), and cTECs and mTECs were further defined as BP1+ and BP1−, respectively (bottom left); isotype Ctrl (shaded), anti-DL1 (dashed line; top right), and anti-DL4 (continuous line; bottom right) are shown. Percentages of DL4+ TECs represent the mean ± SD of five mice. (D) DL1 and DL4 are not detectably expressed on thymocytes. (left) A representative flow cytometric analysis of CD4 versus CD8 of wild-type thymocytes. (right) DL1 and DL4 expression are shown as representative histograms (shaded, isotype Ctrl; dashed line, DL1; continuous line, DL4) after gating on the indicated thymic subpopulations: DN (Lin−, CD45+, CD4−, CD8−), DP (Lin−, CD45+, CD4+, CD8+), CD4SP (Lin−, CD45+, CD4+, CD8−), and CD8SP (Lin−, CD45+, CD4−, CD8+) thymocytes. Percentages of individual thymic subpopulations are indicated within the plots. Data are representative of three independent experiments.
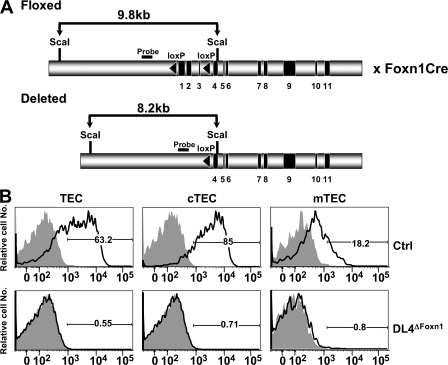
Studying the role of DL4 in hematopoiesis and, more specifically, during T lineage commitment by a conventional loss-of-function approach is hampered by the fact that DL4 gene-targeted mice are embryonic lethal (24, 30). We therefore generated conditional gene-targeted mice for DL4. These mice carry loxP sites flanking the first three coding exons of the DL4 gene (DL4lox/lox). Because the gene expression and antibody studies suggested that DL4 expression in TECs might be critical for T cell lineage commitment, we crossed the DL4lox/lox (Ctrl) mice to knock-in mice expressing the Cre recombinase under the control of the Foxn1 gene (31), which is highly expressed in TECs, to generate DL4lox/lox&Foxn1-Cre (DL4ΔFoxn1) mice (Fig. 2 A). Staining of DL4ΔFoxn1 TECs with anti-DL4 and -DL1 mAbs confirmed the complete loss of DL4 protein in the gene-targeted mice compared with Ctrl animals (Fig. 2 B). Interestingly, DL1 expression on TECs derived from DL4ΔFoxn1 mice was not detectable, indicating that there is no functional compensation by DL1 as a consequence of loss of DL4 (Fig. 2 B).
Figure 2.
Specific targeting of the DL4 gene in TECs by the Foxn1-Cre recombinase. (A) A schematic representation of the genomic organization of the DL4lox/lox locus is shown. Exons 1–3 are flanked by loxP sequences (black triangles). DL4lox/lox mice were crossed to mice in which the Cre recombinase was knocked into the Foxn1 locus (Foxn1-Cre) to obtain DL4lox/loxFoxn1-Cre mice (DL4ΔFoxn1). (B) Histograms of total TECs (PanCyt+CD45−), cTECs (PanCyt+CD45−BP1+), and mTECs (PanCyt+CD45−BP1−) from Ctrl (DL4lox/lox) or DL4ΔFoxn1 mice stained for DL1 (shaded) and DL4 (continuous lines). DL1 staining was indistinguishable from Ctrl isotype staining (not depicted). Data are representative of five Ctrl and three DL4ΔFoxn1 mice aged 2–3 wk from three independent experiments. Percentages of cells positively staining for DL4 are indicated in the histograms.
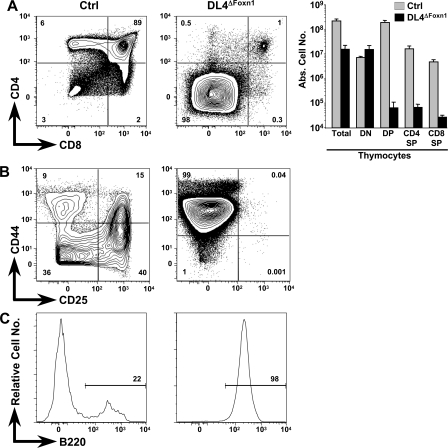
Side-by-side analysis of littermate thymi derived from Ctrl and DL4ΔFoxn1 mice revealed a 14-fold decrease in absolute thymocyte numbers in DL4ΔFoxn1 compared with Ctrl mice (Fig. 3 A). Flow cytometric analysis of CD4 and CD8 expression on thymocytes derived from DL4ΔFoxn1 mice shows nearly complete loss (<1%) of the CD4SP, the CD8SP, and the DP thymocyte subsets, whereas all of the major subsets are present in the expected frequency in Ctrl mice. _DL4ΔFoxn1_-derived thymi were almost exclusively populated with cells falling into the DN gate (Fig. 3 A). In absolute numbers, DP, CD4SP, and CD8SP subsets were reduced by 180–3,000-fold in DL4ΔFoxn1 mice, whereas DN cells were slightly increased (twofold). Whether the small residual population of T lineage cells in the thymus of DL4ΔFoxn1 mice results from a failure to delete DL4 efficiently in a subset of TECs (which stain below the threshold of detection with our anti-DL4 mAb) or, alternatively, represents a minor pathway of DL4-independent (and hence, presumably Notch-independent) T cell development remains to be investigated.
Figure 3.
Complete block in T cell development and accumulation of thymic B cells in DL4ΔFoxn1 mice. (A) Representative flow cytometric analysis of thymocytes derived from Ctrl and DL4ΔFoxn1 14-d-old mice. Total thymocytes were stained with anti-CD4 and -CD8 antibodies. Absolute cell numbers for total thymocytes and indicated subsets are shown as bar diagrams on a logarithmic scale. The bar diagrams represent mean values ± SD (n = 5 for Ctrl and 6 for DL4ΔFoxn1). (B) Representative FACS analysis of CD4−CD8−lin− thymocytes stained with anti-CD44 and -CD25 antibodies. (C) Representative histograms of B220 staining gated on lin−CD44+CD25− cells. Data are representative of three independent experiments. Percentages of positively stained cells are indicated within the contour plots and histograms.
Further analysis of the DN thymic compartment of DL4ΔFoxn1 mice for CD44 and CD25 expression reveals the presence of only CD44+CD25− DN1 cells, whereas all immature thymocyte subsets (DN1–4) were present in normal numbers in Ctrl animals (Fig. 3 B). Thus, the thymic phenotype of DL4ΔFoxn1 mice is very similar to the one previously published for Notch1lox/loxMx-Cre (N1ΔMx) mice (4, 5). In the case of N1ΔMx mice, accumulating DN1 cells in the thymus were found to be ectopically developing B cells (4, 5). Similarly, essentially all CD44+CD25− DN cells in DL4ΔFoxn1 mice expressed B220, whereas only a small fraction of DN1 cells in Ctrl thymi were B220+ (Fig. 3 C). To examine this phenotypic similarity in greater detail, we studied side-by-side B cells within the thymi of 2-wk-old Ctrl, DL4ΔFoxn1, and N1ΔMx mice 2 wk after N1 inactivation and compared them with BM B cells.
A small number of thymic B cells are present in Ctrl mice, and the majority have a mature IgM+B220+ phenotype. In contrast, a much larger population of B cells was found in DL4ΔFoxn1 and N1ΔMx mice, and these B cells express heterogeneous levels of IgM and B220. Moreover, DL4ΔFoxn1 and N1ΔMx B cells also express CD93 and BP1, which are typical markers for immature B cells, normally only present in the BM (Fig. 4 A). All B220+IgM− cells within the thymi of DL4ΔFoxn1 and N1ΔMx mice were CD19+, confirming that these are indeed immature B cells (unpublished data). In absolute numbers, immature thymic B cells were increased 340- and 30-fold over Ctrl values in DL4ΔFoxn1 and N1ΔMx mice, respectively (Fig. 4 B). In conclusion, the immature B cell phenotype observed in the thymi of N1ΔMx mice is largely recapitulated in DL4ΔFoxn1 mice, strongly suggesting that DL4 is the natural ligand for N1 during thymic T cell lineage commitment.
Figure 4.
Comparative phenotypic analysis of B cells within thymi of DL4ΔFoxn1 and N1ΔMx mice. (A) Representative flow cytometric analysis of B cells in the DN thymus compartment from Ctrl, N1ΔMx, and DL4ΔFoxn1 mice stained for B220 and IgM (top) or BP1 and CD93 (bottom). B220/IgM staining is electronically gated on lineage-negative CD44+CD25− cells, whereas BP1/CD93 staining is gated on lineage-negative B220+ cells. (left) A comparative analysis of normal BM B cells to highlight the phenotypic similarity. Percentages of populations staining positively for the indicated markers are shown in the contour plots. (B) Bar graphs show the absolute numbers ± SD of immature (B220+IgM−) B cells per thymus derived from Ctrl, Notch1ΔMx, and DL4ΔFoxn1 mice (n = 6 mice per sample group). Note the logarithmic scale. All mice used were between 2–3 wk old, and data are representative of three independent experiments.
Although DL4ΔFoxn1 and N1ΔMx mice exhibit qualitatively similar thymic phenotypes, it is clear that enhancement of immature B cell development (Fig. 4 B) is more pronounced in the DL4ΔFoxn1 background. This quantitative difference between DL4ΔFoxn1 and N1ΔMx mice most likely reflects the consequence of inactivation of the floxed DL4 and N1 genes using two different Cre systems. The Mx-Cre transgene inducibly inactivates floxed genes preferentially in hematopoietic cells in the BM (32). Thus, postnatal inactivation of N1 will generate N1-deficient BM progenitors that will migrate to the thymus and develop into B cells that will have to compete with residual thymocytes for the availability of thymic niches, growth factors, and cytokines. In contrast, constitutive _Foxn1-Cre_–mediated DL4 gene inactivation in TECs occurs already during embryogenesis (31). Thus, the first wave of incoming lymphoid progenitors will encounter an empty DL4-deficient thymic epithelium. Although these precursors cannot be instructed toward the T cell lineage, they can develop into the B cell lineage and efficiently expand in the absence of competition, as all necessary cytokines and growth factors are abundantly present in the neonatal thymus.
Although Foxn1-Cre expression within the thymus should be restricted to TECs (31) and wild-type thymocytes do not stain detectably with anti-DL4 mAbs (Fig. 1 D), it remains formally possible that rare DL4-expressing hematopoietic cells might be involved in T cell lineage commitment in the thymus. In this scenario, loss of DL4 expression on these cells could theoretically contribute to the generation of immature B cells in the DL4ΔFoxn1 thymus. To exclude this possibility, we generated DL4lox/loxMx-Cre (DL4ΔMx) mice to inducibly inactivate DL4 in BM progenitors. Because the Mx-Cre transgene is also active to some extent in TECs (16), BM from DL4ΔMx or Ctrl mice (both CD45.2+) was transplanted into CD45.1+ wild-type hosts to critically evaluate whether DL4ΔMx hematopoietic cells could contribute to the phenotype. Ctrl and DL4ΔMx BM chimeras were analyzed 8 wk after transplantation. The reconstitution efficiency of CD45.2+ cells for both Ctrl and DL4ΔMx chimeras was >95%. A portion of the BM cells of the corresponding BM chimeras was analyzed by Southern blot analysis to ensure that reconstitution was not caused by rare cells that might have escaped deletion of the DL4 locus (Fig. 5 E). As expected, Ctrl and DL4ΔMx BM progenitors generated all major blood lineages in the BM and spleen (unpublished data). More importantly, all major mature and immature thymocyte subsets were generated normally from Ctrl and DL4ΔMx BM progenitors (Fig. 5, A and B), and only very small numbers of immature B cells were found in the thymus (Fig. 5, C and D). These data exclude a significant role for putative DL4-expressing hematopoietic cells in thymic T cell lineage commitment.
Figure 5.
Normal T cell development and absence of immature thymic B cells in BM chimeras reconstituted with DL4ΔMx BM. Ctrl or DL4ΔMx CD45.2+ BM was transplanted into lethally irradiated CD45.1+ hosts and analyzed 8 wk after reconstitution. The reconstitution efficiency for both Ctrl and DL4ΔMx BM chimeras was >95% (not depicted). (A) Representative flow cytometric analysis of CD45.2+ thymocytes stained with anti-CD4 and -CD8. (B) CD45.2+ lineage-negative DN thymocytes were analyzed for the expression of CD44 and CD25. (C and D) Lineage-negative thymocytes were analyzed for the presence of B cells expressing B220 and IgM, or BP1 and CD93. Data in A–D are representative of three individual chimeras per group with virtually identical results, and relative percentages are indicated in the contour plots. Two independent experiments were performed. (E) Southern blot analysis of _ScaI_-digested genomic DNA derived from BM cells of Ctrl and DL4ΔMx chimeras showing the floxed alleles in Ctrl chimeras and the completely deleted DL4 locus in DL4ΔMx chimeras. The targeting strategy and size of the restriction fragments are as described in Fig. 2 A.
Collectively, we have shown that DL4 is expressed by TECs, whereas DL1 is not detectable. Moreover, DL4ΔFoxn1 mice exhibit a loss of DL4 expression in TECs, which correlates with a complete block in T cell development and the accumulation of immature B cells within the thymus. DL4 deficiency within hematopoietic progenitors seems not to contribute to this phenotype, as assessed by BM chimeras, suggesting that the loss of DL4 on TECs is causative. A similar accumulation of immature thymic B cells is found in mice in which the N1 receptor is inactivated in hematopoietic progenitors. Collectively, these data demonstrate that DL4 expression on TECs is essential for thymic T cell development. Furthermore, they strongly suggest that N1-expressing T cell progenitors only commit to the T cell lineage after entry into the thymus.
Although in vitro DL1 and DL4 are functionally redundant in their ability to promote T cell development from BM precursors when expressed on OP9 cells, in vivo DL1 and DL4 mediate nonredundant functions within the hematopoietic system by binding to specific Notch receptors. Our data demonstrate that DL4 and N1 constitute within the thymus a nonredundant ligand–receptor pair essential for T cell lineage commitment. Similarly, DL1 pairs with N2 in the spleen to specify marginal zone B cell development in a nonredundant manner (16, 33). Differential expression of Notch ligands has also been reported to influence T helper cell differentiation in peripheral CD4 T cells (34). Thus, all of these examples are consistent with the possibility that specificity of Notch signaling within the hematopoietic system might be generally regulated by tissue-specific expression of Notch ligands.
MATERIALS AND METHODS
Mice: generation of DL4 gene-targeted floxed mice.
We generated the conditional Dl4 loss-of-function mutant mice by flanking the first three exons of the DL4 allele with two loxP sites. Mouse DL4 genomic clones were isolated from a 129/Sv genomic library. One loxP site was inserted into the 5′ untranslated region in the XhoI restriction site, 75 bp downstream of the transcription initiation site. The other loxP site, together with the flipase recognition target (FRT) site–flanked PGK-neomycin selection cassette, was inserted in the EcoRI restriction site of intron 3. Upon Cre-mediated recombination, the translation initiation site and the coding sequence for the first 132 amino acids of the protein were removed, creating a null allele. Electroporation and selection of embryonic stem (ES) cells (R1) was performed using standard protocols. Homologous recombination of the targeted DL4 locus in ES cells was confirmed first by PCR and then by ScaI restriction digest of the genomic tail DNA and Southern blotting. A 400-bp ApaI-ApaI genomic fragment localized upstream of the 5′ arm of the targeting vector was used as a probe (Fig. 2 A). Blastocyst injection of the targeted ES cells was performed according to standard protocols. To remove the FRT site–flanked PGK-neomycin selection cassette, germline chimeras were crossed with ACTB-Flpe mice (35) in a pure C57BL6 background. DL4 homozygous floxed mice were obtained in the expected Mendelian ratio, suggesting normal expression of a functional DL4 protein from the modified allele. Homozygous DL4lox/lox mice were bred to Mx-Cre mice (32) and to Foxn1-Cre mice (31) to generate DL4lox/+&Mx-Cre+/− and DL4lox/+&Foxn1-Cre+/− mice, respectively. These mice were intercrossed to generate DL4lox/lox&Mx-Cre+/− (DL4ΔMx) and DL4lox/lox&Foxn1-Cre+/− (DL4ΔFoxn1) mice and were subsequently backcrossed for at least five generations into C57BL6 mice. Mice were genotyped for homozygosity of the loxP sites and for the presence of the Mx-Cre or FoxN1-Cre transgene by PCR.
Conditional gene-targeted N1 mice were previously described (4). B6.SJL-Ptprca (CD45.1+) mice were originally purchased from the Jackson Laboratory. CD45.1+ recipient mice were used as BM recipients for the chimeras described in the following section. This study has been reviewed and approved by the Service Vétérinaire Cantonal of Etat de Vaud.
BM chimeras.
BM chimeras were generated using T cell–depleted BM cells from polyI-polyC (pI-pC; InvivoGen) Ctrl (DL4lox/lox) or DL4ΔMx (DL4lox/lox&MxCre+/−) mice. Lethally irradiated (1,000 cGy) 10-wk-old recipients (CD45.1+) received 10 × 106 donor BM cells (CD45.2+) i.v. Radiation chimeras were maintained on antibiotic water and analyzed after 8 wk.
Activation of the Mx-Cre recombinase.
3–4-wk-old DL4ΔMx and Ctrl mice received 4 i.p. injections of 2 μg/g body weight pI-pC at 2 d intervals. Mice were killed at the time points indicated in the figures, and genomic DNA was prepared from 10 × 106 BM cells. The deletion efficiency in cells from chimeric, Ctrl, and DL4ΔMx mice was assessed by Southern blot analysis of _ScaI_-digested genomic DNA and hybridized with a 400-bp ApaI-ApaI probe (Figs. 2 and 5). The analysis was quantified using a phosphorimager (FLA3000; Fujifilm).
Staining for β-galactosidase activity and histology.
β-Galactosidase activity was detected as previously described (36). In brief, whole thymi were isolated and fixed for 60 min in 0.2% glutaraldehyde in 100 mM of potassium phosphate buffer, pH 7.4, containing 5 mM EGTA and 2 mM MgCl2. Thymi were washed three times in 0.01% Na-desoxycholate and 0.02% Nonidet P-40 in 100 mM of potassium phosphate buffer with 5 mM EGTA and 2 mM MgCl2. For detection of β-galactosidase activity, thymi were incubated in washing solution containing 0.5 mg/ml X-gal, 10 mM K3Fe(CN)6, and 10 mM K4Fe(CN)6 at 37°C. After staining, thymi were washed again in washing solution and, finally, in 1 × PBS. For histology, thymi were dehydrated in isopropanol, cleared in xylene, and transferred to paraffin. Embedded thymi were sectioned at 7–10 μm and counterstained with eosin according to standard procedures.
TEC enrichment.
TECs from normal and DL4ΔFoxn1 thymi were prepared using a modified protocol from Klein et al. (37). In brief, individual thymi were cut into small pieces and washed three times with DMEM supplemented with 2% FCS (PAA Laboratories) to remove thymocytes. TECs were then extracted from the thymic fragments sequentially using collagenase D and DNaseI digestions (both from Roche), followed by TrypLE Express (Invitrogen) digestion. Finally, TECs were recovered at the interface of a 38–60% Percoll gradient, stained with the mAbs indicated in the figures, and analyzed by FACS.
mAbs and flow cytometry.
Single-cell suspensions from BM, thymus, and TECs were prepared and stained using standard protocols, as previously described (5). FACS analyses (6- and 8-color) were performed using a FACSCanto flow cytometer (Becton Dickinson) and a CyAn ADP flow cytometer (Dako). Data were analyzed using FlowJo software (Tree Star, Inc.) or FACS Aria software (Becton Dickinson). The following antibodies were used to analyze hematopoietic cells and TECs, and the subsequent mAb conjugates were purchased from eBioscience: TCRβ (H57-597)-FITC; CD4 (GK1.5)-PECy5, -PECy5.5, and -PECy7; CD8α (53.6.7)-PE and -PECy7; CD11b (M1/70)-PECy7; Gr1 (Ly6G, RB6-8C5)-PECy7; Ter119 (Ly76)-PECy7; B220 (RA3-6B2)-FITC and -PECy5; CD44 (Pgp-1, IM7)-PECy7; CD117 (c-kit, 2B8)-allophycocyanin (APC); CD25 (IL-2R, PC61.5)–APC–Alexa Fluor 750; BP-1 (6C3)–PE; IgM (eB121-15F9)-APC; CD93 (AA4.1, C1qRp)-FITC; CD45 (30-F11)-PECy5.5; CD45.1 (A20.1)-PE and –APC–Alexa Fluor 750. CD45.2 (104)–PacificBlue was purchased from BioLegend. TCRγδ (GL3)-FITC, CD8α (53.6.7)-FITC and –Alexa Fluor 647, CD4 (GK1.5)-FITC, CD3 (145-2C11)-FITC, CD11b (M1/70)-FITC, Gr1 (Ly6G, RB6-8C5)-FITC, and Ter119 (Ly76)-FITC were purified from hybridoma supernatants and conjugated in our laboratory according to standard protocols. Alexa Fluor 647 conjugates were prepared using the appropriate Alexa Fluor protein labeling kits (Invitrogen). Intracellular staining of TECs was performed with a PanCyt mAb (C11) from Sigma-Aldrich.
Rat mAbs were generated against the extracellular domains of DL1 and DL4. Details of the immunization and screening will be presented in a future paper (unpublished data). Purified mAbs were biotinylated in our laboratory according to standard protocols and revealed with streptavidin-PE. Both mAbs were assayed for specificity on OP-9 stromal cells engineered to express DL1 or DL4 combined with enhanced GFP (OP-9–DL1–EGFP or OP-9–DL4–EGFP; Fig. 1 B).
Acknowledgments
We thank Olivier Randin and Pierre Dubied for assistance in preparation of the figures.
This work was in part supported by the Swiss National Science Foundation, the Swiss Cancer League, and a Marie Heim-Vögtlein Fellowship (PMP DB-110307/1 to E. Fiorini).
The authors have no conflicting financial interests.
References
- 1.Rothenberg, E.V. 2000. Stepwise specification of lymphocyte developmental lineages. Curr. Opin. Genet. Dev. 10:370–379. [DOI] [PubMed] [Google Scholar]
- 2.Godfrey, D.I., and A. Zlotnik. 1993. Control points in early T-cell development. Immunol. Today. 14:547–553. [DOI] [PubMed] [Google Scholar]
- 3.Maillard, I., T. Fang, and W.S. Pear. 2005. Regulation of lymphoid development, differentiation, and function by the Notch pathway. Annu. Rev. Immunol. 23:945–974. [DOI] [PubMed] [Google Scholar]
- 4.Radtke, F., A. Wilson, G. Stark, M. Bauer, J. van Meerwijk, H.R. MacDonald, and M. Aguet. 1999. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 10:547–558. [DOI] [PubMed] [Google Scholar]
- 5.Wilson, A., H.R. MacDonald, and F. Radtke. 2001. Notch 1–deficient common lymphoid precursors adopt a B cell fate in the thymus. J. Exp. Med. 194:1003–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han, H., K. Tanigaki, N. Yamamoto, K. Kuroda, M. Yoshimoto, T. Nakahata, K. Ikuta, and T. Honjo. 2002. Inducible gene knockout of transcription factor recombination signal binding protein-J reveals its essential role in T versus B lineage decision. Int. Immunol. 14:637–645. [DOI] [PubMed] [Google Scholar]
- 7.Wolfer, A., A. Wilson, M. Nemir, H.R. MacDonald, and F. Radtke. 2002. Inactivation of Notch1 impairs VDJbeta rearrangement and allows pre-TCR-independent survival of early alpha beta lineage thymocytes. Immunity. 16:869–879. [DOI] [PubMed] [Google Scholar]
- 8.Hoflinger, S., K. Kesavan, M. Fuxa, C. Hutter, B. Heavey, F. Radtke, and M. Busslinger. 2004. Analysis of Notch1 function by in vitro T cell differentiation of Pax5 mutant lymphoid progenitors. J. Immunol. 173:3935–3944. [DOI] [PubMed] [Google Scholar]
- 9.Felli, M.P., M. Maroder, T.A. Mitsiadis, A.F. Campese, D. Bellavia, A. Vacca, R.S. Mann, L. Frati, U. Lendahl, A. Gulino, and I. Screpanti. 1999. Expression pattern of notch1, 2 and 3 and Jagged1 and 2 in lymphoid and stromal thymus components: distinct ligand-receptor interactions in intrathymic T cell development. Int. Immunol. 11:1017–1025. [DOI] [PubMed] [Google Scholar]
- 10.Schmitt, T.M., and J.C. Zuniga-Pflucker. 2002. Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity. 17:749–756. [DOI] [PubMed] [Google Scholar]
- 11.Harman, B.C., E.J. Jenkinson, and G. Anderson. 2003. Entry into the thymic microenvironment triggers Notch activation in the earliest migrant T cell progenitors. J. Immunol. 170:1299–1303. [DOI] [PubMed] [Google Scholar]
- 12.Schmitt, T.M., M. Ciofani, H.T. Petrie, and J.C. Zuniga-Pflucker. 2004. Maintenance of T cell specification and differentiation requires recurrent notch receptor–ligand interactions. J. Exp. Med. 200:469–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang, R., Y. Lan, H.D. Chapman, C. Shawber, C.R. Norton, D.V. Serreze, G. Weinmaster, and T. Gridley. 1998. Defects in limb, craniofacial, and thymic development in Jagged2 mutant mice. Genes Dev. 12:1046–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mancini, S.J., N. Mantei, A. Dumortier, U. Suter, H.R. Macdonald, and F. Radtke. 2005. Jagged1-dependent Notch signaling is dispensable for hematopoietic stem cell self-renewal and differentiation. Blood. 105:2340–2342. [DOI] [PubMed] [Google Scholar]
- 15.Jaleco, A.C., H. Neves, E. Hooijberg, P. Gameiro, N. Clode, M. Haury, D. Henrique, and L. Parreira. 2001. Differential effects of Notch ligands Delta-1 and Jagged-1 in human lymphoid differentiation. J. Exp. Med. 194:991–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hozumi, K., N. Negishi, D. Suzuki, N. Abe, Y. Sotomaru, N. Tamaoki, C. Mailhos, D. Ish-Horowicz, S. Habu, and M.J. Owen. 2004. Delta-like 1 is necessary for the generation of marginal zone B cells but not T cells in vivo. Nat. Immunol. 5:638–644. [DOI] [PubMed] [Google Scholar]
- 17.Besseyrias, V., E. Fiorini, L.J. Strobl, U. Zimber-Strobl, A. Dumortier, U. Koch, M.L. Arcangeli, S. Ezine, H.R. Macdonald, and F. Radtke. 2007. Hierarchy of Notch–Delta interactions promoting T cell lineage commitment and maturation. J. Exp. Med. 204:331–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan, X.Q., U. Sarmiento, Y. Sun, G. Huang, J. Guo, T. Juan, G. Van, M.Y. Qi, S. Scully, G. Senaldi, and F.A. Fletcher. 2001. A novel Notch ligand, Dll4, induces T-cell leukemia/lymphoma when overexpressed in mice by retroviral-mediated gene transfer. Blood. 98:3793–3799. [DOI] [PubMed] [Google Scholar]
- 19.de La Coste, A., E. Six, N. Fazilleau, L. Mascarell, N. Legrand, M.P. Mailhe, A. Cumano, Y. Laabi, and A.A. Freitas. 2005. In vivo and in absence of a thymus, the enforced expression of the notch ligands delta-1 or delta-4 promotes T cell development with specific unique effects. J. Immunol. 174:2730–2737. [DOI] [PubMed] [Google Scholar]
- 20.Wolfer, A., T. Bakker, A. Wilson, M. Nicolas, V. Ioannidis, D.R. Littman, P.P. Lee, C.B. Wilson, W. Held, H.R. MacDonald, and F. Radtke. 2001. Inactivation of Notch 1 in immature thymocytes does not perturb CD4 or CD8 T cell development. Nat. Immunol. 2:235–241. [DOI] [PubMed] [Google Scholar]
- 21.Tanigaki, K., M. Tsuji, N. Yamamoto, H. Han, J. Tsukada, H. Inoue, M. Kubo, and T. Honjo. 2004. Regulation of alphabeta/gammadelta T cell lineage commitment and peripheral T cell responses by Notch/RBP-J signaling. Immunity. 20:611–622. [DOI] [PubMed] [Google Scholar]
- 22.Mohtashami, M., and J.C. Zuniga-Pflucker. 2006. Three-dimensional architecture of the thymus is required to maintain delta-like expression necessary for inducing T cell development. J. Immunol. 176:730–734. [DOI] [PubMed] [Google Scholar]
- 23.Hrabe de Angelis, M., J. McIntyre II, and A. Gossler. 1997. Maintenance of somite borders in mice requires the Delta homologue DII1. Nature. 386:717–721. [DOI] [PubMed] [Google Scholar]
- 24.Duarte, A., M. Hirashima, R. Benedito, A. Trindade, P. Diniz, E. Bekman, L. Costa, D. Henrique, and J. Rossant. 2004. Dosage-sensitive requirement for mouse Dll4 in artery development. Genes Dev. 18:2474–2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rossi, S.W., A.P. Chidgey, S.M. Parnell, W.E. Jenkinson, H.S. Scott, R.L. Boyd, E.J. Jenkinson, and G. Anderson. 2007. Redefining epithelial progenitor potential in the developing thymus. Eur. J. Immunol. 37:2411–2418. [DOI] [PubMed] [Google Scholar]
- 26.Gray, D.H., N. Seach, T. Ueno, M.K. Milton, A. Liston, A.M. Lew, C.C. Goodnow, and R.L. Boyd. 2006. Developmental kinetics, turnover, and stimulatory capacity of thymic epithelial cells. Blood. 108:3777–3785. [DOI] [PubMed] [Google Scholar]
- 27.Tsukamoto, N., M. Itoi, M. Nishikawa, and T. Amagai. 2005. Lack of Delta like 1 and 4 expressions in nude thymus anlages. Cell. Immunol. 234:77–80. [DOI] [PubMed] [Google Scholar]
- 28.Benedito, R., and A. Duarte. 2005. Expression of Dll4 during mouse embryogenesis suggests multiple developmental roles. Gene Expr. Patterns. 5:750–755. [DOI] [PubMed] [Google Scholar]
- 29.Lehar, S.M., J. Dooley, A.G. Farr, and M.J. Bevan. 2005. Notch ligands Delta 1 and Jagged1 transmit distinct signals to T-cell precursors. Blood. 105:1440–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gale, N.W., M.G. Dominguez, I. Noguera, L. Pan, V. Hughes, D.M. Valenzuela, A.J. Murphy, N.C. Adams, H.C. Lin, J. Holash, et al. 2004. Haploinsufficiency of delta-like 4 ligand results in embryonic lethality due to major defects in arterial and vascular development. Proc. Natl. Acad. Sci. USA. 101:15949–15954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gordon, J., S. Xiao, B. Hughes III, D.M. Su, S.P. Navarre, B.G. Condie, and N.R. Manley. 2007. Specific expression of lacZ and cre recombinase in fetal thymic epithelial cells by multiplex gene targeting at the Foxn1 locus. BMC Dev. Biol. 7:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuhn, R., F. Schwenk, M. Aguet, and K. Rajewsky. 1995. Inducible gene targeting in mice. Science. 269:1427–1429. [DOI] [PubMed] [Google Scholar]
- 33.Saito, T., S. Chiba, M. Ichikawa, A. Kunisato, T. Asai, K. Shimizu, T. Yamaguchi, G. Yamamoto, S. Seo, K. Kumano, et al. 2003. Notch2 is preferentially expressed in mature B cells and indispensable for marginal zone B lineage development. Immunity. 18:675–685. [DOI] [PubMed] [Google Scholar]
- 34.Amsen, D., J.M. Blander, G.R. Lee, K. Tanigaki, T. Honjo, and R.A. Flavell. 2004. Instruction of distinct CD4 T helper cell fates by different notch ligands on antigen-presenting cells. Cell. 117:515–526. [DOI] [PubMed] [Google Scholar]
- 35.Rodriguez, C.I., F. Buchholz, J. Galloway, R. Sequerra, J. Kasper, R. Ayala, A.F. Stewart, and S.M. Dymecki. 2000. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat. Genet. 25:139–140. [DOI] [PubMed] [Google Scholar]
- 36.Beckers, J., A. Clark, K. Wunsch, M. Hrabe De Angelis, and A. Gossler. 1999. Expression of the mouse Delta1 gene during organogenesis and fetal development. Mech. Dev. 84:165–168. [DOI] [PubMed] [Google Scholar]
- 37.Klein, L., M. Klugmann, K.A. Nave, V.K. Tuohy, and B. Kyewski. 2000. Shaping of the autoreactive T-cell repertoire by a splice variant of self protein expressed in thymic epithelial cells. Nat. Med. 6:56–61. [DOI] [PubMed] [Google Scholar]