The Biosynthetic Pathway for Synechoxanthin, an Aromatic Carotenoid Synthesized by the Euryhaline, Unicellular Cyanobacterium Synechococcus sp. Strain PCC 7002 (original) (raw)
Abstract
The euryhaline, unicellular cyanobacterium Synechococcus sp. strain PCC 7002 produces the dicyclic aromatic carotenoid synechoxanthin (χ,χ-caroten-18,18′-dioic acid) as a major pigment (>15% of total carotenoid) and when grown to stationary phase also accumulates small amounts of renierapurpurin (χ,χ-carotene) (J. E. Graham, J. T. J. Lecomte, and D. A. Bryant, J. Nat. Prod. 71:1647-1650, 2008). Two genes that were predicted to encode enzymes involved in the biosynthesis of synechoxanthin were identified by comparative genomics, and these genes were insertionally inactivated in Synechococcus sp. strain PCC 7002 to verify their function. The cruE gene (SYNPCC7002_A1248) encodes β-carotene desaturase/methyltransferase, which converts β-carotene to renierapurpurin. The cruH gene (SYNPCC7002_A2246) encodes an enzyme that is minimally responsible for the hydroxylation/oxidation of the C-18 and C-18′ methyl groups of renierapurpurin. Based on observed and biochemically characterized intermediates, a complete pathway for synechoxanthin biosynthesis is proposed.
Carotenoids are multifunctional, C40 isoprenoid pigments that are found in all chlorophototrophs, all retinalophototrophs, and many nonphototrophic organisms (24). In particular, oxygenated carotenoids, known as xanthophylls, play crucial roles in quenching reactive oxygen species. The cyclic carotenoids of cyanobacteria contain mostly β-rings, although ɛ-rings occur in the Prochlorococcus spp., in which α-carotene is the principal carotene (42). Cyanobacterial xanthophylls include compounds with various oxidative modifications of their β-rings: hydroxyl groups may occur at the C-2, C-3, or C-4 positions, and/or keto groups may occur at the C-4 positions (37, 44). Additionally, some cyanobacteria produce the monocyclic xanthophyll myxoxanthophyll or the acyclic xanthophyll oscillaxanthin. These are compounds in which the ψ ends of the molecules are hydroxylated at the C-1 and C-2 positions and an additional double bond is added at the C-3,4 position; this produces either 12 or 13 conjugated double bonds in the resulting carotenoid, respectively (23, 35).
Carotenoids are important and integral components of both photosystem I and photosystem II. β-Carotene is the primary carotenoid associated with the photosystems in most cyanobacteria (3, 17), and the cyanobacterial cytochrome b6f complex also contains one carotenoid molecule (4, 47, 50). Carotenoids are also found in the water-soluble orange carotenoid protein (20), which binds the xanthophyll 3′-hydroxy-echinenone in Synechocystis sp. strain PCC 6803 (49). The orange carotenoid protein functions in nonphotochemical quenching of energy transfer phycobilisomes (19, 48). Aside from their roles in various photosynthetic complexes, the xanthophylls of cyanobacteria are located in the thylakoid, cytoplasmic, and outer membranes of these gram-negative organisms. Anacystis nidulans (now Synechococcus sp. strain PCC 6301) contains primarily β-carotene and zeaxanthin in its thylakoid membranes (30). In contrast, the cell envelope (outer membrane and cell wall) and cytoplasmic membranes of this organism contain zeaxanthin and nostoxanthin in similar proportions (30, 32). In Synechocystis sp. strain PCC 6714, β-carotene is found primarily in the thylakoid fractions (where photosystem I and II occur), and zeaxanthin occurs primarily in the cytoplasmic membranes (33). However, the outer membrane fractions of Synechocystis sp. strain PCC 6714 are bright orange-red and predominantly contain myxoxanthophyll as well as other carotenoids (18). Thus, cyanobacterial carotenoids play diverse roles, and these differing roles may in part explain the diversity of carotenoids found in these organisms and their asymmetric distribution within the cell.
We have recently established that the cyanobacterium Synechococcus sp. strain PCC 7002 produces a novel xanthophyll, which is a bicyclic, aromatic carotenoid diacid (χ,χ-caroten-18,18′-dioic acid) and which we have given the common name synechoxanthin (16). This discovery has further increased the chemical diversity of carotenoids produced by cyanobacteria, and the discovery of synechoxanthin has prompted us to study its biosynthesis. Previous studies of the biosynthesis of aryl carotenoids had suggested that these compound are synthesized from lycopene through monocyclic (β,ψ) or dicyclic (β,β) carotene intermediates (22, 29). The enzyme CrtU was subsequently characterized from Streptomyces griseus as a novel carotene desaturase/methyltransferase that converts β rings to φ rings (21). A gene in the green sulfur bacterium Chlorobaculum tepidum that encodes an enzyme with sequence similarity to S. griseus CrtU was characterized (12). A C. tepidum crtU mutant accumulated γ-carotene instead of the aromatic carotenoid chlorobactene.
Frigaard et al. (12) had observed that cyanobacterial genomes encode proteins apparently homologous to CrtU (γ-carotene-desaturase/methyltransferase) from C. tepidum. However, until recently, cyanobacteria were believed to be unable to synthesize aromatic carotenoids (9, 12, 27). For this reason, Frigaard et al. proposed that these CrtU homologs might be the then-unknown lycopene cyclases in those cyanobacteria that lack CrtL-type lycopene cyclases (12). However, the subsequent discovery of the CruA/CruP family of lycopene cyclases by Maresca et al. identified the “missing” carotene cyclases in chlorophototrophs (26). The recent discovery of synechoxanthin, an aromatic carotenoid produced by Synechococcus sp. strain PCC 7002, increased the likelihood that the cyanobacterial CrtU homologs were involved in the aromatization of carotenoids, and this prompted us to study the function of these crtU homologs in cyanobacteria (26).
In this study two cruE genes, which encode the cyanobacterial homologs of CrtU in Synechococcus sp. strain PCC 7002 and Synechocystis sp. strain PCC 6803, were mutated and shown to be required for the synthesis of aromatic carotenoids in these organisms. The cruH gene, which encodes a second enzyme involved in the biosynthesis of synechoxanthin in Synechococcus sp. strain PCC 7002, was also identified by comparative genomics, and the function of its product in synechoxanthin synthesis was also verified by insertional inactivation. The characterization of these cruE and cruH mutants reveals how aromatic carotenoids are synthesized in cyanobacteria and provides insights into the synthesis of χ,χ-carotenoids in other organisms.
MATERIALS AND METHODS
Strains and growth conditions.
Synechococcus sp. strain PCC 7002 wild type and mutants were routinely grown under standard growth conditions (38°C, continuous illumination at 250 μmol photons m−2 s−1, bubbling with 1% [vol/vol] CO2) in 25-ml test tube cultures in medium A supplemented with 1 mg NaNO3 ml−1 (medium A+) (41). Media were supplemented with 100 μg kanamycin ml−1 or 100 μg spectinomycin ml−1 as required. Freshwater strains, including Synechocystis sp. strain PCC 6803, Nostoc sp. strain MAC, Nostoc sp. strain PCC 7120, and Synechococcus sp. strain PCC 7942, were grown in B-HEPES medium or on solid B-HEPES medium supplemented with 15 g Bacto agar liter−1 (Difco, Voigt Global Distribution, Lawrence, KS). B-HEPES medium was prepared by supplementing BG-11 medium with 4.6 mM HEPES-KOH and 18 mg liter−1 ferric ammonium citrate (39). Plates were incubated at 30°C at a light intensity of 150 μmol photons m−2 s−1. Synechococcus sp. strain C9 was grown in medium D supplemented with 10 mM HEPES buffer, pH 7.6, at 45°C in 30-ml test tube cultures or in 12-liter carboys (8). Other cyanobacterial cell materials were the kind gifts from other laboratories.
Identification of genes and phylogenetic comparisons.
Analyses of cyanobacterial genomes were conducted using databases, the Blastp algorithm (1), and the phylogenetic profiling tools at the Integrated Microbial Genomes website (Joint Genome Institutes, Walnut Grove, CA; http://img.jgi.doe.gov/cgi-bin/pub/main.cgi). For construction of phylogenetic trees, amino acid sequences were aligned with the ClustalW tool in MacVector 9.0 (MacVector, Inc., Cary, NC), and neighbor-joining trees were generated from the resulting alignments with PAUP 4.0 (Sinauer Associates, Inc., Sunderland, MA).
Inactivation of genes by interposon mutagenesis.
The upstream and downstream flanking sequences of genes selected for insertional inactivation were amplified by PCR by using genomic DNA templates isolated from wild-type Synechococcus sp. strain PCC 7002 or wild-type Synechocystis sp. strain PCC 6803 with the primers described in Table S1 in the supplemental material. The reverse primers for the left flank and the forward primers for the right flank contained BamHI, EcoRI, or HindIII restriction sites as indicated in Table S1 in the supplemental material. PCR products were digested with the appropriate restriction enzymes and purified from agarose gels by use of an Eppendorf Perfectprep gel cleanup kit (catalog number 0032 007.759; Eppendorf, Westbury, NY). For insertional activation of SYNPCC7002_A1248, the aphAII gene, conferring resistance to kanamycin, was excised from plasmid pRL170 (13) with HindIII and purified by electrophoresis on an agarose gel. For insertional inactivation of sll0254 and SYNPCC7002_A2246, the aadA gene, conferring resistance to streptomycin and spectinomycin, was excised from plasmid pSRA2 (14) with EcoRI or BamHI, respectively, and purified in the same manner. The fragments were mixed at a left flank/antibiotic resistance cassette/right flank ratio of 3:1:3 and ligated by incubating with T4 DNA ligase. The ligation products were separated on agarose gels, and the desired products were excised, purified as described above, and used to transform Synechococcus sp. strain PCC 7002 or Synechocystis sp. strain PCC 6803 as previously described by Frigaard et al. (13).
HPLC analysis of pigment extracts.
Wild-type and mutant cells of Synechococcus sp. strain PCC 7002 were harvested in mid-exponential phase (optical density at 730 nm [OD730] of ∼1.0) or stationary phase (OD730 of ∼14). Wild-type and mutant cells of Synechocystis sp. strain PCC 6803 were harvested in mid-exponential phase (OD730 of ∼1.0). Pigments were extracted from whole cells by sonication in 7:2 acetone-methanol, or in methanol alone when indicated, and the cellular debris was removed by centrifugation. The supernatant was filtered through a 0.2-μm polytetrafluoroethylene syringe filter (6783-0402; Whatman, Clifton, NJ). Filtered extracts were analyzed immediately without further manipulation. The high-pressure liquid chromatography (HPLC) system consisted of an analytical 5-μm Discovery C18 column (25 cm by 4.6 mm; Supelco, Bellefonte, PA) fitted to a binary pump (model G1312A) and solvent degasser (model 1379A) (1100 series; Agilent Technologies, Palo Alto, CA). Eluates were monitored with a 1,024-element diode array detector (model G1315B, 1100 series; Agilent Technologies, Palo Alto, CA), and the system was controlled with Agilent ChemStation software for HPLC. Solvent A was methanol-acetonitrile-water (21:16.5:62.5, vol/vol/vol) and contained 10 mM ammonium acetate; solvent B was methanol-acetonitrile-ethyl acetate (50:20:30, vol/vol/vol). The solvent gradient program (time [min], %B, flow rate [ml min−1]) employed was as follows: 0, 20, 0.750; 10, 70, 1.0; 40, 100, 1.0; and 60, 100, 1.0.
Pigment purification and analysis.
For larger-scale purification of pigments, a semipreparative 5-μm Discovery C18 column (25 cm by 10 mm; Supelco, Bellefonte, PA) was used with the same HPLC equipment and gradient as described above except that the flow rate was increased to 3.5 ml min−1. Pigment-containing fractions were collected and dried under a stream of nitrogen gas. Chemical analyses of compounds was performed as previously described (11). To test for the presence of hydroxyl groups, a small (200-ng) sample of the carotenoid was dissolved in anhydrous pyridine and treated with Sigma-Sil-A (10 to 100 μl) (trimethylchlorosilane-1,1,1,3,3,3-hexamethyldisilazane-pyridine, 1:3:9) (Sigma, St. Louis, MO). Reactions were quenched with excess methanol, and the solution was dried under nitrogen and resuspended in methanol prior to HPLC analysis. Reduction of carbonyls by sodium borohydride or lithium aluminum hydride was performed as previously described, except that in some cases the starting material was initially solubilized in tetrahydrofuran or anhydrous pyridine rather than diethyl ether (11). The production of methyl esters from acidic carotenoids in the presence of trimethylsilyl-diazomethane and methanol was used as a diagnostic reaction for the presence of carboxylic acid moieties. Methanol (100 μl) was added to a dried carotenoid sample (200 ng), trimethylsilyl-diazomethane (50 μl of a 2 M stock solution prepared in hexane) was added in excess, and the mixture was incubated for 1 h prior to quenching the reaction with excess ethyl acetate. The sample was then dried under nitrogen and resuspended in methanol for further analyses. Eric Snyder and Li Zhang at the Proteomics and Mass Spectrometry Core Facility of the Huck Institutes of the Life Sciences, The Pennsylvania State University, performed the mass spectrometric analyses.
RESULTS
Identification of candidate genes and phylogenetic analyses.
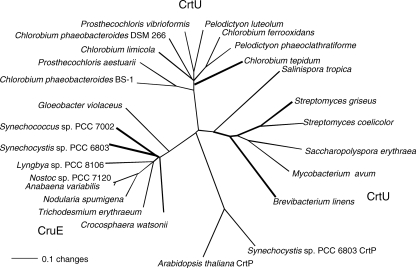
As previously noted by Frigaard et al. (12), genes encoding cyanobacterial enzymes with strong sequence similarity to CrtU carotene desaturases are found in some but not all cyanobacteria whose genomes have been sequenced. CrtU homologs are not found in some sequenced strains, including Nostoc punctiforme strain PCC 73102, Thermosynechococcus elongatus strain BP-1, Synechococcus sp. strain JA-2-3B′a(2-13), and Synechococcus sp. strain JA-3-3Ab, which also lack sequences similar to those of CrtL-type lycopene cyclases. Thus, because all cyanobacteria produce ring-containing carotenes, these CrtU homologs are unlikely to be carotenoid cyclases. The cyanobacterial CrtU-like proteins form a clade that is distinct from the CrtU enzymes of both nonphotosynthetic bacteria and the green sulfur bacteria (Fig. 1). As will be shown below, the enzymatic activity attributed to the products of these cyanobacterial genes is also distinct from that of these other bacteria, and therefore we have given these genes a distinctive gene locus designation, cruE. The open reading frame that carries cruE in Synechococcus sp. strain PCC 7002 is SYNPCC7002_A1248, and that in Synechocystis sp. strain PCC 6803 is sll0154. CruE sequences resemble the CrtU sequences of green sulfur bacteria by having a Rieske iron/sulfur cluster domain inserted into the C terminus of a flavin-binding, pyridine nucleotide-disulfide oxidoreductase superfamily domain. The latter domain is similar to domains that are characteristic of flavin-containing amine oxidases and CrtP-type phytoene desaturases. The CrtU sequences of Streptomyces spp. and other, related organisms lack the domain for binding a Rieske iron/sulfur cluster and are likely to have a different mechanism from those enzymes that have such domains.
FIG. 1.
Neighbor-joining distance tree of aryl-carotenoid-forming β-ring carotene desaturase/methyltransferase enzymes, CrtU and CruE. The CrtP sequences of Arabidopsis thaliana and Synechocystis sp. strain PCC 6803 are included as an outgroup. The bold lines indicate biochemically or genetically characterized enzymes.
Phylogenetic profiling (34) was employed to identify a second enzyme that is involved in synechoxanthin biosynthesis (see below). This enzyme, encoded by open reading frame SYNPCC7002_A2246 in Synechococcus sp. strain PCC 7002 and here named CruH, is related to the phenylpropionate dioxygenases, a family of ring-hydroxylating enzymes that contain domains which ligate Rieske iron-sulfur clusters. The distribution of CruH among sequenced cyanobacteria precisely matched the distribution of CruE. Furthermore, in two cases, Gloeobacter violaceus strain PCC 7421 and Trichodesmium erythraeum strain IMS101, the cruH gene was found in the same gene neighborhood as cruE. CruH from Synechococcus sp. strain PCC 7002 is 63% identical and 78% similar to its ortholog, Sll1869, in Synechocystis sp. strain PCC 6803, and it is 57% identical and 74% similar to its ortholog (CwatDRAFT_4939) in Crocosphaera watsonii.
Characterization of mutant strains.
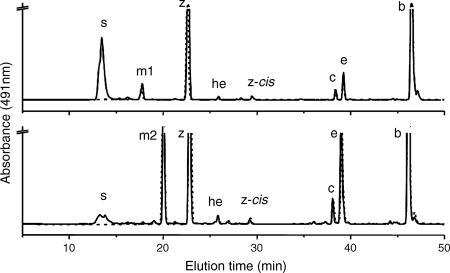
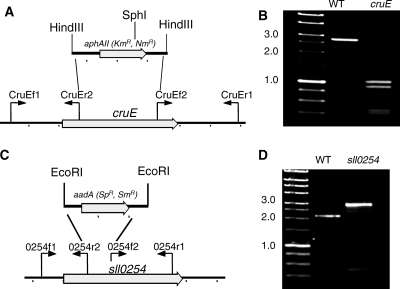
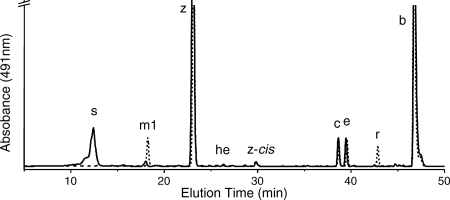
When carotenoids were isolated from exponentially growing wild-type Synechococcus sp. strain PCC 7002, seven major carotenoids were resolved (not counting geometrical isomers) (Fig. 2). Based on previous characterization studies as well as mass spectrometric analyses, these compounds corresponded to synechoxanthin, myxol-2′-fucoside, zeaxanthin, 3′-hydroxy-echinenone, cryptoxanthin, echinenone, and β-carotene (15, 16). These carotenoids were identical to those found in Synechocystis sp. strain PCC 6803, except that this organism synthesizes myxol-2′-dimethylfucoside instead of myxol-2′-fucoside (5, 15, 43). A fully segregated cruE disruption mutant (SYNPCC7002_A1248) was obtained in Synechococcus sp. strain PCC 7002 under standard growth conditions. The mutant was constructed using the primer pairs CruEf1/CruEr2 and CruEf2/CruEr1 (see Table S1 in the supplemental material), and segregation was verified using the primer pair CruEf1/CruEr1 (see Table S1 in the supplemental material). In this mutant the aphAII cartridge from pRL171 replaces 1,307 bp of the coding region of cruE (Fig. 3A). Digestion of the PCR amplicon from the mutant strain with SphI produced two fragments as expected and showed that the resulting mutant strain was homozygous (Fig. 3B).
FIG. 2.
HPLC elution profiles for pigments for two cyanobacteria. (A) HPLC elution profile for pigments extracted from the wild-type (solid line) and cruE (dotted line) mutant strains of Synechococcus sp. strain PCC 7002. (B) HPLC elution profile for pigments extracted from wild-type (solid line) and sll0254_/cruE_ (dotted line) mutant strains of Synechocystis sp. strain PCC 6803. s, synechoxanthin; m1, myxol-2′-fucoside; m2, myxol-2′-dimethylfucoside; z, zeaxanthin; he, 3′-hydroxy-echinenone; z-_cis, cis_-zeaxanthin; c, cryptoxanthin; e, echinenone; b, β-carotene.
FIG. 3.
Restriction maps and PCR verification of cruE mutants of two cyanobacteria. (A and C) Restriction maps showing construction of disruption mutations of the cruE gene of Synechococcus sp. strain PCC 7002 (A) and the sll0254_/cruE_ gene of Synechocystis sp. strain PCC 6803 (C). (B and D) Agarose gel electrophoresis of PCR amplicons from the wild-type (WT) and cruE mutant strains of Synechococcus sp. strain PCC 7002 (B) and wild-type and sll0254/cruE mutant strains of Synechocystis sp. strain PCC 6803 (D). In panel B the PCR amplicons were both digested with SphI to demonstrate the difference, because the inserted drug resistance cartridge was nearly identical in size to the DNA fragment that had been deleted. The data show that both mutant strains are completely segregated. Size markers are indicated at the left of panels B and D, and selected sizes in kb are indicated.
Although a previous study reported that sll0254 (cruE) was essential for viability and that a homozygous mutant with a mutation at this locus could not be obtained (27), we readily obtained a fully segregated sll0254 (cruE) disruption mutant of Synechocystis sp. strain PCC 6803 under standard growth conditions. The mutant was generated by using primer pairs 0254f1/0254r2 and 0254f2/0254r1, and segregation was verified by PCR amplification from genomic DNA using the primer pair 0254f1/0254r1 (see Table S1 in the supplemental material). In this mutant, the aadA cartridge from pSRA2, which lacks the strong transcription termination sequences of the Ω cartridge (14), was inserted via EcoRI restriction sites into the coding sequence, and it replaces 398 bp in the 5′ portion of the gene (Fig. 3C). Complete segregation was verified by PCR amplification from genomic DNA (Fig. 3D).
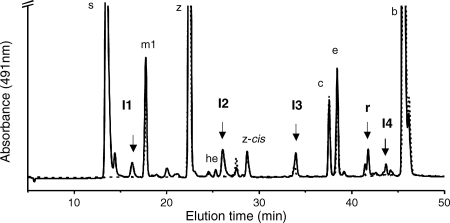
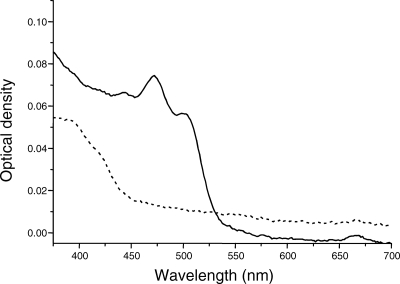
Mutants disrupted at the cruE loci in Synechococcus sp. strain PCC 7002 and Synechocystis sp. strain PCC 6803 did not produce detectable synechoxanthin, but otherwise these mutants had essentially normal carotenoid contents (Fig. 2). Furthermore, the growth rates of these mutants under standard conditions were indistinguishable from those of the corresponding wild-type strains (data not shown). When wild-type Synechococcus sp. strain PCC 7002 was grown to high cell density (OD730 of ∼14) and cells were harvested in stationary phase, several minor compounds that had absorption spectra similar to those of synechoxanthin and renierapurpurin could be identified, and all of these compounds were also missing from the carotenoids of the cruE mutant (Fig. 4). Thus, the cruE mutant was presumed to be incapable of producing χ,χ-carotenes. The spent growth medium of stationary-phase cultures of Synechococcus sp. strain PCC 7002 (OD730 of ∼10 to 14) accumulated an orange pigment (Fig. 5; see Fig. S1 in the supplemental material). This pigment was missing from the spent growth medium of stationary-phase cultures of the cruE mutant (Fig. 5). When the growth medium from wild-type cells was evaporated, extracted with methanol, and analyzed by HPLC, synechoxanthin was the major pigment from the extract (see Fig. S2 in the supplemental material).
FIG. 4.
HPLC elution profile of pigments extracted from stationary-phase cells of the wild-type (solid line) and cruE mutant (dotted line) strains of Synechococcus sp. strain PCC 7002. I1, I2, I3, and I4 indicate intermediates in the synthesis of synechoxanthin (s) (see text for details). r, renierapurpurin. The abbreviations used to identify other carotenoids are the same as in Fig. 2.
FIG. 5.
Absorption spectra of methanolic extracts of spent growth media recovered from stationary-phase cultures of the wild-type (solid line) and cruE mutant (dotted line) strains of Synechococcus sp. strain PCC 7002.
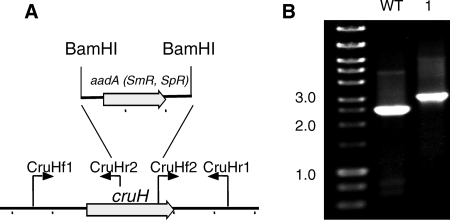
A cruH mutant of Synechococcus sp. strain PCC 7002 was constructed by interposon mutagenesis by using the primer pairs CruHf1/CruHr2 and CruHf2/CruHr1 (Fig. 6; see Table S1 in the supplemental material). Segregation was verified by PCR of genomic DNA using the primers CruHf1/CruHr1 (see Table S1 in the supplemental material; Fig. 6). Methanol extracts of the cruH mutant were analyzed by HPLC, and the mutant also failed to produce synechoxanthin but accumulated renierapurpurin (Fig. 7). This observation suggests that renierapurpurin is the substrate for CruH. Thus, CruH is postulated to act as the C-18 hydroxylase in at least the first step in the oxidation of renierapurpurin to synechoxanthin. It is possible that CruH performs sequential hydroxylations at the C-18 position to produce a carboxylic acid in the same manner that thymine hydroxylase performs three successive hydroxylation/oxidations (45).
FIG. 6.
Restriction map and PCR verification of the cruH mutant of Synechococcus sp. strain PCC 7002. (A) Restriction map showing the construction of the cruH mutant of Synechococcus sp. strain PCC 7002. (B) Agarose gel electrophoresis of amplicons from the cruH locus for the wild type (WT) and a cruH::aadA mutant (lane 1). The data show that the cruH and cruH::aadA alleles are fully segregated in the mutant strain.
FIG. 7.
HPLC elution profile of pigments extracted from stationary-phase cells of the wild-type (solid line) and cruH mutant (dotted line) strains of Synechococcus sp. strain PCC 7002. s, synechoxanthin; r, renierapurpurin. The designations of other compounds is the same as for Fig. 2.
Identification of synechoxanthin intermediates.
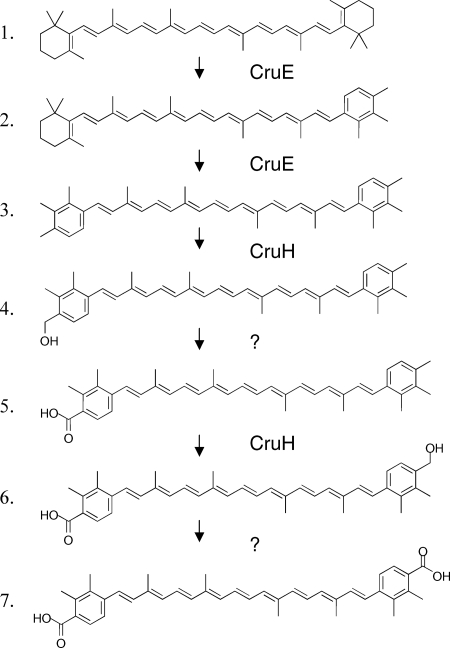
Including renierapurpurin, five compounds whose absorption spectra and elution times corresponded to rational intermediates for synechoxanthin biosynthesis were identified (Fig. 4 and 7). These intermediates are here referred to, in order of decreasing polarity, as I1, I2, I3, R (renierapurpurin), and I4. These intermediates were purified by HPLC and subjected to chemical and HPLC-mass spectrometry (HPLC-MS) analyses. Intermediate I1 had an absorbance spectrum and elution time that were identical to those of the partial reduction product of synechoxanthin (18′-hydroxy-χ,χ-caroten-18-oic acid) (see Fig. S3 and S4 in the supplemental material). I1 was unmodified by treatment with sodium borohydride as well as by saponification (data not shown). Intermediate I1 could, however, be modified by Sigma-Sil-A and was reduced by lithium aluminum hydride to a compound with a retention time and spectrum identical to those of the complete reduction product of synechoxanthin (χ,χ-carotene-18, 18′-diol) (see Fig. S4 in the supplemental material). A methyl ester could be formed from intermediate I1 (see Fig. S4 in the supplemental material). These observations indicated that intermediate I1 was identical to the partial reduction product of synechoxanthin and thus was 18′-hydroxy-χ,χ-caroten-18-oic acid. This was confirmed by HPLC-MS analysis, which showed that the molecular mass for this compound was 574 Da. Intermediate I2 was unmodified by treatment with sodium borohydride, by saponification, and by Sigma-Sil-A. Compound I2 could, however, be reduced with lithium aluminum hydride to form a compound with an elution time and spectrum identical to those of intermediate I3 (see Fig. S5 in the supplemental material). The molecular mass for intermediate I2 that was determined by HPLC-MS was 558 Da. Intermediate I3 was not modified by treatment with lithium aluminum hydride or sodium borohydride or by saponification, but this compound was modified by Sigma-Sil-A (see Fig. S6 in the supplemental material). Thus, intermediate I2 is χ,χ-caroten-18-oic acid, and intermediate I3 is 18-hydroxy-renierapurpurin (χ,χ-carotene-18-ol). Renierapurpurin (16) had an elution time of about 42 min and eluted ∼4 min earlier than β-carotene. Intermediate I4 had an elution time roughly midway between those of renierapurpurin and β-carotene (see Fig. S7 in the supplemental material); the spectrum of this compound had absorption maxima intermediate between those of renierapurpurin and β-carotene, and its elution time was similar to that of β,φ-carotene (see Fig. S7 in the supplemental material). Thus, we concluded that intermediate I4 was β,χ-carotene. Based on the observed intermediates, the biosynthetic pathway of synechoxanthin can be postulated to occur as follows (Fig. 8). CruE acts sequentially on the two rings of β-carotene to form β,χ-carotene and renierapurpurin (χ,χ-carotene). CruH is responsible for hydroxylation at the C-18 and C-18′ positions. Subsequently, an unidentified enzyme oxidizes 18-hydroxyrenierapurpurin to the carboxylic acid (χ,χ-caroten-18-oic acid), or CruH acts sequentially to oxidize the methyl group to the carboxylic acid as noted above. CruH then presumably acts on the second χ ring to form 18′-hydroxy-χ,χ-caroten-18-oic acid, which is subsequently oxidized to form synechoxanthin (Fig. 8). No intermediate corresponding to the diol (18,18′-dihydroxy-renierapurpurin) was found. This suggested that the end groups are sequentially oxidized in this cyanobacterium. Furthermore, no C-18 aldehyde derivatives were identified. This observation suggested either that the oxidation of the C-18 hydroxyl group to the carboxylic acid occurs in a single step or that the aldehyde intermediate is very rapidly converted to the corresponding carboxylic acid.
FIG. 8.
Proposed pathway for the conversion of β-carotene into synechoxanthin. The roles of CruE and CruH are indicated. For additional details, see the text. It should be noted that it is possible that CruH also carries out the hydroxylation/oxidation steps indicated by the two question marks.
Identification of synechoxanthin and synechoxanthin intermediates in other strains.
Based on elution time and absorption spectrum, synechoxanthin was identified by HPLC in the pigment extracts of several cyanobacteria. Synechoxanthin was found in Nostoc sp. strain MAC, Nostoc sp. strain PCC 7120, Synechococcus sp. strain C9, and Synechocystis sp. strain PCC 6308. Only small amounts of synechoxanthin accumulated in Synechococcus sp. strain C9 and G. violaceus strain PCC 7421. However, these two cyanobacteria accumulated compounds with elution times and spectra identical to those of compounds I2 (18′-hydroxy-χ,χ-caroten-18-oic acid) and I3 (χ,χ-caroten-18-oic acid) from Synechococcus sp. strain PCC 7002. These compounds were isolated from Synechococcus sp. strain C9 and analyzed by similar chemical means as described above, and it was found that the compounds had masses and chemical behaviors identical to those of the corresponding intermediate compounds isolated from Synechococcus sp. strain PCC 7002. Exponentially growing Anabaena variabilis strain ATCC 29413 did not contain significant amounts of synechoxanthin, although nearly stationary-phase cells did (15) (see Fig. S8 in the supplemental material). Similarly, Nostoc sp. strain PCC 7120 produced very small amounts of synechoxanthin when growing exponentially, but nearly stationary-phase cultures contained large amounts of synechoxanthin (15) (see Fig. S8 in the supplemental material). Although the genome of T. erythraeum strain IMS101 has homologs of cruE and cruH, extracts of this organism did not contain detectable amounts of synechoxanthin or synechoxanthin intermediates. These observations suggest that the production of synechoxanthin might occur only under specific growth conditions in some organisms that have the capacity to produce it.
DISCUSSION
The identification of genes involved in the production of synechoxanthin in Synechococcus sp. strain PCC 7002 establishes the biochemical basis for the production of aromatic carotenoids in cyanobacteria. The characterization of Synechococcus sp. strain PCC 7002 strains with mutations of cruE (SYNPCC7002_A1248) and of Synechocystis sp. strain PCC 6803 strains with mutations of its homolog, sll0254, demonstrated a role for the products of these genes in the aromatization of carotenoid β-rings. Although they were unable to produce a fully segregated mutant with a mutation at the sll0254 locus, Mohamed and Vermaas (27) had previously proposed that the product of this gene was a bifunctional lycopene cyclase/dioxygenase required for myxoxanthophyll biosynthesis in Synechocystis sp. strain PCC 6803. We previously showed that CruA and CruP and not Sll0254 are lycopene cyclases in Synechococcus sp. strain PCC 7002 and Synechocystis sp. strain PCC 6803 and that myxoxanthophyll is produced in cruE mutants of both strains (26). The failure of Mohamed and Vermaas (27) to obtain a fully segregated sll0254 mutant was likely due to a polarity effect on the downstream gene sll1586. In this work, the aadA cartridge from the plasmid PSRA2 was used (14). This plasmid is derived from pHP45Ω but has the strong transcriptional terminators removed and has been used repeatedly without causing polarity effects (12, 13). By eliminating the transcription terminators that flank the aadA gene of the Ω cartridge, we were able to obtain homozygous mutants with mutations at the same locus with the same drug resistance marker used by Mohamed and Vermaas (27). The product of the downstream sll1586 gene has strong sequence similarity to AsmA, which is important for outer membrane biogenesis in E. coli (10). This might also explain the membrane-related phenotypes described by Mohamed and Vermaas (10, 27). Pigment loss was likely due to cell death caused by increasing antibiotic concentration rather than loss of carotenoid cyclase activity as they suggested.
The complete loss of synechoxanthin and synechoxanthin intermediates in the cruE mutant strains of both Synechococcus sp. strain PCC 7002 and Synechocystis sp. strain PCC 6803 suggests that CruE acts at the branch point that leads to synechoxanthin biosynthesis. Unfortunately, attempts to demonstrate CruE activity directly by expressing the cruE gene in an Escherichia coli strain producing β-carotene have not yet been successful. However, the similarity of CruE to other carotenoid β-ring desaturase/methyltransferase enzymes, whose activities have been demonstrated biochemically (21), strongly supports its role in forming renierapurpurin from β-carotene. CruE is the first member of the carotene desaturase family that is involved in the synthesis of carotenoids with χ rings, which have methyl groups at the 1, 2, and 3 positions of the aryl ring. CrtU-type carotene desaturase/methyltransferases produce carotenoids with φ rings, which have methyl groups at the 1, 2, and 5 positions of the aryl rings. This difference presumably arises because the methytransferase activities of CrtU and CruE differ mechanistically.
The loss of synechoxanthin production in the cruH mutant, as well as the accumulation of renierapurpurin (χ,χ-carotene), establishes a role for CruH in the initial oxidation of renierapurpurin at the C-18 and C-18′ positions. Because CruH belongs to the phenylpropionate dioxygenase family of ring-hydroxylating dioxygenases, it is probable that CruH is the enzyme that performs this oxidation rather than a protein that only regulates this activity. Thus, CruH is probably the first enzyme capable of oxygenating the χ rings of carotenoids. Other organisms, including Thiocystis gelatinosa, can oxygenate carotenoids with χ rings at the C-18 position, but the enzymes that perform this reaction have not yet been identified (2).
Synechocystis sp. strain PCC 6803, which is one of the best-studied cyanobacteria, produces significant quantities of synechoxanthin (Fig. 2), and several other cyanobacteria also produce significant quantities of this carotenoid. Nevertheless, this compound went mostly undetected or at least unidentified for decades. This is probably due to the extremely polar and unusual chemical nature of this carotenoid. Bramley and Sandmann claimed to have systematically identified all of the carotenoids produced by Synechocystis sp. strain PCC 6803 as early as 1985 (5), but those authors did not describe any compound with the properties of synechoxanthin. Synechoxanthin is probably identical to the unidentified acidic carotenoid that was described by Omata and Murata and that was found in the “cell wall” fraction of Synechocystis sp. strain PCC 6714 (33). Furthermore, an extremely polar carotenoid was found to be a major pigment of the outer membrane of Synechocystis sp. strain PCC 6714, although it was absent from inner (cytoplasmic) and thylakoid membranes (18). Although they assigned the earliest-eluting carotenoid compound in wild-type Synechocystis sp. strain PCC 6803 as myxol, synechoxanthin is almost certainly the early-eluting compound shown in the chromatograms of Mohamed and Vermaas (28). In our hands, the only major carotenoids that eluted earlier than myxol-2′-dimethylfucoside from Synechocystis sp. strain PCC 6803 were synechoxanthin and myxol (from A. variabilis strain ATCC 29413). The latter had an elution time similar to that of myxol-2′-dimethylfucoside (19.5 min versus 20.0 min), albeit with a different HPLC system (15).
If synechoxanthin is specifically localized in the cell wall and outer membranes of Synechocystis spp., this localization suggests that this carotenoid likely plays some unique role in cyanobacterial physiology. This distribution would presumably be the same in Synechococcus sp. strain PCC 7002, and the presence of synechoxanthin in spent growth medium is consistent with this conclusion and further suggests that there must be some mechanism for the secretion of this compound. Although other roles cannot be excluded, the likely occurrence of synechoxanthin in cell walls suggests that this compound might act as a protective “sunscreen” for cells growing in high light intensity. Indeed, Synechococcus sp. strain PCC 7002 has the highest levels of synechoxanthin of all the cyanobacteria that we have analyzed, and this organism is known to be remarkably tolerant to high light intensity (31). Physiological studies, which will be reported elsewhere, indicate that the cruE and cruH mutants are more sensitive to high light intensity than the wild type and accumulate higher intracellular levels of reactive oxygen species after a 30-min exposure to high light intensity. For cyanobacteria in stationary phase, “turnover” methods of protection from oxidative stress are reduced, and synechoxanthin could provide an additional level of protection from radicals and lipid peroxides, which can otherwise induce catastrophic cell death through oxidation (36). Moreover, the nearly water-soluble nature of synechoxanthin should allow this compound to function as an antioxidant in molecular environments where few other carotenoids could be localized. Thus, synechoxanthin may provide a “chemical” solution to the problem of placing carotenoids into aqueous environments. This would be in contrast with “biochemical” solutions, e.g., the water-soluble, carotenoid-binding proteins (20), which could be an energetically more expensive solution to this problem for cells in stationary phase. Although the mechanism for this increase will require further study, the increased production of synechoxanthin is likely to be a common adaptive mechanism for cyanobacteria that produce this compound. It is not yet clear whether this increase is governed by high-density cell populations, by nutrient limitation, or by other physiological pressures exerted in stationary-phase laboratory cultures.
The production and secretion of synechoxanthin by cyanobacteria may in part explain the aromatic carotenoids of marine sponges. Sponges, like all other animals, are incapable of the de novo synthesis of carotenoids and must obtain these compounds from microbial symbionts or through modification of dietary carotenoids (7, 24). Marine sponges have an astounding array of associated cyanobacteria, including members of the genera Synechococcus, Prochlorococcus, and Oscillatoria among others (40). Therefore, it is possible that cyanobacterial symbionts are one source of the χ,χ-carotenes found in marine sponges, especially in light of the fact that cyanobacteria apparently can secrete synechoxanthin.
The distribution of CruE, or β-carotene desaturase/methyltransferase, in a variety of cyanobacteria, including the very early diverging strain G. violaceus strain PCC 7421, suggests that this enzyme has probably been inherited vertically in most strains that have the gene for this enzyme. If this is the case, then it is likely that the common ancestor of present-day cyanobacteria was capable of producing aromatic carotenoids. It has previously been assumed that χ,χ-carotene is produced by purple sulfur bacteria and that the diagenetic reduction products of this compound are thus reliable geochemical biomarkers for these organisms and for anoxic, sulfidic environments (6). However, the production of significant quantities of synechoxanthin by oxygen-evolving cyanobacteria in mid-exponential-phase cultures disproves this assumption and further suggests that the interpretation of χ,χ-carotanes in the geological record will be more complicated than initially presumed. In fact, many purple sulfur bacteria do not produce χ-carotenes at all and instead produce only acyclic carotenoids of the spirilloxanthin/spheroidene series (46). Given the coexistence of the myxoxanthophyll biosynthesis pathway and the synechoxanthin pathway in a variety of cyanobacteria, it is certainly possible that some cyanobacteria produce monocyclic χ-carotenoids (25). Although such compounds have not yet been reported, it should be noted that their diagenesis products would be similar to those produced from okenone, an important biomarker for purple sulfur bacteria. The interpretation of χ-ringed carotanes in the geological record should be very carefully evaluated. It may still be possible to distinguish diagenetic products of cyanobacteria and purple sulfur bacteria through specific carbon isotope ratios or distinct diagenetic pathways (7, 38). However, if alternate diagenesis pathways were to exist for synechoxanthin and renierapurpurin, this would also have significant implications for the interpretation of the biomarker signatures relating to these compounds. For instance, demethylation of the ring in renierapurpurin diagenetic products should be random, whereas demethylation of synechoxanthin products would likely be targeted because of the possibility of decarboxylation at the C-4 position (7). Attention to such details may increase the utility of χ,χ-carotenes as biomarkers and possibly help in answering which types of organisms have produced these compounds and at what times in evolutionary history. Obviously, it is impossible to verify which organisms had CrtU or CruE homologs billions of years ago. Because very few sequenced genomes of purple sulfur bacteria are currently available, homologs of CrtU and/or CruE from these organisms are unfortunately not yet known. Ongoing sequencing projects for cyanobacteria will probably reveal more CruE homologs. This will contribute new information on the distribution of aromatic carotenoids in nature and will possibly help to reveal the evolutionary history of these compounds. The discovery of the CruH oxygenase also raises some interesting possibilities. Under anoxic conditions, this enzyme presumably could not function, and thus the synechoxanthin pathway should shift toward the production of renierapurpurin. CruH is also likely to have homologs in purple sulfur bacteria such as Thiocystis gelatinosa, which produces carotenoids that are aryl esters (2). Further biochemical and genetic studies of a broader range of phototrophic prokaryotes should resolve these remaining questions.
Supplementary Material
[Supplemental material]
Acknowledgments
This research was supported by National Science Foundation grants MCB-0077586 and MCB-0519743 to D.A.B.
We gratefully acknowledge Eric Snyder and Li Zhang (The Pennsylvania State University) for assistance with the mass spectrometry. We thank Richard W. Castenholz (University of Oregon), John B. Waterbury (Woods Hole Oceanographic Institution), Theresa Thiel (University of Missouri-St. Louis), and Carlos Gomez-Lojero (Centro de Investigación y Estudios Avanzados del IPN, Mexico), who provided cyanobacterial cells or strains for analyses.
Footnotes
▿
Published ahead of print on 10 October 2008.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215403-410. [DOI] [PubMed] [Google Scholar]
- 2.Andrews, A. G., and S. Liaaen-Jensen. 1972. Carotenoids of Thiorhodaceae. 9. Stuctural elucidation of five minor carotenoids from Thiothece gelatinosa. Acta Chem. Scand. 262194-2204. [DOI] [PubMed] [Google Scholar]
- 3.Barber, J., and S. Iwata. 2005. Refined X-ray structure if photosystem II and its implications, p. 469-489. In T. Wydrzynski, and K. Satoh (ed.), Advances in photosynthesis and respiration, vol. 22. Photosystem II: the light driven water:plastiquinone oxidoreductase. Springer, Dordrecht, The Netherlands. [Google Scholar]
- 4.Boronowsky, U., S.-O. Wenk, D. Schneider, C. J.äger, and M. Rögner. 2001. Isolation of membrane protein subunits in their native state: evidence for selective binding of chlorophyll and carotenoid to the _b_6 subunit of the cytochrome b6f complex. Biochim. Biophys. Acta 150655-66. [DOI] [PubMed] [Google Scholar]
- 5.Bramley, P., and G. Sandmann. 1985. In vitro and in vivo biosynthesis of xanthophylls by the cyanobacterium Aphanocapsa. Phytochemistry 242919-2922. [Google Scholar]
- 6.Brocks, J., G. Love, R. E. Summons, A. H. Knoll, and G. A. Logan. 2005. Biomarker evidence for green and purple sulfur bacteria in a stratified paleoproterozoic sea. Nature 437866-870. [DOI] [PubMed] [Google Scholar]
- 7.Brocks, J. J., and P. Schaeffer. 2008. Okenane, a biomarker for purple sulfur bacteria (Chromatiaceae), and other new carotenoid derivatives from the 1640 Ma Barney Creek Formation. Geochem. Cosmochem. Acta 721396-1414. [Google Scholar]
- 8.Castenholz, R. W. 1981. Isolation and cultivation of thermophilic cyanobacteria, p. 236-246. In M. P. Starr, H. Stolp, H. G. Trüper, A. Balows, and H. G. Schlegel (ed.), The prokaryotes: a handbook on habitats, isolation and identification of bacteria. Springer, Berlin, Germany.
- 9.Cheng, Q. 2006. Structural diversity and functional novelty of new carotenoids biosynthesis genes. J. Ind. Microbiol. Biotechnol. 33552-559. [DOI] [PubMed] [Google Scholar]
- 10.Danese, P. N., and T. J. Sihavy. 1998. Targeting and assembly of periplasmic and outer membrane proteins in Escherichia coli. Annu. Rev. Genet. 3259-94. [DOI] [PubMed] [Google Scholar]
- 11.Eugester, C. 1995. Chemical derivitization: microscale tests for the prescence of common functional groups in carotenoids, p. 71-80. In G. Britton, S. Liaan-Jensen, and H. Pfander (ed.), Carotenoids, vol. 1A. Isolation and analysis. Birkhaueser Verlag, Basel, Switzerland. [Google Scholar]
- 12.Frigaard, N.-U., J. A. Maresca, C. E. Yunker, A. D. Jones, and D. A. Bryant. 2004. Genetic manipulation of carotenoid biosynthesis in the green sulfur bacterium Chlorobium tepidum. J. Bacteriol. 1865210-5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frigaard, N.-U., Y. Sakuragi, and D. A. Bryant. 2004. Gene inactivation in the cyanobacterium Synechococcus sp. PCC 7002 and the green sulfur bacterium Chlorobium tepidum using in vitro made DNA constructs and natural transformation. Methods Mol. Biol. 274324-330. [DOI] [PubMed] [Google Scholar]
- 14.Frigaard, N.-U., G. D. Voigt, and D. A. Byant. 2002. Chlorobium tepidum mutant lacking bacteriochlorophyll c made by inactivation of the bchK gene, encoding bacteriochlorophyll c synthase. J. Bacteriol. 1843368-3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graham, J. E. 2008. Carotenoid biosynthesis in Synechococcus sp. PCC 7002: identification of the enzymes and the carotenoids. Dissertation. The Pennsylvania State University, University Park, PA.
- 16.Graham, J. E., J. T. Lecomte, and D. A. Bryant. 2008. Synechoxanthin: an aromatic C40 xanthophyll is a major carotenoid in the cyanobacterium Synechococcus sp. PCC 7002. J. Nat. Prod. 711647-1650. [DOI] [PubMed] [Google Scholar]
- 17.Jordan, P., P. Fromme, H. T. Witt, O. Klukas, W. Sanger, and N. Krauss. 2001. Three-dimensional structure of cyanobacterial photosystem I at 2.5 Å resolution. Nature 411909-911. [DOI] [PubMed] [Google Scholar]
- 18.Jurgens, U. J., and J. Weckesser. 1985. Carotenoid-containing outer membrane of Synechocystis sp. strain PCC6714. J. Bacteriol. 164384-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karapetyan, N. V. 2007. Non-phtochemical quenching of fluorescence in cyanobacteria. Biochemistry (Moscow) 721127-1135. [DOI] [PubMed] [Google Scholar]
- 20.Kerfeld, C. A. 2004. Water-soluble carotenoid proteins of cyanobacteria. Arch. Biochem. Biophys. 4302-9. [DOI] [PubMed] [Google Scholar]
- 21.Krugel, H., P. Krubasik, K. Weber, H. Saluz, and G. Sandmann. 1999. Functional analysis of genes from Streptomyces griseus involved in the synthesis of isorenieratene, a carotenoid with aromatic end groups, revealed a novel type of carotenoid desaturase. Biochim. Biophys. Acta 143957-64. [DOI] [PubMed] [Google Scholar]
- 22.Leutwiler, L. S., and D. J. Chapman. 1981. Biosynthesis of aryl carotenoids: studies on the nature of the precyclization intermediate. Plant Cell Phys. 22781-787. [Google Scholar]
- 23.Liaaen-Jensen, S., and A. G. Andrews. 1972. Microbial carotenoids. Annu. Rev. Microbiol. 26225-248. [DOI] [PubMed] [Google Scholar]
- 24.Liaaen-Jensen, S., B. Renstrom, T. Ramdahl, M. Hallenstvet, and P. Berquist. 1982. Carotenoids of marine sponges Biochem. Syst. Ecol. 10167-174. [Google Scholar]
- 25.Maresca, J. A., J. E. Graham, and D. A. Bryant. 2008. The biochemical basis for structural diversity in the carotenoids of chlorophototrophic bacteria Photosynth. Res. 97121-140. [DOI] [PubMed] [Google Scholar]
- 26.Maresca, J. A., J. E. Graham, M. Wu, J. A. Eisen, and D. A. Bryant. 2007. Identification of a fourth family of lycopene cyclases in photosynthetic bacteria. Proc. Natl. Acad. Sci. USA 10411784-11789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mohamed, H. E., and W. F. Vermaas. 2006. Sll0254 (CrtLdiox) is a bifunctional lycopene cyclase/dioxygenase in cyanobacteria producing myxoxanthophyll. J. Bacteriol. 1883337-3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mohamed, H. E., and W. F. Vermaas. 2004. Slr1293 in Synechocystis sp. strain PCC 6803 is the C-3′,4′ desaturase (CrtD) involved in myxoxanthophyll bioynthesis. J. Bacteriol. 1865621-5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moshier, S. E., and D. J. Capman. 1973. Biosynthetic studies on aromatic carotenoids. Biochem. J. 136395-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murata, N., N. Sato, and Y. Kuwabara. 1981. Separation and characterization of thylakoid and cell envelop of the blue-green alga (cyanobacterium) Anacystis nidulans. Plant Cell Phys. 22855-866. [Google Scholar]
- 31.Nomura, C. T., T. Sakamoto, and D. A. Bryant. 2006. Roles for heme-copper oxidases in extreme high-light and oxidative stress response in the cyanobacterium Synechococcus sp PCC 7002. Arch. Microbiol. 185471-479. [DOI] [PubMed] [Google Scholar]
- 32.Omata, T., and N. Murata. 1983. Isolation and characterization of the cytoplasmic membranes from the blue-green alga (cyanobacterium) Anacystis nidulans. Plant Cell Phys. 241101-1112. [Google Scholar]
- 33.Omata, T., and N. Murata. 1984. Isolation and characterization of three types of membranes from the cyanobacterium Synechocystis PCC 6714. Arch. Microbiol. 139113-116. [Google Scholar]
- 34.Pellegrini, M., E. M. Marcotte, M. J. Thompson, D. Eisenberg, and T. O. Yeates. 1999. Assigning protein functions by comparative genome analysis: protein phylogenetic profiles Proc. Natl. Acad. Sci. USA 964285-4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pfander, H. 1976. Carotenoid glycosides. Pure Appl. Chem. 47121-128. [Google Scholar]
- 36.Sakamoto, T., V. B. Delgaizo, and D. A. Bryant. 1998. Growth on urea can trigger death and peroxidation of the cyanobacterium Synechococcus sp. strain PCC 7002. Appl. Environ. Microbiol. 642361-2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sandmann, G. 1994. Carotenoid biosynthesis in microoganisms and plants. Eur. J. Biochem. 2237-24. [DOI] [PubMed] [Google Scholar]
- 38.Scheaffer, P., P. Adam, P. Wehrung, and P. Albrecht. 1997. Novel aromatic carotenoid derivatives from sulfur photosynthetic bacteria in sediments. Tetrahedron Lett. 388413-8416. [Google Scholar]
- 39.Stanier, R., R. Kunisawa, M. Mandel, and G. Cohen-Bazire. 1971. Purification and properties of unicellular blue-green algae (order Chroococcales). Bacteriol. Rev. 35171-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steindler, L., D. Huchon, A. Avni, and M. Ilan. 2005. 16S rRNA phylogeny of sponge associated cyanobacteria. Appl. Environ. Microbiol. 714127-4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stevens, D., and R. Porter. 1980. Transformation of Agmenellum quadruplicatum. Proc. Natl. Acad. Sci. USA 776052-6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stickforth, P., S. Steiger, W. R. Hess, and G. Sandmann. 2003. A novel type of lycopene ɛ-cyclase in the marine cyanobacterium Prochlorococcus marinus MED4. Arch. Microbiol. 179409-415. [DOI] [PubMed] [Google Scholar]
- 43.Takaichi, S., Maoka, T., and K. Masamoto. 2001. Myxoxanthophyll in Synechocystis sp. PCC 6803 is myxol 2′-dimethyl-fucoside, (3R,2S)-myxol 2′-(2,4-di-O-methyl-α-L-fucoside), not rhamnoside. Plant Cell Physiol. 42756-762. [DOI] [PubMed] [Google Scholar]
- 44.Takaichi, S., M. Mochimaru, and T. Maoka. 2006. Presence of free myxol and 4-hydroxymyxol and the absence of myxol glycosides in Anabaena variabilis ATCC29413, and proposal of a biosynthetic pathway of carotenoids. Plant Cell Physiol. 47211-216. [DOI] [PubMed] [Google Scholar]
- 45.Thornburg, L. D., M.-T. Lai, J. S. Wishnok, and J. Stubbe. 1993. A non-heme iron protein with heme tendancies: an investigation of the substrate specificity of thymine hydroxylase. Biochemistry 3214023-14033. [DOI] [PubMed] [Google Scholar]
- 46.Toropygina, O. A., Z. K. Makhneva, and A. A. Moshalenko. 2005. Reconstituion of okenone into light harvesting complexes from Allochomatium minutissimum. Biochemistry (Moscow) 701231-1237. [DOI] [PubMed] [Google Scholar]
- 47.Wenk, S. O., D. Schnneider, U. Boronowsky, C. Jager, C. Klughammer, F. L. de Weerd, H. Roon, W. F. Vermaas, J. P. Dekker, and M. Rogner. 2005. Functional implications of pigments bound to a cyanobacterial b6f complex. FEBS J. 272582-592. [DOI] [PubMed] [Google Scholar]
- 48.Wilson, A., G. Ajlani, J. Verbatz, I. Vass, C. A. Kerfeld, and D. Kirilovsky. 2006. A soluble carotenoid protein involved in phycobilisome-related energy dissipation in cyanobacteria. Plant Cell 18992-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu, Y. P., and D. W. Krogman. 1997. The orange carotenoid protein of Synechocystis PCC 6803. Biochim. Biophys. Acta 13221-7. [DOI] [PubMed] [Google Scholar]
- 50.Zhang, H., D. Huang, and W. A. Cramer. 1999. Stoichiometrically bound β-carotene in the cytochrome b6f complex of oxygenic photosynthesis protects against oxygen damage. J. Biol. Chem. 2741581-1587. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental material]