Worldwide Distribution of HIV Type 1 Epitopes Recognized by Human Anti-V3 Monoclonal Antibodies (original) (raw)
Abstract
Epitopes, also known as antigenic determinants, are small clusters of specific atoms within macromolecules that are recognized by the immune system. Such epitopes can be targeted with vaccines designed to protect against specific pathogens. The third variable loop (V3 loop) of the HIV-1 pathogen's gp120 surface envelope glycoprotein can be a highly sensitive neutralization target. We derived sequence motifs for the V3 loop epitopes recognized by the human monoclonal antibodies (mAbs) 447-52D and 2219. Searching the HIV database for the occurrence of each epitope motif in worldwide viruses and correcting the results based on published WHO epidemiology reveal that the 447-52D epitope we defined occurs in 13% of viruses infecting patients worldwide: 79% of subtype B viruses, 1% of subtype C viruses, and 7% of subtype A/AG sequences. In contrast, the epitope we characterized for human anti-V3 mAb 2219 is present in 30% of worldwide isolates but is evenly distributed across the known HIV-1 subtypes: 48% of subtype B strains, 40% of subtype C, and 18% of subtype A/AG. Various assays confirmed that the epitopes corresponding to these motifs, when expressed in the SF162 Env backbone, were sensitively and specifically neutralized by the respective mAbs. The method described here is capable of accurately determining the worldwide occurrence and subtype distribution of any crystallographically resolved HIV-1 epitope recognized by a neutralizing antibody, which could be useful for multivalent vaccine design. More importantly, these calculations demonstrate that globally relevant, structurally conserved epitopes are present in the sequence variable V3 loop.
Introduction
An effective HIV vaccine with a B cell-mediated (humoral) component will present one or more epitopes (or epitope mimics) capable of eliciting broadly neutralizing antibodies from the naive host immune system. HIV-1 isolates have previously been classified genotypically into subtypes based on the nucleic acid sequences of HIV genes or the complete HIV genome. However, B cell epitopes are often formed from a few amino acids at discontinuous positions in the linear sequence of a protein. Therefore, sequence analysis does not necessarily reveal all, or even most, epitopes recognized by antibodies. In addition, multiple neutralization epitopes may occur within the same sequence region, but a single virus strain cannot belong to more than one genotype. Thus, genotype does not necessarily correlate with serotype, and this has previously been noted.1,2 To date, the only relationship that has been observed between virus genotype and neutralization sensitivity to various sera is that between subtypes B and E.3
From the point of view of developing a protective vaccine, reclassifying viruses according to the presence in their proteome of broadly neutralizing antibody epitopes is highly informative. Conversely, understanding the distribution of the epitope recognized by a particular neutralizing antibody among viruses causing the worldwide HIV-1 pandemic can help to establish the value of that particular antibody for vaccine design. Several broadly neutralizing monoclonal antibodies (mAbs) have been isolated and characterized in an effort to understand the molecular basis of broad neutralization. The epitopes recognized by several of these mAbs have been defined, and some epitopes have been resolved at the atomic level by X-ray crystallography.4
The V3 loop of gp120 contains several epitopes capable of inducing broadly neutralizing antibodies.5,6 Of all available anti-V3 mAbs, mAb 447-52D is the best characterized and exhibits both broadly binding7 and broadly neutralizing activity.8,9 Here, we attempted to precisely define the epitope “sequence motifs” of mAb 447-52D and 2219 according to their 3D structure. Bioinformatics was used to assess the presence of this sequence motif in the global population of HIV-1 sequences.
Materials and Methods
Overall method
Estimating the occurrence of the epitope recognized by a given mAb (in this case mAbs 447-52D and 2219) in the diversity of HIV-1 viruses infecting patients worldwide consists of three steps:
- Definition of the sequence motif of the epitope from the crystallographic structure: An epitope sequence motif can be derived from the 3D structure of the complex of a V3 peptide with the neutralizing antibody. This sequence motif is then tested for biologic relevance using a neutralization assay against V3 chimeric SF162 pseudoviruses (psVs) to determine whether the derived sequence motif, in the SF162 Env background, is neutralization-relevant. Based on the crystallographic and neutralization data, the presence of the defined sequence motif in any V3 sequence from any strain indicates that the virus strain contains the neutralization epitope for the antibody in question.
- Calculation of the occurrence of the sequence motif in the Los Alamos National Laboratory (LANL) database: The derived epitope sequence motif recognized by the mAb in question is used to search the LANL database of HIV-1 viral sequences to establish the percentage of recorded HIV-1 sequences that contains the epitope.
- Calculation of the worldwide occurrence of the epitope sequence motif: Since the distribution of subtypes in the LANL database is biased toward subtype B and does not match the actual distribution10–12 of subtypes causing the HIV pandemic, the percentage calculated in step 2 is converted to a more realistic estimate of the worldwide distribution of the epitope, and to a more realistic estimate of the distribution of the epitopes within each subtype.
Derivation of the sequence motifs from the crystal structures of mAbs 447-52D and 2219
The relevant amino acid positions were identified by analysis of the atomic contacts in the 3D structure of the V3MN peptide complexed with mAb 447-52D6 and mAb 2219.13 Graphic analysis and contact map extraction from the two crystal structures were performed using ICM-Pro (Molsoft LLC, La Jolla, CA).
Analysis of neutralization of psVs carrying the relevant sequence motifs of the epitopes recognized by mAbs 447-52D and 2219
For studies of psV neutralization sensitivity, each chimeric psV was constructed to contain a different V3 loop sequence grafted in to replace the V3 loop in the SF162 Env, where the V3 loop is relatively accessible (“unmasked”).14 Thus, the observed differences in psV neutralization elicited by each mAb map to differences in the V3 loop sequences inserted into the neutralization-sensitive SF162 envelope backbone where the V3 loop is relatively accessible (“unmasked”). A panel of 58 V3 chimeric SF162 psVs was constructed with V3 mutations introduced randomly in the consensus subtype B or consensus subtype C V3 loop, or V3 chimeric SF162 psVs were constructed in which the SF162 V3 loop was replaced with the V3 loop consensus sequences from subtypes B, C, F, H, or CRF01_AE (A/E). The neutralization by mAbs 447-52D and 2219 of each of these 58 psVs was assessed using methods previously described.15 Briefly, neutralizing activity was determined with a single-cycle infectivity assay using psVs generated with the _env_-defective luciferase-expressing pNL4-3.Luc.R-E- plasmid16 pseudotyped with the SF162 V3 variants described above. The psVs were incubated with serial dilutions of mAbs for 1.5 h at 37°C, and then added to CD4+CCR5+ U87 target cells plated in 96-well plates in the presence of polybrene (10 mg/ml). After 24 hrs, cells were re-fed with RPMI medium containing 10% FBS and 10 μg/ml polybrene, followed by an additional 24–48 h of incubation. Luciferase activity was determined 48–72 h postinfection with a microtiter plate luminometer (HARTA, Inc.) using assay reagents from Promega, Inc. Geometric mean titers for 50% neutralization (GMT50) were determined by interpolation from neutralization curves and are averages of at least three independent assays.
Statistical 3D structure tests of the predictive value of epitope sequence motifs
To estimate what percentage of LANL sequences that contain the epitope sequence motif was structurally compatible with each mAb, we modeled the 3D structures of the range of LANL V3 loop sequence variations into the 447-52D and 2219 mAb combining sites. A positive control set of V3 sequences containing the 447-52D-defined sequence motif and known experimentally to bind 447-52D (SBIND)7,17,18 was compared to a set of 100 sequences (SLANL) representative of the natural variation in V3 sequences from the LANL HIV sequence database (www.hiv.lanl.gov).19 SBIND was derived from random phage display studies18 or ELISA V3 loop peptide binding data.7,17 SLANL consists of the 100 most common sequences chosen with the “one sequence per patient” restriction applied to limit sampling bias. Our approach takes advantage of the fact that the phage display sequences included in SBIND are not constrained by viral fitness but exhaustively sample the mAb 447-52D combining site. We can thus compare the sample of unconstrained 447-52D bound phage display sequences to the sample of naturally occurring viral sequences found in LANL. Only the portion of the V3 loop sequence exhibiting contact with mAb 447-52D in the crystal structure was considered in the comparison.
The comparison was performed as follows: 3D homology models of the complex of mAb 447-52D with each of the SBIND and SLANL peptide sequences were constructed. Each model was energy minimized as previously described,20 and the distribution of binding energies for the two sets was compared. In each case, the energy of the uncomplexed peptide and mAb was separately calculated as the common reference state so that the energies could be compared across different complexes. _E_complex − (_E_free peptide + _E_free antibody) equaled the binding energy score used in our analysis. Terms for van der Waals contacts, hydrogen bonding, electrostatics, entropy, and solvation were included as previously described.21 This comparison resulted in two distributions of energy scores: one for known 447-52D binders and one unknown or test set of scores representing the LANL diversity. If the two distributions are similar, then the LANL set may be inferred statistically to be 447 binders. If the distributions are different, the LANL set may be inferred not to bind the antibody. The LANL set may also be the integration of two different populations, in which case the distribution will be similar to the known binders, but differ in one or the other tail of the distribution. This last scenario was the case, so we plotted the distributions to estimate the relative sizes of the two populations: a larger set of 447 binders and a smaller set of non-447 binders found in the tail of the distribution.
Phylogenetic test of the predictive value of epitope sequence motifs
Independently, we compared by sequence alignment the “width” or spread of SBIND and SLANL. A single multiple sequence alignment of all sequences in SBIND and SLANL was constructed using the pairwise alignment algorithm of Needleman and Wunsch22 with the standard gap open penalty of 2.4 and gap extension penalty of 0.15 to generate an initial alignment. This alignment was then adjusted according to the structure and the locations of the highly conserved GPG turn at the tip of the V3 loop and certain N-terminal conserved residues in order to ensure that divergent sequences were assigned to the correct residue and to the correct structural position. A phylogenetic tree was then constructed via the neighbor-joining method.23
There are two clear outcomes that are possible from this analysis. If the tree representing the diversity of SBIND has a larger “spread” within the tree than that representing the diversity of SLANL, then mAb 447-52D binds to a wider range of sequence variation than the that exhibited by the LANL HIV-1 sequences, indicating that 447 should bind all naturally occurring GPGR sequences. If, on the other hand, the spread of SLANL is wider than SBIND, then there are sequences that occur in nature containing a GPGR motif that may not bind to mAb 447-52D.
Results
Identification of the epitope “signature motif ” recognized by mAb 447-52D
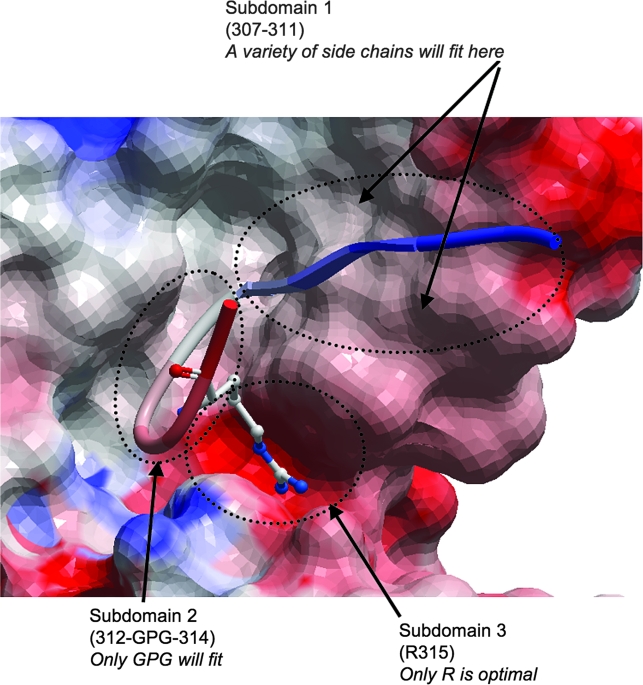
The protein interaction surface seen in the crystallographic structure of mAb 447-52D in complex with a V3 loop peptide from the MN strain of HIV-16 can be divided into three subdomains in terms of how snugly V3 loop side chains are bound by the antibody and how likely that side chain is to vary among HIV-1 viruses (Fig. 1). Subdomain 1 is a side-by-side β-strand hydrogen bonding interaction between the backbone atoms of the N-terminal β-strand of the V3 crown and the backbone atoms of a β-strand in the CDR3 of the mAb; in this subdomain, the area occupied by the side chains of the V3 residues is loose and will accommodate extensive sequence variation. Indeed, the sequence in this region of the V3 loop varies extensively. Subdomain 2 consists of the GPG β-turn, which fits snugly in the structure, but the GPG motif is nearly universal in HIV-1 isolates. Subdomain 3 consists of arginine 315 (R315) at the tip of the V3 loop. This arginine exhibits a snug shape and electrostatic complementarity with a deep pocket on the surface of the 447-52D antibody. Substitutions to any other side chain here would be expected to weaken the V3 loop:mAb interaction, perhaps substantially. This arginine is present in only a subset of viruses (mainly subtype B), so some viruses may be able to escape this antibody due to substitutions at this position. Therefore, the specific epitope sequence motif suggested by the mAb 447-52D complex structure with the V3 loop is R315.
FIG. 1.
Electrostatic surface of mAb 447-52D (red = negatively-charged surface, blue = positively-charged surface). The V3 crown peptide shown in ribbon depiction is colored from the N-terminus 307 position (blue) to the C-terminal 316 position (red) in a smooth gradient. R315 is shown in stick depiction in situ in the 447-52D electrostatic protein surface. The V3:mAb interaction surface is divided into three subdomains in terms of complementarity and how sequence variation would affect the interaction. Subdomain 1 is a side-by-side β-strand interaction between V3 and the mAb, and the area occupied by the V3 loop side chains (not shown, but pointing up and down perpendicular to the β-strand, see arrows) in this subdomain is loose and accommodating, so sequence variation in this subdomain is highly tolerated. Conversely, the surface enclosing the side chains of subdomains 2 and 3 demonstrates a tight shape and electrostatic complementarity, indicating poor tolerance for sequence substitution (which results in a side chain change) at these sites. However, the sequence in subdomain 2 (GPG) is nearly universal in HIV-1 isolates so the tight complementarity does not constrain the mAb to specific virus strains. The R315 of subdomain 3 is present in only a subset of viruses, and the shape and electrostatic complementarity of the pocket enclosing R315 are seen to be tight and unforgiving. Therefore the sequence-specific epitope motif of mAb 447-52D based on this structural analysis is R315.
Broad applicability of this definition of the epitope sequence motif to any documented V3 loop sequence
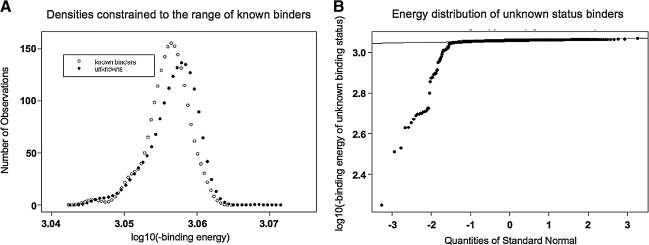
How many of the naturally occurring sequence variations recorded in LANL within the first subdomain of the V3 crown cannot fit into the 447-52D antibody-combining site? We compared peptide sequences that are known experimentally to bind 447-52D (SBIND) to a representative set of LANL sequences (SLANL) in order to make this determination. SBIND exhibits a normal (Gaussian) distribution of values. The same normal distribution with a similar mean is present in SLANL, suggesting the presence of a large subset of SLANL that is a true positive binder to the mAb (Fig. 2A). However, a transformation of the plot identifies an additional small population of subdomain 1 sequences in SLANL (Fig. 2B). This second distribution represents 7% of the LANL sequences that do not fit the mAb well, probably due to van der Waals clashes, backbone strain, or electrostatic incompatability. This 7% of LANL sequences may or may not bind the mAb, but cannot be predicted by this statistical method to bind. Thus, with statistical and 3D structural confidence, 93% of sequences with R315 in the LANL database fit the antigen-binding site of mAb 447-52D. It has yet to be determined whether the outlier 7% of V3 sequences represents artificial or biologically relevant sequences or whether, despite deviating energy scores, they still do bind the mAb efficiently, but this analysis establishes statistically that any naturally occurring V3 loop sequence containing the epitope motif, i.e., R315, has at least a 93% chance of containing a 447-52D-compatible structure in subdomain 1.
FIG. 2.
(A) Histogram plot of the logarithm of the binding energies (_x_-axis) calculated for peptides modeled into the 447-52D antibody surface of subdomain 1. Filled circles: peptide structures corresponding to the LANL sequence variation in subdomain 1. Open circles: peptide structures corresponding to the phage display sequence variation and in vitro ELISA tested peptides proven experimentally to bind to mAb 447-52D. Both distributions primarily form a normal or Gaussian distribution about a similar mean binding energy value. (B) Plot of expected observations occurring along a standard normal distribution (solid line) centered on the observed mean from (A), as plotted by standard deviations (_x_-axis). A tail of scores on the left indicates the presence of a subpopulation in LANL that deviates from the normal/Gaussian distribution. This subpopulation is predicted to be LANL subdomain 1 sequences that cannot be predicted statistically to bind mAb 447-52D and represents approximately 7% of R315 LANL sequences. This sequence population is characterized by the presence of phenylalanine, glutamate, glycine, and proline amino acids in subdomain 1.
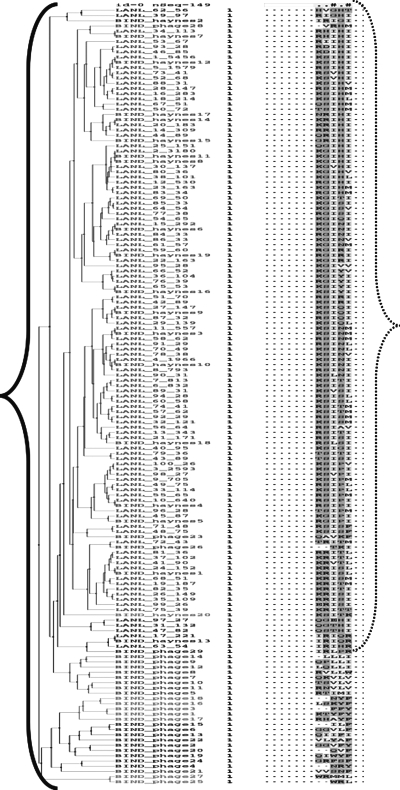
An independent phylogenetic comparison indicated that 100% of the naturally occurring sequences within subdomain 1 in strains carrying R315 are compatible with the 3D structure of the 447-52D antibody-combining site, because SLANL is distributed evenly and is completely contained within SBIND in a combined phylogenetic tree (Fig. 3). This finding suggests that the ability of mAb 447-52D to accommodate peptides carrying Arg at position 315 exceeds the diversity seen in naturally occurring isolates, whose sequence variation is subject to the constraints of infectivity and immune evasion. Thus, by sequence comparison, mAb 447-52D can accommodate 100% of all R315 V3 loop sequences.
FIG. 3.
Phylogenetic tree demonstrating the relationship between the sequences in subdomain 1 of SBIND and SLANL. SBIND members consist of phage display sequences from a previously published study18 and mAb 447-52D binding sequences,7,17 and are denoted by a prefix of “BIND_phage” and “BIND_,” respectively. SLANL are denoted by the pattern “LANL_ x_ y,” where x is the rank of the sequence by occurrence (1–100) and y is the number of times the sequence occurs in the database. Brackets (solid = SBIND; dashed =SLANL) indicate the “width” of evolutionary distance for each of the two sets. The names within the earliest or root 10 branches of the tree are colored alternatively and repetitively in shades of gray to highlight the branch groupings. Similarly, various shades of gray indicate amino acid conservation in the alignment.
Qualitatively, the convergent statistical/structural (93%) and phylogenetic (100%) studies indicate that any HIV-1 isolate that contains an arginine at position 315 (position 18 of the V3 loop) contains the epitope of mAb 447-52D regardless of sequence variation at other positions in the V3 loop (see Materials and Methods). So, the crystallographic structure suggests that the epitope motif is specific, and the bioinformatics suggest that the epitope motif is sensitive.
Functional a_ssessment of the neutralization relevance of the epitope sequence motif recognized by mAb 447-52D_
Binding does not necessarily correlate with neutralization, even in the absence of masking of the epitope, possibly due to a threshold affinity effect or artificial in vitro binding conditions. The presence of the 447-52D epitope sequence motif in a given virus strain even in the controlled SF162 background of this study does not necessarily mean it can be neutralized by mAb 447-52D. We thus sought to determine the neutralization relevance of the 447-52D epitope signature motif we have defined. A diverse set of V3 chimeric psVs was constructed using the SF162 Env. These V3 chimeric psVs varied only at residues in the V3 region and could be divided into two groups: one carrying R315 and the other carrying Q315 (almost every HIV-1 isolate that does not have arginine at this position has glutamine at this position). These V3 chimeric psVs were tested for their ability to be neutralized by mAb 447-52D. psVs with R315 were neutralized very well on average by mAb 447-52D, with a concentration range of 0.000078 to 0.067 μg/ml, while psVs with Q315 were neutralized much more weakly on average by mAb 447-52D in a concentration range of 0.025 to >20 μg/ml (Table 1). Statistically, the two sets are dramatically different (p = 0.00000002) indicating that R315 plays a sensitive and specific role in determining neutralization sensitivity to mAb 447-52D. This extends earlier studies of the effects on neutralization by mAb 447-52D of mutations at V3 position 315.24 Thus, the 3D structural information correlates very well with independent neutralization patterns. In combination with the statistical and phylogenetic analysis, the occurrence of R315 in any naturally occurring V3 loop sequence sensitively and specifically indicates the presence of a neutralization epitope recognized by mAb 447-52D.
Table 1.
A V3 Chimeric SF 162 psV in Which the V3 of SF 162 V3 Is Replaceda,b
| | Sequence change from consensus | 447 | | | ------------------------------------------------------- | ----------------- | ------- | | R315 | Consensus B | 0.00061 | | | Consensus B-R306A | 0.00050 | | | | Consensus B-K307A | 0.00038 | | | | Consensus B-K307E | 0.00033 | | | | Consensus B-I309M | 0.00037 | | | | Consensus B-I309V | 0.00021 | | | | Consensus B-H310P | 0.00048 | | | | Consensus B-H310P/I311M | 0.00078 | | | | Consensus B-I311L | 0.00026 | | | | Consensus B-I311M | 0.00026 | | | | Consensus B-N295V (no N-terminal glycan) | 0.000078 | | | | Consensus B-R298A | 0.00027 | | | | Consensus B-R298Q | 0.00037 | | | | Consensus B-N303I (no internal glycan) | 0.00027 | | | | Consensus B-P313A | 0.0011 | | | | Consensus B-R315K | 0.0068 | | | | Consensus B-T317A | 0.0013 | | | | Consensus B-D326N | 0.00027 | | | | Consensus B-D326A | 0.00026 | | | | Consensus B-T317A/E320D | 0.0024 | | | | Consensus B-T317A/Q327K | 0.00049 | | | | Consensus B-H308R/T317A/E320D | 0.0011 | | | | Consensus B-I307V/H308R/T317A/E320D | 0.00025 | | | | Consensus B-H308T/T317A/E320D (SF 162) | 0.067 | | | Q315 | Consensus B-R315Q | 0.40 | | | Consensus C | >20 | | | | Consensus C-R306A | >20 | | | | Consensus C-R306A/R301G | >20 | | | | Consensus C-R306E | >20 | | | | Consensus C-R306I/I309M/R310G | >20 | | | | Consensus C-R306I/K307Q/I309M/R310G | >20 | | | | Consensus C-K307A | 0.025 | | | | Consensus C-K307A/I311M | 6.5 | | | | Consensus C-K307E | 1.0 | | | | Consensus C-K307R/R310P/I311V | >20 | | | | Consensus C-S308G/R310G | >20 | | | | Consensus C-I309M | 0.55 | | | | Consensus C-I309M/I311L (N295V-no N-terminal glycan) | >20 | | | | Consensus C-I309M/R310P/T316A | >20 | | | | Consensus C-I309M/R310S/T316A | >20 | | | | Consensus C-I309M (N295V-no N-terminal glycan) | 0.13 | | | | Consensus C-I309V | 0.76 | | | | Consensus C-R310N/I311F/T316A | >20 | | | | Consensus C-R310P/T316A | >20 | | | | Consensus C-R310S/T316A | >20 | | | | Consensus C-R310T/I311F/T316A | >20 | | | | Consensus C-R310T/I311L/T316A | >20 | | | | Consensus C-I311F | 0.96 | | | | Consensus C-I311L | >20 | | | | Consensus C-I311L (N295V-no N-terminal glycan) | >20 | | | | Consensus C-I311M | 12.1 | | | | Consensus C-I311V | 0.45 | | | | Consensus C-T316A | 0.58 | | | | Consensus C-N295V (no N-terminal glycan) | 7.7 | | | | Consensus C-H308T/R313Q/A314T/T317A/E320D | >20 | | | | Consensus F | 0.27 | | | | Consensus H | 15.9 | | | | Consensus A/E | 1.2 | |
The sequence motif for the neutralization epitope recognized by mAb 447-52D is therefore confirmed to be the following:
If position 315 in gp120 (or position 18 in the V3 loop) is equal to arginine, a neutralization-sensitive 447-52D epitope is present in the third variable loop of the isolate. Otherwise, the neutralization of the isolate will be considerably weaker, and the isolate may qualitatively be classified as lacking the 447-52D neutralization epitope.
We searched the LANL database with the 447-52D neutralization epitope sequence motif and found that 79% of subtype B sequences contain the R315 sequence motif along with 1% of subtype C and 7% of subtype A/AG viruses. It is highly unlikely that these LANL sequences are biased toward or against R315 as subtype assignment in the LANL HIV database is made independently based on the complete viral genome sequence. These percentages are thus predictive of the occurrence of this epitope in the worldwide distribution of each subtype.
Osmanov et al.25 estimated that the year 2000 worldwide proportion of HIV-1 viruses that are subtypes B, C, or A/AG was 12%, 47%, and 27%, respectively. These three subtypes therefore represented 86% of the viruses causing the HIV-1 pandemic in the year 2000. We therefore estimated the occurrence of the 447-52D epitope sequence motif in these three subtypes as a proxy for the epitope's occurrence throughout the true global distribution of strains: (79% of the 12% of global viruses that are subtype B) + (1% of the 47% of the global viruses that are subtype C) + (7% of the 27% of the global viruses that are subtype A/AG).
According to this calculation, a total of 12% of subtypes A, B, and C HIV-1 viruses contain the 447-52D neutralization-relevant epitope sequence motif, consisting primarily of subtype B viruses. Extrapolating to all subtypes (12%/86%) gives a 14% occurrence of this epitope in worldwide isolates. Since a minimum of 93% of these should bind the mAb (see above), the final calculation is that the neutralization-relevant epitope sequence motif of mAb 447-52D is present in approximately 13% of worldwide isolates.
Identification of the epitope sequence motif recognized by mAb 2219 and functional a_ssessment of the accuracy of this epitope sequence motif_
The same analysis was performed to identify the epitope sequence motif recognized by mAb 2219. This demonstrates the following:
- Analysis of the structure of the complex (not shown) suggests that the sequence motif of the epitope recognized by mAb 2219 is a lysine at position 307 (K307), an isoleucine at position 309 (I309), and a tyrosine at position 318 (Y318). The isoleucine at position 311 also appears to restrict the epitope; however, it is replaced by a leucine in one of the peptides in the published crystallographic complex at this position. Energetic analyses also suggest that several amino acids could fit in this position (not shown). Therefore, for this prototype analysis, we have excluded position 311 from the motif.
- V3 chimeric SF162-based psVs containing the epitope sequence motif recognized by mAb 2219 are neutralized on average by mAb concentrations well below 0.05 μg/ml (ranging from 0.00042 to 20 μg/ml) while those deviating from the motif are mostly not neutralized at all (range from 0.054 to 20 μg/ml; p = 0.000000062; Table 2). So, the defined 2219 epitope motif is neutralization relevant.
- In the LANL database 48% of subtype B, 40% of subtype C, and 18% of subtype A/AG viruses have the K307-I309-Y318 mAb 2219 epitope sequence motif.
Table 2.
A Chimeric V3 Chimeric SF 162 psV as Described for Table 1a,b
| | Sequence change from consensus | 2219 | | | ---------------------------------------------- | ----------------------------- | ------ | | K10/I12/Y21 | Consensus B | 0.0011 | | | Consensus B-R306A | 0.0007 | | | | Consensus B-I309M | 0.012 | | | | Consensus B-I309V | 0.0012 | | | | Consensus B-I311L | 0.00066 | | | | Consensus B-I311M | 0.001 | | | | Consensus B-N295V, no N-terminal glycan | 0.00071 | | | | Consensus B-R298A | 0.00053 | | | | Consensus B-R298Q | 0.00053 | | | | Consensus B-N303I (removed internal glycan) | 0.00042 | | | | Consensus B-P313A | 0.00056 | | | | Consensus B-R315K | 0.00076 | | | | Consensus B-R315Q | 0.0012 | | | | Consensus B-T317A | 0.0023 | | | | Consensus B-D326N | 0.00084 | | | | Consensus B-D326A | 0.0012 | | | | Consensus B-T317A; E320D | 0.0041 | | | | Consensus B-T317A; Q327K | 0.00061 | | | | Consensus B-H308R; T317A; E320D | 0.001 | | | | Consensus B-I307V; H308R; T317A; E320D | 0.00052 | | | | Consensus B-H308T; T317A; E320D (SF 162) | 0.0044 | | | | Consensus C | 0.15 | | | | Consensus F | 0.0034 | | | | Consensus H | 0.21 | | | | Consensus A/E | 0.23 | | | | Consensus C-R310P/T316A | 5 | | | | Consensus C-R310S/T316A | 1.2 | | | | Consensus C-T316A | 0.029 | | | | Consensus C-N295V, no N-terminus glycan | 0.035 | | | | Consensus C-H308T R313Q;A314T;T317A; E320D | 6.6 | | | | Consensus C-R310N/I311F/T316A | 0.036 | | | | Consensus C-R310T/I311F/T316A | 0.075 | | | | Consensus C-R310T/I311L/T316A | 18.9 | | | | Consensus C-I311F | 0.1 | | | | Consensus C-I311L | 2.3 | | | | Consensus C-I311L; (N295V) | 0.4 | | | | Consensus C-I311M | 0.034 | | | | Consensus C-I311V | 0.01 | | | | Consensus C-S308G/R310G | >20 | | | | Consensus C-R306A/R310G | >20 | | | Non-K10/I12/Y21 | Consensus C-R306I/I309M/R310G | >20 | | | Consensus C-R9I/K10Q/I12M/R13G | >20 | | | | Consensus C-K307A | >20 | | | | Consensus C-K307A/I311M | >20 | | | | Consensus C-K307E | >20 | | | | Consensus C-K307R/R1310P/I311V | >20 | | | | Consensus C-I309M | 0.23 | | | | Consensus C-I309M/I311L; (N295V) | >20 | | | | Consensus C-I309M/R310P/T316A | >20 | | | | Consensus C-I309M/R310S/T316A | >20 | | | | Consensus C-I309M; (N295V) | 0.14 | | | | Consensus C-I309V | 0.054 | |
Thus, the worldwide occurrence of the neutralization epitope recognized by mAb 2219, calculated the same way as that for mAb 447-52D, is 30%. However, in this case, the epitope is relatively evenly distributed across the subtypes.
Assessment of the occurrence and distribution of the signature motifs recognized by mAbs 447-52D and 2219 combined in the global HIV-1 population
Using the same analyses as described above, 37% of worldwide isolates were found to contain either the 447-52D or the 2219 epitope signature motifs: 90% of subtype B isolates, 40% of subtype C isolates, and 24% of subtype A/AG isolates.
Discussion
According to the calculations described herein, V3 loop neutralization epitopes recognized by either mAbs 447-52D or 2219 are conserved in 37% of HIV-1 viruses infecting patients worldwide despite the sequence variation observed in the V3 loop. This implies that a vaccine capable of generating both the 447-52D- and 2219-like antibodies in humans would potentially be capable of neutralizing 37% of viruses worldwide, across all subtypes. However, realizing this potential would mean that all HIV-1 isolates worldwide would present the V3 loop to the human antibody response with accessibility that is comparable to SF162. In reality, the accessibility of the V3 loop in worldwide isolates is highly variable and, unfortunately, most of the time V3 is less available (more masked) than it is in the SF162 envelope, at least partly due to glycosylation of the envelope. Thus, the actual percentage of worldwide isolates that could be neutralized by these antibodies is likely to be much lower than that represented by these calculations. It remains to be seen to what extent these masking effects can be overcome by the induction of relatively high levels of these antibodies and/or the induction of antibodies with higher affinities. Nevertheless, we cannot neutralize an epitope that isn't there, so this study establishes a rational baseline for the comparative utility of these antibodies, and the method described can be applied to other epitopes recognized by neutralizing mAbs.
Thus, using data generated from bioinformatics, crystallography, epidemiology, and viral neutralization studies, we developed an approach for measuring the degree of conservation of neutralization epitopes in a variable region of the gp120 envelope glycoprotein of diverse, globally relevant strains of HIV-1. This first version of the method depends on the fidelity of several techniques. The percentages we have calculated are only first estimates that may very well underestimate or overestimate the breadth of 447 and 2219, and the accuracy of the method may be improved in subsequent versions.
First, although a major advantage of this method is that it reduces a complex structural interaction to a sequence motif, V3 loop crown structures may contain a sequence motif, but due to backbone folding, may not fit the combining site of some antibodies, and vice versa (ones that do not contain the sequence motif but fold into a perfect shape for binding through weaker contacts). Indeed, some of the outlier sequences in Table 2 have been determined to be resistant to neutralization due to folding effects despite the presence of the sequence motif (data not shown). Methods to incorporate backbone effects may improve the estimates.
Second, the method depends on neutralization assays and correlates 3D structural observations directly with neutralization patterns without using potentially noisy V3 loop-antibody binding data as an intermediary. However, some Q315 V3 loop peptides have been observed to bind 447-52D,7 and a few are neutralized relatively well compared to the average for Q315 viruses. A better understanding of the relationship between V3 loop-antibody binding observations and V3 loop-mediated neutralization may provide further refinement of the motif to include some Q315 viruses in the definition of the 447-52D epitope. In addition, although the SF162 chimeric pseudovirus system used partitioned the data sufficiently in this case, global interactions between non-V3 and V3 positions in the gp120 monomer and trimer may still have biased these results. A better understanding of these effects may improve the results.
Third, the LANL database is a biased representation of the worldwide distribution of HIV-1 isolates. Improvements in this database to reduce sampling bias may increase the precision of the estimates derived by our method.
Fourth, the method depends on the accuracy of epidemiologic estimates of the global distribution of subtypes. As these estimates become more precise and/or change over time due to virus evolution,12 the epidemiologic relevance of these calculations may improve. Indeed, greater detail may result from calculating the distribution in every defined subtype or from the subtype distribution in specific geographic regions instead of extrapolating from just the three subtypes (A/AG, B, and C) that currently make up 86% of the worldwide pandemic as we did here.
Fifth, the method depends on our statistical and phylogenetic techniques to precisely assess how well all the LANL sequences corresponding to the derived 3D motif fit the mAb under study. Novel methods to make this assessment may improve the precision of the calculation.
Finally, the method also depends on the quality of the crystal structures and psV neutralization assay data. With additional structural and viral data, the precision and relevance of the calculations may be improved.
The method described here is applicable to any crystallographically resolved mAb/peptide epitope complex—including those in sequence variable, surface exposed regions on any pathogen—and it clearly clusters known HIV-1 viruses into vaccine-relevant groups that bear little or no relationship to the subtype designations of viral groups based on genotyping (Fig. 4). This in silico serotyping is relevant to vaccine design because it rapidly allows comparison of promising neutralizing antibodies that can be studied for the rational engineering of protective antibody responses. Upon application to epitopes located in several regions of gp120, this method may serve as a tool for the rational design of multivalent neutralizing antibody-based vaccines, which will protect against the maximum proportion of HIV-1 strains while targeting the minimum number of epitopes.
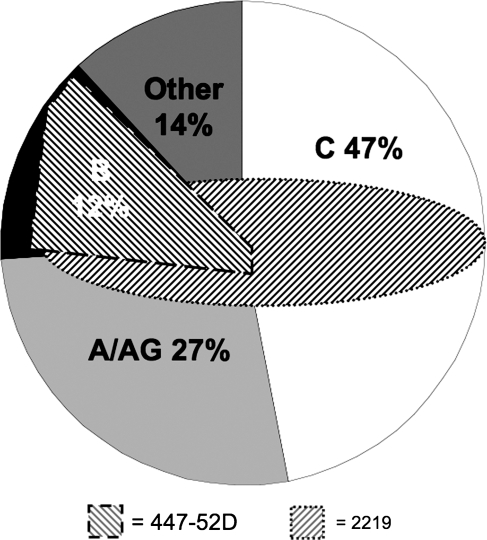
FIG. 4.
Worldwide HIV-1 clade distribution. This diagram depicts the estimated global distribution of HIV-1 genetic subtypes in the year 2000.25 Hatched shape overlays illustrate the theoretical coverage of the two mAbs (447-52D and 2219) calculated in this work. The areas do not correspond exactly to the numbers calculated and are for illustrative purposes only.
Acknowledgments
The authors would like to thank Dr. Xiang-Peng Kong and Dr. Catarina Hioe for helpful discussions. Dr. Jennifer Fuller provided helpful comments in forming and editing the manuscript. The work was supported by grants from the Bill and Melinda Gates Foundation (#38631), the NIH, including DP2 OD004631 (TC), AI36085 (SZP), HL59725 (SZP), AI46283 (AP), and AI27742 (NYU Center for AIDS Research), and research funds from the Department of Veterans Affairs.
Disclosure Statement
No competing financial interests exist.
References
- 1.Barin F. Lahbabi Y. Buzelay L. Lejeune B. Baillou-Beaufils A. Denis F. Mathiot C. M'Boup S. Vithayasai V. Dietrich U. Goudeau A. Diversity of antibody binding to V3 peptides representing consensus sequences of HIV type 1 genotypes A to E: An approach for HIV type 1 serological subtyping. AIDS Res Hum Retroviruses. 1996;12(13):1279–1289. doi: 10.1089/aid.1996.12.1279. [DOI] [PubMed] [Google Scholar]
- 2.Nyambi PN. Nadas A. Mbah HA. Burda S. Williams C. Gorny MK. Zolla-Pazner S. Immunoreactivity of intact virions of human immunodeficiency virus type 1 (HIV-1) reveals the existence of fewer HIV-1 immunotypes than genotypes. J Virol. 2000;74(22):10670–10680. doi: 10.1128/jvi.74.22.10670-10680.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mascola JR. Louwagie J. McCutchan FE. Fischer CL. Hegerich PA. Wagner KF. Fowler AK. McNeil JG. Burke DS. Two antigenically distinct subtypes of human immunodeficiency virus type 1: Viral genotype predicts neutralization serotype. J Infect Dis. 1994;169(1):48–54. doi: 10.1093/infdis/169.1.48. [DOI] [PubMed] [Google Scholar]
- 4.Pantophlet R. Burton DR. GP120: Target for neutralizing HIV-1 antibodies. Annu Rev Immunol. 2006;24:739–769. doi: 10.1146/annurev.immunol.24.021605.090557. [DOI] [PubMed] [Google Scholar]
- 5.Gorny MK. Xu JY. Karwowska S. Buchbinder A. Zolla-Pazner S. Repertoire of neutralizing human monoclonal antibodies specific for the V3 domain of HIV-1 gp120. J Immunol. 1993;150(2):635–643. [PubMed] [Google Scholar]
- 6.Stanfield RL. Gorny MK. Williams C. Zolla-Pazner S. Wilson IA. Structural rationale for the broad neutralization of HIV-1 by human monoclonal antibody 447-52D. Structure. 2004;12(2):193–204. doi: 10.1016/j.str.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 7.Zolla-Pazner S. Gorny MK. Nyambi PN. VanCott TC. Nadas A. Immunotyping of human immunodeficiency virus type 1 (HIV): An approach to immunologic classification of HIV. J Virol. 1999;73(5):4042–4051. doi: 10.1128/jvi.73.5.4042-4051.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gorny MK. Conley AJ. Karwowska S. Buchbinder A. Xu JY. Emini EA. Koenig S. Zolla-Pazner S. Neutralization of diverse human immunodeficiency virus type 1 variants by an anti-V3 human monoclonal antibody. J Virol. 1992;66(12):7538–7542. doi: 10.1128/jvi.66.12.7538-7542.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gorny MK. Williams C. Volsky B. Revesz K. Cohen S. Polonis VR. Honnen WJ. Kayman SC. Krachmarov C. Pinter A. Zolla-Pazner S. Human monoclonal antibodies specific for conformation-sensitive epitopes of V3 neutralize human immunodeficiency virus type 1 primary isolates from various clades. J Virol. 2002;76(18):9035–9045. doi: 10.1128/JVI.76.18.9035-9045.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Osmanov S. Pattou C. Walker N. Schwardlander B. Esparza J. Estimated global distribution and regional spread of HIV-1 genetic subtypes in the year 2000. J Acquir Immune Defic Syndr. 2002;29(2):184–190. doi: 10.1097/00042560-200202010-00013. [DOI] [PubMed] [Google Scholar]
- 11.Buonaguro L. Tornesello ML. Buonaguro FM. Human immunodeficiency virus type 1 subtype distribution in the worldwide epidemic: Pathogenetic and therapeutic implications. J Virol. 2007;81(19):10209–10219. doi: 10.1128/JVI.00872-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hemelaar J. Gouws E. Ghys PD. Osmanov S. Global and regional distribution of HIV-1 genetic subtypes and recombinants in 2004. AIDS. 2006;20(16):W13–23. doi: 10.1097/01.aids.0000247564.73009.bc. [DOI] [PubMed] [Google Scholar]
- 13.Stanfield RL. Gorny MK. Zolla-Pazner S. Wilson IA. Crystal structures of human immunodeficiency virus type 1 (HIV-1) neutralizing antibody 2219 in complex with three different V3 peptides reveal a new binding mode for HIV-1 cross-reactivity. J Virol. 2006;80(12):6093–6105. doi: 10.1128/JVI.00205-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krachmarov CP. Honnen WJ. Kayman SC. Gorny MK. Zolla-Pazner S. Pinter A. Factors determining the breadth and potency of neutralization by V3-specific human monoclonal antibodies derived from subjects infected with clade A or clade B strains of human immunodeficiency virus type 1. J Virol. 2006;80(14):7127–7135. doi: 10.1128/JVI.02619-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krachmarov CP. Kayman SC. Honnen WJ. Trochev O. Pinter A. V3-specific polyclonal antibodies affinity purified from sera of infected humans effectively neutralize primary isolates of human immunodeficiency virus type 1. AIDS Res Hum Retroviruses. 2001;17(18):1737–1748. doi: 10.1089/08892220152741432. [DOI] [PubMed] [Google Scholar]
- 16.Connor RI. Chen BK. Choe S. Landau NR. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995;206(2):935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 17.Haynes BF. Ma B. Montefiori DC. Wrin T. Petropoulos CJ. Sutherland LL. Scearce RM. Denton C. Xia SM. Korber BT. Liao HX. Analysis of HIV-1 subtype B third variable region peptide motifs for induction of neutralizing antibodies against HIV-1 primary isolates. Virology. 2006;345(1):44–55. doi: 10.1016/j.virol.2005.08.042. [DOI] [PubMed] [Google Scholar]
- 18.Keller PM. Arnold BA. Shaw AR. Tolman RL. Van Middlesworth F. Bondy S. Rusiecki VK. Koenig S. Zolla-Pazner S. Conard P, et al. Identification of HIV vaccine candidate peptides by screening random phage epitope libraries. Virology. 1993;193(2):709–716. doi: 10.1006/viro.1993.1179. [DOI] [PubMed] [Google Scholar]
- 19.Leitner T. Foley B. Hahn B. Marx P. McCutchan FE. Mellors J. Wolinsky S. Korber B. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory; Los Alamos, NM: 2005. HIV Sequence Compendium. LA-UR 04-7420. [Google Scholar]
- 20.Cardozo T. Totrov M. Abagyan R. Homology modeling by the ICM method. Proteins. 1995;23(3):403–414. doi: 10.1002/prot.340230314. [DOI] [PubMed] [Google Scholar]
- 21.Abagyan R. Totrov M. Biased probability Monte Carlo conformational searches and electrostatic calculations for peptides and proteins. J Mol Biol. 1994;235(3):983–1002. doi: 10.1006/jmbi.1994.1052. [DOI] [PubMed] [Google Scholar]
- 22.Needleman SB. Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48(3):443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 23.Saitou N. Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 24.Zolla-Pazner Zhong P. Revesz K, et al. The cross-clade neutralizing activity of a human monoclonal antibody is determined by the GPGR V3 motif of HIV type 1. AIDS Res Hum Retroviruses. 2004;20(11):1254–1258. doi: 10.1089/aid.2004.20.1254. [DOI] [PubMed] [Google Scholar]
- 25.Osmanov S. Pattou C. Walker N, et al. Estimated global distribution and regional spread of HIV-1 genetic subtypes in the year 2000. J Acquir Immune Defic Syndr. 2002;29(2):184–190. doi: 10.1097/00042560-200202010-00013. [DOI] [PubMed] [Google Scholar]