Aurora-A Expression Is Independently Associated with Chromosomal Instability in Colorectal Cancer (original) (raw)
Abstract
AURKA (the official symbol for Aurora-A, STK15, or BTAK) regulates the function of centrosomes, spindles, and kinetochores for proper mitotic progression. AURKA overexpression is observed in various cancers including colon cancer, and a link between AURKA and chromosomal instability (CIN) has been proposed. However, no study has comprehensively examined AURKA expression in relation to CIN or prognosis using a large number of tumors. Using 517 colorectal cancers in two prospective cohort studies, we detected AURKA overexpression (by immunohistochemistry) in 98 tumors (19%). We assessed other molecular events including loss of heterozygosity (LOH) in 2p, 5q, 17q, and 18q, the CpG island methylation phenotype (CIMP), and microsatellite instability (MSI). Prognostic significance of AURKA was evaluated by Cox regression and Kaplan-Meier method. In both univariate and multivariate logistic regressions, AURKA overexpression was significantly associated with CIN (defined as the presence of LOH in any of the chromosomal segments; multivariate odds ratio, 2.97; 95% confidence interval, 1.40–6.29; P = .0045). In multivariate analysis, AURKA was associated with cyclin D1 expression (P = .010) and inversely with PIK3CA mutation (_P_=.014), fatty acid synthase expression (_P_=.028), and family history of colorectal cancer (P = .050), but not with sex, age, body mass index, tumor location, stage, CIMP, MSI, KRAS, BRAF, BMI, LINE-1 hypomethylation, p53, p21, β-catenin, or cyclooxygenase 2. AURKA was not significantly associated with clinical outcome or survival. In conclusion, AURKA overexpression is independently associated with CIN in colorectal cancer, supporting a potential role of Aurora kinase-A in colorectal carcinogenesis through genomic instability (rather than epigenomic instability).
Introduction
Chromosomal instability (CIN) in cancer is characterized by frequent chromosomal abnormalities including translocations, gains, and losses of chromosomes or their segments. Chromosomal instability promotes carcinogenesis through loss of tumor suppressors and copy number gains of oncogenes [1]. Causes of CIN are still poorly understood but possibly include mitotic spindle checkpoint gene (e.g., BUB1 and BUB1B) deregulation [2], DNA checkpoint gene (e.g., TP53) mutation [3], cell cycle regulator (e.g., FBXW7) inactivation [4], telomere dysfunction [5], and abnormal centrosome number and function [6–8]. Centrosome dysfunction causes abnormal centrosome segregation during mitosis, which may lead to CIN [6–8].
AURKA (the official symbol for Aurora-A, also known as STK15/BTAK) is a member of the Aurora family of cell cycle-regulating serine/threonine kinases and functions in centrosome regulation and mitotic spindle formation [9,10]. Activation of AURKA in experimental systems confers malignant phenotype by inducing centrosome amplification and genomic instability, indicating AURKA as an oncogene [11–13]. AURKA expression has been reported in various human cancers [14–20], including colorectal cancers [16]. AURKA amplification correlates with CIN in colorectal cancer [21]. However, to our knowledge, no study has comprehensively examined the relation between AURKA protein overexpression and CIN in colorectal cancer or whether the relation is independent of other related molecular features including microsatellite instability (MSI) and the CpG island methylator phenotype (CIMP).
In this study using a large number (N = 517) of colorectal cancers, we examined AURKA overexpression in relation to CIN and patient survival. Because our tumor database included other related molecular events, we were able to assess whether there was an independent relation between AURKA and CIN. Our current data support AURKA as one of potential contributing factors for CIN in colorectal cancer.
Materials and Methods
Study Group
We used the databases of two large prospective cohort studies; the Nurses' Health Study (NHS; N = 121,700 women followed since 1976) [22,23], and the Health Professionals Follow-up Study (HPFS; N = 51,500 men followed since 1986) [23]. Data on height and weight were obtained by biennial questionnaire. A subset of the cohort participants developed colorectal cancers during prospective follow-up. Previous studies on the NHS and HPFS have described baseline characteristics of cohort participants and incident colorectal cancer cases and confirmed that our colorectal cancers were good representatives of a population-based sample [22,23]. Data on tumor location and stage were obtained through medical record review. We collected paraffin-embedded tissue blocks from hospitals where patients had undergone resections of colorectal cancers. On the basis of availability of adequate tissue specimens and results, a total of 517 colorectal cancers were included. Written informed consent was obtained from all study subjects. Among our cohort studies, there was no significant difference in demographic features between cases with tissue available and those without available tissue [23]. This current analysis represents a new analysis of AURKA on the existing colorectal cancer database that have been previously characterized for CIMP, MSI, p53, KRAS, BRAF, PIK3CA, long interspersed nucleotide element 1 (LINE-1), cyclooxygenase 2 (COX-2), and clinical outcome [23–26], which is analogous to novel studies using the well-described cell lines or animal models. In any of our previous studies, we have not examined AURKA expression or the relations of AURKA with clinical outcome and other molecular events. This study represents a unique novel study in term of 1) a large sample size analyzed for AURKA; 2) the comprehensive clinical and tissue molecular database used, including the long-term follow-up outcome data, CIMP, MSI, KRAS, BRAF, PIK3CA, p53, β-catenin, LINE-1 methylation, and COX-2; and 3) a number of molecular correlates that have been analyzed. Tissue collection and analyses were approved by the Harvard School of Public Health and Brigham and Women's Hospital Institutional Review Boards.
Histopathologic Evaluations
Hematoxylin and eosin-stained tissue sections were examined by a pathologist (S.O.) unaware of other data. The tumor grade was categorized as low (≥50% gland formation) versus high (<50% gland formation). The presence and extent of extracellular mucin were categorized as 0% (no mucin), 1% to 49%, or ≥50% of the tumor volume [27]. The presence and extent of signet ring cells were categorized as absent (0%) or present (≥1%) [27].
Sequencing of KRAS, BRAF, and PIK3CA
Genomic DNA was extracted from tumor and polymerase chain reaction (PCR) and Pyrosequencing targeted for KRAS (codons 12 and 13) [28], BRAF (codon 600) [29], and PIK3CA (exons 9 and 20) [30] were performed as previously described.
Microsatellite Instability Analysis
Microsatellite instability analysis was performed, using 10 microsatellite markers (D2S123, D5S346, D17S250, BAT25, BAT26, BAT40, D18S55, D18S56, D18S67, and D18S487) [27]. MSI-high was defined as the presence of instability in ≥30% of the markers. MSI-low was defined as instability in <30% of the markers, and microsatellite stable (MSS) tumors were defined as tumors without an unstable marker [27].
Loss of Heterozygosity Analysis
For loss of heterozygosity (LOH) analysis using microsatellite markers (D2S123, D5S346, D17S250, D18S55, D18S56, D18S67, and D18S487), we duplicated PCR in each sample to exclude allele drop-outs of one of two alleles [27,31]. Loss of heterozygosity at each locus was defined as ≥40% reduction of one of two allele peaks in tumor DNA relative to normal DNA. Chromosomal instability (CIN) positivity was defined as the presence of LOH in any of the chromosomal segments among 2p, 5q, 17q, and 18q. CIN negativity was defined as the absence of LOH in any of the chromosomal segments with the presence of at least two informative segments.
Real-time PCR for Quantitative DNA Methylation Analysis
Sodium bisulfite treatment on genomic DNA and subsequent realtime PCR (MethyLight) were validated and performed as previously described [32]. We quantified DNA methylation in eight CIMP-specific promoters [CACNA1G, CDKN2A (p16), CRABP1, IGF2, MLH1, NEUROG1, RUNX3, and _SOCS1_] [24,33,34]. CIMP-high was defined as the presence of six or more of eight methylated promoters, CIMP-low as the presence of one to five of eight methylated promoters, and CIMP-0 as the absence of methylated promoters, according to the previously established criteria [24,35].
Pyrosequencing to Measure LINE-1 Methylation
To accurately quantify relatively high methylation levels in LINE-1 repetitive elements, we used Pyrosequencing as previously described [25,36].
Immunohistochemistry for AURKA, p53, p21, Cyclin D1, β-Catenin, COX-2, and Fatty Acid Synthase
Tissue microarrays were constructed as previously described [23,37]. Methods of immunohistochemical procedures and examples of staining patterns can be found in the previous reports as follows: p53 [38], p21 (CDKN1A) [39,40], cyclin D1 [41], β-catenin [42], COX-2 [27,43], and fatty acid synthase (FASN) [27,44]. For AURKA, antigen retrieval was performed, and deparaffinized tissue sections in Antigen Retrieval Citra Solution (Biogenex Laboratories, San Ramon, CA) were treated with microwave in a pressure cooker for 25 minutes. Tissue sections were incubated with 3% H2O2 (30 minutes) to block endogenous peroxidase (Dako Cytomation, Carpinteria, CA). A primary antibody [mouse monoclonal to AURKA (ab13824), 1:100 dilution; Abcam Inc, Cambridge, MA] was applied, and the slides were maintained at 4°C overnight, followed by mouse secondary antibody (Vector Laboratories, Burlingame, CA) for 60 minutes, an avidin-biotin complex conjugate (Vector Laboratories) for 60 minutes, diaminobenzidine (5 minutes) and methyl-green counterstain. Nuclear AURKA expression was recorded as no expression, weak expression, moderate expression, or strong expression (Figure 1). AURKA overexpression was defined as moderate to strong expression in any portion of tumor cells or at least 50% of tumor cells with weak staining. Appropriate positive and negative controls were included in each run of immunohistochemistry. Each immunohistochemical maker was interpreted by one of the investigators (AURKA by Y.B.; cyclin D1 and β-catenin by K.N.; p53, p21, COX-2, and FASN by S.O.) unaware of other data. A random selection of 117 cases was examined for AURKA by a second observer (K.S.) unaware of other data, and concordance between the two observers was 0.85 (κ = 0.62, P < .0001), indicating substantial agreement. For each of the other immunohistochemical markers, a second observer (S.O. for β-catenin; K.S. for cyclin D1 and p21; K.N. for p53, COX-2, and FASN) examined a random selection of more than 100 tumors unaware of other data, and the concordance rate between the two observers was always greater than 0.82 [all _κ_ > 0.61 (except for FASN, κ = 0.57), all P < .0001], indicating generally substantial agreement.
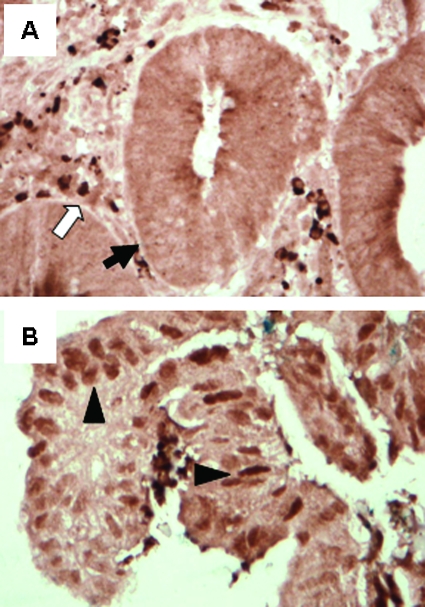
Figure 1.
AURKA (Aurora-A) expression in colorectal cancer cells. (A) Negative for nuclear AURKA overexpression in colorectal cancer cells (arrow). Inflammatory cells serve as an internal positive control for AURKA positivity (white arrow). (B) Positive for nuclear AURKA overexpression in colorectal cancer cells (arrowheads).
Statistical Analysis
All statistical analyses used SAS program (Version 9.1; SAS Institute, Cary, NC). All P values were two-sided, and statistical significance was set at P ≤ .05. For categorical data, the _χ_2 test was performed, and odds ratio (OR) with 95% confidence interval (CI) was computed. To compare mean LINE-1 methylation levels, the t test assuming unequal variances was performed. The κ coefficient was calculated to assess an agreement between the two interpreters in immunohistochemical analyses. To assess independent relations of AURKA overexpression with CIN, a multivariate logistic regression analysis was performed. Odds ratio was adjusted for sex, age (continuous), body mass index (BMI, <30 vs ≥30 kg/m2), family history of colorectal cancer (present vs absent), tumor location (right colon vs left colon vs rectum), tumor stage (I–II vs III–IV), tumor grade (low vs high), mucinous component (0 vs ≥1%), signet ring cell component (0 vs ≥1%), CIMP status (high vs CIMP-low/0), MSI status (high vs low/MSS), LINE-1 methylation (continuous), BRAF, KRAS, PIK3CA, p53, p21, cyclin D1, β-catenin, COX-2, and FASN.
For survival analysis, Kaplan-Meier method was used to assess survival time distribution according to AURKA status, and log-rank test was used to test significance of a deviation from the null hypothesis. For the analyses of colorectal cancer-specific mortality, death as a result of colorectal cancer was the primary end point and deaths as a result of other causes were censored. To assess independent effect of AURKA on mortality, we constructed a multivariate, stage-matched conditional Cox proportional hazard model to compute a hazard ratio (HR) according to AURKA status, adjusted for sex, age, year of diagnosis (continuous), BMI, family history of colorectal cancer, tumor location, stage, grade, CIMP, MSI, CIN, BRAF, KRAS, PIK3CA, LINE-1 methylation, p53, p21, cyclin D1, β-catenin, COX-2, and FASN.
Results
AURKA Overexpression in Colorectal Cancer
Among the 517 colorectal cancers in this study, 98 (19%) showed nuclear AURKA overexpression (i.e., AURKA-positive; Figure 1). Table 1 shows the frequencies of AURKA overexpression according to various clinical and pathologic features. AURKA overexpression was inversely associated with family history of colorectal cancer (OR, 0.49; 95% CI, 0.26–0.89; P = .018).
Table 1.
Frequency of AURKA Overexpression in Colorectal Cancer.
| Clinical or Pathologic Feature | Total N | AURKA (+) | Univariate OR (95% CI) | P |
|---|---|---|---|---|
| All cases | 517 | 98 (19%) | ||
| Gender | ||||
| Men | 203 | 35 (17%) | 1 | |
| Women | 314 | 63 (20%) | 1.20 (0.76–1.90) | |
| Age (years) | ||||
| ≤59 | 118 | 20 (17%) | 1 | |
| 60–69 | 233 | 43 (18%) | 1.10 (0.62–1.99) | |
| ≥70 | 166 | 35 (21%) | 1.31 (0.71–2.41) | |
| BMI (kg/m2) | ||||
| <25 | 209 | 43 (21%) | 1 | |
| 25–30 | 192 | 36 (19%) | 0.89 (0.54–1.46) | |
| ≥30 | 87 | 12 (14%) | 0.62 (0.31–1.24) | |
| Family history of colorectal cancer | ||||
| (-) | 396 | 84 (21%) | 1 | Referent |
| (+) | 121 | 14 (12%) | 0.49 (0.26–0.89) | .018 |
| Tumor location | ||||
| Right colon (cecum to transverse colon) | 243 | 53 (22%) | 1 | |
| Left colon (splenic flexure to sigmoid) | 165 | 25 (15%) | 0.64 (0.38–1.08) | |
| Rectum | 89 | 14 (16%) | 0.66 (0.35–1.28) | |
| Stage | ||||
| I | 106 | 18 (17%) | 1 | |
| II | 158 | 30 (19%) | 1.14 (0.60–2.18) | |
| III | 138 | 19 (14%) | 0.78 (0.39–1.57) | |
| IV | 71 | 15 (21%) | 1.31 (0.61–2.81) | |
| Tumor grade | ||||
| Low | 456 | 82 (18%) | 1 | |
| High | 45 | 11 (24%) | 1.48 (0.72–3.03) | |
| Mucinous component | ||||
| 0% | 267 | 43 (16%) | 1 | |
| 1%–49% | 116 | 27 (23%) | 1.58 (0.92–2.71) | |
| ≥50% | 64 | 16 (25%) | 1.74 (0.90–3.34) | |
| Signet ring cell component | ||||
| 0% | 387 | 72 (19%) | 1 | |
| ≥1% | 29 | 7 (24%) | 1.39 (0.57–3.38) |
AURKA, CIN, and Other Molecular Features in Colorectal Cancer
Table 2 summarizes the frequencies of AURKA overexpression according to status of CIN and other molecular features in colorectal cancer. Chromosomal instability (CIN) was defined as the presence of LOH in any of the chromosomal segments among 2p, 5q, 17q, and 18q. AURKA overexpression was significantly associated with CIN (OR, 2.17; 95% CI, 1.09–4.32; P = .024).
Table 2.
Frequency of AURKA Overexpression in Colorectal Cancer According to Various Molecular Features.
| Molecular Feature | Total N | AURKA (+) | Univariate OR (95% CI) | P |
|---|---|---|---|---|
| CIN* | ||||
| (-) | 105 | 11 (10%) | 1 | Referent |
| (+) | 291 | 59 (20%) | 2.17 (1.09–4.32) | .024 |
| CIMP status (no. of methylated CIMP markers) | ||||
| CIMP-0 (0) | 224 | 43 (19%) | 1 | |
| CIMP-low (1–5) | 202 | 30 (15%) | 0.73 (0.44–1.22) | |
| CIMP-high (6–8) | 77 | 21 (27%) | 1.58 (0.86–2.88) | |
| MSI status | ||||
| MSS | 376 | 66 (18%) | 1 | |
| MSI-low | 52 | 11 (21%) | 1.26 (0.62–2.58) | |
| MSI-high | 87 | 21 (24%) | 1.49 (0.86–2.61) | |
| BRAF mutation | ||||
| (-) | 437 | 80 (18%) | 1 | |
| (+) | 67 | 17 (25%) | 1.51 (0.83–2.77) | |
| KRAS mutation | ||||
| (-) | 327 | 72 (22%) | 1 | Referent |
| (+) | 189 | 26 (14%) | 0.56 (0.35–0.92) | .021 |
| PIK3CA mutation | ||||
| (-) | 397 | 86 (22%) | 1 | Referent |
| (+) | 62 | 4 (6%) | 0.25 (0.09–0.71) | .0050 |
| LINE-1 methylation | ||||
| ≥70% | 61 | 9 (15%) | 1 | |
| 50%–70% | 360 | 66 (18%) | 1.30 (0.61–2.76) | |
| <50% | 76 | 19 (25%) | 1.93 (0.80–4.63) | |
| p53 expression | ||||
| (-) | 311 | 67 (22%) | 1 | |
| (+) | 202 | 31 (15%) | 0.66 (0.41–1.05) | |
| p21 | ||||
| Expressed | 104 | 24 (23%) | 1 | |
| Lost | 403 | 73 (18%) | 0.73 (0.44–1.24) | |
| Cyclin D1 expression | ||||
| (-) | 142 | 21 (15%) | 1 | |
| (+) | 349 | 75 (21%) | 1.57 (0.93–2.68) | |
| β-Catenin† | ||||
| Inactive (score 0–2) | 294 | 62 (21%) | 1 | Referent |
| Active (score 3–5) | 175 | 24 (14%) | 0.60 (0.36–0.99) | .046 |
| COX-2 expression | ||||
| (-) | 90 | 23 (26%) | 1 | |
| (+) | 426 | 74 (17%) | 0.61 (0.36–1.05) | |
| FASN expression | ||||
| (-) | 350 | 70 (20%) | 1 | |
| (+) | 65 | 7 (11%) | 0.48 (0.21–1.10) |
AURKA overexpression was inversely associated with KRAS mutation (OR, 0.56; 95% CI, 0.35–0.92; P = .021), PIK3CA mutation (OR, 0.25; 95% CI, 0.09–0.71; P = .0050), and β-catenin activation (OR, 0.60; 95% CI, 0.36–0.99; P = .046; Table 2).
Multivariate Analysis to Assess Independent Relations with AURKA
We performed multivariate logistic regression analysis to examine whether AURKA was independently associated with CIN (Table 3). AURKA overexpression was significantly associated with CIN (multivariate OR 2.97; 95% CI, 1.40–6.29; P = .0045) independent of other variables. In addition, AURKA seemed to be associated with cyclin D1 expression (P = .012) and inversely with PIK3CA mutation (P = .010), FASN expression (P = .032), and family history of colorectal cancer (P = .036; Table 3); however, considering multiple hypotheses testing and these P values between .05 and .01, any of these additional associations might simply be a chance event.
Table 3.
Multivariate Logistic Regression to Show Independent Relationship between CIN and AURKA Overexpression in Colorectal Cancer.
| Variable Independently Associated with AURKA | Multivariate OR (95% CI) | P |
|---|---|---|
| CIN* | 2.97 (1.40–6.29) | .0045 |
| Other significant variables | ||
| PIK3CA mutation | 0.24 (0.08–0.70) | .010 |
| Cyclin D1 expression | 1.93 (1.15–3.24) | .012 |
| FASN expression | 0.37 (0.15–0.92) | .032 |
| Family history of colorectal cancer | 0.50 (0.26–0.96) | .036 |
AURKA and Patient Survival
We assessed the influence of AURKA overexpression on clinical outcome of patients with stage I to IV colorectal cancer and adequate follow-up. We have previously shown that clinical outcome data in our two independent cohort studies are valid and reliable in detecting significant molecular predictors of patient survival [25,26,43,44]. There were a total of 216 deaths, including 124 colorectal cancer-specific deaths. In Kaplan-Meier analysis, AURKA was not significantly associated with colorectal cancer-specific (log rank, P = .67) or overall survival (log rank, P = .78). We also performed Cox regression analysis to assess patient mortality according to AURKA status (Table 4). For both cancer-specific and overall mortality, AURKA overexpression was not significantly related with patient outcome in univariate analysis, stage-matched analysis, or multivariate analysis. When we assessed patients with colon cancers, AURKA remained unrelated with patient mortality.
Table 4.
AURKA Expression and Patient Mortality in Colorectal Cancer.
| Total N | Cancer-Specific Mortality | Overall Mortality | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Deaths/Person-Years | Univariate HR (95% CI) | Stage-matched HR (95% CI) | Multivariate HR (95% CI) | Deaths/Person-Years | Univariate HR (95% CI) | Stage-matched HR (95% CI) | Multivariate HR (95% CI) | ||
| Colon and rectal cancers | |||||||||
| AURKA (-) | 398 | 103/3180 | 1 (referent) | 1 (referent) | 1 (referent) | 179/3180 | 1 (referent) | 1 (referent) | 1 (referent) |
| AURKA (+) | 89 | 21/688 | 0.90 (0.57–1.45) | 0.98 (0.60–1.60) | 0.97 (0.57–1.65) | 37/688 | 0.95 (0.67–1.36) | 1.00 (0.69–1.45) | 1.03 (0.69–1.53) |
| Colon cancers | |||||||||
| AURKA (-) | 324 | 78/2648 | 1 (referent) | 1 (referent) | 1 (referent) | 144/2648 | 1 (referent) | 1 (referent) | 1 (referent) |
| AURKA (+) | 75 | 19/569 | 1.07 (0.65–1.77) | 1.13 (0.66–1.93) | 0.89 (0.49–1.60) | 31/569 | 0.99 (0.67–1.46) | 1.00 (0.66–1.50) | 0.99 (0.63–1.55) |
Discussion
We conducted this study to examine the relationship between AURKA overexpression and CIN in colorectal cancer. In addition, we assessed the relationship of AURKA with clinical, pathologic, and other molecular features and with patient survival. We found that AURKA was associated with CIN, independent of any of the clinical, pathologic, and molecular variables examined. Our rich tumor database allowed us to examine whether there is an independent relationship of AURKA with CIN as well as with clinical outcome. Our data support the hypothesis that AURKA overexpression is one of the contributing factors for CIN during colorectal cancer development.
Studying molecular alterations is important in cancer research [45–56]. Our resource of a large number (N = 517) of colorectal cancers derived from the two independent prospective cohort studies has enabled us to precisely estimate the frequency of colorectal cancers with a specific molecular feature (such as AURKA overexpression, MSI, etc.). The large number of cases has also provided us with a sufficient power in the multivariate logistic regression analysis and survival analysis. Thus, in survival analysis, we can conclude that AURKA expression is not significantly associated with patient survival.
Accumulating evidence suggests that AURKA activation is related to cancer development through CIN [11–13,57]. AURKA is mainly localized at spindle poles and the mitotic spindle during mitosis, where it regulates the function of centrosomes, spindles, and kinetochores, all of which are required for proper mitosis progression [9,10]. AURKA protein expression has also been related with TERT (telomerase) activity, thus possibly related with telomere function [16]. AURKA gene amplification [58,59] and AURKA gene overexpression [58] have been reported in colorectal cancer. Another study using colon cancer cell lines and a small number (N = 48) of human colorectal cancer tissues has shown that high copy number of AURKA is associated with CIN and AURKA protein overexpression [21]. However, the discrepancy in the frequencies of AURKA amplification and AURKA protein overexpression has been reported in several other cancers [13,19,60]. To the best of our knowledge, a significant relationship between AURKA protein overexpression and CIN has not been shown using a large number of colorectal cancer tissues. Molecular correlates with AURKA protein overexpression are important for the better understanding of genetic and epigenetic alterations during the colorectal carcinogenic process, especially in relation to CIN.
It remains controversial whether AURKA expression level correlates with malignant phenotype or patient prognosis in human cancers. Studies on several types of human cancers have indicated that AURKA overexpression may be related with poor prognosis [15,17,18,61] or with higher recurrence rate [62]. However, other studies have shown that AURKA expression is not associated with patient prognosis [20,63]. Another study has shown that activation of AURKA is associated with an early stage disease in ovarian cancer [19]. In a previous study on colorectal cancer (N = 200) [16], AURKA overexpression was associated with high-grade tumor, but the relationship between AURKA and clinical outcome was unclear. In our current study (N = 517), AURKA overexpression was not associated with tumor grade. This discrepancy might be due to a difference in the patient cohorts or the methods to assess AURKA overexpression or simply due to a chance variation between different studies. In addition, our analysis found that AURKA overexpression was not associated with prognosis of patients with stage I to IV colorectal cancer. Our findings suggest that AURKA overexpression may not mark an aggressive type of colorectal cancer.
Interestingly, we did observe inverse relations between AURKA and PIK3CA mutation as well as FASN expression in colorectal cancer, independent of clinical, pathologic, and other molecular variables. In various cancers including colorectal cancer, mutant PIK3CA stimulates the PI3K-AKT signaling pathway [64]. Fatty acid synthase overexpression has been associated with the PI3K-AKT pathway activation in some cancers [65,66]. A study using human cancer cell lines [67] has shown that AKTis likely responsible for the up-regulation of AURKA for mitotic progression. In contrast, another study using mouse oocytes [68] has shown that the activation of AURKA on microtubule-organizing centers is independent of the PI3K-AKT pathway. As to the relationship between AURKA and the PI3K-AKT pathway, further studies are needed. We also demonstrated that AURKA overexpression was associated with nuclear cyclin D1 expression. A previous study using a mouse model [11] has reported that AURKA overexpression cause nuclear accumulation of cyclin D1, which is in agreement with our current data.
Recently, Aurora kinases have been targeted for cancer therapy, and a new class of drugs known as Aurora kinase inhibitors has been undergoing preclinical and clinical assessments [69,70]. Among them, VX-680 has shown promising results in animal studies, inhibiting tumor growth in a range of xenograft models and leading to regression of colon tumor [71]. VX-680 is already undergoing the clinical study, but there is no biomarker for selecting patients to benefit for clinical trials of this drug. Hereafter, AURKA expression in the resected specimens might attract increasing attention as a biomarker for patient selection. In this respect, our findings may have clinical implications.
In particular, a possible inverse relation between AURKA and family history of colorectal cancer merits discussion. An association between a molecular change and family history of colorectal cancer implies a genetic factor (and/or a shared environmental factor) that may contribute to the development of the given molecular change. The inverse association between AURKA and family history likely supports genetic predisposition to molecular changes (such as MSI) alternative to the AURKA-CIN pathway. Clarification of this issue by future research is important considering AURKA inhibitors as potential targeted therapy against colorectal cancers, including familial cases.
In conclusion, using a large number of colorectal cancers, we have shown that AURKA overexpression is independently associated with CIN. Our data support the hypothesis that AURKA may contribute to colorectal carcinogenesis through CIN.
Acknowledgments
The authors thank the NHS and HPFS cohort participants who have generously agreed to provide biological specimens and information through responses to questionnaires. The authors thank Frank Speizer, Walter Willett, Susan Hankinson, Graham Colditz, Meir Stampfer, and many other staff members who implemented and have maintained the cohort studies.
Abbreviations
AURKA
Aurora-A
BMI
body mass index
CI
confidence interval
CIN
chromosomal instability
CIMP
CpG island methylator phenotype
FASN
fatty acid synthase
HPFS
Health Professionals Follow-up Study
HR
hazard ratio
LOH
loss of heterozygosity
MSI
microsatellite instability
MSS
microsatellite stable
NHS
Nurses' Health Study
OR
odds ratio
Footnotes
1
This work was supported by US National Institutes of Health (NIH) grants P01 CA87969, P01 CA55075, P50 CA127003 (to C.S.F.), and K07 CA122826 (to S.O.) and in part by grants from the Bennett Family Fund and from the Entertainment Industry Foundation through the National Colorectal Cancer Research Alliance (NCCRA). K.N. was supported by a fellowship grant from the Japan Society for Promotion of Science. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or NIH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. No conflicts of interest exist.
References
- 1.Grady WM, Carethers JM. Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology. 2008;135:1079–1099. doi: 10.1053/j.gastro.2008.07.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cahill DP, Lengauer C, Yu J, Riggins GJ, Willson JK, Markowitz SD, Kinzler KW, Vogelstein B. Mutations of mitotic checkpoint genes in human cancers. Nature. 1998;392:300–303. doi: 10.1038/32688. [DOI] [PubMed] [Google Scholar]
- 3.Gualberto A, Aldape K, Kozakiewicz K, Tlsty TD. An oncogenic form of p53 confers a dominant, gain-of-function phenotype that disrupts spindle checkpoint control. Proc Natl Acad Sci USA. 1998;95:5166–5171. doi: 10.1073/pnas.95.9.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rajagopalan H, Jallepalli PV, Rago C, Velculescu VE, Kinzler KW, Vogelstein B, Lengauer C. Inactivation of hCDC4 can cause chromosomal instability. Nature. 2004;428:77–81. doi: 10.1038/nature02313. [DOI] [PubMed] [Google Scholar]
- 5.Maser RS, DePinho RA. Connecting chromosomes, crisis, and cancer. Science. 2002;297:565–569. doi: 10.1126/science.297.5581.565. [DOI] [PubMed] [Google Scholar]
- 6.Fukasawa K. Oncogenes and tumour suppressors take on centrosomes. Nat Rev Cancer. 2007;7:911–924. doi: 10.1038/nrc2249. [DOI] [PubMed] [Google Scholar]
- 7.Pihan GA, Purohit A, Wallace J, Knecht H, Woda B, Quesenberry P, Doxsey SJ. Centrosome defects and genetic instability in malignant tumors. Cancer Res. 1998;58:3974–3985. [PubMed] [Google Scholar]
- 8.Lingle WL, Barrett SL, Negron VC, D'Assoro AB, Boeneman K, Liu W, Whitehead CM, Reynolds C, Salisbury JL. Centrosome amplification drives chromosomal instability in breast tumor development. Proc Natl Acad Sci USA. 2002;99:1978–1983. doi: 10.1073/pnas.032479999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu J, Bian M, Jiang Q, Zhang C. Roles of Aurora kinases in mitosis and tumorigenesis. Mol Cancer Res. 2007;5:1–10. doi: 10.1158/1541-7786.MCR-06-0208. [DOI] [PubMed] [Google Scholar]
- 10.Marumoto T, Zhang D, Saya H. Aurora-A—a guardian of poles. Nat Rev Cancer. 2005;5:42–50. doi: 10.1038/nrc1526. [DOI] [PubMed] [Google Scholar]
- 11.Wang X, Zhou YX, Qiao W, Tominaga Y, Ouchi M, Ouchi T, Deng CX. Overexpression of aurora kinase A in mouse mammary epithelium induces genetic instability preceding mammary tumor formation. Oncogene. 2006;25:7148–7158. doi: 10.1038/sj.onc.1209707. [DOI] [PubMed] [Google Scholar]
- 12.Bischoff JR, Anderson L, Zhu Y, Mossie K, Ng L, Souza B, Schryver B, Flanagan P, Clairvoyant F, Ginther C, et al. A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J. 1998;17:3052–3065. doi: 10.1093/emboj/17.11.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou H, Kuang J, Zhong L, Kuo WL, Gray JW, Sahin A, Brinkley BR, Sen S. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat Genet. 1998;20:189–193. doi: 10.1038/2496. [DOI] [PubMed] [Google Scholar]
- 14.Jeng YM, Peng SY, Lin CY, Hsu HC. Overexpression and amplification of Aurora-A in hepatocellular carcinoma. Clin Cancer Res. 2004;10:2065–2071. doi: 10.1158/1078-0432.ccr-1057-03. [DOI] [PubMed] [Google Scholar]
- 15.Landen CN, Jr, Lin YG, Immaneni A, Deavers MT, Merritt WM, Spannuth WA, Bodurka DC, Gershenson DM, Brinkley WR, Sood AK. Overexpression of the centrosomal protein Aurora-A kinase is associated with poor prognosis in epithelial ovarian cancer patients. Clin Cancer Res. 2007;13:4098–4104. doi: 10.1158/1078-0432.CCR-07-0431. [DOI] [PubMed] [Google Scholar]
- 16.Lam AK, Ong K, Ho YH. Aurora kinase expression in colorectal adenocarcinoma: correlations with clinicopathological features, p16 expression, and telomerase activity. Hum Pathol. 2008;39:599–604. doi: 10.1016/j.humpath.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 17.Tanaka E, Hashimoto Y, Ito T, Okumura T, Kan T, Watanabe G, Imamura M, Inazawa J, Shimada Y. The clinical significance of Aurora-A/STK15/BTAK expression in human esophageal squamous cell carcinoma. Clin Cancer Res. 2005;11:1827–1834. doi: 10.1158/1078-0432.CCR-04-1627. [DOI] [PubMed] [Google Scholar]
- 18.Nadler Y, Camp RL, Schwartz C, Rimm DL, Kluger HM, Kluger Y. Expression of Aurora A (but not Aurora B) is predictive of survival in breast cancer. Clin Cancer Res. 2008;14:4455–4462. doi: 10.1158/1078-0432.CCR-07-5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gritsko TM, Coppola D, Paciga JE, Yang L, Sun M, Shelley SA, Fiorica JV, Nicosia SV, Cheng JQ. Activation and overexpression of centrosome kinase BTAK/Aurora-A in human ovarian cancer. Clin Cancer Res. 2003;9:1420–1426. [PubMed] [Google Scholar]
- 20.Kurai M, Shiozawa T, Shih HC, Miyamoto T, Feng YZ, Kashima H, Suzuki A, Konishi I. Expression of Aurora kinases A and B in normal, hyperplastic, and malignant human endometrium: Aurora B as a predictor for poor prognosis in endometrial carcinoma. Hum Pathol. 2005;36:1281–1288. doi: 10.1016/j.humpath.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 21.Nishida N, Nagasaka T, Kashiwagi K, Boland CR, Goel A. High copy amplification of the Aurora-A gene is associated with chromosomal instability phenotype in human colorectal cancers. Cancer Biol Ther. 2007;6:525–533. doi: 10.4161/cbt.6.4.3817. [DOI] [PubMed] [Google Scholar]
- 22.Colditz GA, Hankinson SE. The Nurses' Health Study: lifestyle and health among women. Nat Rev Cancer. 2005;5:388–396. doi: 10.1038/nrc1608. [DOI] [PubMed] [Google Scholar]
- 23.Chan AT, Ogino S, Fuchs CS. Aspirin and the risk of colorectal cancer in relation to the expression of COX-2. N Engl J Med. 2007;356:2131–2142. doi: 10.1056/NEJMoa067208. [DOI] [PubMed] [Google Scholar]
- 24.Ogino S, Kawasaki T, Kirkner GJ, Kraft P, Loda M, Fuchs CS. Evaluation of markers for CpG island methylator phenotype (CIMP) in colorectal cancer by a large population-based sample. J Mol Diagn. 2007;9:305–314. doi: 10.2353/jmoldx.2007.060170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ogino S, Nosho K, Kirkner GJ, Kawasaki T, Chan AT, Schernhammer ES, Giovannucci EL, Fuchs CS. A cohort study of tumoral LINE-1 hypomethylation and prognosis in colon cancer. J Natl Cancer Inst. 2008;100:1734–1738. doi: 10.1093/jnci/djn359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogino S, Nosho K, Kirkner GJ, Kawasaki T, Meyerhardt JA, Loda M, Giovannucci EL, Fuchs CS. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009;58:90–96. doi: 10.1136/gut.2008.155473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ogino S, Brahmandam M, Cantor M, Namgyal C, Kawasaki T, Kirkner G, Meyerhardt JA, Loda M, Fuchs CS. Distinct molecular features of colorectal carcinoma with signet ring cell component and colorectal carcinoma with mucinous component. Mod Pathol. 2006;19:59–68. doi: 10.1038/modpathol.3800482. [DOI] [PubMed] [Google Scholar]
- 28.Ogino S, Kawasaki T, Brahmandam M, Yan L, Cantor M, Namgyal C, Mino-Kenudson M, Lauwers GY, Loda M, Fuchs CS. Sensitive sequencing method for KRAS mutation detection by pyrosequencing. J Mol Diagn. 2005;7:413–421. doi: 10.1016/S1525-1578(10)60571-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ogino S, Kawasaki T, Kirkner GJ, Loda M, Fuchs CS. CpG island methylator phenotype-low (CIMP-low) in colorectal cancer: possible associations with male sex and KRAS mutations. J Mol Diagn. 2006;8:582–588. doi: 10.2353/jmoldx.2006.060082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nosho K, Kawasaki T, Ohnishi M, Suemoto Y, Kirkner GJ, Zepf D, Yan L, Longtine JA, Fuchs CS, Ogino S. PIK3CA mutation in colorectal cancer: relationship with genetic and epigenetic alterations. Neoplasia. 2008;10:534–541. doi: 10.1593/neo.08336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nosho K, Shima K, Kure S, Irahara N, Baba Y, Chen L, Kirkner GJ, Fuchs CS, Ogino S. °C virus T-antigen in colorectal cancer is associated with p53 expression and chromosomal instability, independent of CpG island methylator phenotype. Neoplasia. 2009;11:87–95. doi: 10.1593/neo.81188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogino S, Kawasaki T, Brahmandam M, Cantor M, Kirkner GJ, Spiegelman D, Makrigiorgos GM, Weisenberger DJ, Laird PW, Loda M, et al. Precision and performance characteristics of bisulfite conversion and real-time PCR (MethyLight) for quantitative DNA methylation analysis. J Mol Diagn. 2006;8:209–217. doi: 10.2353/jmoldx.2006.050135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ogino S, Cantor M, Kawasaki T, Brahmandam M, Kirkner GJ, Weisenberger DJ, Campan M, Laird PW, Loda M, Fuchs CS. CpG island methylator phenotype (CIMP) of colorectal cancer is best characterised by quantitative DNA methylation analysis and prospective cohort studies. Gut. 2006;55:1000–1006. doi: 10.1136/gut.2005.082933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, Kang GH, Widschwendter M, Weener D, Buchanan D, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 35.Nosho K, Irahara N, Shima K, Kure S, Kirkner GJ, Schernhammer ES, Hazra A, Hunter DJ, Quackenbush J, Spiegelman D, et al. Comprehensive biostatistical analysis of CpG island methylator phenotype in colorectal cancer using a large population-based sample. PLoS ONE. 2008;3:e3698. doi: 10.1371/journal.pone.0003698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ogino S, Kawasaki T, Nosho K, Ohnishi M, Suemoto Y, Kirkner GJ, Fuchs CS. LINE-1 hypomethylation is inversely associated with microsatellite instability and CpG island methylator phenotype in colorectal cancer. Int J Cancer. 2008;122:2767–2773. doi: 10.1002/ijc.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogino S, Brahmandam M, Kawasaki T, Kirkner GJ, Loda M, Fuchs CS. Combined analysis of COX-2 and p53 expressions reveals synergistic inverse correlations with microsatellite instability and CpG island methylator phenotype in colorectal cancer. Neoplasia. 2006;8:458–464. doi: 10.1593/neo.06247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ogino S, Kawasaki T, Kirkner GJ, Yamaji T, Loda M, Fuchs CS. Loss of nuclear p27 (CDKN1B/KIP1) in colorectal cancer is correlated with microsatellite instability and CIMP. Mod Pathol. 2007;20:15–22. doi: 10.1038/modpathol.3800709. [DOI] [PubMed] [Google Scholar]
- 39.Ogino S, Meyerhardt JA, Cantor M, Brahmandam M, Clark JW, Namgyal C, Kawasaki T, Kinsella K, Michelini AL, Enzinger PC, et al. Molecular alterations in tumors and response to combination chemotherapy with gefitinib for advanced colorectal cancer. Clin Cancer Res. 2005;11:6650–6656. doi: 10.1158/1078-0432.CCR-05-0738. [DOI] [PubMed] [Google Scholar]
- 40.Ogino S, Kawasaki T, Kirkner GJ, Ogawa A, Dorfman I, Loda M, Fuchs CS. Down-regulation of p21 (CDKN1A/CIP1) is inversely associated with microsatellite instability and CpG island methylator phenotype (CIMP) in colorectal cancer. J Pathol. 2006;210:147–154. doi: 10.1002/path.2030. [DOI] [PubMed] [Google Scholar]
- 41.Nosho K, Kawasaki T, Chan AT, Ohnishi M, Suemoto Y, Kirkner GJ, Fuchs CS, Ogino S. Cyclin D1 is frequently overexpressed in microsatellite unstable colorectal cancer, independent of CpG island methylator phenotype. Histopathology. 2008;53:588–598. doi: 10.1111/j.1365-2559.2008.03161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kawasaki T, Nosho K, Ohnishi M, Suemoto Y, Kirkner GJ, Dehari R, Meyerhardt JA, Fuchs CS, Ogino S. Correlation of beta-catenin localization with cyclooxygenase-2 expression and CpG island methylator phenotype (CIMP) in colorectal cancer. Neoplasia. 2007;9:569–577. doi: 10.1593/neo.07334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ogino S, Kirkner GJ, Nosho K, Irahara N, Kure S, Shima K, Hazra A, Chan AT, Dehari R, Giovannucci EL, et al. Cyclooxygenase-2 expression is an independent predictor of poor prognosis in colon cancer. Clin Cancer Res. 2008;14:8221–8227. doi: 10.1158/1078-0432.CCR-08-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ogino S, Nosho K, Meyerhardt JA, Kirkner GJ, Chan AT, Kawasaki T, Giovannucci EL, Loda M, Fuchs CS. Cohort study of fatty acid synthase expression and patient survival in colon cancer. J Clin Oncol. 2008;26:5713–5720. doi: 10.1200/JCO.2008.18.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roschke AV, Glebov OK, Lababidi S, Gehlhaus KS, Weinstein JN, Kirsch IR. Chromosomal instability is associated with higher expression of genes implicated in epithelial-mesenchymal transition, cancer invasiveness, and metastasis and with lower expression of genes involved in cell cycle checkpoints, DNA repair, and chromatin maintenance. Neoplasia. 2008;10:1222–1230. doi: 10.1593/neo.08682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Toma MI, Grosser M, Herr A, Aust DE, Meye A, Hoefling C, Fuessel S, Wuttig D, Wirth MP, Baretton GB. Loss of heterozygosity and copy number abnormality in clear cell renal cell carcinoma discovered by high-density Affymetrix 10K single nucleotide polymorphism mapping array. Neoplasia. 2008;10:634–642. doi: 10.1593/neo.08160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang S, Liu H, Ren L, Pan Y, Zhang Y. Inhibiting colorectal carcinoma growth and metastasis by blocking the expression of VEGF using RNA interference. Neoplasia. 2008;10:399–407. doi: 10.1593/neo.07613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ahlquist T, Bottillo I, Danielsen SA, Meling GI, Rognum TO, Lind GE, Dallapiccola B, Lothe RA. RAS signaling in colorectal carcinomas through alteration of RAS, RAF, NF1, and/or RASSF1A. Neoplasia. 2008;10:680–686. doi: 10.1593/neo.08312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chung YL, Troy H, Kristeleit R, Aherne W, Jackson LE, Atadja P, Griffiths JR, Judson IR, Workman P, Leach MO, et al. Noninvasive magnetic resonance spectroscopic pharmacodynamic markers of a novel histone deacetylase inhibitor, LAQ824, in human colon carcinoma cells and xenografts. Neoplasia. 2008;10:303–313. doi: 10.1593/neo.07834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Derks S, Postma C, Carvalho B, van den Bosch SM, Moerkerk PT, Herman JG, Weijenberg MP, de Bruine AP, Meijer GA, van Engeland M. Integrated analysis of chromosomal, microsatellite and epigenetic instability in colorectal cancer identifies specific associations between promoter methylation of pivotal tumour suppressor and DNA repair genes and specific chromosomal alterations. Carcinogenesis. 2008;29:434–439. doi: 10.1093/carcin/bgm270. [DOI] [PubMed] [Google Scholar]
- 51.Henkhaus RS, Roy UK, Cavallo-Medved D, Sloane BF, Gerner EW, Ignatenko NA. Caveolin-1-mediated expression and secretion of kallikrein 6 in colon cancer cells. Neoplasia. 2008;10:140–148. doi: 10.1593/neo.07817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ueno K, Hiura M, Suehiro Y, Hazama S, Hirata H, Oka M, Imai K, Dahiya R, Hinoda Y. Frizzled-7 as a potential therapeutic target in colorectal cancer. Neoplasia. 2008;10:697–705. doi: 10.1593/neo.08320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiong H, Zhang ZG, Tian XQ, Sun DF, Liang QC, Zhang YJ, Lu R, Chen YX, Fang JY. Inhibition of JAK1, 2/STAT3 signaling induces apoptosis, cell cycle arrest, and reduces tumor cell invasion in colorectal cancer cells. Neoplasia. 2008;10:287–297. doi: 10.1593/neo.07971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gorringe KL, Choong DY, Williams LH, Ramakrishna M, Sridhar A, Qiu W, Bearfoot JL, Campbell IG. Mutation and methylation analysis of the chromodomain-helicase-DNA binding 5 gene in ovarian cancer. Neoplasia. 2008;10:1253–1258. doi: 10.1593/neo.08718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Potter N, Karakoula A, Phipps KP, Harkness W, Hayward R, Thompson DN, Jacques TS, Harding B, Thomas DG, Palmer RW, et al. Genomic deletions correlate with underexpression of novel candidate genes at six loci in pediatric pilocytic astrocytoma. Neoplasia. 2008;10:757–772. doi: 10.1593/neo.07914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kawasaki T, Nosho K, Ohnishi M, Suemoto Y, Kirkner GJ, Fuchs CS, Ogino S. IGFBP3 promoter methylation in colorectal cancer: relationship with microsatellite instability, CpG island methylator phenotype, and p53. Neoplasia. 2007;9:1091–1098. doi: 10.1593/neo.07760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Meraldi P, Honda R, Nigg EA. Aurora-A overexpression reveals tetraploidization as a major route to centrosome amplification in p53-/- cells. EMBO J. 2002;21:483–492. doi: 10.1093/emboj/21.4.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Carvalho B, Postma C, Mongera S, Hopmans E, Diskin S, van de Wiel MA, van Criekinge W, Thas O, Matthai A, Cuesta MA, et al. Multiple putative oncogenes at the chromosome 20q amplicon contribute to colorectal adenoma to carcinoma progression. Gut. 2009;58:79–89. doi: 10.1136/gut.2007.143065. [DOI] [PubMed] [Google Scholar]
- 59.Killian A, Di Fiore F, Le Pessot F, Blanchard F, Lamy A, Raux G, Flaman JM, Paillot B, Michel P, Sabourin JC, et al. A simple method for the routine detection of somatic quantitative genetic alterations in colorectal cancer. Gastroenterology. 2007;132:645–653. doi: 10.1053/j.gastro.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 60.Sakakura C, Hagiwara A, Yasuoka R, Fujita Y, Nakanishi M, Masuda K, Shimomura K, Nakamura Y, Inazawa J, Abe T, et al. Tumour-amplified kinase BTAK is amplified and overexpressed in gastric cancers with possible involvement in aneuploid formation. Br J Cancer. 2001;84:824–831. doi: 10.1054/bjoc.2000.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ogawa E, Takenaka K, Katakura H, Adachi M, Otake Y, Toda Y, Kotani H, Manabe T, Wada H, Tanaka F. Perimembrane Aurora-A expression is a significant prognostic factor in correlation with proliferative activity in non-small-cell lung cancer (NSCLC) Ann Surg Oncol. 2008;15:547–554. doi: 10.1245/s10434-007-9653-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Comperat E, Camparo P, Haus R, Chartier-Kastler E, Radenen B, Richard F, Capron F, Paradis V. Aurora-A/STK-15 is a predictive factor for recurrent behaviour in non-invasive bladder carcinoma: a study of 128 cases of non-invasive neoplasms. Virchows Arch. 2007;450:419–424. doi: 10.1007/s00428-007-0383-x. [DOI] [PubMed] [Google Scholar]
- 63.Royce ME, Xia W, Sahin AA, Katayama H, Johnston DA, Hortobagyi G, Sen S, Hung MC. STK15/Aurora-A expression in primary breast tumors is correlated with nuclear grade but not with prognosis. Cancer. 2004;100:12–19. doi: 10.1002/cncr.11879. [DOI] [PubMed] [Google Scholar]
- 64.Samuels Y, Velculescu VE. Oncogenic mutations of PIK3CA in human cancers. Cell Cycle. 2004;3:1221–1224. doi: 10.4161/cc.3.10.1164. [DOI] [PubMed] [Google Scholar]
- 65.Van de Sande T, Roskams T, Lerut E, Joniau S, Van Poppel H, Verhoeven G, Swinnen JV. High-level expression of fatty acid synthase in human prostate cancer tissues is linked to activation and nuclear localization of Akt/PKB. J Pathol. 2005;206:214–219. doi: 10.1002/path.1760. [DOI] [PubMed] [Google Scholar]
- 66.Wang HQ, Altomare DA, Skele KL, Poulikakos PI, Kuhajda FP, Di Cristofano A, Testa JR. Positive feedback regulation between AKT activation and fatty acid synthase expression in ovarian carcinoma cells. Oncogene. 2005;24:3574–3582. doi: 10.1038/sj.onc.1208463. [DOI] [PubMed] [Google Scholar]
- 67.Liu X, Shi Y, Woods KW, Hessler P, Kroeger P, Wilsbacher J, Wang J, Wang JY, Li C, Li Q, et al. Akt inhibitor a-443654 interferes with mitotic progression by regulating aurora a kinase expression. Neoplasia. 2008;10:828–837. doi: 10.1593/neo.08408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Saskova A, Solc P, Baran V, Kubelka M, Schultz RM, Motlik J. Aurora kinase A controls meiosis I progression in mouse oocytes. Cell Cycle. 2008;7:2368–2376. doi: 10.4161/cc.6361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Agnese V, Bazan V, Fiorentino FP, Fanale D, Badalamenti G, Colucci G, Adamo V, Santini D, Russo A. The role of Aurora-A inhibitors in cancer therapy. Ann Oncol. 2007;18(Suppl 6):vi47–vi52. doi: 10.1093/annonc/mdm224. [DOI] [PubMed] [Google Scholar]
- 70.Gautschi O, Heighway J, Mack PC, Purnell PR, Lara PN, Jr, Gandara DR. Aurora kinases as anticancer drug targets. Clin Cancer Res. 2008;14:1639–1648. doi: 10.1158/1078-0432.CCR-07-2179. [DOI] [PubMed] [Google Scholar]
- 71.Harrington EA, Bebbington D, Moore J, Rasmussen RK, Ajose-Adeogun AO, Nakayama T, Graham JA, Demur C, Hercend T, Diu-Hercend A, et al. VX-680, a potent and selective small-molecule inhibitor of the Aurora kinases, suppresses tumor growth in vivo. Nat Med. 2004;10:262–267. doi: 10.1038/nm1003. [DOI] [PubMed] [Google Scholar]
- 72.Jass JR, Biden KG, Cummings MC, Simms LA, Walsh M, Schoch E, Meltzer SJ, Wright C, Searle J, Young J, et al. Characterisation of a subtype of colorectal cancer combining features of the suppressor and mild mutator pathways. J Clin Pathol. 1999;52:455–460. doi: 10.1136/jcp.52.6.455. [DOI] [PMC free article] [PubMed] [Google Scholar]