Identification of small gains and losses in single cells after whole genome amplification on tiling oligo arrays (original) (raw)
Abstract
Clinical DNA is often available in limited quantities requiring whole-genome amplification for subsequent genome-wide assessment of copy-number variation (CNV) by array-CGH. In pre-implantation diagnosis and analysis of micrometastases, even merely single cells are available for analysis. However, procedures allowing high-resolution analyses of CNVs from single cells well below resolution limits of conventional cytogenetics are lacking. Here, we applied amplification products of single cells and of cell pools (5 or 10 cells) from patients with developmental delay, cancer cell lines and polar bodies to various oligo tiling array platforms with a median probe spacing as high as 65 bp. Our high-resolution analyses reveal that the low amounts of template DNA do not result in a completely unbiased whole genome amplification but that stochastic amplification artifacts, which become more obvious on array platforms with tiling path resolution, cause significant noise. We implemented a new evaluation algorithm specifically for the identification of small gains and losses in such very noisy ratio profiles. Our data suggest that when assessed with sufficiently sensitive methods high-resolution oligo-arrays allow a reliable identification of CNVs as small as 500 kb in cell pools (5 or 10 cells), and of 2.6–3.0 Mb in single cells.
INTRODUCTION
Many clinical applications, such as pre-implantation and non-invasive prenatal diagnosis, would benefit from the ability to characterize the entire genome of individual single cells by high resolution. Furthermore, in specific cancer research applications, such as the investigation of disseminated tumor cells (micrometastases) in bone marrow or circulating tumor cells in blood, often only single cells or very small cell numbers are available for analyses. The same applies to precancerous lesions, such as cells with dysplasia or early adenomas. In addition, due to the discovery that the genome of all humans has copy-number variations (CNVs) (1–3) and that these may contribute to phenotype variability and disease susceptibility (4), screening of whole genomes for CNVs represents one of the most fascinating areas in human genetics at present. More recently, evidence was reported that CNVs may arise in somatic cells resulting in somatic CNV mosaicism in differentiated human tissues (5,6). The prospect that the presence of some CNVs may be limited to confined somatic areas and their potential impact on physiological processes further fuels the need for reliable CNV screening approaches in small cell amounts.
Comparative genomic hybridization (CGH) allows scanning of the whole genome for CNVs. However, CGH is usually performed with DNA extracted from thousands of cells and thus measures the average copy number of a large population of cells. Accordingly, CGH is sensitive to CNV heterogeneity within the cell population. Without preceding special, unbiased whole genome amplification, CGH is not amenable to single cell or few cell analyses.
Recently, first results were published describing the hybridization of single cell amplification products to various array platforms. Initial studies reported resolution limits of entire chromosomes (7) or of 34 Mb at best (8) and thus failed to demonstrate a significant improvement compared with conventional methodologies. By using the GenomePlex library technology for DNA amplification (GenomePlex Single Cell Whole Genome Amplification Kit, Sigma-Aldrich), we reported that copy number changes as small as 8.3 Mb in single cells can be detected reliably (9). Another group employed a 3.000 BAC array and achieved the detection of about 60% of gains, losses and interspersed normal regions ‘smaller than 20 Mb’ (10). Therefore, to the best of our knowledge, even the most advanced published single cell array-CGH technologies have resolution limits which represent only a slight improvement as compared to conventional CGH on metaphase spreads.
These earlier studies do not offer a detailed map of how robust a genome with CNVs is represented when whole genome amplification products are applied to oligo tiling arrays. To this end, we specifically selected clinical samples from some individuals in which previous analyses had revealed defined deletions on chromosome 22. We performed analyses on oligo tiling array platforms, which possess the highest density of oligonucleotides at present, i.e. NimbleGen's Chromosome 22 Tiling array (HG18 CHR22 FT) covering 385.210 oligos resulting in a median probe spacing of 65 bp and to a custom made chromosome 22 array (Agilent) with 241.700 oligo probes and a median probe spacing of 104 bp. In addition, we employed the NimbleGen Whole Genome Tiling Array (HG18 WG Tiling 2.1M CGH v2.0D) consisting of 2.1-million oligo probes, resulting in a median probe spacing of 1169 bp. During the evaluation of these cells, we noted that standard array CGH-evaluation programs are not suited for the evaluation of single cell amplification products and we therefore developed a new algorithm. In order to test the robustness of this algorithm and to start to address specific biological questions, we analyzed single cells from two cancer cell lines and polar bodies on a 244K whole genome array (Agilent).
As reported previously multiple displacement amplification with Φ29 polymerase results in different amplification of regions in relation to the GC content (11). The same applies to a linker adaptor whole genome amplification approach (12), because when these amplification products were hybridized to a BAC array GC rich regions on chromosome 19 had to be excluded from analysis (10). As we did not observe any nucleotide related amplification bias when applying the GenomePlex library technology to a tiling BAC array (9), we applied this amplification method to all experiments described here.
MATERIALS AND METHODS
Samples from clinical cases, cancer cell lines and polar bodies
We used cells from two probands (P1 and P2) with mental retardation and dysmorphic features in whom previous analyses performed on the whole genome 44K Agilent array had shown deletions on chromosome 22 with sizes of 2.8 Mb (P1) and 3 Mb and 1.2 Mb (both P2), respectively. Furthermore, we prepared new cells from the stable female renal cell carcinoma cell line 769P, because we are very familiar with this cell line from previous analyses (9) and the colorectal cancer cell line HT29, which is known to be chromosomally instable (13). For polar body analyses oocyte collection and processing were done according to standard protocols.
Isolation of single cells and whole genome amplification
Cultured cells were centrifuged at 700 g for 10 min, re-suspended in 1×PBS and transferred onto a polyethylene-naphthalate (PEN) membrane covered microscope slide (Zeiss, Austria) by cyto-centrifugation at 120 g for 3 min. After removing the supernatant, slides were air dried at room temperature overnight. Isolation of single cells and cell pools was carried out using a laser microdissection and pressure catapulting system (LMPC; P.A.L.M., Zeiss, Austria). Single cells and cells pools were randomly selected and directly catapulted into the cap of a 200 µl Eppendorf tube containing 10 µl digestion mix.
We performed whole genome amplification of the single cells and cell pools according to our recently published protocol (9,14). In brief, we employed the GenomePlex Single Cell Whole Genome Amplification Kit (#WGA4; Sigma-Aldrich, Germany) according to the manufacturer's instructions with some modifications. In a final volume of 10 µl, the specimens were centrifuged at 20.800 g for 10 min at 4°C. After cell lysis and Proteinase K digest, the DNA was fragmented and libraries were prepared. Amplification was performed by adding 7.5 µl of 10× Amplification Master Mix, 51 µl of nuclease-free water and 1.5 µl Titanium Taq DNA Polymerase (#639208; Takara Bio Europe/Clontech, France). Samples were amplified using an initial denaturation of 95°C for 3 min followed by 25 cycles, each consisting of a denaturation step at 94°C for 30 s and an annealing/extension step at 65°C for 5 min. After purification using the GenElute PCR Clean-up Kit (#NA1020; Sigma-Aldrich, UK), DNA concentration was determined by a Nanodrop spectrophotometer. Amplified DNA was stored at −20°C.
The quality of the amplification was evaluated using a multiplex PCR approach (15) and samples with four bands on an agarose gel were selected for further array-CGH analysis.
Array-comparative genomic hybridization (array-CGH)
We carried out array-CGH using various oligonucleotide microarray platforms as outlined in the text. For the analysis of amplified DNA samples, reference DNA amplified with the same protocol as described above was used.
Agilent platform
Samples were labeled with the Bioprime Array CGH Genomic Labeling System (#18095-12, Invitrogen, Carlsberg, CA) according to the manufacturer's instructions. Briefly, 500 ng test DNA and reference DNA were differentially labeled with dCTP-Cy5 or dCTP-Cy3 (#PA53021 and #PA55021, GE Healthcare, Piscataway, NJ). Slides were scanned using a microarray scanner (#G2505B; Agilent Technologies, Santa Clara, CA).
NimbleGen platform
Hybridizations on the 2.1 M whole genome array (HG18 WG Tiling 2.1M CGH v2.0D) and the chromosome 22 specific 385K array (HG18 CHR22 FT, both Roche NimbleGen Systems, Reykjavik, Iceland) were performed at service from Roche NimbleGen.
Array-CGH evaluation platform
Data normalization and calculation of ratio values were conducted employing NimbleGen's NimbleScan software package and the Feature Extraction software 9.1 from Agilent Technologies, respectively. The algorithm developed for this study focuses on detecting which ratio values differ significantly [two times standard deviation (SD)] from the ratio profile's mean and should therefore be considered as over- or underrepresented. The concept of the algorithm includes the employment of running means with different window sizes and analyses at progressively greater levels of smoothing and then combining these analyses.
The algorithm is implemented in ‘R’ (version 2.7.0) (16) and addresses three specific issues (i.e. location of windows, window size and threshold selection), which have a significant impact on the identification of very small CNVs in noisy CGH-profiles.
Positioning of windows
Consecutive data points are combined and their mean ratio values are presented in graphs of array-CGH results. The algorithm iterates through the profile by changing window positions, employing a sliding window approach.
The positioning of such windows may have an impact on the ability to detect small CNVs: the scheme in Supplementary Figure 1a illustrates a heterozygous deletion (black), the windows (red) used for the calculation of mean ratio values, and their calculated ratio profiles (blue). In the example on the left side, one window (light red) is located directly inside the deletion, thus the mean ratio value characterizing this region will reflect the actual DNA loss. In addition, the size of the deletion is shown correctly in the ratio profile. On the right side of Supplementary Figure 1a, the windows are positioned in such a way that two windows cover deleted and undeleted regions by half. As a result, these two windows are assigned mean ratio values generated in equal parts from balanced and lost regions. Therefore, the decrease of the ratio value will be lower and the region displayed in the profile (i.e. the size of the two windows) will be larger than the actual deletion.
Taking this into account, the algorithm calculates the mean ratio value for each window and assigns it only to the center of the respective window (Supplementary Figure 1b, blue dots).
As a consequence, CNVs do not appear with a sharp transition border at the location of breakpoints but as a more or less steep slope. For example, Supplementary Figure 1c shows the ratio profiles of the non-amplified DNA (upper panel) in comparison with the averaged ratio profile of the 10-cell pool (lower panel) obtained with DNA of proband P2. The 10-cell pool ratio was generated with a window size of 5.000 oligos (corresponding to 325 kb). Iterative calculations were made with windows of the same size, each moved by 1000 oligos. Note that the three largest CNVs (i.e. deletions with sizes of 3 and 1.2 Mb, and duplication of 532 kb) have already been correctly identified and are therefore shown in green and red, respectively. However, the ratio profile of the 10-cell pool shows no sharp change of the ratio values at the breakpoints.
Window size and threshold selection
The mean ratio value is calculated for each window based on the ratio values it contains. Assuming that a window's ratio values are distributed normally, we estimate the SD by considering the outmost value that is within ±34.1% of the mean. In our previous single cell experiments performed on BAC-arrays, we defined thresholds as ±1.5 times the SD (9). Due to the higher noise on oligo-arrays as compared to BAC-arrays, thresholds had to be defined more stringently as ±2 times the SD.
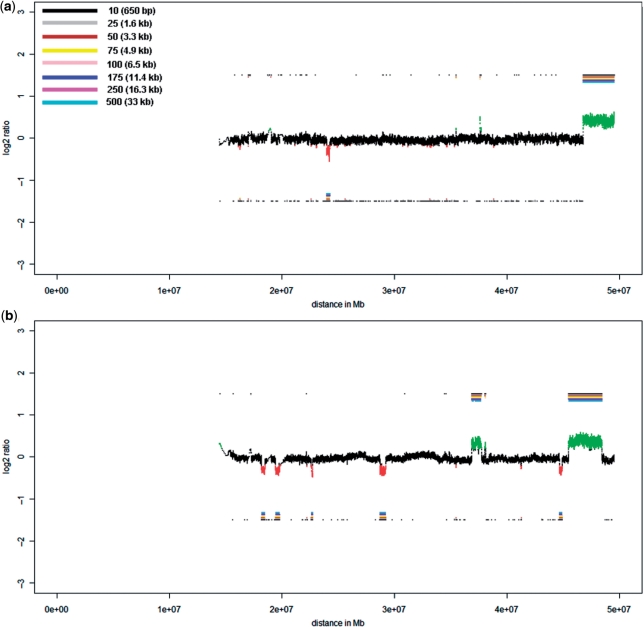
Importantly, when testing calculations with various window sizes we noted that different regions may be called over- or underrepresented. Supplementary Figure 2a and b illustrate again two calculations of the 10-cell pool of proband P2. Both calculations were made with fixed window sizes of 500 oligos (corresponding to 32.5 kb) (Supplementary Figure 2a) and 2.500 oligos (162.5 kb) (Supplementary Figure 2b). In each case, the mean ratio for the entire window and not only the center position is shown. When using the 500 oligo size windows, many of the respective mean ratio values at the chromosomal locations of the three largest CNV regions are above or below the thresholds and are therefore displayed in green and red. However, within these regions there are also many windows which are neither significantly increased nor decreased (black colored regions), and are therefore impeding the distinct identification of CNVs. On the other hand, there are no false positive calls. When using larger windows, e.g. 2.500 oligos, there are more regions within the three largest CNVs which are significantly increased or decreased (Supplementary Figure 2b). However, also some false positive regions are now identified which were not observed with the 500 oligo windows.
These data suggest that a simple increase of the window size alone may not be efficient for improvements of CNV identification. At the same time the observation that different window sizes identify different regions as over- or underrepresented suggests that real CNVs should show specific patterns if the calculations are repeated with various window sizes. Furthermore, these patterns should enable to distinguish between false positive calls and real existing CNVs as illustrated in Supplementary Figure 2c. Panel (1) shows four different calculations, each with a different window size and threshold, as a different SD exists for every calculation. If a window shows a significantly increased or decreased mean ratio value, the mean position of that window will be displayed above or below the respective region of the ratio profile [panel (2)]. Depending on the window size it will be labeled with a different color and distance to the _X_-axis. The thus generated color bar code facilitates the estimation of the size of a CNV because the smaller the CNV the less color bars will be generated [panel (3); compare for example Figure 2a and b]. For more detailed size estimations the algorithm generates a table with all localizations of significant calls which allows the estimation of the CNV size very accurately.
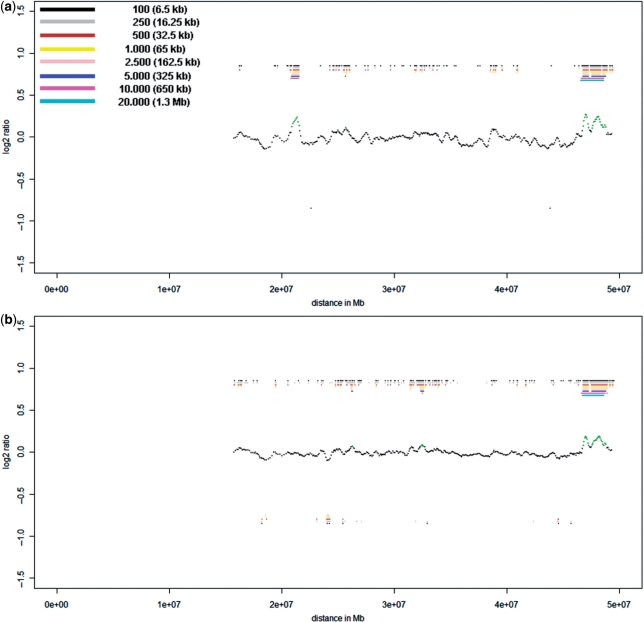
Figure 2.
This figure displays the same ratio profiles as in Figure 1a and b, i.e. the ratio profiles of probands P1 (a) and P2 (b), now calculated with the algorithm described in this manuscript. The center profile is based on calculations with window sizes of 100 adjacent oligos (corresponding to 6.5 kb). A color bar code presents the window size (each in adjacent oligos and the respective physical size) for which calculations have been conducted. In the case of non-amplified DNA we selected very small window sizes, in the other cases with whole genome amplification products the window sizes were larger.
A correctly identified CNV should show the smallest sized windows and also larger windows (depending on the size of the CNV) which have been determined as significant gains or losses [panel (2)]. Other bar code patterns should not occur as they suggest that regions identified as decreased or increased are more likely to be artifacts [panel (4)]: an example of this would be that no gains and losses are identified using the smallest windows but noted at larger window sizes [panel (4), left; for further examples see Supplementary Figures 6c and 7]. Due to the noisy CGH-pattern our algorithm does not require all windows to be detected as CNVs; although the majority of windows of a given size should be identified as gained or lost. Windows detected as CNVs should be continuous, thus no gap between the identification of two different window sizes should occur [panel (4), center]. A single call at any window size, except the smallest window size, is certainly an artifact [panel (4), right]. Therefore the pattern of identified regions with significant deviations from the mean ratio value can help to distinguish between true and false positives. This iterative color bar code generation avoids that a user has to adjust the window size for an individual experiment, therefore preventing the introduction of user bias.
The only user-defined option to interfere with the data representation is the selection which of the ratio profiles should be shown in the center.
RESULTS
Cells from clinical cases (probands P1 and P2) and establishment of their CNV status
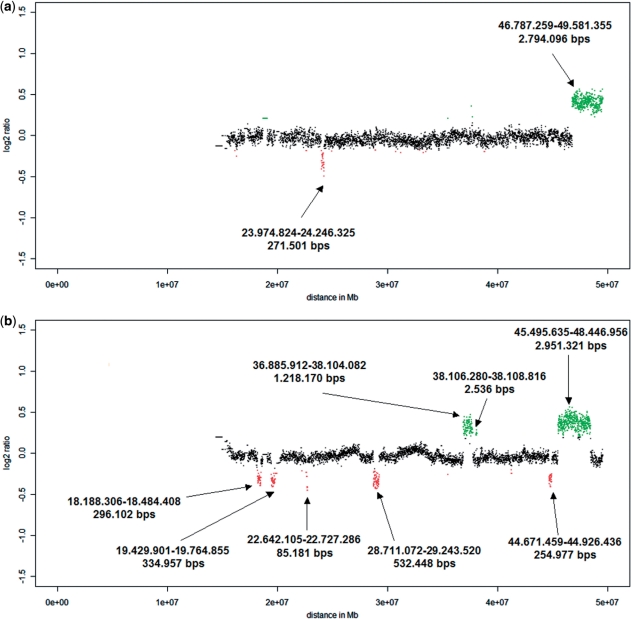
We used cells from two probands (P1 and P2). Previous analyses performed on the whole genome 44K Agilent array had shown deletions on chromosome 22 with sizes of 2.8 Mb (P1) and 3 Mb and 1.2 Mb (both P2), respectively. When hybridizing non-amplified DNA to the NimbleGen Chromosome 22 Tiling array, we observed additional CNVs below the resolution limits of the 44K Agilent array. Proband P1 had an additional duplication of 272 kb (Figure 1a), whereas in proband P2 one additional deletion (size: 2.5 kb) and five duplications of various sizes (532, 335, 296, 255 and 85 kb) (Figure 1b) were observed. These additional CNVs, which had been unknown to us when we designed the experiments, turned out to be very useful for the estimation of resolution limits.
Figure 1.
Ratio profiles of non-amplified DNA of probands P1 (a) and P2 (b) on the NimbleGen Chromosome 22 Tiling array. The calculation of these ratio profiles was based on a classical approach, using a window size of 100 adjacent oligos (corresponding to 6.5 kb) thresholds were simply determined as ±2 times SD. On the NimbleGen arrays losses are illustrated in green above the _X_-axis, whereas gains are shown in red below the _X_-axis. The sizes of observed CNVs are displayed at the respective locations.
For each proband, we prepared cell pools, each consisting of 5 and 10 cells. In addition, we prepared one single cell from P2 and three different single cells from P1. Cell isolation by laser microdissection and subsequent hybridization were performed as previously (14). All experiments were conducted on the NimbleGen Chromosome 22 Tiling array (HG18 CHR22 FT), all amplification products of proband P2 were hybridized to the Agilent custom-made chromosome 22 array and the samples of proband P1 were additionally hybridized to the Whole Genome Tiling Array (HG18 WG Tiling 2.1M CGH v2.0D).
Evaluation of CNVs of probands P1 and P2 in noisy ratios in whole genome amplification products
As expected from our previous experience (9), amplification products yielded significantly noisier ratio profiles on the oligo-arrays than non-amplified DNA did. SDs are a reliable estimate of this noise (9). On the NimbleGen arrays the SDs of non-amplified DNA were in the range of about 0.3, whereas for amplified single-cell or cell-pool material they increased to values ranging from 0.45 to 0.7 (Table 1). By contrast, the SDs on the Agilent arrays were generally lower, i.e. about 0.1 for non-amplified DNA and 0.35–0.66 for amplification products (Table 1). When trying to evaluate these noisy ratios with currently used CGH-programs, such as those available on CGHweb (http://compbio.med.harvard.edu/CGHweb; e.g. CBS, CGHseg, cghFLasso), CNVs were not detected and/or the rate of false positive calls was high (data not shown). This reflects that present CGH programs are not designed for the evaluation of noisy ratio profiles.
Table 1.
Summary of the standard deviations determined for each experiment on the various array-platforms
| Proband | Sample | NimbleGen | Agilent | |
|---|---|---|---|---|
| Chromosome 22 array | Whole genome array | Chromosome 22 array | ||
| P1 | Non-amplified DNA | 0.29 | 0.29 | ND |
| Pool 10 cells | 0.45 | 0.50 | ND | |
| Pool 5 cells | 0.42 | 0.68 | ND | |
| Single cell #1 | 0.59 | 0.75 | ND | |
| Single cell #2 | 0.80 | 0.89 | ND | |
| Single cell #3 | 0.66 | 1.05 | ND | |
| P2 | Non-amplified DNA | 0.30 | ND | 0.11 |
| Pool 10 cells | 0.51 | ND | 0.30 | |
| Pool 5 cells | 0.59 | ND | 0.35 | |
| Single cell #1 | 0.87 | ND | 0.66 |
We therefore developed a new CGH evaluation algorithm. New features of this algorithm include that the entire evaluation is conducted in an automated way without user interaction in order to avoid that selection of thresholds or sliding window sizes are influenced by user bias. The algorithm iteratively calculates values above or below thresholds for various window sizes, analyses the data at progressively greater levels of smoothing and then combines the data. These calculations result in a pattern distribution of regions identified as imbalanced, which allows to distinguish between artifacts and real imbalances and also to estimate the size of CNVs (details in ‘Materials and Methods’ section).
In a first step, we reevaluated the array-CGH profiles of the non-amplified DNA, shown in Figure 1, with this algorithm. As expected, all previously observed gains and losses could be identified again (Figure 2a and b). In addition, we evaluated the DNA of proband P2 on the custom-made Agilent Chromosome 22 Tiling array (Supplementary Figure 3), yielding an almost identical ratio profile as on the NimbleGen array.
Results obtained with cell samples from proband P2
Analyses of amplification products obtained with 5- and 10-cell pools
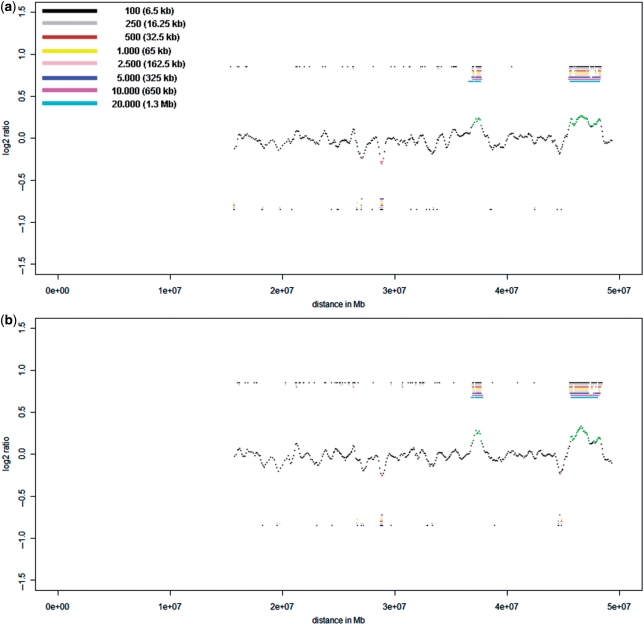
The NimbleGen Chromosome 22 Tiling array comprises 385.210 oligonucleotides and has a median probe spacing of 65 bp. On this array platform, we detected the three largest CNVs of 3, 1.2 Mb and 532 kb with ease (Figure 3a and b) with both amplification products of the cell pools (5 or 10 cells). However, smaller CNVs could not be identified.
Figure 3.
Cell-pool results obtained for proband P2 on the NimbleGen Chromosome 22 Tiling array. (a) Evaluation of the 10-cell pool on the NimbleGen Chromosome 22 Tiling array. The profile shown in the center was obtained with a window size of 5.000 oligos (corresponding to 325 kb). The two largest CNVs show bar codes from black to cyan, demonstrating that the size of the CNVs is in the range of 1.3 Mb or larger (actual sizes: 3 and 1.2 Mb, respectively; compare Figure 1b). In contrast, the largest duplication has a bar code ranging only from black to blue, showing that the size of this CNV is somewhere between 325 and 650 kb (the actual size is 532 kb, Figure 1b). To the left side of this duplication another region at position 26.5 Mb appears to be potentially duplicated. However, the calls are not uninterrupted from black to blue, as there is no pink bar revealing that this CNV call is likely to be an artifact [compare panel (4) in Supplementary Figure 2c]. (b) Hybridization of the 5-cell pool from proband P2 on the NimbleGen Chromosome 22 Tiling array resulted in a CNV recognition pattern similar to that of the 10-cell pool. The algorithm shows the presence of the 255 kb large duplication at position of about 44.7–44.8 (compare Figure 1b), however, the larger 296 and 335 kb duplications were not identified.
The custom-made Agilent array consists of 241.700 oligo probes with a median probe spacing of 104 bp. When applying the 5- and 10-cell pools to this array platform, we identified only the 3 Mb deletion in each case, but no other CNVs (Supplementary Figures 4a and b).
These results suggest that probe density on the array platform may have an important impact on resolution limits. Thus, depending on the array platform resolution limits for the CNV, detection in cell pools consisting of 5–10 cells are in the range of about 500 kb.
Analyses of amplification products obtained with a single cell
As expected, noise of the single cell amplification products was increased, which is also reflected in the SD (Table 1), and resulted in a poorer resolution. On the NimbleGen Chromosome 22 Tiling array, we clearly detected the 3 Mb-deletion, whereas smaller CNVs could not be identified (Figure 4). Similarly, this deletion was also detected on the Agilent Chromosome 22 Tiling array (Supplementary Figure 5).
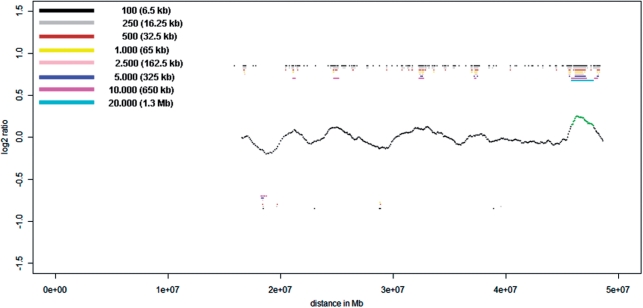
Figure 4.
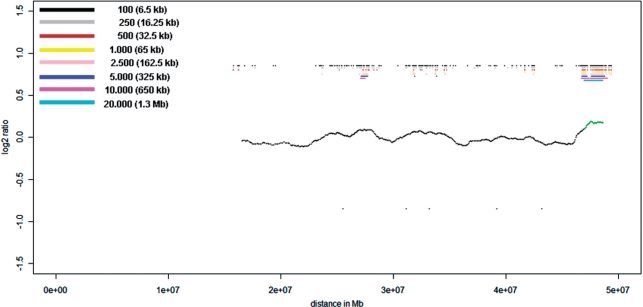
Chromosome 22 profile for proband P2 obtained with a single cell amplification product on the NimbleGen Chromosome 22 Tiling array. Beside the 3 Mb deletion, the bar code pattern displays a possible presence of two smaller deletions at positions 34 and 38 Mb with sizes between 650 kb and 1.3 Mb. The deletion at position 38 Mb corresponds to the location of the real existing 1.2 Mb deletion. However, the second putative deletion at position 34 Mb is false positive, demonstrating that CNVs with a size of <2 Mb cannot be reliably detected in a single cell. Here the center profile was obtained with a 20.000 oligo sliding window (1.3 Mb).
These results suggest that CNVs in single cells with a size of 3 Mb can be detected on appropriate array platforms.
Results obtained with cell samples from proband P1
Analyses of amplification products obtained with 5- and 10-cell pools
In general, the hybridization patterns with the cell samples from proband P1 appeared to be noisier on the NimbleGen Chromosome 22 Tiling array as compared to proband P2. This is not reflected in the SDs (Table 1), which may be due to the fact that the SDs of proband P2 are increased as a result of the unexpected large number of CNVs on chromosome 22. When applying our evaluation algorithm, this increased noise is reflected in the multiple regions above the threshold, which could only be identified with small window sizes (Figure 5). In both cell pools (5 or 10 cells), we detected the deletion of 2.8 Mb, but not the duplication of 271 kb (Figure 5a and b). However, the 10-cell pool also identified a 650 kb large deletion at position 21 Mb (Figure 5a). As shown below, when the same amplification product was hybridized to another array platform, i.e. the NimbleGen Whole Genome Array, this deletion was not visible suggesting that this copy number change is a false positive result and was probably caused by a hybridization artifact rather than by an amplification artifact.
Figure 5.
Cell-pool results obtained for proband P1 on the NimbleGen Chromosome 22 Tiling array. (a) Hybridization of the 10-cell pool clearly identified the 2.8 Mb-deletion. The algorithm also identified another deletion with a size of about 650 kb at position 21 Mb. This deletion is likely to be an artifact (compare Supplementary Figure 6b and details in text). (b) The 5-cell pool of proband P1 also allowed precise identification of the 2.8 Mb-deletion. In addition, at positions 27 and 32 Mb, the algorithm shows the possible presence of two further deletions, each with a size below the 500 kb limit for reliable CNV identification in cell pools. At position 23–24 Mb some bar codes reveal a duplication, which in fact corresponds to the real 272 kb duplication. In both cases the center profile was obtained with a sliding window of 5.000 oligos (325 kb).
For proband P1, we could also compare the ratio profiles of the NimbleGen Chromosome 22 Tiling array with the NimbleGen Whole Genome Tiling Array. On the latter array, chromosome 22 is represented with 26.718 clones, corresponding to median probe spacing of 937 bp. We first compared the ratio profiles obtained with non-amplified DNA on both array platforms and found that these were nearly identical (Supplementary Figure 6a). With the amplification products of the 5- and 10-cell pools, we again detected the 2.8 Mb deletion in each case (Supplementary Figure 6b and c).
In this case, there were no significant resolution differences between the two array-platforms. In fact, the hybridization patterns on the whole genome tiling array appeared to be less noisy as compared to the chromosome 22 tiling array (compare Figure 5a and b with Supplementary Figure 6b and c). In summary, our results suggest that resolution limits for the CNV detection in cell pools consisting of 5–10 cells are in the range of ∼500 kb.
Analyses of amplification products obtained with single cells
We hybridized three different single cell amplification products from proband P1 to the NimbleGen Chromosome 22 Tiling array. However, only in one of the three single cells (‘Single cell #1’) of proband P1, we were able to identify the 2.8 Mb deletion (Figure 6).
Figure 6.
Identification of the 2.8 Mb deletion in a single cell (‘#1’) of proband P1 on the NimbleGen Chromosome 22 Tiling array. The center profile was generated using a 20.000 oligo sliding window (1.3 Mb).
When repeating the single cell analyses of cells on NimbleGen's Whole Genome Tiling Array, we made the same observation, i.e. we discovered the 2.8 Mb-deletion only with the same amplification product from the cell which had allowed us to identify the deletion on the chromosome 22 tiling array (Supplementary Figure 7).
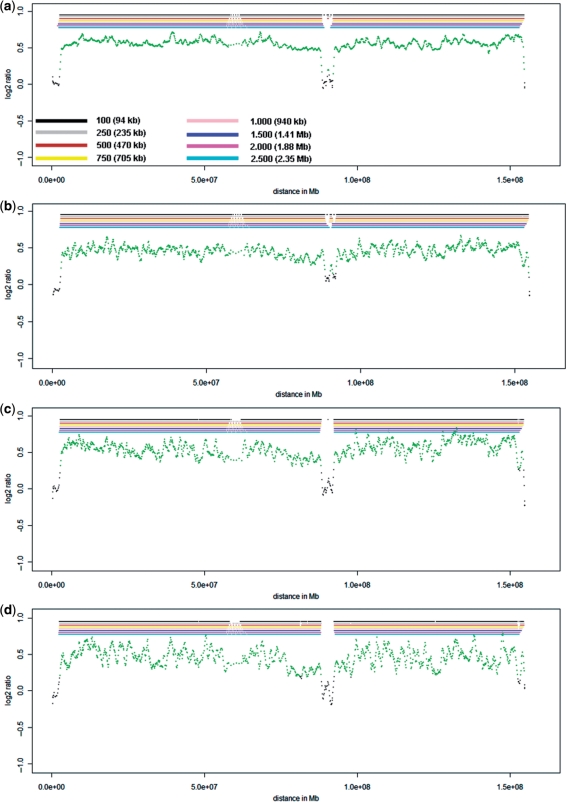
In order to get a more detailed insight whether CNVs with the size of 2.8–3.0 Mb are only borderline-detectable, we also evaluated well-known landmarks on the X-chromosome for hybridizations performed on the NimbleGen 2.1 M Whole Genome Tiling Array. The X-chromosome is represented by 106.458 oligos on this array. Proband P1 is male and the hybridization was carried out with female reference DNA. Due to the different sexes of proband and reference DNA certain landmarks regions on the X-chromosome should show a balanced profile, whereas other regions should show decreased ratio values. The balanced regions include the first pseudoautosomal region (PAR1; size: 2.6 Mb) at chromosome Xp22.3, the XY homology region (XY-HR; size: 4 Mb) at chromosome Xq21.3, and the second pseudoautosomal region (PAR2; size: 320 kb) at chromosome Xq28 (Supplementary Figure 8a). This expected hybridization pattern was indeed observed with non-amplified DNA (Figure 7a). Moreover, both the PAR1 and XY-HR were reliably detected in the cell pool hybridizations (Figure 7b and c) and even in all three single cells (Figure7d and Supplementary Figure 8b and c).
Figure 7.
Ratio profiles of the X-chromosome. (a) Evaluation of the X-chromosome with non-amplified DNA. All X-chromosome landmark regions, i.e. PAR1, PAR2 and the XY-homology region (compare Supplementary Figure 8a) are identified. (b) X-chromosome evaluation of the 10-cell pool, which results in a similar ratio profile as obtained with the non-amplified DNA. (c) X-chromosome evaluation of the 5-cell pool, again with a similar ratio profile. (d) X-chromosome evaluation of the single cell ‘#1’ from proband P1. For this cell the deletion on chromosome 22 was also identified.
Analysis of single cells from two cancer cell lines
In order to further examine how reliably our new algorithm works, we tested single cells from two cancer cell lines on a 244K whole-genome array (Agilent). The first cell line was the female renal cell carcinoma cell line 769P. This cell line is chromosomally very stable as shown by our own previous analyses (9) and by other extensive studies employing M-FISH and array-CGH (17,18). Therefore, we expected that all analyzed cells should show an almost identical CGH-profile. The second cell line was colorectal cancer cell line HT29, which has a good level of chromosomal instability (CIN) with a highly reproducible modal chromosome number (13). Therefore, in this case, we estimated that these cells could show some cell-to-cell variation. Thus, in addition to testing our algorithm's robustness, we could also address the phenomenon of CIN, which is frequently observed in cancer and which is characterized by cell-to-cell variability (19).
In cell line 796P areas of copy number change identified by hybridization of non-amplified DNA could also be detected with the single cell products. To test the reproducibility of the algorithm we compared the ratio profile of non-amplified DNA (Supplementary Figure 9a) with four single cells which met our described quality criteria. For example, chromosome 1 harbors the equivalent of a single copy deletion on the p-arm covering a region of ∼30 Mb and the equivalent of a single copy gain on the q-arm of ∼90 Mb (9). 769P also has a small single copy deletion on chromosome 9 of 6.3 Mb (genomic position 16.7–23.0 Mb) (9). These regions of copy number change were easily identified in single-cell amplified material and non-amplified DNA (Supplementary Figure 9b). We indeed always discovered the same numerical aberrations and, notably, the ∼6.3 Mb deletion on chromosome 9p was detected in each cell.
Cell line HT29 is near triploid and, according to previous publications, shows relative excess of chromosome arms 8q, 13q, 19q and 20q, relative deficiency of 8p, 14q, 17p, 18q and 21q, and pronounced intermetaphase variation (13). To the best of our knowledge, no high-resolution array-CGH profile of this cell line has been published yet. However, a partial high-resolution profile is available on the Agilent web-page (http://www.servicexs.com/blobs/Agilent/Agilent_CGH_brochure.pdf). Our array-CGH profile obtained with non-amplified DNA was consistent with previously published numerical aberrations (13) and with gains and losses described on the aforementioned Agilent web-page (Supplementary Figure 10a). This cell line also harbors two small homozygous deletions on 16p (size: 1.29 Mb; genomic position 6.0–7.3 Mb) and on 20p (size: 1.81 Mb; genomic position 14.2–16.0 Mb). The aforementioned larger numerical changes were easily observed in all four different single cells shown in Supplementary Figure 10b–e. Interestingly, we could even unequivocally identify the small deletions on 16p and 20p in three cells (16p deletion: Supplementary Figure 10c–e; 20p deletion: Supplementary Figure 10b, d and e). In the other cells the ratios at the respective regions were decreased, yet they did not exceed the threshold. Thus, it may be especially easy to detect very small (<2 Mb) homozygous deletions in single cell amplification products.
As expected from the previously reported intermetaphase variation (13), we also observed some alterations not present in all cells. The best example is the deletion of the distal part of 6q. This deletion is easily visible with non-amplified DNA, however, the decrease of the ratio values is not as pronounced as e.g. for 3p or the distal region of 4q (Supplementary Figure 10a), suggesting that this numerical change may be present as mosaic. In fact, in two (Supplementary Figure 10c and d) of the four analyzed single cells, we observed a balanced ratio profile for the entire chromosome 6. In one cell (Supplementary Figure 10b) there was no gain of 18p, which was otherwise visible in all other cells and also in cells from non-amplified DNA. Furthermore, in another cell we observed a large, balanced region within an area on chromosome 7, which was overrepresented in all other cells (Supplementary Figure 10c). This suggests that CIN is in this cell line not only caused by whole-chromosome changes but also by structural rearrangements resulting in segmental aneuploidies. Applying single-cell array-CGH, we had previously made similar observations with the colorectal cell line HCT116 (9).
Analysis of polar bodies
Polar bodies represent an interesting model as chromosomal gains and losses observed in the first and second polar body should complement one another to a large extent. For example, a gain of a certain chromosome in the first polar body leaves two options for this chromosome for the second polar body: first the same chromosome could be lost, indicating a balanced status for this chromosome in the oocyte, or it could be balanced, indicating a loss of this chromosome in the oocyte. However, the gain of a certain chromosome should never be observed in both the first and the second polar body and the same applies for the loss of a chromosome. In preimplantation genetic diagnosis, we focus on polar bodies as Austrian legislation prohibits the analyses of blastomeres.
By now, we have analyzed by CGH 231 polar bodies, including 170 matching first and polar bodies demonstrating that our approach is highly reliable even for the analyses of haploid genomes (manuscript in preparation). Here we present an particularly interesting pair of first and second polar bodies showing complementary gains and losses for chromosomes 1, 9, 10, 13, 18, 20 and 21 (Supplementary Figure 11a and b). However, the first polar body had in addition a gain of chromosome 14 (Supplementary Figure 11a), whereas the second polar body had additional gains of chromosomes 16 and17 and losses of chromosomes 2, 3, 4, 6, 7, 11 and 15 (Supplementary Figure 11b). Thus, the corresponding oocyte should be unbalanced.
Inspection of the ratio profiles revealed another interesting phenomenon: in each polar body ratio, values were at four different levels. For example, in the first polar body (Supplementary Figure 11a), we observed chromosomes with average ratio profiles of about 1 (i.e. chromosomes 10, 14 and 19), 0 (i.e. chromosomes 2, 3, 4, 6, 7, 11, 15, 22), −0.3 (i.e. chromosomes 5, 8, 12, 16, 17), and −1.5 (i.e. chromosomes 1, 9, 13, 18, 20, 21). These different ratio levels are indicated on the right side of each figure (‘1–4’; Supplementary Figure 11). If the two meiotic divisions proceed without any errors, the first polar body should receive 23 chromosomes, each consisting of two chromatids, whereas the second polar body should get 23 chromosomes, each consisting of one chromatid. The four different levels of ratio values we observed in this and other (manuscript in preparation) polar body pairs most likely reflects that meiotic segregation errors even during meiosis I may involve not only chromosomes but also single chromatids. This pair of polar bodies and results from other polar bodies (our unpublished data) demonstrate that high rates of chromosome segregation errors may occur during female meiosis.
DISCUSSION
In this study, we evaluated the performance of amplification products from cell pools or single cells on oligo tiling path arrays. Our results suggest that the use of arrays with a sufficient density of oligos allows the reliable detection of CNVs with a size of 3 Mb (P2) or 4 Mb (size of XY-HR). However, below 3 Mb, the detection of CNVs in single cells becomes critical as we missed a deletion of 2.8 Mb in 2 of 3 cells, whereas we identified the PAR1 region of 2.6 Mb on the X-chromosome in all of these three analyzed single cells. This indicates that reliable detection of CNVs with a size range of below 3 Mb is already at the resolution limit of present protocols for single cell analysis. In contrast, both robustness and resolution increase if only 5 or 10 cells are being analyzed, as we were able to identify CNVs as small as 500 kb in such cell pools.
Confirming our previous observations (9) our results again demonstrate that CGH-profiles from single cells or from a few cells are significantly noisier than those from non-amplified DNA. Amplification of the entire genome of a single cell most likely includes multiple stochastic amplification events due to the low amount of template DNA. Thus, while whole genome amplification products appear to be ‘unbiased’ at low resolution, e.g. if hybridized to metaphase chromosomes [as shown for example by (12) or (20)], variant amplification becomes more obvious on oligo tiling arrays and affects the detection sensitivity of small CNVs.
This requires particularly sensitive methods for data interpretation. Currently available array-CGH programs have been developed for the evaluation of ratio profiles with limited noise, which are usually achieved when non-amplified DNA is used.
In previous experiments when we (9) or others (8,10) tested the performance of amplified DNA on array-platforms, the standard procedure involved a comparison of ratio profiles obtained with amplified DNA versus a baseline profile usually generated with non-amplified DNA. Resolution is then estimated based on the concordance between the two ratio profiles. During these comparisons users will presumably adjust parameters, such as window smoothing or thresholds, until the best correlation between the profiles is achieved. However, since whole genome amplification of single cells or few cells involves a number of stochastic events, CGH-evaluation parameters, which may be optimal for a particular single cell experiment, may be less suited in the next experiment. Accordingly, lacking the option of a comparison with a baseline-ratio profile, the user will not know which parameters are optimal for a most sensitive CNV identification. In fact, in most scenarios performing single cell/few cell analyses reliable baseline profiles are not available for comparison, because otherwise there would be no need for an elaborate single cell analysis. Accordingly, our tests with various standard array-CGH programs revealed in fact that these had not been developed for noisy CGH-patterns and therefore they are not suited for the identification of very small changes in extremely noisy CGH ratio patterns.
For these reasons we developed a new algorithm with the specific aim of detecting small CNVs in very noisy ratio profiles. For the aforementioned reasons the algorithm excludes user interaction. Instead, ratios are iteratively calculated at progressively greater levels of smoothing and the analyses are then combined. This generates a pattern of regions gained or lost. Based on such a pattern the algorithm determines regions of significant ratio deviation. Thus, the main advantages over and differences from other CGH-programs include (i) no user interaction and thus avoidance of user bias; (ii) identification of small CNVs in noisy ratio profiles; (iii) distinction between real CNVs and artifacts; and (iv) reliable size estimates for CNVs based on color coding and tables listing positions of over- and underrepresented regions.
Our comparisons of the ratio profiles between different chromosome 22 tiling array platforms and other oligo tiling arrays suggest that probe density on the array may have an important impact on the resolution limits. Furthermore, as demonstrated in our cell pool experiments, stochastic amplification artifacts are already reduced if only 5 or 10 cells are amplified, resulting in a drastic improvement of both robustness and resolution. This will pave the way for the establishment of detailed CNV-maps from small cell numbers. In addition, we demonstrated that specific biological questions can now be addressed with unprecedented resolution such as CIN in biological samples including cancer cells or polar bodies.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Commission (DISMAL project, contract no. LSHC-CT-2005-018911 and GENINCA, contract no. HEALTH-F2-2008-202230), the FWF Austrian Science Fund (P19455), the Österreichische Nationalbank (12569) and the Austrian Ministry for Science and Research (Project GEN-AU BIN); PhD-Program Molecular Medicine of the Medical University of Graz (to A.C.O.). Funding for open access charge: Medical University of Graz.
Conflict of interest statement. None declared.
Supplementary Material
[Supplementary Data]
ACKNOWLEDGEMENTS
We are grateful to Mag. Maria Langer-Winter for critically reading the manuscript.
Footnotes
The authors wish it to be known that, in their opinion, the first two authors should be regarded as joint First Authors.
REFERENCES
- 1.Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Detection of large-scale variation in the human genome. Nat. Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 2.Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Maner S, Massa H, Walker M, Chi M, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 3.Tuzun E, Sharp AJ, Bailey JA, Kaul R, Morrison VA, Pertz LM, Haugen E, Hayden H, Albertson D, Pinkel D, et al. Fine-scale structural variation of the human genome. Nat. Genet. 2005;37:727–732. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- 4.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruder CE, Piotrowski A, Gijsbers AA, Andersson R, Erickson S, de Stahl TD, Menzel U, Sandgren J, von Tell D, Poplawski A, et al. Phenotypically concordant and discordant monozygotic twins display different DNA copy-number-variation profiles. Am. J. Hum. Genet. 2008;82:763–771. doi: 10.1016/j.ajhg.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Piotrowski A, Bruder CE, Andersson R, de Stahl TD, Menzel U, Sandgren J, Poplawski A, von Tell D, Crasto C, Bogdan A, et al. Somatic mosaicism for copy number variation in differentiated human tissues. Hum. Mutat. 2008;29:1118–1124. doi: 10.1002/humu.20815. [DOI] [PubMed] [Google Scholar]
- 7.Hu DG, Webb G, Hussey N. Aneuploidy detection in single cells using DNA array-based comparative genomic hybridization. Mol. Hum. Reprod. 2004;10:283–289. doi: 10.1093/humrep/gah038. [DOI] [PubMed] [Google Scholar]
- 8.Le Caignec C, Spits C, Sermon K, De Rycke M, Thienpont B, Debrock S, Staessen C, Moreau Y, Fryns JP, Van Steirteghem A, et al. Single-cell chromosomal imbalances detection by array CGH. Nucleic Acids Res. 2006;34:e68. doi: 10.1093/nar/gkl336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fiegler H, Geigl JB, Langer S, Rigler D, Porter K, Unger K, Carter NP, Speicher MR. High resolution array-CGH analysis of single cells. Nucleic Acids Res. 2007;35:e15. doi: 10.1093/nar/gkl1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuhrmann C, Schmidt-Kittler O, Stoecklein NH, Petat-Dutter K, Vay C, Bockler K, Reinhardt R, Ragg T, Klein CA. High-resolution array comparative genomic hybridization of single micrometastatic tumor cells. Nucleic Acids Res. 2008;36:e39. doi: 10.1093/nar/gkn101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lage JM, Leamon JH, Pejovic T, Hamann S, Lacey M, Dillon D, Segraves R, Vossbrinck B, Gonzalez A, Pinkel D, et al. Whole genome analysis of genetic alterations in small DNA samples using hyperbranched strand displacement amplification and array-CGH. Genome Res. 2003;13:294–307. doi: 10.1101/gr.377203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klein CA, Schmidt-Kittler O, Schardt JA, Pantel K, Speicher MR, Riethmuller G. Comparative genomic hybridization, loss of heterozygosity, and DNA sequence analysis of single cells. Proc. Natl Acad. Sci. USA. 1999;96:4494–4499. doi: 10.1073/pnas.96.8.4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abdel-Rahman WM, Katsura K, Rens W, Gorman PA, Sheer D, Bicknell D, Bodmer WF, Arends MJ, Wyllie AH, Edwards PA. Spectral karyotyping suggests additional subsets of colorectal cancers characterized by pattern of chromosome rearrangement. Proc. Natl Acad. Sci. USA. 2001;98:2538–2543. doi: 10.1073/pnas.041603298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geigl JB, Speicher MR. Single-cell isolation from cell suspensions and whole genome amplification from single cells to provide templates for CGH analysis. Nat. Protoc. 2007;2:3173–3184. doi: 10.1038/nprot.2007.476. [DOI] [PubMed] [Google Scholar]
- 15.van Beers EH, Joosse SA, Ligtenberg MJ, Fles R, Hogervorst FB, Verhoef S, Nederlof PM. A multiplex PCR predictor for aCGH success of FFPE samples. Br. J. Cancer. 2006;94:333–337. doi: 10.1038/sj.bjc.6602889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.R: A Language and Environment for Statistical Computing. Vienna, Austria.: R Foundation for Statistical Computing; 2008. R Development Core Team. ISBN 3-900051-07-0, URL http://www.R-project.org. [Google Scholar]
- 17.Fiegler H, Redon R, Andrews D, Scott C, Andrews R, Carder C, Clark R, Dovey O, Ellis P, Feuk L, et al. Accurate and reliable high-throughput detection of copy number variation in the human genome. Genome Res. 2006;16:1566–1574. doi: 10.1101/gr.5630906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fiegler H, Carr P, Douglas EJ, Burford DC, Hunt S, Scott CE, Smith J, Vetrie D, Gorman P, Tomlinson IP, et al. DNA microarrays for comparative genomic hybridization based on DOP-PCR amplification of BAC and PAC clones. Genes Chromosomes Cancer. 2003;36:361–374. doi: 10.1002/gcc.10155. [DOI] [PubMed] [Google Scholar]
- 19.Geigl JB, Obenauf AC, Schwarzbraun T, Speicher MR. Defining ‘chromosomal instability’. Trends Genet. 2008;24:64–69. doi: 10.1016/j.tig.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 20.Gangnus R, Langer S, Breit E, Pantel K, Speicher MR. Genomic profiling of viable and proliferative micrometastatic cells from early-stage breast cancer patients. Clin. Cancer Res. 2004;10:3457–3464. doi: 10.1158/1078-0432.CCR-03-0818. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplementary Data]