Getting to First Base in Proteasome Assembly (original) (raw)
. Author manuscript; available in PMC: 2013 Nov 12.
Published in final edited form as: Cell. 2009 Jul 10;138(1):10.1016/j.cell.2009.06.035. doi: 10.1016/j.cell.2009.06.035
Abstract
Assembly of complex structures such as the eukaryotic 26S proteasome requires intricate mechanisms that ensure precise subunit arrangements. Recent studies have shed light on the pathway for ordered assembly of the base of the 19S regulatory particle of the 26S proteasome by identifying new precursor complexes and four dedicated chaperones involved in its assembly.
The formation of biological structures—from multimeric enzymes to large molecular machines—relies primarily on the assembly of protein subunits with complementary surfaces. When a structure is composed of identical or a few subunits, self-assembly can drive the formation of large structures such as polyhedral viruses. However, when a structure consists of many diverse components that must associate in a definite order and can have potentially detrimental nonproductive interactions, specific assembly factors are required for its formation. These factors act as process-specific molecular chaperones that prevent incorrect subunit associations. Recent studies (Funakoshi et al., 2009; Kaneko et al., 2009; Park et al., 2009; Roelofs et al., 2009; Saeki et al., 2009) have revealed an ordered pathway to assemble the 19S component of the 26S proteasome, the primary site for protein degradation in eukaryotic cells. In this assembly pathway, four chaperones ensure efficient formation of this complex structure.
The 26S proteasome is a 2.5 MDa proteolytic machine composed of 33 distinct subunits that are highly conserved among eukaryotes (Figure 1). Its primary function is to rapidly degrade proteins marked for destruction by ubiquitination. Consequently, the proteasome has many essential homeostatic functions, including protecting against the accumulation of misfolded polypeptides and controlling diverse processes through the regulated destruction of critical enzymes or transcription factors. Protein substrates are digested within the 20S core particle, a hollow cylinder composed of four stacked rings. The two outer rings contain seven homologous α subunits. The two inner rings consist of seven homologous β subunits and enclose a central compartment containing six proteolytic sites that are the targets for the proteasome inhibitors widely used in research and cancer therapy (Goldberg, 2007). Substrates enter the proteasome through a gated pore. This gate is formed by the interlacing N termini of the α subunits and prevents the nonspecific degradation of cellular proteins. Consequently, proteolysis is regulated by activating complexes that cause gate opening and allow substrate entry. In the 26S proteasome, gate opening is controlled by the 19S regulatory particle (PA700), which caps one or both ends of the 20S particle (Figure 1). This complex catalyzes several critical ATP-stimulated processes, including binding of ubiquitinated proteins, disassembly of ubiquitin chains, and unfolding of globular polypeptides and facilitation of their entry into the 20S core.
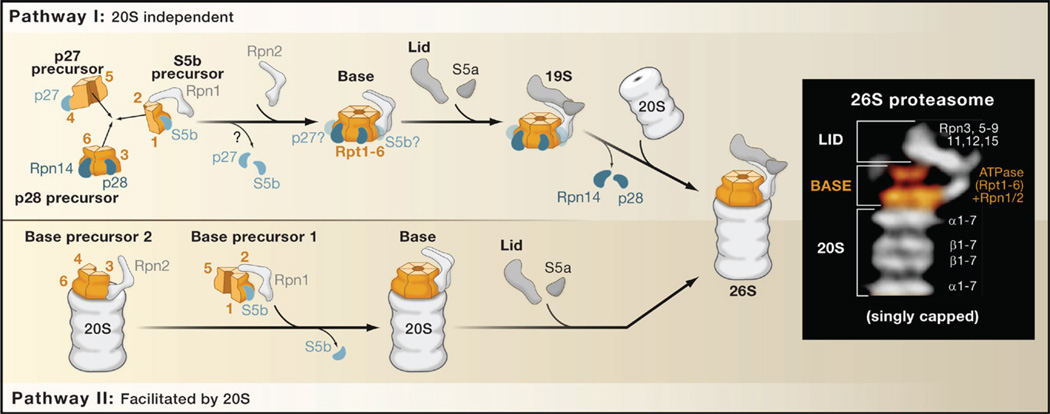
Figure 1. Pathways of Proteasome Assembly.
The eukaryotic 26S proteasome consists of base, lid, and 20S particles (inset, electron micrographs of structures and subunit composition). The 20S particle is composed of 14 different subunits, and its assembly requires five cofactors (not shown). The proteasome base contains six homologous ATPase subunits (Rpt1–6) that assemble into a hexameric ring with Rpn1 and Rpn2. Base assembly is mediated by four chaperones called p27, S5b, p28, and Rpn14 in mammals and Nas2, Hsm3, Nas6 and Rpn14 in yeast (the mammalian nomenclature is used in the figure). Each chaperone interacts with one or two Rpt subunits (indicated as 1–6) and guides their assembly into the base. The exact sequence of events during assembly is unclear. In one possible pathway (pathway I) favored by Funakoshi et al. (2009),Kaneko et al. (2009), and Murata et al. (2009), chaperone-bound subcomplexes coalesce to form a hexameric base that includes Rpn1, Rpn2, Rpn14, and p28. In this complex, the S5b precursor likely releases S5b, and Rpn2 displaces p27. The lid assembles independently by an unknown mechanism and completes the 19S along with S5a (Rpn10 in yeast). The 20S particle then displaces p28 and Rpn14 to form the active 26S proteasome. In an alternative pathway (pathway II) favored by Park et al. (2009) and Roelofs et al. (2009), the 20S particle serves as a template on which Rpn2, Rpt4, Rpt6, and Rpt3 (base precursor 2) assemble with the assistance of p28 and Rpn14 to form a subcomplex. This subcomplex is then joined by the S5b precursor and Rpt5 (base precursor 1) as well as the lid and S5a to form the 26S proteasome.
Despite major advances in our knowledge of the 26S proteasome, many questions remain unanswered. The structure of the 20S particle is known with atomic resolution, and the pathway for its ordered assembly has been elucidated (Murata et al., 2009). However, the spatial organization of 19S subunits and their precise roles in substrate degradation are still mysterious. A full understanding of proteasomal mechanisms will also require a high-resolution structure of the 19S complex. Nevertheless, important progress has been made recently in defining the intricate pathway for assembly of the 19S regulatory particle.
Eukaryotic Proteasome Formation Requires Multiple Assembly Factors
Archaea and certain actinomycetes harbor simple forms of the 20S proteasome where the four-ring, 28 subunit complex is composed of one type of α and β subunit. Consequently, proteasome formation in these organisms can occur by self-assembly. In contrast, the α ring of the eukaryotic proteasome is composed of seven distinct α subunits and requires the heterodimeric protein PAC1/2 (Pba1/2 in yeast) for assembly (Murata et al., 2009). The α ring acts as a platform for the arrangement of seven β subunits to form a two-ring “half-proteasome.” This process is facilitated by another heterodimeric cofactor, PAC3/4 (Pba3/4 in yeast). In addition to ensuring correct spatial organization of the α and β subunits, this assembly process prevents the exposure of proteolytic sites that could cause nonspecific destruction of cellular components. For the proteolytic sites to become active, a leader sequence must be cleaved autocatalytically from the β subunits. This process is inhibited by the maturation factor, Ump1 (Pomp in mammalian cells), which is degraded when the two half-proteasomes coalesce into the four-ring cylinder (Murata et al., 2009).
The assembly pathway of the 19S regulatory particle is more complex than that of the 20S particle. The 19S is composed of two subcomplexes—the lid and the base—whose association is stabilized by the S5a/Rpn10 ubiquitin-binding subunit (Figure 1). The simultaneous efforts of five laboratories now shed light on how the base of the regulatory particle is formed. Supported by an earlier study (Le Tallec et al., 2009), Funakoshi et al. (2009) and Saeki et al. (2009), reporting in Cell, and Park et al. (2009) and Roelofs et al. (2009), reporting in Nature, describe multistep pathways for 19S base assembly in budding yeast that require four chaperone-like cofactors—Nas2, Hsm3, Nas6, and Rpn14.Kaneko et al. (2009) also report in Cell a similar pathway for 19S base assembly in mammals that involves homologs of these yeast assembly factors. All of these chaperones were previously described as integral 26S subunits or proteins associated with the proteasome. However, these recent studies show that they are not components of the mature 26S complex but are associated only with free 19S particles or their precursors.
The base of the 19S regulatory particle is composed of a hexameric ring of homologous ATPases (Rpt1–6) whose C termini dock in specific pockets of the α ring of the 20S particle (Smith et al., 2007). The base also contains two larger subunits, Rpn1 and Rpn2, as well as a small ubiquitin-binding subunit, Rpn13. As demonstrated by all six recent studies, the 19S base forms independently of the lid through a process involving four assembly factors that differ markedly in structure from one another. Each of these proteins—p27, S5b (Hsm3 in yeast), p28 (Nas6 in yeast), and Rpn14—specifically interacts with the C-terminal domain of a different ATPase (Rpt) subunit (Park et al., 2009; Roelofs et al., 2009; Saeki et al., 2009). Each ATPase subunit (except Rpt2, which binds to the base subunit Rpn1, and Rpt4), associates with one of these assembly factors (Figure 1). During base assembly, S5b interacts with Rpt1, p28 interacts with Rpt3, Rpn14 interacts with Rpt6, and p27 interacts with Rpt5, as demonstrated by coimmunoprecipitation and mass spectrometric analyses.
Chaperones Guide 19S Base Assembly via Precursor Complexes
Two models emerge from these recent studies to explain how each Rpt subunit finds its correct position within the ATPase ring during base assembly (Figure 1). In one model, Funakoshi et al., Kaneko et al., and Saeki et al. present evidence that chaperones support the stepwise formation of precursor subcomplexes that then assemble into the 19S particle and join with a 20S particle to form the 26S proteasome (Figure 1). These investigators suggest that complementary interfaces on the Rpt subunits allow the four chaperone-associated ATPases (Rpt1, 3, 5, 6), Rpt2 (in association with Rpn1), and Rpt4 to undergo dimerization to form three precursor subcomplexes— Rpn1-Rpt2-Rpt1-S5b (S5b precursor), p28-Rpt3-Rpt6-Rpn14 (p28 precursor), and Rpt4-Rpt5-p27 (p27 precursor). In addition, Rpn2 may also associate with Rpn13 (Funakoshi. et al., 2009; Kaneko. et al., 2009). The assembly of these precursor subcomplexes is elegantly documented by Kaneko et al. through a combination of RNA interference and glycerol gradient analysis. They show that p28 in the p28 precursor facilitates the association of Rpt3 with Rpt1 in the S5b precursor to release S5b and to form a complex containing four ATPase subunits.
The incorporation of the p27 precursor into the complex containing the four ATPase subunits completes the hexameric ATPase ring of the base. This structure then associates with the remaining base subunit Rpn2 (probably together with Rpn13) and releases p27, which seems to inhibit the premature entry of Rpn2 into the base (Kaneko et al., 2009). The assembled base associates with the 19S lid through an as yet undefined mechanism that is likely to be facilitated by S5a/Rpn10. Once the lid and base are joined together, the 20S particle is able to associate, triggering the release of Rpn14 and p28 (Figure 1). Thus, in this pathway of 19S assembly, formation of the 19S base appears to be guided by two sets of seemingly redundant chaperones—p27/S5b and p28/Rpn14. Of these factors, p27 and S5b seem primarily involved in assembling the base, whereas p28 and Rpn14 seem to coordinate base attachment to the 20S. However, each chaperone is likely to also have distinct functions as their sequence motifs differ considerably. Although the exact sequence of events in base formation remains unclear, the general scheme of chaperone-directed base formation is strongly supported by two-hybrid analysis in yeast (Le Tallec et al., 2009; Saeki et al., 2009), as well as by coimmunoprecipitation experiments and mass spectrometric analysis of assembly intermediates.
The 20S Particle Can Be a Template in Base Assembly
Evidence for an alternative mechanism of base assembly is presented by Park et al. and Roelofs et al. (Figure 1). These investigators show that the 20S particle can serve as a template for assembling the base, as was proposed in a previous study (Kusmierczyk et al., 2008). They presented evidence that half of the base—a complex they call base precursor 2 (BP2) that includes Rpn2, Rpt4, Rpt6, and possibly Rpt3— assembles on top of the 20S particle. This structure is then joined by a protein complex they term base precursor 1 (BP1), which resembles the S5b-Rpt1-Rpt2-Rpn1 precursor but also contains Rpt5 (Park et al., 2009; Roelofs et al., 2009). These investigators show that the chaperones S5b, Rpn14, and p28 interact specifically with the ultimate C-terminal residues of the ATPases. The outer ring of the 20S particle contains binding pockets for the C termini of each of the ATPases (Smith et al., 2007). The C termini of Rpt2 and Rpt5 contain the conserved HbYX motif, which docks into specific intersubunit pockets of the α ring and triggers gate opening (Rabl et al., 2008; Smith et al., 2007). Interestingly, Rpt4 and Rpt6 assemble first on the 20S particle (as part of BP2), but because they lack the HbYX motif, their binding should not cause premature gate opening and proteolysis. Deletion of the C-terminal residues of either Rpt4 or Rpt6 disrupts base assembly, providing strong support for the template model (Park et al., 2009).
Park et al. and Roelofs et al. also reveal important structural insight into the base assembly process by showing that these precursor complexes shield the C termini of the ATPases (to prevent premature activation) and also guide the ordered arrangement of the precomplexes on the 20S. Using the recently reported crystal structure of PAN (Zhang et al., 2009), the homologous proteasome-regulatory complex in archaea (Goldberg, 2007; Smith et al., 2007), these investigators model the Rpt3-p28 interaction and show that p28 prevents the C terminus of Rpt3 from reaching its docking site on the 20S α ring. Both the chaperones p28 and Rpn14 appear to be released from the regulatory particle when either the complete 19S (Figure 1, pathway I) or possibly base precursor 2 (Figure 1, pathway II) docks onto the 20S. During ring assembly, p28 and Rpn14 seem to be brought together through their associations with the adjacent Rpt3 and Rpt6 subunits. The presence of either chaperone is sufficient to ensure 19S formation as the loss of either protein has only minor phenotypic consequences.
These two seemingly distinct models for 19S base assembly may actually represent two stages of a single process. A role for the 20S as a template in base assembly is not addressed in the studies of Funakoshi et al., Kaneko et al., and Saeki et al., which focus on the earlier steps in the pathway. Similarly, Park et al. and Roelofs et al. do not investigate the assembly of base precursor 1 and 2. Possibly, the 20S particle promotes the interaction of the p27 precursor (p27-Rpt4-Rpt5) with the p28 precursor (p28-Rpt3-Rpt6-Rpn14) to form base precursor 2 (Figure 1). These models could merge if Rpn2 also plays a key role in this interaction through its ability to bind Rpt4 and Rpt6 (Park et al., 2009), as well as the 20S particle (Rosenzweig et al., 2008).
Questions for Future Study
A complete understanding of the roles of the four assembly factors will require more knowledge of the structures and interactions of subunits within the 19S base. One outstanding issue emphasized by these studies concerns the roles of the large 19S base subunits, Rpn1 and Rpn2. The Rpn1/2 heterodimer can bind to the 20S particle, and it has been proposed that the six ATPases form a ring enclosing Rpn1/2 (Rosenzweig et al., 2008). Such a model seems difficult to reconcile with the new images of the ring structure of PAN (Zhang et al., 2009) and the finding (in all of the new studies) that Rpn1 is incorporated into a precursor complex at an initial stage in base assembly. Unfortunately, the location of Rpn1/2 in the assembled particle is still uncertain, and the mode for the incorporation of Rpn2 remains controversial. However, because assembly of the base is blocked by an N-terminal deletion in Rpn2 (Isono et al., 2007), the Rpn1/2 heterodimer might, through its ring-like structure (Effantin et al., 2009), still serve as an alternative scaffold for the attachment of the ATPase subunits in the absence of specific chaperones.
Now that the general pathway and cofactors for 19S base assembly have been elucidated, it will be important to define the pathway for lid formation. The lid resembles the COP9 signalosome complex and the eIF3 translation-initiating complex in subunit composition. It is the least-studied proteasome component, and even its role in substrate degradation is unclear. Although the general chaperone Hsp90 as well as the protein Int6/eIF3e have been implicated in lid formation (Murata et al., 2009), their exact roles are uncertain. Whether additional chaperones catalyze lid formation and how this process is coordinated with assembly of the base needs to be investigated.
As nearly all proteasomal subunits are essential for viability, it is surprising that deletion of any of the four base chaperones only produces minor phenotypic effects under normal conditions. There are clearly two sets of redundant assembly factors with partially overlapping roles—p27/S5b (Nas2/Hms3 in yeast) and p28/Rpn14 (Nas6/Rpn14 in yeast)—such that in the absence of any one chaperone, a less efficient alternative pathway for base assembly can function. Future studies should clarify how the abundance and activities of these chaperones are regulated. The cell’s need for new proteasomes and possibly also for assembly factors increases in situations such as heat shock when cells generate large amounts of misfolded proteins that must be rapidly degraded. Intriguingly, Roelofs et al. report that in yeast, cells lacking Nas6, Hms3, or Rpn14 have a growth defect at elevated temperatures that is enhanced by the loss of the transcription factor Rpn4. Rpn4 controls the expression of 26S subunits and is activated when proteasome capacity is insufficient to handle substrate load (Hanna and Finley, 2007). Thus, increased proteasome production can compensate for inadequate proteasome function. This capacity to produce and assemble new proteasomes may also determine cell survival in many conditions, such as the exposure to proteasome inhibitors during cancer therapy (although no Rpn4-like factor has yet been identified in higher eukaryotes). If the supply of these chaperones is rate limiting in proteasome formation, one might predict that their expression is also regulated (by Rpn4 in yeast and by an analogous factor in mammals) to ensure efficient 26S assembly when proteasome production is induced.
Three of the chaperones involved in 20S particle formation (Ump1 and PAC1/2) are degraded by the proteasome after they function in assembly. Kaneko et al. observe that base assembly factors are stable, which is consistent with these factors functioning catalytically in multiple rounds of base assembly. They also observe that if a chaperone is downregulated, the ATPase subunit bound by that factor becomes unstable and fails to accumulate. Thus, base assembly factors seem to also function like classic chaperones to protect their client proteins from premature destruction.
These chaperones may also have roles other than as intermediates in 26S proteasome assembly. In fact, p28 (also called gankyrin) was isolated independently of its association with the 19S complex as an oncoprotein that is overexpressed in hepatocellular carcinoma. p28 binds to the tumor suppressors pRb and p53 and enhances their degradation. The relationship, if any, between these oncogenic properties and the role of p28 in 19S assembly clearly merits further study (Dawson et al., 2006). Also, the p27-Rpt4-Rpt5 precursor was originally isolated and shown to promote association between the 19S and 20S particles (Adams et al., 1997). By controlling the interactions between the base and the 20S particle, these chaperones may not only regulate the formation of the 26S particle, but could also influence the level of free 19S in cells. Several intriguing studies have implicated the 19S base in 20S-independent regulation of transcription (Collins and Tansey, 2006). In addition, alternate tissue-specific forms of the 19S complex may exist, as they do for the 20S particle (for example, the immunoproteasome and thymoproteasome that function in immune surveillance) (Murata et al., 2009). These newly identified assembly factors could have additional roles in regulating the assembly of tissue-specific forms of the 19S.
In identifying new precursors and chaperones involved in 26S assembly, these important studies should lead to a fuller understanding of the overall regulation of intracellular proteolysis. They may also reveal targets for the development of inhibitors that reduce proteasome abundance and thus may have therapeutic applications.
REFERENCES
- Adams GM, Falke S, Goldberg AL, Slaughter CA, DeMartino GN, Gogol EP. J. Mol. Biol. 1997;273:646–657. doi: 10.1006/jmbi.1997.1334. [DOI] [PubMed] [Google Scholar]
- Collins GA, Tansey WP. Curr. Opin. Genet. Dev. 2006;16:197–202. doi: 10.1016/j.gde.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Dawson S, Higashitsuji H, Wilkinson AJ, Fujita J, Mayer RJ. Trends Cell Biol. 2006;16:229–233. doi: 10.1016/j.tcb.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Effantin G, Rosenzweig R, Glickman MH, Steven AC. J. Mol. Biol. 2009;386:1204–1211. doi: 10.1016/j.jmb.2009.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funakoshi M, Tomko RJ, Jr, Kobayashi H, Hochstrasser M. Cell. 2009;137:887–899. doi: 10.1016/j.cell.2009.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AL. Biochem. Soc. Trans. 2007;35:12–17. doi: 10.1042/BST0350012. [DOI] [PubMed] [Google Scholar]
- Hanna J, Finley D. FEBS Lett. 2007;581:2854–2861. doi: 10.1016/j.febslet.2007.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isono E, Nishihara K, Saeki Y, Yashiroda H, Kamata N, Ge L, Ueda T, Kikuchi Y, Tanaka K, Nakano A, et al. Mol. Biol. Cell. 2007;18:569–580. doi: 10.1091/mbc.E06-07-0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko T, Hamazaki J, Iemura S, Sasaki K, Furuyama K, Natsume T, Tanaka K, Murata S. Cell. 2009;137:914–925. doi: 10.1016/j.cell.2009.05.008. [DOI] [PubMed] [Google Scholar]
- Kusmierczyk AR, Kunjappu MJ, Funakoshi M, Hochstrasser M. Nat. Struct. Mol. Biol. 2008;15:237–244. doi: 10.1038/nsmb.1389. [DOI] [PubMed] [Google Scholar]
- Le Tallec B, Barrault MB, Guerois R, Carre T, Peyroche A. Mol. Cell. 2009;33:389–399. doi: 10.1016/j.molcel.2009.01.010. [DOI] [PubMed] [Google Scholar]
- Murata S, Yashiroda H, Tanaka K. Nat. Rev. Mol. Cell Biol. 2009;10:104–115. doi: 10.1038/nrm2630. [DOI] [PubMed] [Google Scholar]
- Park S, Roelofs J, Kim W, Robert J, Schmidt M, Gygi SP, Finley D. Nature. 2009;459:866–870. doi: 10.1038/nature08065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabl J, Smith DM, Yu Y, Chang SC, Goldberg AL, Cheng Y. Mol. Cell. 2008;30:360–368. doi: 10.1016/j.molcel.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelofs J, Park S, Haas W, Tian G, McAllister FE, Huo Y, Lee BH, Zhang F, Shi Y, Gygi SP, et al. Nature. 2009;459:861–865. doi: 10.1038/nature08063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenzweig R, Osmulski PA, Gaczynska M, Glickman MH. Nat. Struct. Mol. Biol. 2008;15:573–580. doi: 10.1038/nsmb.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeki Y, Toh EA, Kudo T, Kawamura H, Tanaka K. Cell. 2009;137:900–913. doi: 10.1016/j.cell.2009.05.005. [DOI] [PubMed] [Google Scholar]
- Smith DM, Chang SC, Park S, Finley D, Cheng Y, Goldberg AL. Mol. Cell. 2007;27:731–744. doi: 10.1016/j.molcel.2007.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Hu M, Tian G, Zhang P, Finley D, Jeffrey PD, Shi Y. Mol. Cell. 2009;34:473–484. doi: 10.1016/j.molcel.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]