Quantifying cellular interaction dynamics in 3-D fluorescence microscopy data (original) (raw)
. Author manuscript; available in PMC: 2012 Sep 16.
Published in final edited form as: Nat Protoc. 2009 Aug 20;4(9):1305–1311. doi: 10.1038/nprot.2009.129
Abstract
The wealth of information available from advanced fluorescence imaging techniques used to analyze biological processes with high spatial and temporal resolution calls for high-throughput image analysis methods. Here, we describe a fully automated approach to analyzing cellular interaction behavior in 3-D fluorescence microscopy images. As example application we present the analysis of drug-induced and S1P1-knock-out-related changes in bone-osteoclast interactions. Moreover, we apply our approach to images showing the spatial association of dendritic cells with the fibroblastic reticular cell network within lymph nodes and to microscopy data about T-B lymphocyte synapse formation. Such analyses that yield important information about the molecular mechanisms determining cellular interaction behavior would be very difficult to perform with approaches that rely on manual/semi-automated analyses. This protocol integrates adaptive threshold segmentation, object detection, adaptive color channel merging and neighborhood analysis and permits rapid, standardized, quantitative analysis and comparison of the relevant features in large data sets.
INTRODUCTION
The availability and usage of advanced fluorescence imaging techniques such as confocal and multi-photon microscopy have dramatically increased and enabled researchers to investigate biological processes with a high degree of spatial and temporal resolution. This includes, but is not limited to, studies ranging from static detection of the subcellular localization of proteins to dynamic tracking of fluorescent probes in single cells to intravital imaging of cell behavior in complex tissues and organs. These developments have been especially fruitful for fields like immunology, cancer research, and neuroscience where the system behavior is largely governed by dynamic cell interactions, for instance, by the interactions between lymphocytes and antigen-presenting cells in lymph nodes1–3, lymphocyte recruitment to and interaction with tumors4, and synapse-glia dynamics in the brain5. Quantitative analysis of cell interaction behavior can considerably increase the information we can gain about the molecular mechanisms governing cellular communication processes or may be used to assess and quantify the efficacy of drugs. However, the development of computational high-throughput methods for automated and standardized quantitative analysis of the resulting 3-D image data has lagged behind the experimental advances6.
Here, we describe a protocol for the quantitative investigation of cell-cell, cell-tissue or cell-pathogen interactions that uses a fully automated, high-throughput image analysis method. The approach involves four steps that are performed automatically without user intervention: First, the actual (true-positive) signal is separated from background (false-positive image elements) in each fluorescent image channel. Second, individual image objects are detected in the output data of the first step, which permits acquiring object numbers and size statistics and allows for removal of image artifacts based on prior information about, for instance, minimum cell size. The third step merges the different fluorescent color channels to obtain an unambiguous segmentation. This step is especially important for interaction analyses because interfacing objects of different type (i. e., different color) usually show spatial image overlap that an accurate interface analysis must account for. Finally, in the last step, the interface areas are computed.
Advantages and Disadvantages of our method
Previous approaches to interaction image analysis relied on semi-quantitative estimations using manual measurements of such features. For example, in conventional bone-osteoclast interaction analysis, data sets are sent to commercial labs and processed by personnel manually delineating cellular boundaries and interfaces7–11. In contrast, the method presented here enables investigators to perform rapid and standardized analyses that do not require operator interpretation in the vast majority of cases and are especially suited to the quantification of differences between experimental groups in large 3-D data sets.
To assess the limitations and the usability of our protocol for different data types and image features we tested different types of typical confocal/two-photon image data (see details below) acquired by different experimenters using different microscopy platforms. Moreover, we performed sensitivity analyses by assessing the impact of the variation of the threshold selection parameter and the introduction of artifical image noise on the interface analysis. These tests showed that our approach works equally well for all data types we tested and that parameter/quality variations did not significantly impact the interface analysis results. However, we tested typical image data and noise. A limitation of the protocol (and segmentation approaches in general) is that excessive noise and/or high background signal (getting close to the intensity of the object signal) may lead to a failure of the ability of the method to separate noise/background from actual signal which subsequently generates incorrect interface quantification results. As a rule of thumb the protocol can be applied to image data whose intensity histogram meet the shape characteristics we describe. Vice versa, the quality of the analysis can be expected to deteriorate if e. g. the peak in the lower intensity region of the histogram representing the background `collapses' into the higher intensity `signal' region. However, we would like to point out that the main application of our protocol is the comparison of different experimental groups (see following section for examples) and that for this purpose (small) segmentation errors are acceptable as long as they occur consistently which is guaranteed by our approach.
Example applications
The software we use in this protocol was initially developed for quantifying osteoclast-bone interactions in order to analyze how a S1P1 (sphingosine-1-phosphate)-receptor knock-out mutant and the immunomodulatory drug FTY720 (Fingolimod, an S1P1 agonist) that affect a lipid phosphate chemosensing pathway alter osteoclast activity in situ and thereby affect bone homoeostasis12. We present this as our main example application and then demonstrate the versatility of the approach by describing its application to two other types of data. In the second example we show how our approach can be used to analyze image data to quantify the extent to which dendritic cells decorate the fibroblastic reticular cell (FRC) networks on which T lymphocytes migrate within lymph nodes. The last example shows the applicability of the protocol for the investigation of interaction phenomena on a smaller scale: We analyze image data from recently published experiments13 about the influence of a mouse T cell signaling mutation in the gene encoding SAP (signaling lymphocyte activation molecule (SLAM) associated protein, the cause of X-linked lymphoproliferative syndrome (XLP)) on cell membrane contacts of T and B lymphocytes (this SAP mutation is known to affect the development of humoral immunity by influencing the stability of interactions between these T and B cells).
All these examples have in common that their analysis requires a quantitative assessment of either the size and/or the spatial localization of interfaces between cells or spatial regions with certain properties relevant for the biological question at hand but differ in terms of size and shape of the cells and tissue structures.
The applications demonstrate the advantages of using an automated and standardized analysis method to obtain unbiased comparisons of data sets obtained under different experimental conditions. As illustrated by the images in Figures 1 and 2, background fluorescence and the overlap between different color channels can make manual segmentation a challenging and subjective task. Under such conditions, variations in background signal/noise levels between data sets or between color channels in a single data set can easily mask subtle but significant biological differences or artificially create the impression of such differences. The potentially clinically relevant results obtained for the quantification of osteoclast-bone interaction robustly demonstrated the ability of the immunomodulatory drug FTY720 to reduce the adhesion of osteoclasts to bone surfaces based on multiple data sets displaying typical variations in image quality. This effect was not evident from visual inspection of the microscopy images and could be rigorously quantified only because of the adaptive standardization our protocol offers.
Figure 1.
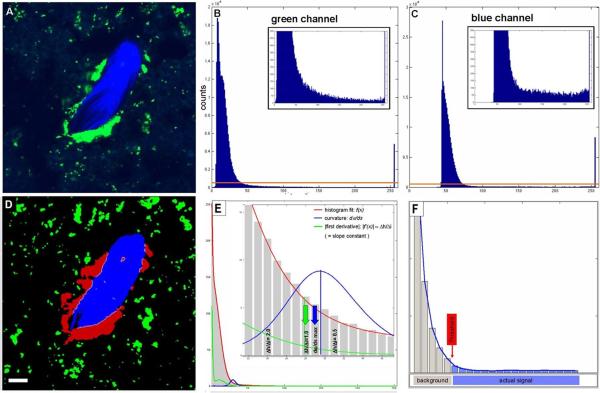
Automated threshold segmentation. Example original (A) fluorescence two-photon image and intensity histograms showing the characteristic shape (B,C). Histograms can be divided into 1) a “peak” region (lower intensity values) and 2) a “constant” region stretching from the end of the “peak” region to higher intensity values. B) osteoclasts (green) and C) bone tissue (blue). D) Resulting segmented image after automated thresholding using maximum curvature estimation method for threshold selection illustrated in E and F. E) The curvature of the graph of the histogram fit (red) is shown in blue. Selection of the threshold at the curvature maximum is indicated by blue arrow. The approximation of the maximum curvature computation by the slope constant criterion f'(x)≈Δh/Δi=1 is depicted by the green curve and arrow. Scale bar represents 10μm. All animal procedures used in this study have been approved by the Animal Care and Use Committee, NIAID, NIH.
Figure 2.
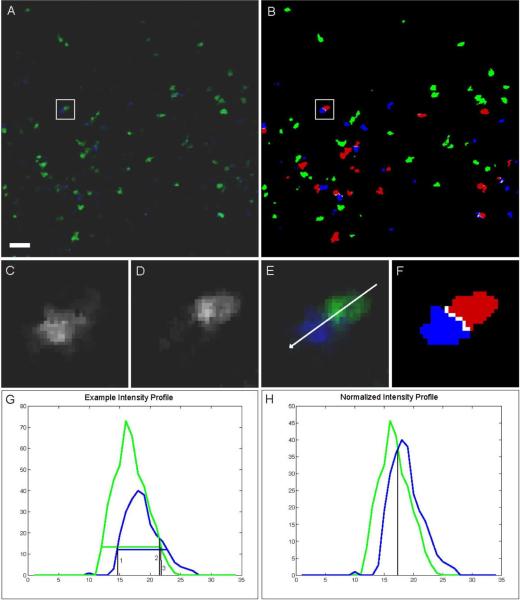
Adaptive channel merging. A, Original image data of T (green) and B (blue) cells. B, segmentation result, interacting T cells (red), interface (white). Magnification of blue (C) and green (D) channels with overlay (E) and segmentation result (F) from inset in A, B. Intensity profile along arrow in E is shown in G. Horizontal green and blue lines in G indicate computed threshold. Black vertical lines (1, 2, and 3) show the selection alternatives of the border between the blue and green channels under the condition that (1) either the blue or (3) the green object is given priority. (2) is the result of the border selection when comparing absolute intensities of competing channels. H, normalized intensity profile allows for balanced identification of the border between color channels (result shown in F). Scale bar represents 10 μm. All animal procedures used in this study have been approved by the Animal Care and Use Committee, NIAID, NIH.
Background to the image analysis program
The image analysis program performs the following steps automatically:
First module: Signal-Background separation by automated adaptive threshold segmentation
The critical step in image segmentation is the separation of signal from background, which includes discrimination of different signals (colors) from one another when considering multicolor fluorescence datasets. The first module of the image processing software that is used here performs an automated adaptive intensity threshold segmentation based on the characteristics of fluorescence confocal or two photon image data. Such intensity histograms show a peak in the lower intensity region that represents the background signal, while the actual image signal is – relative to the form of the background part – approximately uniformly distributed over the (higher) intensity spectrum. We found this to be characteristic of all two photon and confocal fluorescence microscopy images we analyzed and distinct from most conventional (non-fluorescent) image data (as in14) (Figure 1). Because the exact shape and location of the peak and constant region vary depending on the image acquisition features, a stable and reliable segmentation method capable of yielding comparable results has to adapt to these variations that may occur between different as well as within individual data sets. The segmentation algorithm used here accomplishes this by defining the transition point between background and actual signal (i.e., the threshold) as the intensity value at which the curvature of the histogram graph is maximal (see section “Automatic adaptive thresholding: Maximum curvature estimation” for details). In contrast to fully (3-D)-global thresholding methods with single constant or user-selected cut-offs for the whole 3-D image data set, our approach computes the threshold for each image z-slice separately and is therefore capable of compensating for intensity inhomogeneities (or “attenuation”) along the z axis (for instance, in deep-tissue 2-photon imaging in vivo).
Automatic adaptive thresholding: Maximum curvature estimation
In the case of photographic images histograms often have a bimodal shape, with one peak representing background and the other signal portion of the image (Supplementary Figure 1). In such cases it is obvious that the threshold selection criterion is to find the minimum between the two peaks. There is no such criterion that is similarly obvious and also physically plausible in case of two-photon/confocal microscopy image data and the transition between background and signal cannot be defined as a precise location similar to that in case of bimodal histograms. It can only be defined with some remaining degree of fuzziness in the region of the transition between the strongly decaying part of the peak in the lower intensity spectrum and the region with the relatively flat slope. We chose the maximum curvature and its approximation by using a derivative criterion for normalized histograms as threshold selection criterion because it represents a mathematically definable, reasonable estimate of the transition point at which the strongly negative slope of the background peak changes into the relatively flat slope associated with the signal region and moreoever, because it is computationally efficient (alternative threshold selection criteria are discussed in 14).
To compute the curvature of the (discrete) histogram in the relevant region a Gaussian model may be used to fit the histogram in the relevant region, i. e., between global maximum (or local maximum with highest intensity value in the background region) and intensity 254. The maximum intensity histogram bin 255 is omitted in all calculations because it is significantly higher than the other “true positive” intensity values. This is due to the aggregation of all intensities ≥ 255 in this bin. We also omit the 0 bin, because in the case of image data of a low overall intensity (and thus many 0-intensity voxels) its inclusion may interfere with the generation of a normalized histogram that exhibits the shape features required for threshold computation.
With the angle α, f (x) the histogram fit function and the line element ds given by ds=1+(f′(x))2dx the curvature of the graph of f (x) is given by dαds=dαdx⋅dxds=f′′(x)(1+f′(x)2)3∕2 (Figure 1). In our implementation we approximated this process by using the (discretized) derivative of the histogram h(i): To determine the intensity threshold used for the segmentation, the derivative Δ_h_/Δ_i_ is computed stepwise starting at _i_=254 with decreasing i until the transition from the constant image to the non-constant background region is encountered. Prior to the threshold determination, the histogram is smoothed using a Gaussian filter (0 bin is the maximum bin of intensity histogram) scaled by the histogram integral and multiplied by a factor of 255 in order to normalize for different cell/bone tissue coverage of the image area. Because of the common histogram shape features present in all data sets, this scaling/normalization procedure resulted in the value of the discrete derivative ΔhΔi=h(i−Δi)−h(i)Δi to be approximately 1 at the maximum curvature point (Figure 1E). Our sensitivity analysis of the precise location of the selected threshold (see section “Robustness assessment: Impact of variations of the slope constant on the interface area results” for details) shows that this approximation is valid. Supplementary Figure 2 shows an exemplary threshold segmentation result overlaid with the original image.
Second module: Object detection using connected component analysis
Following the automated thresholding procedure, our approach uses a 3-D connected component labeling algorithm (see 15 and references within) to identify individual image objects. The connected component labeling permits computation of the numbers of objects (such as cells) and the corresponding object features such as volume, surface area, and location. It is also required for obtaining detailed information on cell-cell interaction behavior (e.g., the number of cells of type A attached to cells of type B) and for comparing features of potential interest of interacting vs. non-interacting cells. In addition to the utility of the connected component analysis for the quantification of object features, our software also uses this analysis to improve image quality by applying a threshold filter for cell volumes, thus allowing removal from the processed dataset of small pieces of cell debris or bright specks resulting from artifactual dye labeling.
Third and fourth modules: Channel merging and interface area computation using normalized intensity comparison and voxel-neighborhood analysis
The third step of the analysis pipeline finalizes the segmentation by adaptively merging the previously separate segmentations of individual color channels. This is a pivotal step because most current image data involve objects (cells, pathogens, tissue components) that are labeled with different colors and whose fluorescent signals overlap when those objects are in close proximity, resulting in overlap in the segmentations of the individual color channels. Disregarding this issue may lead to distorted interaction results and even to the accidental removal of small objects, especially if one channel is significantly dimmer than the other. Our channel merging approach accounts for spatial overlap between neighboring or interacting image objects by combining the single channel thresholding results with an adaptive intensity comparison of the original image data based on normalization of each channel with its intensity profile. After the initial threshold segmentation (performed for each channel separately) each voxel that has been assigned both class labels (`overlap voxel') is evaluated by comparing the original image intensities in each channel normalized with the corresponding maximum intensity values. The voxel is then assigned to the final segmentation class that represents the channel with the higher relative intensity at that particular voxel. This approach therefore allows for an accurate estimate of the actual interfaces (see Fig. 2 and Supp. Fig. 3).
Finally, the interface areas are computed by identifying those voxels in channel one that are direct neighbors of voxels in channel two. We use the sum of the surface voxels as an estimate of the total contact area. This is sufficient because the analysis result is provided in terms of ratios of interface areas to total cell or tissue surface areas or in terms of comparisons of such areas between different experimental groups.
Robustness assessment: Impact of variations of the slope constant and noise on the interface area results
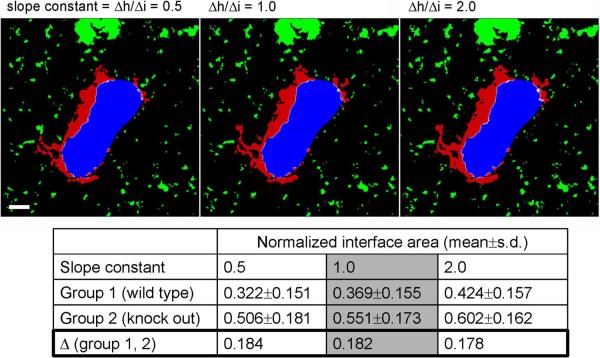
The robustness of an image segmentation and analysis method towards variations of pre-set parameters and image quality is an important measure of its utility. Therefore, we performed a sensitivity analysis to evaluate the impact of variations in the slope value used to identify the boundary between background and true image data. This analysis showed that the average ratio of the interface area / total surface area in two groups (bone-osteoclast gene knock-out vs. wild-type data) with 20 data sets each (each data set consisting of ~10 x–y images of 512×512 pixels) changes only about 10% even when varying the slope value by a factor of 2 in both directions for both channels. More importantly, the differences of the average area ratios between the control and knock-out groups – which is the relevant result in this kind of analysis – show a negligible dependence on the variation of the characteristic slope (change of difference 1.1%; see Figure 3). These results show that the particular choice of the slope value is less important than using the same constant throughout the data analysis, which subsequently generates the proper adaptive threshold value. This ensures that variations of image quality within and between data sets are normalized, thus permitting the consistent comparison of different data sets.
Figure 3.
Analysis of the impact of variations in the derivative constant on the normalized interface area (ratio interface area / total bone surface area) and on the difference between “wild-type” and “knock-out” groups. Mean values are based on 20 data sets each in each group. While changing the derivative constant by a factor of 2 in both directions relative to 1.0 results in average normalized interface area changes of ~10%, the differences change only 1.1% (0.5→1.0) and 2.2% (1.0→2.0). Scale bar represents 10 μm. See also Figure 5. (For more information on the experiments see Ishii et al., Nature, in press). All animal procedures used in this study have been approved by the Animal Care and Use Committee, NIAID, NIH.
In addition to testing our approach with different types of typical confocal and two-photon microscopy data we assessed the robustness of the analysis under the influence of a variety of noise levels. We generated different levels of artifical noise for an exemplary data set (image noise follows a Poisson distribution 16). The comparison of the analysis results yielded interface area differences of ≤5% between different typical Poisson noise levels (see Supplementary Figure 4 for details). However, due to the combination of noise and background signal and the diversity of possible features of fluorescence microscopy data we cannot provide straightforward rules to determine a priori which data our protocol will work with. Low data quality may lead to significant misclassification (see Supplementary Figure 5) and we therefore advise the user to perform tests and examine test results to estimate the suitability of the automated analysis using this protocol and/or to optimize the user-definable parameters for the particular data. However, for the main application of this protocol, the comparative analysis of interaction behavior, it is above all important that data processing is standardized and that the data sets that are to be compared are of roughly similar quality. In that case segmentation errors within reasonable limits are not problematic for comparative analyses using our protocol because they occur consistently.
MATERIALS
Reagents
Images for analysis, see REAGENT SETUP for further information about required format and Supplementary Method for information on how images were acquired for our example application.
Equipment
Software (see Equipment setup for details)
Standary 32bit hardware
The example analyses shown in this protocol were performed on an Intel Xeon Dual Core 3.0 GHz workstation with 16GB memory running 64bit SuSE Linux. CRITICAL While the analyses can in principle be performed on standard 32bit hardware, the memory requirements may go beyond 4GB depending on image size and number of biological objects in the image data and 64bit workstations with 8GB or more memory may be required.
Reagent Setup
Image data preparation
The analysis module of the software we present and use in this protocol requires image data to be in 3-color (RGB) Portable Network Graphics (`.png') format with the z-slices in files consecutively numbered and assigned a defined filename prefix, for instance, `img001.png, img002.png, img003.png …'. Each z-stack has to be saved in a single directory that does not contain any other entries other than the image files. If a 4-D data set is analyzed, each timestep is treated separately and saved in individual directories. Note that because this protocol describes interactions between two types of biological objects, only two color channels are relevant. Here, by convention, we use `green' as the first and `blue' as the second channel. Microscope acquisition software usually permits saving images in TIFF format files with one file per color channel and slice (and in case of 4-D data per timestep) indicated by certain identifiers, for instance `filenameprefix_T000_C0_Z000.tif' where `T', `C' and `Z' indicate timestep, color channel and z slice, respectively. The software `2PISA' offers a module to convert such data into the format described above, if this is not required the protocol should be started at step 10.
Software
The description of the analysis method we provide offers sufficient detail for its implementation with a suitable high-level scripting/programming language (e. g. Matlab). However, we recommend using our implementation of the software `2PISA' (2 Photon Image Segmentation and Analysis), which can be obtained free of charge for non-commercial use from the authors (http://www3.niaid.nih.gov/labs/aboutlabs/psiim/computationalBiology/) and is available for most versions of Linux, MacOS X, and Windows. The software package is written in C/C++ using the Qt library (Nokia, Finland). While the current release of the software supports two-color images, the method can in principle handle an unlimited number of channels as long as each class has a unique color label. An OpenGL-capable system is required for full functionality.
PROCEDURE
Image data preparation
- 1
Start the program `2pisaloader' to prepare the data. - 2
In the menu `Data' select `Convert data'. - 3
If the data you would like to convert is time course data, select yes and enter the single character that identifies the nature of the image parameter of interest. If `T' defines the time step identifier (which is followed by the time step number, for instance: `T001' defines the first time step), enter `T'. Enter the number of digits used to encode the time step: in case of `T001', enter `3'. If the data is not time course data proceed to step 4 - 4
Enter the number of z slices. - 5
Enter the number of digits used for z slice coding. The z stack number must be at the end of the filename before the image format suffix, for instance `…001.png'. - 6
Select the first image file of channel 1 (`green' channel). - 7
Select the first image file of channel 2 (`blue' channel). - 8
Enter the directory in which the converted data will be saved. The software then starts the image file conversion. If time course data are used, directories named according to the time step will be generated within the directory defined and contain the data for each time step. Otherwise, the converted images are saved directly in the directory specified.
Performing data analysis
- 9
Start the software `2pisaloader'. - 10
In the menu `Data', select `Start data analysis'.
TROUBLESHOOTING
- 11
Select the directory that contains the image data. Depending on the content of the selected directory, the software will automatically determine if a single set or multiple data sets are to be analyzed: If the selected directory contains subdirectories with image files, all subdirectories are processed. If no subdirectories are present, the analysis is limited to the files in the selected directory. Because the program counts the number of files in each directory to determine the size of the z stack, it is important that only the relevant images files are in the directory named following the convention described above (see `data preparation'). - 12
Enter the filename prefix. The prefix has to be the same for all images files within the selected directory and subdirectories. Subsequently, the fully automated segmentation and analysis process is started and does not require any further user interaction.
TROUBLESHOOTING
Results
- 13
Locate the segmentation results. These will be saved in the same directories as the original images with the filename prefix `result' followed by the z-slice identifier equivalent to that of the original image, for instance for `originalimage100.png', `result100.png'. In addition to the resulting image files, each directory will contain a file (`Results.txt') providing detailed interaction statistics, such as the number of interfacing cells/tissue components, interface area relative to total surface area, interface area per interfacing component, etc. If multiple data sets/directories are analyzed simultaneously, the top level directory will contain a file (`AllResults.txt') that combines the results of all data sets. - 14
Directlt read these files and/or import to statistics/spreadsheet software for further analysis or visualization.
TROUBLESHOOTING
Step 10: Manual parameter optimization
Even though the default settings for the fully automated processing of the data worked well for all data sets we tested, the user can override the default settings and manually adjust/set the parameters “slope constant”, “threshold value” and “minimal object size” to account for specific data features and optimize the accuracy of the analysis. Manual parameter selection can be enabled in the Settings menu.
Step 12: Memory limitations
If large image data sets containing large numbers of objects are analyzed, the memory of standard 32bit hardware (max. 4GB memory) may be insufficient and the analysis process might be terminated. While it is not possible to precisely determine the memory requirement for a given image size, because it partially depends on the number of image objects processed in the connected component module, a rough estimate is that data sets of size ~250×250×10 (width × length × height) with a number of components similar to that present in the lymphocyte example can in most cases be analyzed on 32bit hardware while datasets equal to or larger than ~500×500×>20 (as is the case for the bone-osteoclast data) are likely to require more memory. As an alternative to using 64bit hardware with sufficient memory (~8–16GB), the data sets can also be split into smaller subsets that are then analyzed separately.
TIMING
The analysis time strongly depends on the size and composition of the data sets and the computer hardware. The approximate duration of the bone-osteoclast interaction analysis with 512×512×~20 data sets on an Intel Xeon 3.0GHz/16GB workstation running 64bit-Linux is in the order of several minutes.
ANTICIPATED RESULTS
Analysis of interactions of osteoclasts and bone tissue
In the first and main example application we show here, the interacting components of interest are bone tissue and osteoclasts (see Figures 1, 3 and 4A for illustrations). The extent of the interface area between osteoclasts and bone provides a measure of osteoclast activity. Generated by fusion of multiple monocyte-derived precursor cells, osteoclasts in their mature state resorb bone tissue, thereby acting as important components for bone and calcium homoeostasis17. To quantify the influence of pathological conditions, therapeutic interventions, or experimental conditions on bone metabolism, previous studies have relied on methods like bone densitometry or the use of manual or semiautomated image analyses7–9, 18 based on a bone histomorphometry standard developed by Parfitt19. Recent data from our laboratory suggested that the lipid mediator S1P and its receptor S1P1 might play important roles in regulating osteoclast generation by controlling the rate of osteoclast precursor detachment from the bone surface before mature osteoclast formation12. To assess the influence of the immunomodulatory drug FTY720 (a blocker of S1P/S1P1-function) on bone metabolism, we developed this protocol to analyze microscopy images of osteoclasts interacting with bone tissue and quantified the osteoclast attachment ratio, which is defined as the ratio of the interface area between osteoclasts and bone tissue and the total bone surface.
Figure 4.
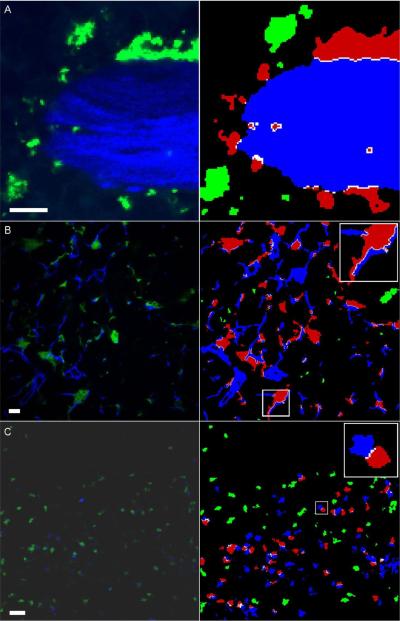
Segmentation example results. Left column: original images; right column: segmentation results. A) Bone-osteoclast interaction (osteoclasts: green; bone: blue; red: interacting osteoclasts). B) Dendritic cells (green/red) and fiber network (blue) in lymph nodes. (white: interface area). Analysis of the whole 3-D data set reveals that 22% of the DC surface is attached to fibers. C) T-B-cell interaction in lymph nodes (T cells: green/red; B cells: blue). Scale bars represent 10 μm. All animal procedures used in this study have been approved by the Animal Care and Use Committee, NIAID, NIH.
We found that this ratio changes from approximately 0.3 in control to 0.6 in ovarectomized mice that develop osteopenia due to unbalanced osteoclast activity and that treatment of these animals with FTY720 reduces the osteoclast attachment to bone almost to normal levels (~0.35, p=0.0006). Evaluation of the influence of S1P1 function on osteoclast function revealed a decrease of the osteoclast attachment ratio from ~0.6 in wild-type to ~0.44 in S1P1−/− knock-out mutants (p=0.0003). (For further details on the experimental design and biological implications of the actual experiments, see12).
Additional example data
While the bone-osteoclast application deals with the interaction of relatively large cells with large tissue structures, the protocol can also be used to analyze interaction behaviour when the relevant dimensions of the biological objects are much smaller. Examples are image data showing dendritic cell - fibroblast reticular network attachment and T - B lymphocyte interaction. Typical results that can be expected from analyzing such data are shown in Figure 4.
Supplementary Material
Supplement
ACKNOWLEDGMENTS
This research was supported by the Intramural Research Program of NIAID, NIH. M.I. was supported by a fellowship grant from International Human Frontier Science Program.
Footnotes
AUTHOR CONTRIBUTION STATEMENT F.K. designed, implemented and tested the method. F.K., M.M.S. and R.N.G. wrote the paper. M.M.S. and R.N.G. supervised the project. M.I., H.Q., M.B., J.E. and F.K. generated and provided experimental data.
COMPETING FINANCIAL INTERESTS STATEMENT The authors declare that they have no competing financial interests.
REFERENCES
- 1.Germain RN, et al. An extended vision for dynamic high-resolution intravital immune imaging. Seminars in immunology. 2005;17:431–441. doi: 10.1016/j.smim.2005.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stoll S, Delon J, Brotz TM, Germain RN. Dynamic imaging of T cell-dendritic cell interactions in lymph nodes. Science (New York, N.Y. 2002;296:1873–1876. doi: 10.1126/science.1071065. [DOI] [PubMed] [Google Scholar]
- 3.Xu X, Meier-Schellersheim M, Yan J, Jin T. Locally controlled inhibitory mechanisms are involved in eukaryotic GPCR-mediated chemosensing. The Journal of cell biology. 2007;178:141–153. doi: 10.1083/jcb.200611096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fisher DT, et al. Hurdles to lymphocyte trafficking in the tumor microenvironment: implications for effective immunotherapy. Immunological investigations. 2006;35:251–277. doi: 10.1080/08820130600745430. [DOI] [PubMed] [Google Scholar]
- 5.Nishida H, Okabe S. Visualization of synapse-glia dynamics. Brain and nerve = Shinkei kenkyu no shinpo. 2007;59:755–761. [PubMed] [Google Scholar]
- 6.Wollman R, Stuurman N. High throughput microscopy: from raw images to discoveries. Journal of cell science. 2007;120:3715–3722. doi: 10.1242/jcs.013623. [DOI] [PubMed] [Google Scholar]
- 7.Yamada C, et al. The murine glucagon-like peptide-1 receptor is essential for control of bone resorption. Endocrinology. 2008;149:574–579. doi: 10.1210/en.2007-1292. [DOI] [PubMed] [Google Scholar]
- 8.Nakamura T, et al. Estrogen prevents bone loss via estrogen receptor alpha and induction of Fas ligand in osteoclasts. Cell. 2007:811–823. doi: 10.1016/j.cell.2007.07.025. 130. [DOI] [PubMed] [Google Scholar]
- 9.Lee SH, et al. v-ATPase V0 subunit d2-deficient mice exhibit impaired osteoclast fusion and increased bone formation. Nature medicine. 2006;12:1403–1409. doi: 10.1038/nm1514. [DOI] [PubMed] [Google Scholar]
- 10.Castellino F, et al. Chemokines enhance immunity by guiding naive CD8+ T cells to sites of CD4+ T cell-dendritic cell interaction. Nature. 2006;440:890–895. doi: 10.1038/nature04651. [DOI] [PubMed] [Google Scholar]
- 11.Henrickson SE, et al. T cell sensing of antigen dose governs interactive behavior with dendritic cells and sets a threshold for T cell activation. Nature immunology. 2008;9:282–291. doi: 10.1038/ni1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishii M, et al. Sphingosine-1-phosphate mobilizes osteoclast precursors and regulates bone homeostasis. Nature. 2009;458:524–528. doi: 10.1038/nature07713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qi H, Cannons JL, Klauschen F, Schwartzberg PL, Germain RN. SAP-controlled T–B cell interactions underlie germinal centre formation. Nature. 2008;455:764–769. doi: 10.1038/nature07345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sezgin H, Sankur B. Survey over image thresholding techniques and quantitative performance evaluation. Journal of Electronic Imaging. 2004;13:146–168. [Google Scholar]
- 15.Suzuki K, Horiba I, Sugie N. Linear-time connected-component labeling based on sequential local operations. Computer Vision and Image Understanding. 2003:1–23. [Google Scholar]
- 16.Kervrann C, Trubuli A. An adaptive window approach for Poisson noise reduction and structure preserving in confocal microscopy. IEEE International Symposium on Biomedical Imaging: Nano to Macro. 2004;1:788–791. 2004. [Google Scholar]
- 17.Bar-Shavit Z. The osteoclast: a multinucleated, hematopoietic-origin, bone-resorbing osteoimmune cell. Journal of cellular biochemistry. 2007;102:1130–1139. doi: 10.1002/jcb.21553. [DOI] [PubMed] [Google Scholar]
- 18.Wada T, et al. The molecular scaffold Gab2 is a crucial component of RANK signaling and osteoclastogenesis. Nature medicine. 2005;11:394–399. doi: 10.1038/nm1203. [DOI] [PubMed] [Google Scholar]
- 19.Parfitt AM, et al. Bone histomorphometry: standardization of nomenclature, symbols, and units. Report of the ASBMR Histomorphometry Nomenclature Committee. J Bone Miner Res. 1987;2:595–610. doi: 10.1002/jbmr.5650020617. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplement