Absolute quantification of microRNAs by using a universal reference (original) (raw)
Abstract
MicroRNAs (miRNAs) are a species of small RNAs ∼21–23-nucleotides long that have been shown to play an important role in many different cellular, developmental, and physiological processes. Accordingly, numerous PCR-, sequencing-, or hybridization-based methods have been established to identify and quantify miRNAs. Their short length results in a high dynamic range of melting temperatures and therefore impedes a proper selection of detection probes or optimized PCR primers. While miRNA microarrays allow for massive parallel and accurate relative measurement of all known miRNAs, they have so far been less useful as an assay for absolute quantification. Here, we present a microarray-based approach for global and absolute quantification of miRNAs. The method relies on the parallel hybridization of the sample of interest labeled with Cy5 and a universal reference of 954 synthetic miRNAs in equimolar concentrations that are labeled with Cy3 on a microarray slide containing probes for all human, mouse, rat, and viral miRNAs (miRBase 12.0). Each single miRNA is quantified with respect to the universal reference canceling biases related to sequence, labeling, or hybridization. We demonstrate the accuracy of the method by various spike-in experiments. Furthermore, we quantified miRNA copy numbers in liver samples and CD34(+)/CD133(−) hematopoietic progenitor cells.
Keywords: miRNA copy number, microarray, liver, hematopoietic progenitor cells
INTRODUCTION
MicroRNAs (miRNAs), short noncoding RNAs of 21–23-nucleotides (nt) in length, regulate target mRNAs post-transcriptionally. They have been shown to play an important role in many different cellular, developmental, and physiological processes as divergent as cell lineage decisions, cell proliferation, apoptosis, morphogenesis, fat metabolism, hormone secretion, neuronal synaptic plasticity, and long-term memory (Aravin and Tuschl 2005). Methods that have been established to identify and quantify miRNAs include Northern blotting (Lim et al. 2003), in situ hybridization (Pena et al. 2009), small RNA library sequencing (Lagos-Quintana et al. 2001), bead arrays (Chen et al. 2008), microarray hybridization (Krichevsky et al. 2003; Barad et al. 2004; Liu et al. 2004; Landgraf et al. 2007), and reverse transcription PCR (Chen et al. 2005). The general methodologies are comparable to well-established conventional mRNA analytics; however, the physicochemical properties of miRNAs that are determined by their short length make it more difficult to generate quantitative data. When compared with mRNAs, a much higher dynamic range of melting temperatures is observed for miRNAs, and a proper selection of hybridization probes or PCR primers is hardly possible. With each miRNA having its specific physicochemical properties, each probe/miRNA pair displays different hybridization efficiencies. Furthermore, true miRNA “housekeeping genes” as established for single mRNA measurements barely exist. Finally, global normalization methods used for massive parallel detection and normalization of the ∼20,000 expressed mRNAs per tissue (Carter et al. 2005a) fail when applied to miRNAs, due to the comparably low number of ∼100 expressed miRNAs per cell type. The strength of miRNA microarrays lay thus far in relative (ratio-based) measurements of known miRNAs in high throughput. In order to optimize the measurement of miRNAs, attempts have been made to adjust the melting temperature or the free energy (Δ_G_) of probe/target pairs by altering the individual probe length or including nucleotide analogs that increase duplex stability, such as 2′OMe modified nucleotides or LNA residues (Castoldi et al. 2006). However, while these approaches narrow the melting temperature differences of miRNA/probe pairs, other effects distorting the microarray signal—such as the labeling efficiency or RNA folding—are not considered. We have developed an miRNA microarray platform (miRXplore) that enabled a semiquantitative description of differential miRNA expression. Recently, we have used our arrays within a large-scale study to identify miRNAs and to assess their expression patterns in >250 small RNA libraries from >26 different organ systems enriched in neuronal as well as normal and malignant hematopoietic cells and tissues (Landgraf et al. 2007). To allow for quantitative miRNA measurement, we have now analyzed different options of probe design, target labeling, and hybridization conditions. We have generated a universal reference (UR), consisting of 954 synthetic human, mouse, rat, and viral miRNAs, with each individual oligoribonucleotide present in equimolar concentrations. Thereby, any single miRNA detected on a microarray can be quantified by directly comparing its signal intensity with the one obtained by the same miRNA sequence present in the UR adjusting for biases related to sequence, labeling, hybridization, or signal detection. In addition, an absolute determination of miRNA copy numbers present in the samples is made possible by using the universal reference as a common quantifiable standard. We found, for example, that mouse liver contains 115,330 copies of miRNAs per cell, and highly purified CD34+/CD133(−) human hematopoietic progenitor cells (HSCs) contain on average 11,587 miRNAs per cell.
RESULTS
Microarray probe design
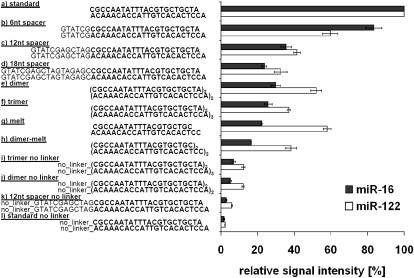
Different probe designs were investigated with respect to sensitivity and selectivity of microarray hybridization. An array with 11 different variants of oligonucleotides for miR-122a and miR-16 was produced (Fig. 1). This included an attempt to normalize the melting temperatures (_T_m) for the probe/miRNA pairs by altering the probe length (Goff et al. 2005). _T_ms were calculated using the oligo analyzer (http://eu.idtdna.com/analyzer/Applications/OligoAnalyzer/Default.aspx?c=EU) and were adjusted to the miRNA probe with the lowest melting temperature (50.2°C ± 0.43°C).
FIGURE 1.
Relative signal intensities of different oligodeoxynucleotide probes for miR-16 and miR-122. The signal intensity of each different oligo variant is displayed in relation to the corresponding standard probe signal intensity, which is set to 100%. All oligodeoxynucleotide probes comprise the reverse complementary sequence of miR-16 and miR-122 and are covalently linked to the slide surface via a C6-amino linker except for the no linker controls. “a),” Standard probe: perfect match reverse complementary miRNA, C6-amino linker; “b)–d).” probe variants with an additional spacer of 6, 12, and 18 random nucleotides on the 5′-end; “e)” and “f),” dimer or trimer probe variants; “g)” and “h),” truncated probe variants; and “i)–l),” probe variants without linker.
Synthetic miR-16 and miR-122a oligoribonucleotides were labeled enzymatically with Cy3 and hybridized to the array.
The DNA probes immobilized via a C6-amino linker gave the highest signal intensities, whereas all other oligonucleotide variants reduced the hybridization efficiency (Fig. 1).
The nine oligo variants with an adjusted _T_m showed a decrease in sensitivity of 40%–93%. Moreover, as also observed for the variation of the other parameters, the effect of adjusted probe length was dependent on the miRNA sequence underlining the difficulty of thorough theoretic _T_m estimation in complex assays. Based on these results, the standard DNA oligonucleotide carrying a C6-amino linker was used for microarray production.
Sensitivity and linear dynamic range of miRNA detection
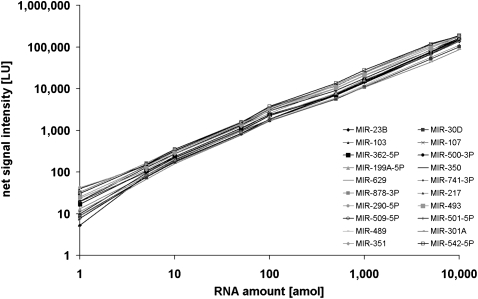
The linear dynamic range and sensitivity of the microarray was measured by hybridizing a dilution series of Universal Reference version 1.0, an equimolar mixture of synthetic miRNAs. The UR was hybridized in stepwise dilutions starting with 10 fmol down to 1 amol of each individual miRNA. Signal intensity was plotted against the amount of 20 selected miRNAs detected in all hybridizations. The slope of the dlog plots was 1.004 (±0.05 SD), with a regression coefficient of >0.98 (Fig. 2). These results show that the linear dynamic range of the microarray exceeded 4 orders of magnitude with an absolute detection sensitivity of <1 amol of target miRNA.
FIGURE 2.
Dynamic range of the miRNA microarrays. An equimolar mixture of miRNAs was labeled and hybridized to microarrays in amounts of 1 amol to 10 fmol per miRNA. Net signal intensities (background subtracted signals) for 20 miRNAs are shown. The slope within the dlog-Plots was 1.004 (±0.05 SD) with a regression coefficient of >0.98.
The influence of different miRNA ligases on miRNA labeling and quantification
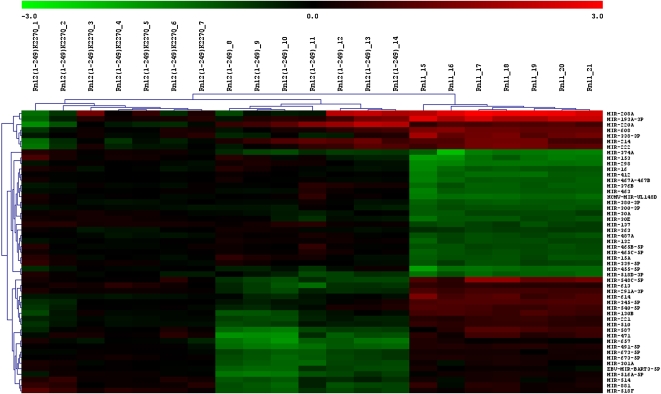
Furthermore, we sought to test whether different ligases and ligation parameters used for the labeling of miRNAs influence the outcome of miRNA quantification upon microarray hybridization. Therefore, we labeled the universal reference (version 0.9) with three ligases—Rnl1, Rnl2(1-249), and Rnl2(1-249)K227Q—and additionally varied the incubation time as well as the amount of labeling dye for the labeling reaction. The influence of labeling time (4–24 h) and amount of labeling dye (2–100 μM) was minuscule (data not shown), and therefore, the data of these arrays were combined. Next, significance analyses of microarrays (SAM) were performed separately for the combined time course experiment (three arrays per ligase) and the dye concentration experiment (four arrays per ligase). The intersection of both analyses led to 51 miRNA candidates [false discovery rate (median): 0.00000%] that were significantly differently labeled by the different ligases. A hierarchical cluster analysis (Fig. 3) of these 51 miRNAs illustrates the potential bias when comparing miRNA data generated using different ligases. A detailed correlation of the labeling efficiency of the 51 miRNAs with their physicochemical properties will help to further characterize the substrate specificity of these RNA ligases and to rationalize the different labeling efficiencies. The influence of the ligases on the labeling efficiency underscores the difficulty of absolute quantification.
FIGURE 3.
Average linkage cluster of miRNAs that are differently labeled by the three tested RNA-ligases. The universal reference (version 0.9) was labeled with Cy5 by three RNA ligases, Rnl1, Rnl2(1-249), and Rnl2(1-249)K227Q (n = 7 for each ligase). Discriminatory gene analysis of the microarray signal intensities by the SAM algorithm resulted in 51 miRNAs that are differentially labeled by the different ligases. These miRNAs were grouped according to similarities using two-dimensional hierarchical clustering (Euclidian distance, average linkage). Log2-transformed expression ratios are indicated from −3.0 (green) to 3.0 (red). (1-7) Sample 1-7 labeled by Rnl2(1-249)K227Q; (8-14) sample 8-14 labeled by Rnl2(1-249); (15-21) sample 15-21 labeled by Rnl1.
Relative quantification of miRNAs across multiple experiments
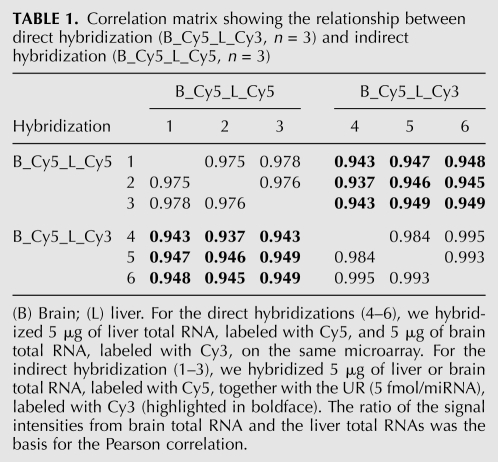
Next, we analyzed to which extent the universal reference can be used as a tool for the relative quantification of miRNAs across multiple experiments. We compared direct hybridizations (i.e., two samples labeled with two different colors on the same array) to indirect hybridizations (one array per sample, labeled with Cy5, with the UR [version 1.0] labeled with Cy3 and hybridized in parallel). For the direct hybridizations, 5 μg of liver total RNA versus 5 μg of brain total RNA (n = 3) were labeled and hybridized to the arrays. The experiments were carried out in triplicates. For the indirect hybridization, 5 μg of liver or brain total RNA and 5 fmol/miRNA of the UR were labeled and the experiments carried out in triplicates. We calculated the so-called re-ratios for the indirect hybridizations by dividing the ratios of sample signal and UR signal of the liver arrays and the brain arrays. These re-ratios were compared with the ratio of the liver and brain sample signals from the direct hybridizations (Supplemental Fig. 1). Pearson correlation coefficients were ∼0.98 for the array replicates and ∼0.95 when comparing the direct hybridizations with the indirect hybridizations (Table 1). Taken together, these results demonstrate that the universal reference can be used as a common reference sample for a relative quantification of miRNA expression measured in independent experiments.
TABLE 1.
Correlation matrix showing the relationship between direct hybridization (B_Cy5_L_Cy3, n = 3) and indirect hybridization (B_Cy5_L_Cy5, n = 3)
Absolute quantification of miRNAs
A direct absolute quantification of miRNAs by measurement of the array signal intensity is impossible due to the sequence-dependent labeling and hybridization efficiencies of miRNAs. In order to test whether the equimolar universal reference (UR) could compensate for sequence-dependent differences and thus allow for an absolute quantification of miRNAs, we measured the amount of spiked-in miRNAs in complex hybridization experiments. We first measured the miRNA expression profiles of mouse liver by microarray hybridization (n = 20) and selected 75 miRNAs that were not detected in this tissue. We then supplemented mouse liver total RNA with 75 synthetic oligoribonucleotides corresponding to these miRNAs. The spiked-in miRNAs were added to 500 ng or 1 μg of liver total RNA in varying amounts ranging from 0.3125 fmol to 20 fmol—with seven miRNAs present in amounts of 0.3125 fmol; five sets of 12 miRNAs containing 0.625, 1.25, 2.5, 5, and 10 fmol per miRNA, respectively; and eight miRNAs with 20 fmol. The liver-RNA including the 75 spike-ins was enzymatically labeled with Cy5 and hybridized together with the UR (version 0.9) and the 75 spike-ins with 2.5 fmol/miRNA labeled with a Cy3 dye. After normalizing the array by calculating the median of the miRControl 3 present in the liver and UR sample, the miRNA amount of the liver spike-ins was calculated with respect to the corresponding miRNA in the UR. The experiments were carried out in quadruplicates.
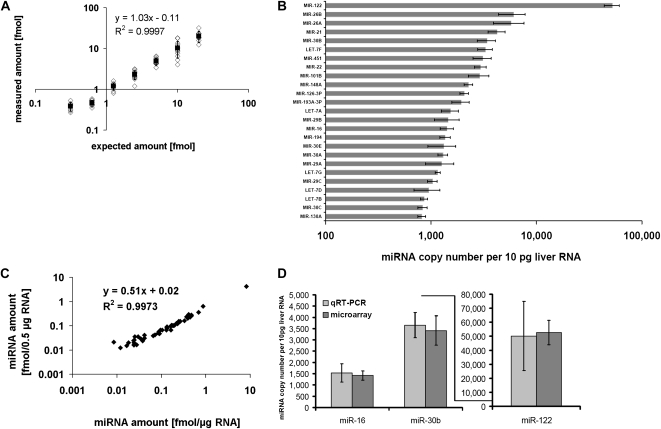
The expected values, and the actually measured values, are shown in Figure 4A and Supplemental Figure 2. Fifty-nine percent (n = 4) of the 75 spike-ins lay within the symmetric 50% interval of the expected value; i.e., 44 ± 5 of the 75 spike-ins showed a deviation of <25%. Furthermore, the absolute amount of miRNA abundance could be quantified with a mean accuracy of 22%–31% (median 17%–25%) for the four arrays. For example, miR-302b* was spiked in with 0.625 fmol (representing the expected value), while the measurement of miR-302b* yielded 0.469 fmol showing a negative deviation of 25%. The absolute measurement of miRNAs across the multiple replicates was highly reproducible, with a mean variation coefficient of 11.5%. Supplemental Table 1 displays the measured amount of the spiked-in miRNAs.
FIGURE 4.
Absolute quantification of miRNAs. (A) Liver total RNA was spiked with 75 synthetic miRNAs not detectable in liver in amounts ranging from 0.3125 to 20 fmol and hybridized versus the universal reference. (Empty black diamonds) The measured and expected amounts for the 75 spike-ins. (Black squares) The measured mean value and its standard deviation. The diagram displays one of four array experiments (complete data are shown in Supplemental Fig. 2). (B) Absolute expression level of the 25 highest expressed miRNAs detected in 1 μg of liver (n = 4). One microgram of liver total RNA was labeled with Cy5 and hybridized together with 2.5 fmol/miRNA of UR. The median miRNA amount calculated from the signal intensities after comparison with the UR was 633 copies per 10 pg of liver total RNA. (C) Dlog-plot showing the calculated amounts for the detected miRNAs in 500 ng and 1 μg of liver RNA. The regression coefficient is 0.997 and the slope 0.51 (expected slope: 0.5). (D) Comparison of microarray and qRT-PCR data for miR-16, miR-30b, and miR-122. One to ten nanograms of liver total RNA were used for the qRT-PCR. Copy numbers per 10 pg of total RNA were calculated using standard curves based on the UR (n = 3). Microarray data are the same as in B.
However, we also observed 17 outliers that reproducibly deviated from the expected value by more than 25% on all four arrays. The measured amount of the worst outlier differed from the expected value by the factor of 2 (Supplemental Fig. 3).
We performed self–self-hybridizations of the UR and compared the labeling of synthetic RNAs with the labeling of synthetic RNAs in the context of total RNA to rule out that the complex mixture of different RNA species present in total RNA interferes with the labeling efficiency. We did not observe significantly differing signal intensities of individual miRNAs for the two different dyes in self–self-hybridizations of the UR. The mean ratio for all UR miRNAs was 1.04 ± 0.27, and the mean ratio for the subset of the spike-ins was 1.05 ± 0.10. Next, we hybridized the spike-ins alone (Cy3) versus the spike-ins plus liver total RNA (Cy5), to compare the labeling of synthetic RNAs with the labeling of synthetic RNAs in the complex background of total RNA. The mean ratio of the 75 spike-ins was 1.06 ± 0.50 (n = 2), pointing to a higher degree of variation as observed in the self–self hybridizations of UR. Taken together, the results of these experiments led to the conclusion that there is no single explanation for the 17 outliers and that, probably, the complexity of a sample has the biggest impact on quantification accuracy.
These experiments show that it is possible to measure the absolute amount of miRNA in a complex sample using microarrays.
Absolute quantification of liver miRNAs
We calculated the amount of miRNAs that was detected in 1 μg of total liver RNA from four independent experiments. The amount of different miRNAs ranged between 0.005 and 8.8 fmol (28–52,567 miRNA copies per 10 pg of total RNA), with the median amount being 0.11 fmol (633 copies per 10 pg total RNA), and the total amount being 19.2 fmol, equivalent to 115,330 copies per 10 pg of RNA (Fig. 4B). As the miRNA proportion in total RNA has been estimated to 0.01% in tissues (Peltier and Latham 2008), one would expect 0.1 ng (15 fmol) of miRNA in 1 μg of total RNA, matching quite well with our calculated overall amount.
Next, we also labeled and hybridized in duplicate 0.5 μg of total liver RNA and compared the calculated miRNA copy numbers with those obtained from 1 μg of total RNA. The regression coefficient between the experiments was 0.996 ± 0.002, indicating no influence of the amount of starting material. The slope of the regression plot was 0.50 ± 0.07, showing that the calculated miRNA amount in 500 ng is half of the amount in 1 μg, as expected (Fig. 4C).
We validated the copy numbers of three selected miRNAs via qRT-PCR. The amounts were calculated with respect to a standard curve based on a serial dilution of the UR. As a control, we also used synthetic miR-30b to generate a standard curve giving similar results.
As shown in Figure 4D, we found that the miRNA copy numbers determined by qRT-PCR for miR-30b, miR-122, and miR-16 agree very well with the amounts measured using microarrays.
Absolute quantification of miRNAs in CD34(+)/CD133(−) hematopoietic progenitor cells
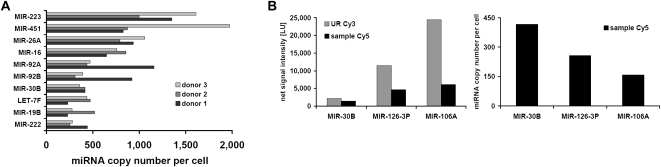
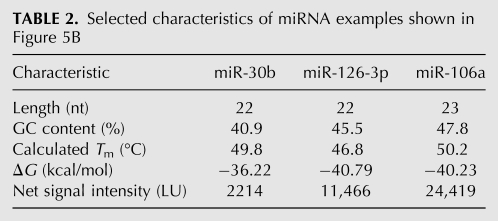
Next, we determined the copy numbers of miRNAs in CD34+ human hematopoietic progenitor cells. We analyzed the RNA extracted from 7 × 105 to 1 × 106 CD34(+)/CD133(−) cells of three different donors. To reduce the biological noise introduced by individual differences between the three independent donors, we focused on strongly expressed miRNA by applying a threshold filter of a signal-to-background ratio of 2. This resulted in 43 miRNAs detected in all three donors. The Spearman correlation coefficients of the miRNA expression profiles were 0.70–0.87 for the three biological replicates. The highest expressed miRNAs were miR-223 and miR-451, with copy numbers of up to 1972 copies per cell. The copy numbers for the 10 most abundant miRNAs are shown in Figure 5A, and the numbers for all 43 detected miRNAs are listed in Supplemental Table 2. The median copy number was 178 copies per cell, and the overall copy number was 11,587 ± 945. The absolute measurement for the three samples had a mean variation coefficient of 39%. Figure 5B displays the net signal intensities on the left and the corresponding miRNA copy number per cell on the right for three different miRNAs. Interestingly, the UR net signal intensities for miR-30B, miR-126-3P, and miR-106A differ notably and do not correlate with the respective Gibbs free energies or melting temperatures (Table 2). This example demonstrates, again, that the signal intensities of different miRNAs are not easily predictable and do not directly correspond with their absolute relation in terms of copy numbers per cell.
FIGURE 5.
MicroRNA copy numbers of CD34(+)/CD133(−) cells prepared from three different donors. (A) The 10 most abundant miRNAs in CD34(+)/CD133(−) cells are shown. The copy numbers per cell for the highest expressed miRNAs miR-223 and miR-451 were determined to be between 828 and 1972. (B, left) Net signal intensities and (right) miRNA copy numbers for miR-30b, miR-126-3p, and miR-106a are shown (donor 2).
TABLE 2.
Selected characteristics of miRNA examples shown in Figure 5B
DISCUSSION
Here, we report a method for the global and absolute quantification of miRNAs. Absolute quantification of miRNAs has so far been difficult due to varying, hardly predictable labeling and hybridization efficiencies of miRNA probe/target pairs or primer/target pairs, respectively. The reasons for this are mainly the high sequence diversity and shortness of miRNAs but also some further properties like the secondary structure whose calculation goes beyond available algorithms for mere primary sequence-based secondary structure prediction. Additionally, when dealing with complex array experiments, solid-phase hybridization of DNA with RNA competes simultaneously with intramolecular hybridization of the respective DNA and RNA, with solid-phase DNA/RNA hybridization of similar sequences and liquid-phase RNA/RNA hybridization of miRNA/miRNA* pairs.
The main approach to deal with some of these problems has been to adjust the melting temperatures of all miRNA probes used in an assay. This is either done by altering the probe length (Goff et al. 2005) or including nucleotide analogs (Castoldi et al. 2006). In addition, different DNA surface attachment strategies have been published to reduce the influence of surface physics and chemistry on the hybridization behavior of probe/target pairs.
We have addressed some of these issues and find that using (1) a simple and straightforward surface attachment procedure, (2) a probe sequence chemistry strictly reflecting the natural paradigm as close as possible, and (3) an extrinsic referencing tool that allows a direct quantification of each single miRNA probe signal is best suited for the representation of the miRNA content by microarrays. Especially, the UR consisting of synthetic oligos of a defined concentration can be used for absolute quantification of the respective target miRNA. Assuming that the different effects influencing microarray signal intensities are comparable for the UR and the sample of interest, miRNA amounts can be calculated by comparing UR versus sample signal intensity.
Our data addressing different probe designs confirm reported observations on important factors influencing binding interactions on microarrays (Shchepinov et al. 1997; Peterson et al. 2002; Luebke et al. 2003). According to Goff et al. (2005), dimer and trimer probes showed enhanced hybridization signal intensity as compared to the monomer. However, we observed a reduction of signal intensity when miRNA sequences are doubled or tripled. These discrepancies are likely explained by the multitude of factors that vary between the different array platforms, such as different attachment and surface chemistries, hybridization buffers, probe complexities, and probe designs. It again underlines the complexity of nucleic acid hybridization reactions. Again, in contrast to Goff et al. (2005), we did not find a uniform hybridization for oligo variants that had been adjusted with respect to their melting temperature but rather a severe loss in sensitivity, and most notably, a variable influence of individual miRNA sequences. Therefore, as also described by Pozhitkov et al. (2006) and Dimitrov and Zuker (2004), we doubt that there is, at least in complex assays, a simple consistency between Gibbs free energy and target sequence.
A few alternative methods have been described for the absolute quantification of miRNAs not relying on altered probe/target design. These approaches, including TaqMan and primer extension assays (Chen et al. 2005; Raymond et al. 2005), an invader assay (Allawi et al. 2004) and a single-molecule analysis method (Neely et al. 2006) are based on the preparation of standard curves for the estimation of the miRNA copy number. However, this calibration curve has to be prepared individually for every miRNA due to the different secondary structure and primer efficiencies. We analyzed 11 randomly selected, synthetic miRNAs and observed CT values differing by up to 4.8 despite the same amount of input material (Supplemental Fig. 4). This corresponds to a 27.9-fold difference of the miRNA amounts calculated without standard curves. Due to the arbitrary selection, we expect even bigger differences for other miRNAs. Therefore, a simple miRNA copy number quantification by comparing CT values or by using one standard curve for all miRNAs is not feasible. However, the Universal Reference can be used to generate standard curves for nearly 1000 miRNAs allowing accurate quantification by qRT-PCR. The accuracy and reproducibility of our absolute microarray-based microRNA quantification surpasses the one reported for mRNA quantification. Due to the mRNA length, it is currently hardly possible to work with a fully synthetic mRNA standard as presented here for miRNAs. Therefore, although the different available microarray platforms for mRNA expression profiling have been extensively evaluated (Canales et al. 2006), there is still no gold standard for quantifying mRNA transcript concentrations by microarrays. The approaches developed so far encompass mathematical modeling (Hekstra et al. 2003; Frigessi et al. 2005), calibrated reference samples (Dudley et al. 2002), exogenous RNA controls (Carter et al. 2005b), and combinations of mathematical modeling and spike measurements (Zhao et al. 2007). The TransCount method (Frigessi et al. 2005) is based on Bayesian statistical modeling and utilizes covariants of the microarray experiment to calculate the concentration from the signal intensity of each probe. A second mathematical approach developed by Hekstra et al. (2003) is based on the Langmuir model. Dudley et al. (2002) used a calibrated reference sample. However, as they use the same reference for each spot, their approach does not control for some important aspects of the microarray system, including sequence-specific hybridization and labeling differences between transcripts. Such aspects are considered if a universal reference consisting of total RNA is used (Novoradovskaya et al. 2004). The disadvantage of current universal references consisting of total RNA is the unknown amount of each single mRNA or miRNA. Therefore, the existing universal reference total RNA is very well suited for the standardization and cross-referencing of microarray experiments but useless for the absolute quantification of mRNAs. In contrast to this, our universal reference consisting of synthetic oligos enables the cross-referencing of experiments and allows in addition for the absolute quantification of miRNAs.
Our measurements of miRNA copy number in liver samples revealed that miR-122 was present with 52,567 copies per 10 pg and both miR-24 and miR-145 with about 600 copies per 10 pg. Thus, the copy numbers are in agreement with estimates of the quantitative primer-extension PCR (Raymond et al. 2005), single-molecule method (Neely et al. 2006), TaqMan (Chen et al. 2005), invader assay (Allawi et al. 2004), and RNase protection assay (Chang et al. 2004). The abundant liver-specific miR-122 has copy numbers 75-fold higher than median copy numbers of liver miRNAs, reflecting its important role in regulation of liver-specific processes, such as cholesterol biogenesis (Krützfeldt et al. 2005). In addition to the copy number of miR-122, Chang et al. (2004) determined also the expression level of the target CAT-1, a high-affinity cationic amino acid transporter. Interestingly, miR-122 is liver-specific, whereas CAT-1 is expressed universally in mammalian tissues with a very low expression in liver compared to other tissues.
In comparison to the liver, the copy numbers of individual miRNAs in CD34+ cells are low, when we assume a general amount of 10 pg of total RNA per cell. As we have only measured miRNAs that are known so far, we cannot exclude that there are additional highly expressed unknown miRNAs that would raise the overall number of miRNAs per CD34+ cell. Interestingly, comparable miRNAs levels were reported by Neilson et al. (2007) for cells representing different stages of T-lymphocyte development. Furthermore, it was demonstrated that global miRNA abundance is higher at high cell densities in diverse animal cell lines (Hwang et al. 2009), so that the cell–cell contact in tissues could also lead to an increased overall miRNA expression.
The principle of absolute quantification presented in our study is not restricted to our microarray platform but can also be applied to other miRNA detection systems, such as Northern blot analysis, RT-PCRs, bead arrays, or sequencing of small RNA libraries. The exact copy number measurement of miRNAs will be of increasing relevance for a detailed analysis of miRNAs and their interaction with other RNA species and proteins, since the expression levels of individual miRNAs influence the targeting properties (Bartel 2009). Also, systems biology approaches and the study of the role of miRNAs in networks as described by Tsang et al. (2007) will gain explanatory power in the context of quantitative data.
MATERIALS AND METHODS
Cell sorting
HSCs were isolated from human bone marrow following written consent from the local Ethics Committee. CD34(+)/CD133(−) cells were enriched using the CliniMACS device (Miltenyi Biotec) by first depleting CD133(+) cells with the CliniMACS CD133 Reagent (clone AC133; Miltenyi Biotec) and subsequent enrichment of CD34(+) cells with the CliniMACS CD34 Reagent (clone QBEND/10; Miltenyi Biotec). The mean purity of the separated cells was 80% as determined by FACS analysis according to ISHAG (Sutherland et al. 1996).
RNA sources
Total RNA was isolated from mouse liver and brain using Trizol (Sigma) and from CD34(+)/CD133(−) cells using Trizol (Sigma) or miRNeasy Mini (QIAGEN). Synthetic 5′ phosphorylated, HPLC-purified oligoribonucleotides were obtained from Sigma Proligo. The quality of each single oligonucleotide was controlled by mass spectroscopy and in addition by PAGE analysis. The concentration of each oligonucleotide was determined by at least two independent photometric measurements. Synthetic oligoribonucleotides were used to prepare a Universal Reference (UR) consisting of a pool of 816 (Universal Reference, version 0.9) (Landgraf et al. 2007) or 954 miRNAs (Universal Reference version 1.0 covering all human, mouse, and rat miRNAs according to miRBase 9.2) (Griffiths-Jones 2006) (miRXplore Universal Reference; Miltenyi), with each individual oligoribonucleotide having a final concentration of 5 fmol/μL.
RNA labeling and hybridization
Four different labeling procedures were investigated, one using the miRCury Power Labeling Kit (Exiqon), and three generic approaches using either Rnl1 (Fermentas), Rnl2(1-249) (Ho et al. 2004), or Rnl2(1-249)K227Q [Plasmid 14072: pET16b-Rnl2(1-249)K227Q, Addgene] to ligate a chemically adenylated Cy3 or Cy5 (AppX) to the 3′-end of miRNAs. In the latter three cases, 2.5 fmol of each individual miRNA (Universal Reference version 0.9) were incubated with the respective ligase in 2 μL of 10× RNA ligation buffer (Pfeffer et al. 2005), 6 μL of 50% DMSO, and 2–100 μM AppX (the synthesis and structure of the dye have been described previously by Pfeffer et al. 2005) in a final volume of 20 μL as indicated in the Results section. The samples were heated for 30 sec at 95°C and immediately placed for 1 min on ice before the ligation reaction that was carried out for 4 h to 24 h.
Hybridization was performed using an automated hybridization instrument following the manufacturer's instructions (a-Hyb Hybridization Station; Miltenyi Biotec). Briefly, the volume of labeled sample was adjusted to 100 μL with nuclease-free water, and 100 μL of 2× Hybridization Solution (Miltenyi) pre-warmed to 42°C was added. After mixing, the solution was pre-heated for 2 min to 70°C prior to application to the a-Hyb Station. Microarray processing in the a-Hyb was performed as follows: incubation in Pre-Hyb Solution (Miltenyi) for 5 min at 42°C, hybridization with the labeled RNAs for 960 min at 42°C, washes with Wash Buffer I (Miltenyi) for 1 min at 10°C (two cycles), and with Wash Buffer II (Miltenyi) for 1 min at 10°C (two cycles). The pump speed at all incubations was set to 1 mL/min. Microarrays were blow-dried and scanned with the Agilent DNA-Microarray Scanner (Agilent).
Microarray design and manufacturing
The design of the microarrays changed slightly during the course of analysis with respect to the newest miRBase sequence database (Griffiths-Jones 2006). The exact configuration of each microarray is deposited the National Center for Biotechnology Information Gene Expression Omnibus (GEO) (accession numbers GSE15831, GSE15832, GSE15833, GSE15834, and GSE15835). In the latest version, each microarray contained 1376 quadruplicate spots of DNA oligonucleotides reverse-complementary to miRNAs from human, mouse, rat, and viruses, and in addition 97 control oligonucleotides. Because some miRNAs are perfectly conserved between species, the 1376 oligonucleotides covered altogether 2063 miRNAs, including 871 human, 634 mouse, 426 rat, and 132 viral miRNAs as deposited in the miRBase sequence database, version 12.0. The 97 positive and negative control probes were chosen as follows: miRControl 1 (four sequences) and miRControl 2 (a single sequence) were used to assess efficiency and sensitivity of labeling, while miRControl 3 (18 sequences) was used to normalize signal intensities across different samples and arrays. All miRControls were artificial sequences that were chosen by bioinformatics means to have no homology with any of the human, mouse, rat, or viral genomes and validated experimentally for cross-hybridization with any of the probes represented on the microarray. A fourth set of positive controls (36 sequences) was composed of probes for other small noncoding RNAs including rRNAs, tRNAs, and U2 and U6 snRNAs. A fifth set of oligonucleotides (13 sequences) were single and double mismatch probes at different positions, antisense and scrambled miRNA sequences, all resembling negative controls for hybridization. The microarrays were manufactured by Miltenyi Biotec. Briefly, 1.2 nL of 10 μM HPLC purified, 5′ amino-modified DNA oligonucleotides (Metabion) were spotted four times on aminopropyltrimethoxysilane/phenylendiisothiocyanate-coated glass slides using a customized piezoelectric spotter.
Microarray data analysis
Signal processing and quantification of the scanned images was done as described before (Landgraf et al. 2007). Depending on the experimental setup, different ratio calculation and normalization methods were applied using an in-house software (miRXplorer). For the direct, relative comparison of two samples—i.e., when two samples were directly compared to each other on the same microarray—a “sample versus sample” modus was chosen. Here, the signal intensities of both channels were normalized either using global normalization methods (e.g., lowess normalization) or according to the median of the Cy5/Cy3 ratios of the miRcontrol 3 probes that were spiked into all experimental and control samples.
For indirect comparisons of multiple experimental samples and absolute quantification, a “sample versus universal reference” modus was chosen. Each sample was hybridized versus a universal reference. The normalization of the sample versus universal reference ratios was according to the spiked-in miRcontrol 3 as described above. As the number of detected signals and the dynamic range differ substantially between the reference and the sample, global normalization methods could not be applied.
Then, signal ratios between sample A and sample B were calculated by multiplying each signal ratio of sample A with the reciprocal value of corresponding miRNA signal ratio of sample B resulting in a so-called re-ratio. The re-ratios were again normalized using standard procedures like lowess normalization. For downstream statistical analysis, ratios were log2 transformed and data were imported in TIGR MeV (Saeed et al. 2003). Two-dimensional hierarchical clustering (Eisen et al. 1998) was done using Euclidean Distance, and SAM was performed using the Tusher et al. (2001) method with at least 1000 permutations per analysis.
Quantitative real-time PCR (qRT-PCR)
The miScript PCR System (QIAGEN) was used to validate the miRNA copy numbers of miR-122, miR-30b, and miR-16. The RT-PCR reaction and the real-time PCR were carried out according to the manufacturer's protocol. For each miRNA quantification, a dilution series of UR ranging from 10 fmol down to 0.01 amol was spiked into a constant amount of carrier RNA, and the CT values were measured. A standard curve was calculated using the linear range of the dilution series (10 fmol down to 10 amol) and used for absolute quantification of the respective miRNAs in liver total RNA. PCR reactions were run on an ABI Prism 7000 SDS Real Time PCR machine (Applied Biosystems) and analyzed using ABI Prism 7000 System SDS software (version 1.1).
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Frank Hübel for support during microarray data analysis; Stephan Preuss, Lena Willnow, and Juliane Stuth for technical support; and Harold Cremer for critical reading of the manuscript.
Footnotes
REFERENCES
- Allawi HT, Dahlberg JE, Olson S, Lund E, Olson M, Ma WP, Takova T, Neri BP, Lyamichev VI. Quantitation of microRNAs using a modified invader assay. RNA. 2004;10:1153–1161. doi: 10.1261/rna.5250604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin A, Tuschl T. Identification and characterization of small RNAs involved in RNA silencing. FEBS Lett. 2005;579:5830–5840. doi: 10.1016/j.febslet.2005.08.009. [DOI] [PubMed] [Google Scholar]
- Barad O, Meiri E, Avniel A, Aharonov R, Barzilai A, Bentwich I, Einav U, Gilad S, Hurban P, Karov Y, et al. MicroRNA expression detected by oligonucleotide microarrays: System establishment and expression profiling in human tissues. Genome Res. 2004;14:2486–2494. doi: 10.1101/gr.2845604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canales RD, Luo Y, Willey JC, Austermiller B, Barbacioru CC, Boysen C, Hunkapiller K, Jensen RV, Knight CR, Lee KY, et al. Evaluation of DNA microarray results with quantitative gene expression platforms. Nat Biotechnol. 2006;24:1115–1122. doi: 10.1038/nbt1236. [DOI] [PubMed] [Google Scholar]
- Carter DE, Robinson JF, Allister EM, Huff MW, Hegele RA. Quality assessment of microarray experiments. Clin Biochem. 2005a;38:639–642. doi: 10.1016/j.clinbiochem.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Carter MG, Sharov AA, VanBuren V, Dudekula DB, Carmack CE, Nelson C, Ko MS. Transcript copy number estimation using a mouse whole-genome oligonucleotide microarray. Genome Biol. 2005b;6:R61. doi: 10.1186/gb-2005-6-7-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castoldi M, Schmidt S, Benes V, Noerholm M, Kulozik AE, Hentze MW, Muckenthaler MU. A sensitive array for microRNA expression profiling (miChip) based on locked nucleic acids (LNA) RNA. 2006;12:913–920. doi: 10.1261/rna.2332406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J, Nicolas E, Marks D, Sander C, Lerro A, Buendia MA, Xu C, Mason WS, Moloshok T, Bort R, et al. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 2004;1:106–113. doi: 10.4161/rna.1.2.1066. [DOI] [PubMed] [Google Scholar]
- Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. Real-time quantification of microRNAs by stem–loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Lozach J, Garcia EW, Barnes B, Luo S, Mikoulitch I, Zhou L, Schroth G, Fan JB. Highly sensitive and specific microRNA expression profiling using BeadArray technology. Nucleic Acids Res. 2008;36:e87. doi: 10.1093/nar/gkn387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov RA, Zuker M. Prediction of hybridization and melting for double-stranded nucleic acids. Biophys J. 2004;87:215–226. doi: 10.1529/biophysj.103.020743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley AM, Aach J, Steffen MA, Church GM. Measuring absolute expression with microarrays with a calibrated reference sample and an extended signal intensity range. Proc Natl Acad Sci. 2002;99:7554–7559. doi: 10.1073/pnas.112683499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigessi A, van de Wiel MA, Holden M, Svendsrud DH, Glad IK, Lyng H. Genome-wide estimation of transcript concentrations from spotted cDNA microarray data. Nucleic Acids Res. 2005;33:e143. doi: 10.1093/nar/gni141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff LA, Yang M, Bowers J, Getts RC, Padgett RW, Hart RP. Rational probe optimization and enhanced detection strategy for microRNAs using microarrays. RNA Biol. 2005;2:93–100. doi: 10.4161/rna.2.3.2059. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S. miRBase: The microRNA sequence database. Methods Mol Biol. 2006;342:129–138. doi: 10.1385/1-59745-123-1:129. [DOI] [PubMed] [Google Scholar]
- Hekstra D, Taussig AR, Magnasco M, Naef F. Absolute mRNA concentrations from sequence-specific calibration of oligonucleotide arrays. Nucleic Acids Res. 2003;31:1962–1968. doi: 10.1093/nar/gkg283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CK, Wang LK, Lima CD, Shuman S. Structure and mechanism of RNA ligase. Structure. 2004;12:327–339. doi: 10.1016/j.str.2004.01.011. [DOI] [PubMed] [Google Scholar]
- Hwang HW, Wentzel EA, Mendell JT. Cell–cell contact globally activates microRNA biogenesis. Proc Natl Acad Sci. 2009;106:7016–7021. doi: 10.1073/pnas.0811523106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA. 2003;9:1274–1281. doi: 10.1261/rna.5980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, Rhoades MW, Burge CB, Bartel DP. The microRNAs of Caenorhabditis elegans. Genes & Dev. 2003;17:991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CG, Calin GA, Meloon B, Gamliel N, Sevignani C, Ferracin M, Dumitru CD, Shimizu M, Zupo S, Dono M, et al. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc Natl Acad Sci. 2004;101:9740–9744. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luebke KJ, Balog RP, Garner HR. Prioritized selection of oligodeoxyribonucleotide probes for efficient hybridization to RNA transcripts. Nucleic Acids Res. 2003;31:750–758. doi: 10.1093/nar/gkg133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neely LA, Patel S, Garver J, Gallo M, Hackett M, McLaughlin S, Nadel M, Harris J, Gullans S, Rooke J. A single-molecule method for the quantitation of microRNA gene expression. Nat Methods. 2006;3:41–46. doi: 10.1038/nmeth825. [DOI] [PubMed] [Google Scholar]
- Neilson JR, Zheng GX, Burge CB, Sharp PA. Dynamic regulation of miRNA expression in ordered stages of cellular development. Genes & Dev. 2007;21:578–589. doi: 10.1101/gad.1522907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoradovskaya N, Whitfield ML, Basehore LS, Novoradovsky A, Pesich R, Usary J, Karaca M, Wong WK, Aprelikova O, Fero M, et al. Universal Reference RNA as a standard for microarray experiments. BMC Genomics. 2004;5:20. doi: 10.1186/1471-2164-5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier HJ, Latham GJ. Normalization of microRNA expression levels in quantitative RT-PCR assays: Identification of suitable reference RNA targets in normal and cancerous human solid tissues. RNA. 2008;14:844–852. doi: 10.1261/rna.939908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena JT, Sohn-Lee C, Rouhanifard SH, Ludwig J, Hafner M, Mihailovic A, Lim C, Holoch D, Berninger P, Zavolan M, et al. miRNA in situ hybridization in formaldehyde and EDC-fixed tissues. Nat Methods. 2009;6:139–141. doi: 10.1038/nmeth.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson AW, Wolf LK, Georgiadis RM. Hybridization of mismatched or partially matched DNA at surfaces. J Am Chem Soc. 2002;124:14601–14607. doi: 10.1021/ja0279996. [DOI] [PubMed] [Google Scholar]
- Pfeffer S, Lagos-Quintana M, Tuschl T. Cloning of small RNA molecules. In: Ausubel FM, et al., editors. Current protocols in molecular biology. Vol. 4. Wiley; New York: 2005. pp. 4.1–4.18. Chap. 26, [DOI] [PubMed] [Google Scholar]
- Pozhitkov A, Noble PA, Domazet-Loso T, Nolte AW, Sonnenberg R, Staehler P, Beier M, Tautz D. Tests of rRNA hybridization to microarrays suggest that hybridization characteristics of oligonucleotide probes for species discrimination cannot be predicted. Nucleic Acids Res. 2006;34:e66. doi: 10.1093/nar/gkl133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond CK, Roberts BS, Garrett-Engele P, Lim LP, Johnson JM. Simple, quantitative primer-extension PCR assay for direct monitoring of microRNAs and short-interfering RNAs. RNA. 2005;11:1737–1744. doi: 10.1261/rna.2148705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. TM4: A free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Shchepinov MS, Case-Green SC, Southern EM. Steric factors influencing hybridisation of nucleic acids to oligonucleotide arrays. Nucleic Acids Res. 1997;25:1155–1161. doi: 10.1093/nar/25.6.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland DR, Anderson L, Keeney M, Nayar R, Chin-Yee I. The ISHAGE guidelines for CD34+ cell determination by flow cytometry. International Society of Hematotherapy and Graft Engineering. J Hematother. 1996;5:213–226. doi: 10.1089/scd.1.1996.5.213. [DOI] [PubMed] [Google Scholar]
- Tsang J, Zhu J, van Oudenaarden A. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol Cell. 2007;26:753–767. doi: 10.1016/j.molcel.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Engelen K, De Moor B, Marchal K. CALIB: A Bioconductor package for estimating absolute expression levels from two-color microarray data. Bioinformatics. 2007;23:1700–1701. doi: 10.1093/bioinformatics/btm159. [DOI] [PubMed] [Google Scholar]