Interactions of Human Mismatch Repair Proteins MutSα and MutLα with Proteins of the ATR-Chk1 Pathway (original) (raw)
Abstract
At clinically relevant doses, chemotherapeutic SN1 DNA methylating agents induce an ATR-mediated checkpoint response in human cells that is dependent on functional MutSα and MutLα. Deficiency of either mismatch repair activity renders cells highly resistant to this class of drug, but the mechanisms linking mismatch repair to checkpoint activation have remained elusive. In this study we have systematically examined the interactions of human MutSα and MutLα with proteins of the ATR-Chk1 pathway using both nuclear extracts and purified proteins. Using nuclear co-immunoprecipitation, we have detected interaction of MutSα with ATR, TopBP1, Claspin, and Chk1 and interaction of MutLα with TopBP1 and Claspin. We were unable to detect interaction of MutSα or MutLα with Rad17, Rad9, or replication protein A in the extract system. Use of purified proteins confirmed direct interaction of MutSα with ATR, TopBP1, and Chk1 and of MutLα with TopBP1. MutSα-Claspin and MutLα-Claspin interactions were not demonstrable with purified proteins, suggesting that extract interactions are indirect or depend on post-translational modification. Use of a modified chromatin immunoprecipitation assay showed that proliferating cell nuclear antigen, ATR, TopBP1, and Chk1 are recruited to chromatin in a MutLα- and MutSα-dependent fashion after _N_-methyl-_N′_-nitro-_N_-nitrosoguanidine treatment. However, chromatin enrichment of replication protein A, Claspin, Rad17-RFC, and Rad9-Rad1-Hus1 was not detected in these experiments. Although our failure to observe enrichment of the latter activities could be due to sensitivity limitations, these observations may indicate a novel mechanism for ATR activation.
Keywords: Cancer, Cell/Checkpoint, Cell/Apoptosis, Cell/Cycle, Chromatin/Immunoprecipitation/ChIP, DNA/Repair, Drug Resistance/Cancer Therapy/Chemotherapy
Introduction
Mismatch repair is a mutation avoidance mechanism that corrects DNA replication errors, inhibits recombination between quasi-homologous DNA sequences, and participates in the early steps of checkpoint and apoptotic responses to several types of DNA damage (reviewed in Refs. 1–4). Inactivation of the human pathway elevates spontaneous mutability, renders cells resistant to certain DNA damaging agents, causes typical and atypical hereditary nonpolyposis colon cancer, and has been implicated in the development of some sporadic tumors.
The most thoroughly studied function of mismatch repair has been its role in replication fidelity. Although the eukaryotic signals that direct repair of DNA biosynthetic errors to the newly synthesized DNA strand have not been identified, a strand-specific nick or gap is sufficient to direct the reaction in mammalian cell extracts (5–7) and in several purified systems (8–11). Analysis of these purified systems has indicated that repair initiation involves activation of a latent endonuclease activity of MutLα (MLH1·PMS2) in a reaction that requires a mismatch, MutSα (MSH2·MSH6), replication factor C (RFC),2 proliferative cell nuclear antigen (PCNA), and ATP (12, 13). Incision by MutLα endonuclease is directed to the heteroduplex strand that contains a pre-existing break, yielding molecules that contain strand discontinuities to either side of the mismatch. These endonucleolytic products serve as substrates for MutSα-activated exonuclease I, which loads at a strand break located 5′ to the mispair and hydrolyzes a DNA segment spanning the mismatch (8, 12). The ensuing gap is filled by replication protein A (RPA) and repaired by DNA polymerase δ in a reaction that also depends on PCNA and RFC (8, 10, 11, 14, 15). Covalent continuity is restored to the repaired strand by the action of a DNA ligase (10).
In addition to their role in replication fidelity, MutSα and MutLα function is required for normal checkpoint and apoptotic responses that occur upon exposure to SN1 DNA methylators, and cells deficient in these activities are highly resistant to killing by these drugs (reviewed in Refs. 1 and 16–18). SN1 methylators, including temozolomide, procarbazine, decarbazine, _N_-methyl-_N_-nitrosourea, and _N_-methyl-_N′_-nitro-_N_-nitrosoguanidine (MNNG), produce several classes of DNA lesion, but _O_6-methylguanine (MeG) is largely responsible for the cytotoxicity of the agents (19, 20). MeG is repaired by _O_6-methylguanine-DNA methyltransferase (MGMT) (18) and pairs with C or T during replication bypass, resulting MeG-C and MeG-T mispairs (21). Although both mismatches activate the repair system, MeG-T is a superior substrate (22–24). The checkpoint response induced by clinically relevant doses of SN1 methylators occurs in the G2 phase of the second cell cycle (25) and appears to be regulated through the ATR-Chk1 pathway with MutSα and MutLα acting upstream of ATR (22, 26–29).
Two models, which are not mutually exclusive, have been proposed to explain the nature of MutSα- and MutLα-dependent damage signaling in response to MeG lesions. The futile repair model invokes mismatch-provoked excision triggered by MeG mispairs at the replication fork. Because the damaged guanine base resides on the template strand and repair targets new DNA, activation of the repair system under these conditions would lead to abortive turnover of the daughter strand, ultimately triggering checkpoint activation (19). The alternate direct signaling model suggests that recruitment of MutSα and MutLα and perhaps other activities to MeG lesions is sufficient to trigger activation of damage signaling and apoptotic pathways (30). Evidence consistent with both models is available. The finding that SN1 methylator-induced checkpoint activation occurs during the second G2 is consistent with the futile repair model, which requires replication past MeG damage (19, 26, 31). Indeed, MeG-dependent abortive turnover that is dependent on a functional mismatch repair system has been documented in nuclear extracts of mammalian cells (24), and in vitro repair synthesis on DNAs containing MeG can lead to persistent single-stranded breaks (24, 32). Electron microscopic analysis has revealed mismatch repair-dependent accumulation of single-stranded DNA gaps in newly replicated DNA in MNNG-treated cells (31), and ATR-dependent checkpoint activation in such cells is accompanied by formation of nuclear foci that contain ATR and RPA, suggesting recruitment of ATR to single-stranded DNA regions (26). Furthermore, exonuclease 1, which participates in the excision step of mammalian mismatch repair, has been implicated in the cytotoxic effects of 6-thioguanine and SN1 DNA alkylators (33, 34).
Although these findings are consistent with a futile repair mechanism, compelling evidence for the direct signaling model is also available. Yoshioka et al. (28) have demonstrated MutSα- and MutLα-dependent ATR activation in mammalian cell extracts using MeG-containing DNAs that are not expected to support mismatch-provoked excision. Additional evidence for such a mechanism has been provided by a mouse Msh6 separation of function mutation that confers a defect in mismatch repair but retains sensitivity to killing by MNNG (35).
MutSα and MutLα are believed to act upstream of the ATR-Chk1 checkpoint pathway in the damage response elicited by MeG. According to current views, ATR-mediated signaling is activated by replication interference or by lesions that stall replication forks, whereas the ATM-Chk2 pathway is activated primarily in response to double-stranded breaks (36). Activation of the ATR-dependent pathway is believed to involve recruitment of ATR-ATRIP, Rad17-RFC, and Rad9-Rad1-Hus1 (9-1-1) to sites of damage or replication fork stress (37), with efficient activation of the system also dependent on TopBP1 and Claspin (38–40). Activated ATR phosphorylates a range of substrates including the Chk1 kinase, which in turn regulates downstream targets such as Cdc25A, Cdc25C, and p53, to inhibit cell cycle progression (36).
Previous studies using co-immunoprecipitation from cell extracts have suggested that MutSα may be capable of interaction with ATR and Chk1 (28, 29, 41), although these experiments suffer from the caveat that the interactions observed could be indirect, mediated by other molecular partners. However, the use of purified proteins has demonstrated that MSH2, in the absence of its MSH6 or MSH3 partners, can directly interact with ATR (29). In an effort to better understand the interplay between mismatch repair and DNA damage checkpoints, we have systematically analyzed interactions between MutSα and MutLα with components of the ATR-Chk1 checkpoint pathway in nuclear extracts, in purified systems, and on chromatin from cells that have been subjected to SN1 methylator damage.
EXPERIMENTAL PROCEDURES
Cell Culture and Reagents
Human lymphoblastoid B-cell lines TK6 and MT1 (19) were grown in RPMI 1640 medium supplemented with 10% (v/v) fetal bovine serum (HyClone). HeLa S3 cells were cultured in RPMI 1640 medium supplemented with 5% fetal bovine serum. The 293T Lα cell line was a gift from Josef Jiricny (University of Zurich) and was cultured as described (42). The cell line was derived from MLH1- and MGMT-deficient human embryonic kidney 293T cells and contains a stably integrated hMLH1 cDNA minigene that is controlled by the Tet-Off expression system. To turn off MLH1 expression in these cells, 50 ng/ml doxycycline (Clontech) was added into the medium every second day. MNNG (Sigma) was dissolved in Me2SO immediately prior to use.
Nuclear Co-immunoprecipitation
Nuclear extracts were prepared at 0–4 °C from TK6 cells and HeLa S3 cells as described (5) with the following modifications. The nuclear pellets were resuspended in 25 mm Hepes-KOH, pH 7.5, 450 mm NaCl, 5 mm MgCl2, 0.1 mm EDTA. 0.15% Triton X-100 (v/v) and then rotated on a ROTO-TORQUE rotator (Cole-Parmer) for 30 min. The samples were centrifuged at 15,000 × g for 20 min, and the supernatant was diluted 3-fold by adding two volumes of 25 mm Hepes-KOH, pH 7.5, 5 mm MgCl2, and 0.1 mm EDTA. Protein concentrations of the resulting nuclear extracts were determined by Bradford assay, and the extracts were used immediately.
For co-immunoprecipitation, 1 mg of nuclear extract was incubated with 5 μg of primary antibody overnight at 4 °C with rotation. The antibodies used were control mouse IgG (Santa Cruz Biotechnology), control rabbit IgG (Santa Cruz Biotechnology), and those directed against MSH6 (BD Biosciences), PMS2 (BD Biosciences), ATR (Santa Cruz Biotechnology), Rad17 (Santa Cruz Biotechnology), Rad9 (Bethyl Laboratories), TopBP1 (Bethyl Laboratories), Claspin (GeneTex), and Chk1 (Santa Cruz Biotechnology). Fifty μl of agarose-protein G beads (50% slurry; Invitrogen) pre-equilibrated with wash buffer (25 mm Hepes-KOH, pH 7.5, 150 mm NaCl, 5 mm MgCl2, 0.1 mm EDTA, 50 μg/ml bovine serum albumin, 0.05% Triton X-100) were then added, and the reactions were rotated at 4 °C for 1 h. The beads were washed three times with wash buffer, resuspended in 30 μl of 2× Laemmli SDS buffer (43), and the samples were subjected to SDS-PAGE and Western blotting.
Purified Protein-Protein Interactions
Preparation of the purified proteins used in this study are described in the supplemental materials.
All of the reactions were carried out at 0 −4 °C. MutSα or MutLα (final concentration, 20 nm) was incubated with 20 nm checkpoint proteins in 500 μl of binding buffer (25 mm Hepes-KOH, pH 7.5, 150 mm NaCl, 5 mm MgCl2, 0.1 mm EDTA, 0.02–0.1% Triton X-100, 50 μg/ml bovine serum albumin, 0.1% phenylmethylsulfonyl fluoride, 1 mm dithiothreitol, and protease inhibitor mixture; Roche Applied Science). After rotation for 2 h, 5 μg of antibody (mouse IgG, anti-MSH2, anti-MSH6, anti-PMS2, or anti-MLH1) was added, and the rotation continued for 1 h. Fifty μl of agarose-protein G beads, pre-equilibrated with binding buffer, were added, and the reactions were rotated for 1 h more. The beads were then washed twice with binding buffer, resuspended in Laemmli buffer, and analyzed as described above.
For GST-tagged proteins (GST-Chk1 and GST-TopBP1) and FLAG-tagged proteins (FLAG-ATR and FLAG-Claspin), tag pull-down assays were also performed. Purified tagged proteins (20 nm in 500 μl of binding buffer) were incubated with 50 μl of glutathione-Sepharose beads (GE Healthcare) or anti-FLAG M2 affinity gel (Sigma) for 1 h, followed by two washes with binding buffer. MutSα or MutLα (20 nm) was then added. After 2 h of incubation, the beads were washed with binding buffer and resuspended in Laemmli SDS buffer as described above.
Modified ChIP Assay
TK6 cells, MT1 cells, and 293T Lα cells (cultured with or without doxycycline) were treated with 0.2 μm MNNG and harvested at 0, 6, 12, 24, 48, and 72 h post-treatment (zero time samples were removed just before MNNG addition). Cross-linking and chromatin preparation were performed by a modification of the procedure of Fousteri et al. (44). Briefly, the cells were treated with 0.5% formaldehyde for 5 min at room temperature, followed by the addition of 0.125 m glycine to quench unreacted formaldehyde. All of the subsequent steps were performed at 0° −4 °C, and all of the buffers contained 0.1 mm EDTA, 0.5 mm EGTA, 1 mm dithiothreitol, and protease inhibitors (0.1% phenylmethylsulfonyl fluoride (saturated in isopropanol), 1 μg/ml leupeptin, 1 μg/ml E-64, 0.5 μg/ml aprotinin, and 5 μg/ml pepstatin A). The cells (1 × 108) were washed with phosphate-buffered saline (pH 7.4) and lysed for 10 min with rotation in 10 ml of 50 mm Hepes-KOH, pH 7.8, 150 mm NaCl, 0.5% Nonidet P-40, 0.25% Triton X-100, 10% (v/v) glycerol. After centrifugation (4,000 × g, 5 min), the pellet was resuspended in 10 ml of 10 mm Tris-HCl, pH 8.0, 200 mm NaCl, rotated 10 min, and centrifuged at 15,000 × g for 10 min. The chromatin pellet was resuspended in 1 ml of 50 mm Tris-HCl, pH 7.9, 5 mm CaCl2 and digested with 500 units of micrococcal nuclease (New England Biolabs) at 37 °C for 10 min. After the addition of EGTA to 10 mm, the samples were centrifuged at 15,000 × g for 20 min, and the supernatant containing cross-linked chromatin was used for measurement of DNA concentration and ChIP assays.
To determine DNA content, 50 μl of chromatin solution were added to 159 μl of 0.25 m NaCl, 62 μg/ml RNase A (Sigma) and incubated for 4 h at 65 °C. Two μl of proteinase K (10 mg/ml; Thermo Fisher Scientific) were added, and the solutions were incubated at 42 °C for 1.5 h. DNA was isolated by phenol/chloroform extraction and ethanol precipitation, and concentration was determined by absorbance at 260 nm. DNA fragment size (weight average size of about 400 bp; mode about 200 bp) was checked by agarose gel electrophoresis.
For ChIP reactions, digested chromatin samples were diluted with radioimmune precipitation assay buffer (10 mm Tris-HCl, pH 8.0, 140 mm NaCl, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS) to 0.5 mg/ml DNA. The diluted samples (1 ml) were immunoprecipitated overnight with 5 μg of anti-MSH6, anti-MLH1, or control mouse IgG. Fifty μl of agarose-protein G beads, pre-equilibrated with radioimmune precipitation assay buffer supplemented with 50 μg/ml bovine serum albumin and 1 mg/ml sonicated calf thymus DNA, were then added, and the reactions were rotated at 4 °C for 1 h. The beads were washed three times with radioimmune precipitation assay buffer and resuspended in 30 μl of 2× Laemmli SDS buffer. The samples were heated at 95 °C for 30 min to reverse cross-links prior to SDS-PAGE and Western blotting.
Western Blotting
Western blot was done as previously described (45). The primary antibodies used were specific for MSH6 and MSH2 (Santa Cruz Biotechnology); PMS2 and MLH1 (Santa Cruz Biotechnology); ATR, Rad17, and Rad9 (Bethyl Laboratories); RPA70, TopBP1, Claspin, Chk1, and β-actin (Novus); poly(ADP-ribose) polymerase-1 (Alexis Biochemicals); and histone H3 (Millipore). Secondary antibodies were anti-mouse or rabbit or goat IgGs conjugated with horseradish peroxidase (Invitrogen). The protein bands were visualized using the ECL Western blotting system (GE Healthcare).
RESULTS
Nuclear Co-immunoprecipitation
Co-immunoprecipitation from cell extracts has indicated that MutSα may interact with ATR and Chk1 (28, 29, 41), but indirect interactions were not excluded in these studies. In addition, the use of purified proteins has indicated that ATR is capable of direct interaction with the MSH2 polypeptide (29), although the latter protein is not known to display biological activity in the absence of its MSH6 or MSH3 partners (46). Here we used both nuclear extracts and purified proteins to systematically examine interactions of MutSα and MutLα with components of the ATR-Chk1 pathway. For nuclear co-immunoprecipitation, HeLa cells and TK6 cells were used, both of which are proficient in mismatch repair and display MutSα- and MutLα-dependent checkpoint responses to SN1 methylators (22, 26, 28, 31).
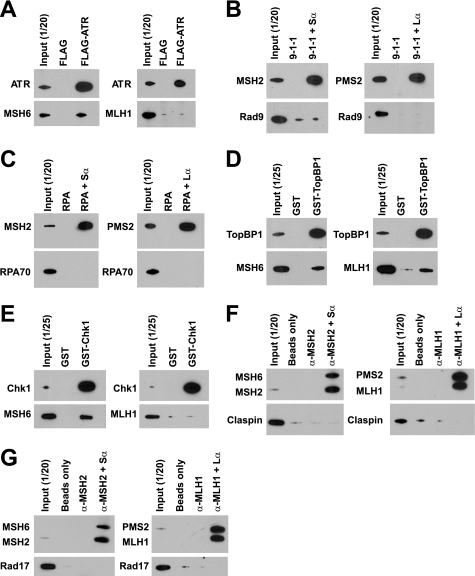
As shown in Fig. 1A (left panel), ATR, TopBP1, Claspin, and Chk1 were co-immunoprecipitated from HeLa and TK6 nuclear extracts by antibodies against the MSH6 subunit of MutSα, indicating direct or indirect interactions between MutSα and these checkpoint activities. Despite the use of several anti-MSH6 antibodies directed against different epitopes, Rad17, Rad9, and RPA were not detected in MSH6 immunoprecipitates. When MutLα was immunoprecipitated by PMS2 antibody, TopBP1, and Claspin were co-immunoprecipitated (Fig. 1A, right panel), but other checkpoint activities were not detected.
FIGURE 1.
Reciprocal nuclear co-immunoprecipitation. A, immunoprecipitation from nuclear extracts of HeLa or TK6 cells was done using anti-MSH6 or anti-PMS2 antibodies as described under “Experimental Procedures.” The lanes marked as Input (1/20) indicate that 120 of total extract was loaded. Mouse IgG was used as a control for MSH6 or PMS2 antibody. The labels on the left indicate proteins co-precipitating with MSH6 or PMS2 as judged by Western blot after SDS-PAGE. Although some ATR signal is evident in the IgG control lane, quantitation of Western results demonstrated that the ATR signal in anti-MSH6 immunoprecipitates is 3–4-fold higher than that in the control sample. B, co-immunoprecipitation from nuclear extracts of TK6 cells was done by using antibodies against the checkpoint proteins indicated at the top of the figure. MutSα and MutLα in immunoprecipitates were evaluated after SDS-PAGE using antibodies directed against MSH2 and MSH6 (upper panel) or MLH1 and PMS2 (lower panel).
Interaction of ATR, TopBP1, Claspin, and Chk1 with MutSα and the interaction of TopBP1 and Claspin with MutLα were confirmed by reciprocal nuclear co-immunoprecipitation using antibodies specific for the checkpoint proteins (Fig. 1B). It is noteworthy that in contrast to input samples, the MSH2 subunit of MutSα was preferentially recovered when co-immunoprecipitated with antibodies directed against TopBP1 or Chk1 (Fig. 1B). This may indicate preferential interaction of these activities with MSH2 and dissociation of MutSα subunits during precipitation.
Interaction of MutSα and MutLα with Checkpoint Proteins in a Purified System
We employed purified proteins to further clarify the nature of these interactions. With the exception of FLAG-ATR and FLAG-Claspin, both of which were ≈80% pure, the proteins used in these experiments were essentially homogeneous (supplemental Fig. S1). When bound to anti-FLAG M2 affinity beads, FLAG-ATR can pull down MSH6 (Fig. 2A), indicating direct interaction of ATR with MutSα. However, interaction between ATR and MutLα was not detected (Fig. 2A, right panel). Consistent with these observations, we have found that immunoprecipitation of MutSα co-precipitates FLAG-ATR, but immunoprecipitation of MutLα does not (data not shown). This is in agreement with nuclear extract results described above and the previous finding of Wang and Qin (29). Similar experiments (Fig. 2, B–G) demonstrated that MutSα interacts directly with Chk1 and TopBP1, but we have been unable to detect MutSα interaction with 9-1-1, Rad17-RFC, RPA, or FLAG-Claspin using pulldown methods. These experiments also demonstrated robust interaction of MutLα with GST-TopBP1, but MutLα interaction with other checkpoint proteins was not detected. Interaction of MutSα with GST-Chk1 and GST-TopBP1 and interaction of MutLα with GST-TopBP1 were confirmed by reciprocal pulldowns using anti-MSH6 or anti-MLH1, respectively (data not shown).
FIGURE 2.
Interactions of MutSα or MutLα with purified checkpoint proteins. A, MutSα or MutLα was incubated with FLAG-ATR as described under “Experimental Procedures.” Anti-FLAG M2 affinity gel was used to pull down FLAG-ATR and its interacting proteins. Incubation with FLAG peptide (20 nm) was used as a control for FLAG-ATR. ATR, MSH6, and MLH1 in the precipitates were determined by Western blot after SDS-PAGE. B, 9-1-1 was incubated alone, or with MutSα or MutLα followed by pulldown with anti-MSH2 or anti-PMS2 bound to protein G beads (“Experimental Procedures”). RAD9, MSH2, and PMS2 in the precipitates were determined by Western blot after SDS-PAGE. C, procedure was as in B, except RPA was used instead of 9-1-1, and RPA70 was probed in the Western blot. D, procedure was as in A except that glutathione-Sepharose beads were used to pull down GST-TopBP1 and its interacting partners. GST was used as a negative control for GST-TopBP1. E, procedure was as in D except GST-Chk1 was used instead of GST-TopBP1. F, after incubation with FLAG-Claspin, MutSα or MutLα was pulled down using anti-MSH2 or anti-MLH1 bound to protein G beads (right lanes). Negative controls included incubation of FLAG-Claspin with protein G beads alone or incubation of FLAG-Claspin with protein G beads and MSH2 or MLH1 antibody in the absence of MutSα or MutLα (lanes marked as α-MSH2 or α-MLH1). G, procedure was as in F except that incubation with Rad17-RFC was substituted for FLAG-Claspin.
Association of MutSα and MutLα with Checkpoint Proteins in Vivo
To evaluate interactions of these proteins in cells upon SN1 methylator treatment, we employed formaldehyde cross-linking in a modified ChIP assay to capture interacting multi-protein-DNA assemblies in vivo (44, 47). Two pairs of mismatch repair proficient/deficient cell lines were used: 293T Lα cells, cultured with or without doxycycline, and TK6 and MT1 cells. 293T Lα cells cultured without doxycycline (referred to as 293T Lα+ cells) express MLH1, are mismatch repair-proficient, and display a G2/M checkpoint response to SN1 methylators, whereas cells cultured in the presence of 50 ng/ml doxycycline (referred to as 293T Lα− cells) shut off MLH1 expression and are defective in mismatch repair and the checkpoint response to SN1 methylators (26, 42). TK6 and MT1 cells are human lymphoblastoid cells with MGMT deficiency. The MT1 cell line was derived from TK6 by single-step selection for high level DNA methylator resistance and is deficient in MutSα due of missense mutations in both alleles of MSH6 (19, 30, 48).
The four cell lines were treated with 0.2 μm MNNG and harvested at 0, 6, 12, 24, 48, and 72 h post-treatment, and DNA-bound protein complexes were cross-linked with formaldehyde. Chromatin fractions were obtained after removal of cytoplasm and soluble nuclear fractions by detergent extraction, which was confirmed by Western blotting using antibodies against marker proteins (supplemental Fig. S2). Chromatin was digested to an average DNA fragment size of about 400 bp with microccoal nuclease, and chromatin-associated MutSα (in 293T Lα+ and 293T Lα− cells) or MutLα (in TK6 and MT1 cells) immunoprecipitated using anti-MSH6 or anti-MLH1, respectively. Proteins present in immunoprecipitates were then analyzed by SDS-PAGE and Western blot after cross-link reversal.
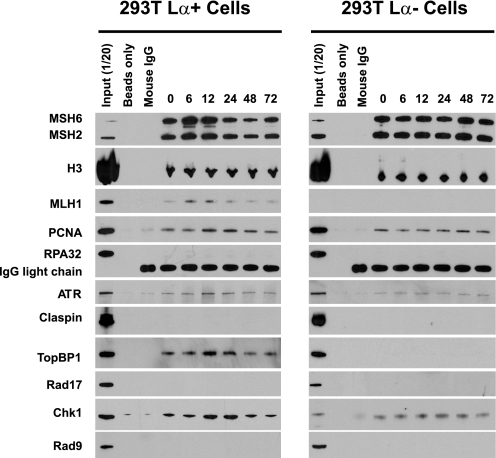
The results of anti-MSH6 immunoprecipitation of equivalent amounts of chromatin fragments (0.50 mg DNA) derived from 293T Lα+ and 293 Lα− cells are shown in Fig. 3 and presented in quantitative form in Fig. 4. As can be seen, a number of activities co-immunoprecipitated with MSH6 in the zero time formaldehyde cross-linked chromatin sample, including MSH2, MLH1, PCNA, ATR, TopBP1, and Chk1 (Fig. 3). Because this sample was removed prior to MNNG treatment, these findings indicate significant association of these repair and checkpoint activities with MSH6-enriched chromatin fractions that have not been subjected to exogenous alkylator damage. The possibility of chromatin enrichment of repair and checkpoint activities was evaluated by normalizing Western blot signals for each protein of interest to that for histone H3 in the sample (Fig. 4). This analysis revealed a transient 1.7–1.8-fold increase in chromatin-associated MutSα that was evident in 6- and 12-h post-treatment samples from 293T Lα+ cells but not observed with chromatin from 293T Lα− cells. The enrichment of MutSα in 293T Lα+ chromatin was accompanied by increases in chromatin-bound MLH1, with both repair activities returning to basal levels by 24 h. PCNA, ATR, TopBP1, and Chk1 were also subjected to transient elevation in MSH6-associated chromatin fractions, but with a delay relative to chromatin enrichment of MutSα and MLH1. Thus, elevation of PCNA, ATR, TopBP1, and Chk1 levels was initially observed in 12-h chromatin samples and persisted to 24 h, returning to near basal levels by 48 h. In contrast to these results obtained with chromatin isolates obtained from 293T Lα+ cells, no significant variation of chromatin association of MutSα or the latter set of activities was observed in samples obtained from 293 Lα− cells (Figs. 3 and 4B), implying that these effects are MLH1-dependent. Interestingly, the basal association of TopBP1 observed in anti-MSH6 zero time chromatin precipitates from 293T Lα+ cells was not observed in otherwise identical samples from 293T Lα− cells (Fig. 3), suggesting that this association is also MLH1-dependent.
FIGURE 3.
Modified ChIP assay using 293T Lα+ and 293T Lα− cells. Immunoprecipitation of micrococcal nuclease-digested formaldehyde-cross-linked chromatin samples with anti-MSH6 was performed as described under “Experimental Procedures.” After cross-link reversal, the samples were resolved by electrophoresis on SDS gels, which were probed by Western blot for proteins indicated on the left side of the figure. Negative controls included incubation of chromatin samples with protein G beads (Beads only) or incubation with protein G beads and mouse IgG (Mouse IgG). In each panel, the numbers above the right six lanes indicate the time after MNNG exposure in hours. Zero time samples were removed immediately prior to MMNG addition. Input (1/20) indicates that 5% of the zero time chromatin sample was loaded.
FIGURE 4.
Quantification of protein bands for modified ChIP assay using 293T Lα+ and 293T Lα− cells. Band intensities of MSH6, histone H3, MLH1, PCNA, ATR, TopBP1, and Chk1 at 0–72 h post MNNG treatment were quantified using Image J (National Institutes of Health) and for each time sample normalized to the corresponding histone H3 signal. The relative intensity was then calculated by dividing the normalized intensity at each time by the zero time sample on the same gel. The results from triplicate experiments are expressed as the means ± S.D. The values that differ significantly from zero time relative intensities are indicated with asterisks (p < 0.05, two-sample t test). Quantification of results obtained with 293T Lα+ and 293T Lα− chromatin samples are shown in A and B, respectively.
Similar results were obtained in comparative studies with TK6 and MT1 cells, which are MSH6-proficient and -deficient, respectively, when cross-linked chromatin fragments were precipitated with anti-MLH1. However, in this case the basal chromatin association of PCNA, ATR, TopBP1, and Chk1 in zero time samples was much reduced as compared with those obtained with 293T cells (Fig. 5). Because of the reduced levels of basal association, the variation in chromatin association of mismatch repair and checkpoint activities was particularly evident in samples from TK6 cells. As observed with 293T Lα+ cells, chromatin enrichment of MutLα and MSH6 was observed in chromatin samples isolated 6 and 12 h after MNNG exposure of TK6 cells, with enrichment for PCNA, ATR, TopBP1, and Chk1 demonstrable only in 12- and 24-h post-exposure isolates. No detectable enrichment for any of these proteins was observed in chromatin samples obtained from MSH6-deficient MT1 cells.
FIGURE 5.
Modified ChIP assay using TK6 and MT1 cells. A, MLH1 antibody was used to immunoprecipitate MutLα. The assay was done as in Fig. 3. B, quantification of protein bands for the modified ChIP assay using TK6 and MT cells. The band intensity of MutLα and MSH2 at 0–72 h post MNNG treatment was quantified and analyzed as for Fig. 4.
It is noteworthy that although we have demonstrated MutSα-, MutLα-, and damage-dependent chromatin association of PCNA, ATR, TopBP1, and Chk1, we have been unable to demonstrate chromatin enrichment of several other activities that are expected to be involved in signaling via the ATR-Chk1 pathway. In particular enrichment of RPA, Rad17, Rad9, or Claspin was not observed in chromatin fractions obtained by precipitation with anti-MSH6 or anti-MLH1 in two different cell line backgrounds, although these activities were readily detected in bulk chromatin samples used as input for immunoprecipitation assay (Figs. 3 and 5).
The enrichment of checkpoint activities that we have observed in anti-MLH1 and anti-MSH6 immunoprecipitates is not a consequence of their nonspecific association with damaged chromatin. Western analysis of bulk chromatin samples used as input for immunoprecipitation assay demonstrated that the levels of ATR, TopBP1, and Chk1 did not vary significantly over the course of the experiment (supplemental Fig. S4). Chromatin-associated RPA did increase noticeably at 48 h post-MNNG treatment, but this protein was not enriched in anti-MLH1 or anti-MSH6 immunoprecipitates.
As noted above, different levels of basal association of PCNA, ATR, TopBP1, and Chk1 were observed when chromatin samples derived from 293T Lα+ were precipitated with anti-MSH6 as compared with TK6 chromatin samples precipitated with anti-MLH1. To address this difference, fragmented chromatin from TK6 cells were precipitated using anti-MSH6. As shown in supplemental Fig. S3, the results obtained were essentially identical to those observed when 293T Lα+ cell chromatin was precipitated with anti-MSH6 (Fig. 3), indicating that this effect is cell line-independent. This finding suggests that the basal association differences observed in these experiments may be due to precipitation of different chromatin subpopulations by anti-MSH6 and anti-MLH1 antibodies.
DISCUSSION
The nature of the DNA damage signaling events triggered by MutSα and MutLα are uncertain, although the consensus view has implicated downstream involvement of the ATR-Chk1 pathway in mismatch repair-dependent responses to SN1 DNA methylators. To better understand the interplay between these two pathways, we have systematically examined interactions of MutSα and MutLα with seven components of the ATR-Chk1 system, including ATR, Chk1, Rad17-RFC, 9-1-1, RPA, TopBP1, and Claspin. Co-immunoprecipitation in nuclear extracts indicated MutSα interaction with ATR, TopBP1, Claspin, and Chk1 and interaction of MutLα with TopBP1 and Claspin. Although purified MSH2 has been previously shown to interact directly with ATR (29), our experiments extend this finding to the functional MSH2·MSH6 (MutSα) form of the protein. We have also demonstrated that MutSα is capable of direct interaction with Chk1 and TopBP1 and that MutLα interacts directly with TopBP1. We were unable to detect MutSα-Claspin and MutLα-Claspin interactions using purified components. Mismatch repair protein-Claspin interactions that occur in nuclear extracts may therefore be indirect or depend on post-translational modification(s) that were not represented in the purified recombinant proteins used for this study. These findings confirm and extend the results of others (29, 41, 49–51), indicating the existence of a complex interaction network involving MutSα, MutLα, and components of the ATR checkpoint pathway. Although RPA, Rad17-RFC, and 9-1-1 have also been implicated in ATR-dependent checkpoint activation (36, 52), we were unable to detect interactions of MutSα or MutLα with these activities by immunological assay either in extracts or by use of purified components.
The significance of the interactions observed with purified proteins was substantiated in formaldehyde-cross-linked chromatin fractions obtained from MNNG-treated cells. Precipitation of chromatin fragments with anti-MSH6 or anti-MLH1 resulted in enrichment for ATR, Chk1, and TopBP1, as well as PCNA. Enrichment in this manner was dependent on prior treatment with MNNG and functionality of MLH1 and MSH6 (Figs. 3–5), which are required for G2 checkpoint activation in response to SN1 DNA methylators. Because the chromatin fragments used in our experiments were relatively small (weight average size, ≈400 bp; mode, ≈200 bp), the enrichment of several activities may indicate MutSα- and MutLα-dependent assembly of a large multi-protein complex during the course of the damage response. Interestingly, recruitment of MutSα and MutLα to MNNG-damaged chromatin precedes the recruitment of PCNA, ATR, Chk1, and TopBP1, but neither the basis nor the implications of this temporal effect are clear. Surprisingly, enrichment of RPA, Rad17, Rad9, or Claspin, all of which have been implicated in ATR-dependent checkpoint events, was not observed in the MNNG-damaged chromatin subpopulations that were selected by precipitation with anti-MSH6 or anti-MLH1. There are several possible explanations for this finding. Although the presence of these proteins was readily demonstrable in the input chromatin samples, it is possible that fraction recruited to the vicinity of MutSα/MutLα-chromatin complexes was simply too small to be detected by the Western blot methods we have used. This possibility is consistent with previous suggestions that DNA-bound RPA and 9-1-1 play important roles in the recruitment of ATR·ATRIP and TopBP1 to single-stranded DNA gaps (53–55).
A second possibility is suggested by the ability of MutSα to interact directly with ATR and TopBP1, and MutLα with TopBP1. Such interactions could provide an alternate pathway for recruitment of ATR and TopBP1 to DNA lesions in a manner that does not require RPA or 9-1-1 (Fig. 6). Such a mechanism would also account for our inability to detect chromatin enrichment of RPA, Rad17-RFC, or 9-1-1 in anti-MSH6 or anti-MLH1 immunoprecipitates. Inasmuch as TopBP1 has been shown to activate ATR (39, 40, 54–59), co-recruitment of these two activities to DNA lesions by MutSα and MutLα might be sufficient to trigger ATR-dependent checkpoint activation. Although such a mechanism could be viewed as consistent with the direct signaling model described above, our experiments do not rule out the futile cycling model. For example, if the mismatch repair DNA synthesis step is fast relative to excision triggered by a MeG lesion, then the steady-state level of the RPA-bound gapped excision intermediate could be very low. Indeed and as noted above, there is excellent evidence implicating Exo1, the primary hydrolytic activity involved in eukaryotic mismatch repair (60), in the cellular response to SN1 DNA methylator damage (34). Unfortunately we have been unable to test for the presence of this activity in chromatin immunoprecipitates because the levels of this activity are so low that it cannot be detected immunologically in cell extracts. The idea that direct recruitment of ATR and TopBP1 by MutSα and MutLα may provide an alternate mechanism for ATR activation makes several testable predictions, and we are pursuing these possibilities.
FIGURE 6.
MutSα and MutLα as a scaffold for recruitment of checkpoint proteins. Based on the results of this study and previous findings of others (29, 41, 49, 61), we suggest that the MutSα·MutLα complex with a DNA lesion may serve as a scaffold for recruitment of components of the ATR-Chk1 pathway.
Acknowledgments
We thank Ravi Iyer, Leonid Dzantiv, and Farid Kadyrov for purified proteins and Elisabeth Penland for cell culture.
*
This work was supported in part by National Institutes of Health Grants GM45190 (to P. M.) and GM32833 (to A. S.).
2
The abbreviations used are:
RFC
replication factor C
PCNA
proliferating cell nuclear antigen
RPA
replication protein A
MNNG
_N_-methyl-_N′_-nitro-_N_-nitrosoguanidine
MeG
_O_6-methylguanine
MGMT
_O_6-methylguanine-DNA methyltransferase
GST
glutathione _S_-transferase
ChIP
chromatin immunoprecipitation.
REFERENCES
- 1.Iyer R. R., Pluciennik A., Burdett V., Modrich P. L. (2006) Chem. Rev. 106, 302–323 [DOI] [PubMed] [Google Scholar]
- 2.Jiricny J. (2006) Nat. Rev. Mol. Cell Biol. 7, 335–346 [DOI] [PubMed] [Google Scholar]
- 3.Li G. M. (2008) Cell Res. 18, 85–98 [DOI] [PubMed] [Google Scholar]
- 4.Hsieh P., Yamane K. (2008) Mech. Aging Dev. 129, 391–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holmes J., Jr., Clark S., Modrich P. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 5837–5841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomas D. C., Roberts J. D., Kunkel T. A. (1991) J. Biol. Chem. 266, 3744–3751 [PubMed] [Google Scholar]
- 7.Iams K., Larson E. D., Drummond J. T. (2002) J. Biol. Chem. 277, 30805–30814 [DOI] [PubMed] [Google Scholar]
- 8.Genschel J., Modrich P. (2003) Mol. Cell 12, 1077–1086 [DOI] [PubMed] [Google Scholar]
- 9.Dzantiev L., Constantin N., Genschel J., Iyer R. R., Burgers P. M., Modrich P. (2004) Mol. Cell 15, 31–41 [DOI] [PubMed] [Google Scholar]
- 10.Zhang Y., Yuan F., Presnell S. R., Tian K., Gao Y., Tomkinson A. E., Gu L., Li G. M. (2005) Cell 122, 693–705 [DOI] [PubMed] [Google Scholar]
- 11.Constantin N., Dzantiev L., Kadyrov F. A., Modrich P. (2005) J. Biol. Chem. 280, 39752–39761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kadyrov F. A., Dzantiev L., Constantin N., Modrich P. (2006) Cell 126, 297–308 [DOI] [PubMed] [Google Scholar]
- 13.Kadyrov F. A., Holmes S. F., Arana M. E., Lukianova O. A., O'Donnell M., Kunkel T. A., Modrich P. (2007) J. Biol. Chem. 282, 37181–37190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Longley M. J., Pierce A. J., Modrich P. (1997) J. Biol. Chem. 272, 10917–10921 [DOI] [PubMed] [Google Scholar]
- 15.Ramilo C., Gu L., Guo S., Zhang X., Patrick S. M., Turchi J. J., Li G. M. (2002) Mol. Cell. Biol. 22, 2037–2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stojic L., Brun R., Jiricny J. (2004) DNA Repair 3, 1091–1101 [DOI] [PubMed] [Google Scholar]
- 17.O'Brien V., Brown R. (2006) Carcinogenesis 27, 682–692 [DOI] [PubMed] [Google Scholar]
- 18.Kaina B., Christmann M., Naumann S., Roos W. P. (2007) DNA Repair 6, 1079–1099 [DOI] [PubMed] [Google Scholar]
- 19.Goldmacher V. S., Cuzick R. A., Jr., Thilly W. G. (1986) J. Biol. Chem. 261, 12462–12471 [PubMed] [Google Scholar]
- 20.Karran P., Bignami M. (1992) Nucleic Acids Res. 20, 2933–2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haracska L., Prakash S., Prakash L. (2000) Mol. Cell. Biol. 20, 8001–8007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duckett D. R., Bronstein S. M., Taya Y., Modrich P. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 12384–12388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duckett D. R., Drummond J. T., Murchie A. I., Reardon J. T., Sancar A., Lilley D. M., Modrich P. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 6443–6447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.York S. J., Modrich P. (2006) J. Biol. Chem. 281, 22674–22683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tominaga Y., Tsuzuki T., Shiraishi A., Kawate H., Sekiguchi M. (1997) Carcinogenesis 18, 889–896 [DOI] [PubMed] [Google Scholar]
- 26.Stojic L., Mojas N., Cejka P., Di Pietro M., Ferrari S., Marra G., Jiricny J. (2004) Genes Dev. 18, 1331–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beardsley D. I., Kim W. J., Brown K. D. (2005) Mol. Pharmacol. 68, 1049–1060 [DOI] [PubMed] [Google Scholar]
- 28.Yoshioka K., Yoshioka Y., Hsieh P. (2006) Mol. Cell 22, 501–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y., Qin J. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 15387–15392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kat A., Thilly W. G., Fang W. H., Longley M. J., Li G. M., Modrich P. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 6424–6428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mojas N., Lopes M., Jiricny J. (2007) Genes Dev. 21, 3342–3355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ceccotti S., Aquilina G., Macpherson P., Yamada M., Karran P., Bignami M. (1996) Curr. Biol. 6, 1528–1531 [DOI] [PubMed] [Google Scholar]
- 33.Schaetzlein S., Kodandaramireddy N. R., Ju Z., Lechel A., Stepczynska A., Lilli D. R., Clark A. B., Rudolph C., Kuhnel F., Wei K., Schlegelberger B., Schirmacher P., Kunkel T. A., Greenberg R. A., Edelmann W., Rudolph K. L. (2007) Cell 130, 863–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klapacz J., Meira L. B., Luchetti D. G., Calvo J. A., Bronson R. T., Edelmann W., Samson L. D. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 576–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang G., Scherer S. J., Shell S. S., Yang K., Kim M., Lipkin M., Kucherlapati R., Kolodner R. D., Edelmann W. (2004) Cancer Cell 6, 139–150 [DOI] [PubMed] [Google Scholar]
- 36.Sancar A., Lindsey-Boltz L. A., Unsal-Kaçmaz K., Linn S. (2004) Annu. Rev. Biochem. 73, 39–85 [DOI] [PubMed] [Google Scholar]
- 37.MacDougall C. A., Byun T. S., Van C., Yee M. C., Cimprich K. A. (2007) Genes Dev. 21, 898–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chini C. C., Chen J. (2003) J. Biol. Chem. 278, 30057–30062 [DOI] [PubMed] [Google Scholar]
- 39.Mordes D. A., Glick G. G., Zhao R., Cortez D. (2008) Genes Dev. 22, 1478–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Navadgi-Patil V. M., Burgers P. M. (2008) J. Biol. Chem. 283, 35853–35859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adamson A. W., Beardsley D. I., Kim W. J., Gao Y., Baskaran R., Brown K. D. (2005) Mol. Biol. Cell 16, 1513–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cejka P., Stojic L., Mojas N., Russell A. M., Heinimann K., Cannavó E., di Pietro M., Marra G., Jiricny J. (2003) EMBO J. 22, 2245–2254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 44.Fousteri M., Vermeulen W., van Zeeland A. A., Mullenders L. H. (2006) Mol. Cell 23, 471–482 [DOI] [PubMed] [Google Scholar]
- 45.Liu Y., Kvaratskhelia M., Hess S., Qu Y., Zou Y. (2005) J. Biol. Chem. 280, 32775–32783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Edelmann W., Umar A., Yang K., Heyer J., Kucherlapati M., Lia M., Kneitz B., Avdievich E., Fan K., Wong E., Crouse G., Kunkel T., Lipkin M., Kolodner R. D., Kucherlapati R. (2000) Cancer Res. 60, 803–807 [PubMed] [Google Scholar]
- 47.Coin F., Oksenych V., Mocquet V., Groh S., Blattner C., Egly J. M. (2008) Mol. Cell 31, 9–20 [DOI] [PubMed] [Google Scholar]
- 48.Szadkowski M., Iaccarino I., Heinimann K., Marra G., Jiricny J. (2005) Cancer Res. 65, 4525–4529 [DOI] [PubMed] [Google Scholar]
- 49.Wang Q., Zhang H., Guerrette S., Chen J., Mazurek A., Wilson T., Slupianek A., Skorski T., Fishel R., Greene M. I. (2001) Oncogene 20, 4640–4649 [DOI] [PubMed] [Google Scholar]
- 50.Quaresima B., Faniello M. C., Baudi F., Crugliano T., Cuda G., Costanzo F., Venuta S. (2006) Hum. Mutat. 27, 715. [DOI] [PubMed] [Google Scholar]
- 51.Cannavo E., Gerrits B., Marra G., Schlapbach R., Jiricny J. (2007) J. Biol. Chem. 282, 2976–2986 [DOI] [PubMed] [Google Scholar]
- 52.Navadgi-Patil V. M., Burgers P. M. (2009) DNA Repair 8, 996–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zou L., Elledge S. J. (2003) Science 300, 1542–1548 [DOI] [PubMed] [Google Scholar]
- 54.Delacroix S., Wagner J. M., Kobayashi M., Yamamoto K., Karnitz L. M. (2007) Genes Dev. 21, 1472–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee J., Kumagai A., Dunphy W. G. (2007) J. Biol. Chem. 282, 28036–28044 [DOI] [PubMed] [Google Scholar]
- 56.Kumagai A., Lee J., Yoo H. Y., Dunphy W. G. (2006) Cell 124, 943–955 [DOI] [PubMed] [Google Scholar]
- 57.Choi J. H., Lindsey-Boltz L. A., Sancar A. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 13301–13306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mordes D. A., Nam E. A., Cortez D. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 18730–18734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Choi J. H., Lindsey-Boltz L. A., Sancar A. (2009) Nucleic Acids Res. 37, 1501–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kadyrov F. A., Genschel J., Fang Y., Penland E., Edelmann W., Modrich P. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 8495–8500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang Y., Cortez D., Yazdi P., Neff N., Elledge S. J., Qin J. (2000) Genes Dev. 14, 927–939 [PMC free article] [PubMed] [Google Scholar]