Deregulation of oncogene-induced senescence and p53 translational control in X-linked dyskeratosis congenita (original) (raw)
Abstract
Defects in ribosome biogenesis and function are present in a growing list of human syndromes associated with cancer susceptibility. One example is X-linked dyskeratosis congenita (X-DC) in which the DKC1 gene, encoding for an enzyme that modifies ribosomal RNA, is found to be mutated. How ribosome dysfunction leads to cancer remains poorly understood. A critical cellular response that counteracts cellular transformation is oncogene-induced senescence (OIS). Here, we show that during OIS, a switch between cap- and internal ribosome entry site (IRES)-dependent translation occurs. During this switch, an IRES element positioned in the 5′untranslated region of p53 is engaged and facilitates p53 translation. We further show that in DKC1m cells, p53 IRES-dependent translation is impaired during OIS ex vivo and on DNA damage in vivo. This defect in p53 translation perturbs the cellular response that counteracts oncogenic insult. We extend these findings to X-DC human patient cells in which similar impairments in p53 IRES-dependent translation are observed. Importantly, re-introduction of wild-type DKC1 restores p53 expression in these cells. These results provide insight into the basis for cancer susceptibility in human syndromes associated with ribosome dysfunction.
Keywords: IRES, p53, RAS, senescence, translation
Introduction
A growing list of inherited human syndromes results from mutations in genes that control different aspects of ribosome synthesis and are characterized by cancer susceptibility (Ruggero and Pandolfi, 2003; Ganapathi and Shimamura, 2008; Alter et al, 2009). X-linked dyskeratosis congenita (X-DC) is invariably associated with mutations of the DKC1 gene encoding for the dyskerin protein, which functions as a pseudouridine synthase that mediates post-transcriptional modification of ribosomal RNA (rRNA) when associated with the H/ACA class of small nucleolar RNAs (Meier, 2005). Other, milder forms of dyskeratosis congenita can arise from mutations in genes affecting other molecular pathways, including components of the telomerase complex that are also found to be associated with dyskerin (Kirwan and Dokal, 2008). Importantly, we have genetically shown by recapitulating X-DC pathogenesis in DKC1 hypomorphic mice (DKC1m) that impairments in rRNA modifications are associated with X-DC pathogenesis when telomeres are of a normal length (Ruggero et al, 2003). Strikingly, more than 50% of DKC1m mice develop tumours with different histological origins, consistent with increased cancer susceptibility present in X-DC patients (Ruggero et al, 2003). DKC1m mice as well as X-DC human patient cells show a selective impairment in the translation of a subset of mRNAs that share a common mode of translation initiation relying on an internal ribosome entry site (IRES) element positioned in their 5′ untranslated region (Yoon et al, 2006). However, the tumour suppressive function of DKC1 is poorly understood, and there is a major gap in our understanding of the molecular events underlying cancer initiation or progression in X-DC.
IRES-dependent translation is a fine-tuning mechanism that regulates the expression of important proteins in specific circumstances when a general decrease in cap-dependent translation is evident, such as during the mitotic phase of the cell cycle, hypoxia, and apoptosis. A balance between cap- and IRES-dependent translation is, therefore, required to maintain accurate cellular homeostasis (Holcik and Sonenberg, 2005). Cellular senescence is an effective and rapid anti-tumour barrier that acts by restraining the uncontrolled proliferation of cells that are carrying potentially oncogenic alterations, such as the H-RasV12 oncogene (Artandi and Attardi, 2005). In this study, we identify a new mechanism by which a selective switch between cap- and IRES-mediated translation modulates gene expression in cells undergoing oncogene-induced senescence (OIS). These findings link IRES-dependent translational control with the earliest tumour suppressive response against oncogenic insult. We further show that p53, which harbours an IRES element (Ray et al, 2006; Yang et al, 2006), is translationally regulated during OIS through this mechanism. Importantly, DKC1m cells display impairments in undergoing cellular senescence after expression of an activated H-RasV12 oncogene and bypass OIS-induced cell cycle arrest, which results in a dramatic increase in their clonogenic potential. At the molecular level, this defect is associated with impaired IRES-dependent translational control of p53 expression during OIS and DNA damage. Moreover, X-DC human patient cells that harbour mutations in the DKC1 gene also show similar defects in p53 IRES-dependent translation, which are associated with a significant decrease in p53 protein expression. Importantly, re-introduction of an exogenous wild-type DKC1 (DKC1WT) into these cells was able to restore p53 expression levels, thus further showing the importance of DKC1 in the regulation of p53 translational control. Altogether, these results provide a novel mode for regulating gene expression during the early steps of cellular transformation, highlighting the importance of p53 translational control and helping to explain how DKC1 functions as a tumour suppressor gene.
Results
A translational switch from cap- to IRES-dependent translation occurs during OIS
Oncogenic Ras triggers premature senescence in primary cells (Serrano et al, 1997). Induction of senescence is a frequent outcome of oncogenic insult and serves as a critical and cell-autonomous tumour suppressive mechanism (Artandi and Attardi, 2005). Although the cellular events during OIS are well established, the molecular mechanisms that regulate gene expression during OIS are poorly understood. Specifically, although the initial effect of oncogenic Ras in normal cells is to trigger proliferation, this aberrant proliferation is not sustained, and after a few doublings, cells enter into a permanent cell cycle arrest indicative of senescence (Serrano et al, 1997). Until now, translational control of gene expression has not been implicated in this process, but may facilitate a more immediate, rapid, and robust mechanism to counteract oncogenic activation. To address this question, we have generated an animal model that expresses a stable, genetically encoded translational reporter for cap- and IRES-dependent translation (CMV-hepatitis C virus (HCV)-IREST). The CMV-HCV-IREST mice harbour a dicistronic mRNA in which the first cistron (Rluc) is translated through cap-dependent translation and the second cistron (Fluc) is translated under the control of the HCV IRES element IREST (Figure 1A). The HCV IRES element was selected because it provides a general readout of IRES-dependent translation activity and, therefore, represents a great tool to quantify differences in overall IRES-dependent translation initiation. Primary cells from this mouse can be isolated, and this provides a means to accurately monitor translational control in distinct cell types such as mouse embryo fibroblasts (MEFs), in which the integrity of the transgenic vector has been carefully characterized (Supplementary Figure S1). Using this genetic reporter, we uncovered that a ‘translational switch' between cap- and IRES-dependent translation occurs during RAS-induced senescence.
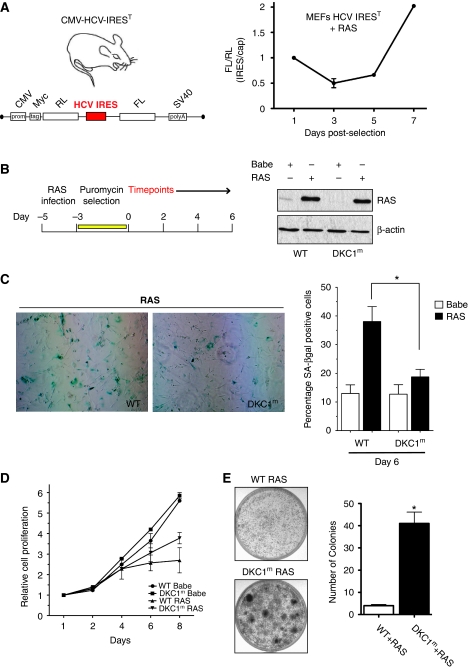
Figure 1.
Induction of OIS is impaired in DKC1m cells. (A) Transgenic CMV-HCV-IREST animals harbouring a translational dicistronic-luciferase reporter (left). Primary mouse embryo fibroblasts (MEFs) prepared from CMV-HCV-IREST mice were infected with RAS and the ratio between HCV IRES-mediated Firefly (FL) and cap-dependent Renilla (RL)-luciferase activities was measured at different time points during OIS (right). (B) Experimental design to study OIS after RAS infection of WT and DKC1m primary MEFs. Western blot analysis of RAS levels in WT and DKC1m retrovirally transduced cells. β-actin was used as loading control. (C) Representative images of WT and DKC1m MEFs stained for SA-β-galactosidase. The number of senescent SA-β-galactosidase-positive cells was determined 6 days post-selection. Graph shows mean±s.e.m. of four independent experiments performed in triplicate *_P_=0.04. (D) Representative growth curve of WT and DKC1m cells transduced with empty vector or RAS retroviruses. Relative number of cells±s.e.m. was determined using crystal violet staining and normalized to the cell number at day 1. (E) Clonogenic potential of WT and DKC1m primary MEFs after RAS infection. Images of RAS-infected WT (top) and DKC1m (bottom) cells plated at clonal density (20 000 cells) in a 6 cm plate and cultured for 3 weeks. The number of colonies was determined using standard crystal violet staining (left). Graph shows mean number of colonies±s.e.m. of three independent experiments performed in triplicate *_P_=0.002.
Early passage HCV IREST MEFs were transduced with retroviruses harbouring an activated H-RasV12 oncogene (herein after referred to as RAS), and IRES- and cap-dependent translation were monitored during the process of OIS (Figure 1A; Supplementary Figure S1A). We observed that while cap-dependent translation is initially upregulated after RAS expression, and is associated with an initial burst in proliferation, it steadily decreases during the process of OIS. In contrast, IRES-mediated translation levels are initially very low, but markedly increase during the process of OIS (Figure 1A; Supplementary Figure S1A). These findings show that a translational switch between cap- and IRES-dependent translation operates during OIS and may be important in modulating gene expression required to counteract oncogenic insult.
OIS is impaired in DKC1m cells
We reasoned that defects in IRES-mediated translation that are present in DKC1m mice and X-DC patient cells (Yoon et al, 2006) might unbalance the synthesis of proteins that are important for OIS induction. Therefore, we first studied OIS in primary DKC1m MEFs. To this end, early passage WT and DKC1m MEFs were transduced with control or RAS viruses and the number of senescent β-galactosidase-positive cells was determined 6 days post-pharmacological selection (Figure 1B and C). Strikingly, we found that in the presence of RAS, the number of DKC1m senescent cells was significantly reduced compared with control cells (Figure 1C; Supplementary Figure S2C). Senescence is characterized by irreversible cell cycle arrest, and overexpression of RAS consistently inhibits the growth of WT cells during this process (Figure 1D). In contrast, DKC1m cells proliferate at a higher rate when compared with RAS-infected controls (Figure 1D). Importantly, defects in OIS induction that are present in DKC1m cells are associated with a dramatic increase in clonogenic potential upon overexpression of only a single oncogene that is more than four-fold higher than that of WT cells (Figure 1E). These results strongly suggest that DKC1 may act as a tumour suppressor gene, at least in part, through control of OIS.
RAS-induced activation of p53 is defective in DKC1m cells
The p53 tumour suppressor gene is a master regulator of OIS (Serrano et al, 1997). Loss of p53 can revert the senescence phenotype elicited by oncogene activation in vivo (Braig et al, 2005; Chen et al, 2005; Collado et al, 2005). Induction of p53 is rapidly observed after expression of oncogenic RAS in primary cells, and p53 null cells are resistant to RAS-induced senescence (Serrano et al, 1997; Lin et al, 1998). This prompted us to examine p53 expression in WT and DKC1m cells during RAS-induced senescence. Strikingly, we found that RAS-dependent p53 accumulation is reduced in DKC1m cells at all time points studied after pharmacological selection of RAS-infected cells (Figure 2A). Induction of p53 activity during senescence is associated with growth arrest, at least in part, through activation of p53 target genes, including cell cycle inhibitors (Serrano et al, 1997; Lin et al, 1998). We tested whether the decrease in p53 expression present in DKC1m cells would manifest itself in changes in downstream targets. We found that p53-dependent activation of the cell cycle inhibitors p21 cip and GADD45 after RAS expression was substantially reduced in DKC1m cells (Figure 2E). Importantly, stable overexpression of an exogenous WT DKC1 in RAS-infected primary DKC1m MEFs was able to rescue p53 protein levels and to restore OIS induction (Supplementary Figure S3A and B).
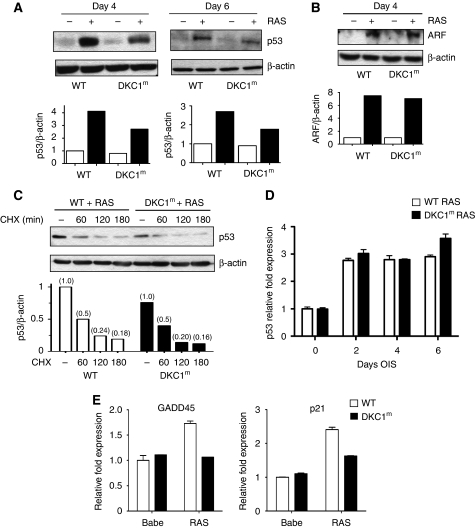
Figure 2.
p53 IRES-mediated translation is active during OIS and is reduced in DKC1m cells. (A) Representative analysis of p53 protein levels measured 4 and 6 days post-selection. Densitometry analysis of p53 protein was normalized over β-actin levels in each sample (bottom). (B) RAS-dependent induction of ARF is not impaired in DKC1m cells. Representative western blots showing ARF levels in empty vector and RAS-infected WT and DKC1m cells 4 days post-selection (top). Densitometry analysis of ARF over β-actin levels in each sample is shown (bottom). (C) Assessment of p53 protein turnover 6 days post-selection. Cells were cultured in the presence of 50 μg/ml of CHX and harvested at the indicated time points. Densitometric analysis of p53 over β-actin protein levels in each sample in WT and DKC1m cells (bottom). Numbers represent relative band intensity normalized to the untreated sample for each genotype. (D) Quantification of p53 mRNA levels in RAS-infected WT (white bars) and DKC1m (black bars) cells during OIS was carried out using real-time Q-PCR; graph shows mean±s.e.m. of a representative experiment performed in duplicate. (E) Determination of p21 cip and GADD45 mRNA levels during OIS in WT and DKC1m cells. p21 and GADD45 mRNA levels were measured 6 days post-selection using Q-PCR and normalized over the amount of β-actin messenger in each sample. Graphs show mean±s.e.m. of a representative experiment performed in triplicate.
We next sought to understand the molecular mechanism by which OIS-dependent p53 induction is impaired in DKC1m cells. One of the main mechanisms by which oncogenic RAS activation engages the p53 pathway in primary murine cells is through induction of the ARF tumour suppressor gene, which positively regulates the stability of the p53 protein (Palmero et al, 1998; Lin and Lowe, 2001; Efeyan et al, 2009). We therefore first analysed the expression of ARF on transduction of oncogenic RAS. ARF expression is similarly induced in WT and DKC1m cells during OIS (Figure 2B). Furthermore, we blocked total protein neosynthesis using cyclohexamide (CHX) to monitor p53 turnover in WT and DKC1m cells during OIS (Figure 2C). In line with the analysis of ARF levels, no significant differences in p53 stability were noticeable between WT and DKC1m cells during OIS (Figure 2C). We next assessed control of p53 transcription in DKC1m cells during OIS. Q-PCR analysis revealed that OIS promotes p53 mRNA transcription to a similar extent in WT and DKC1m cells (Figure 2D). These findings strongly suggest that upstream signals activated during OIS that regulate p53 transcription and/or stability are unperturbed in DKC1m cells and cannot account for the decrease in p53 protein levels evident in these cells.
p53 IRES-mediated translation is active during senescence and reduced in DKC1m cells
We next asked whether impairments in DKC1 might affect p53 translational control during OIS. We therefore directly monitored the association between p53 mRNA and polyribosomes in WT and DKC1m cells in the context of OIS (Figure 3A). Importantly, we found that the amount of p53 mRNA associated with polysomes was markedly augmented during OIS in WT cells. Strikingly, this upregulation in polysomal p53 mRNA levels was completely abrogated in RAS-infected DKC1m cells (Figure 3A, top panels). Notably, the analysis of β-actin mRNA did not reveal significant differences in the polysomal distribution between WT and DKC1m cells, but it did show that the levels of this messenger associated with polysomal fractions were reduced during OIS (Figure 3A, bottom panels). Taken together, these results strongly suggest that a specific defect in p53 translational control is present in DKC1m cells during OIS.
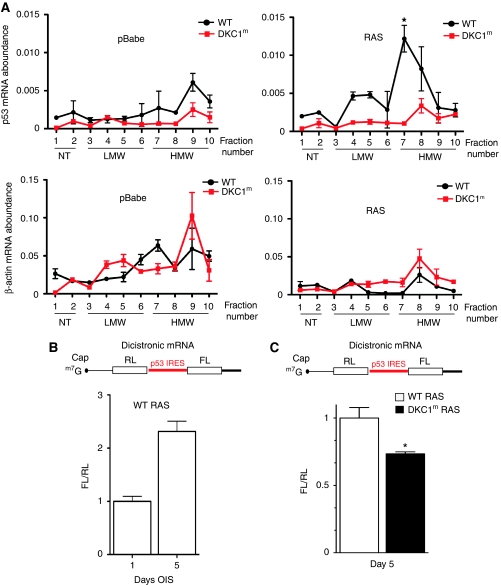
Figure 3.
p53 IRES-mediated translation is impaired in DKC1m cells during OIS. (A) Polysomal analysis of p53 and β-actin mRNAs in empty vector (pBabe)- and RAS-infected WT and DKC1m primary MEFs 6 days post-selection. The polysomal association of p53 and β-actin mRNAs was tested by fractionating cytoplasmic lysates through a sucrose gradient and measuring mRNA abundance by Q–PCR analysis in each of the 10 resulting fractions. Graphs show the abundance of p53 (top) and β-actin (bottom) mRNAs in each gradient fraction from pBabe- (left) and RAS-infected (right) WT and DKC1m cells normalized to the corresponding 18S rRNA levels and corrected for the total mRNA content in one representative experiment. *_P_=0.039 was calculated combining three independent experiments. Fractions in the graphs are indicated as NT (not translated), LMW, and HMW (low and high molecular weight polysomes, respectively). (B) RAS-infected WT cells were transduced with a dicistronic reporter mRNA harbouring the p53 IRES element upstream of the Firefly-luciferase ORF. Graph shows mean±s.e.m. of ratios between Firefly and Renilla-luciferase (FL/RL) activities measured 1 and 5 days post-selection. (C) RAS-infected WT and DKC1m cells transduced with a dicistronic reporter mRNA harbouring the p53 IRES element. Graph shows the ratios between Firefly and Renilla-luciferase (FL/RL) activities measured at day 5. *_P_=0.015 was calculated in four independent experiments carried out in triplicate.
Interestingly, p53 has been shown to possess an IRES element (Ray et al, 2006; Yang et al, 2006); however, p53 IRES activity during the senescence programme has not been earlier characterized. We therefore monitored p53 IRES-dependent activity during OIS. At first, we used the well-established dicistronic expression system in which the first cistron is translated by a cap-dependent initiation mechanism and the second is translated by the preceding p53 IRES element. We transfected WT primary fibroblasts with this RNA reporter construct to monitor p53 IRES activity after selection of RAS-infected cells (Figure 3B; Supplementary Figure S2A and B). We observed that p53 IRES activity is greatly induced as cells undergo senescence (Figure 3B; Supplementary Figure S2A). We next explored the possibility that p53 IRES-mediated translation may be impaired in DKC1m cells. To this end, we compared p53 IRES-dependent translation in WT and DKC1m RAS-infected cells. Strikingly, we found that p53 IRES activity is reduced in DKC1m cells (Figure 3C). Therefore, p53 IRES-dependent translation is active during OIS and is impaired in DKC1m cells, which show reduced p53 protein levels.
p53 IRES-mediated translation and function are impaired in DKC1m cells after DNA damage and mitosis
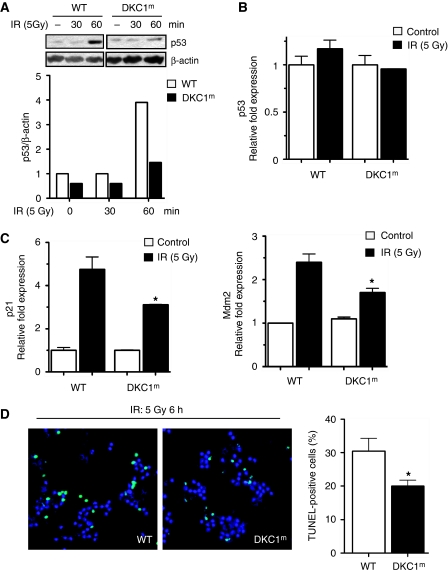
To further analyse the relationship between DKC1 and p53 activation, we measured p53 protein levels during conditions that promote IRES-mediated translation, such as mitosis and DNA damage. Translational control is important in modulating cellular levels of the p53 tumour suppressor protein after DNA damage (Takagi et al, 2005; Halaby and Yang, 2007; Ofir-Rosenfeld et al, 2008) and the p53 IRES element has an important function in stimulating p53 synthesis after treatment with DNA-damaging agents (Ray et al, 2006; Yang et al, 2006). We monitored p53 protein levels after DNA damage, comparing WT and DKC1m primary cells. After 30 min of treatment with etoposide—a DNA-damaging agent that causes double-strand breaks—levels of p53 protein rapidly increase by almost eight-fold in WT cells, but this induction is markedly reduced in DKC1m (Figure 4A). The reduction in p53 protein levels observed in DKC1m cells is not due to changes in p53 transcription or protein stability (Figure 4B and C). On the contrary, we found that on etoposide treatment, p53 IRES-dependent translation is increased in WT, but not in DKC1m cells (_P_=0.0005; Figure 4D; Supplementary Figure S4). Importantly, the specific defect in p53 IRES-mediated translation in DKC1m cells perturbs p53 function. Indeed, we found that the transactivation of p53 target genes, namely p21 cip and Mdm2, was markedly reduced in DKC1m cells after DNA damage (Figure 4E). Consistent with these findings, the percentage of DKC1m apoptotic cells was significantly decreased as compared with control cells after treatment with etoposide (Figure 4F).
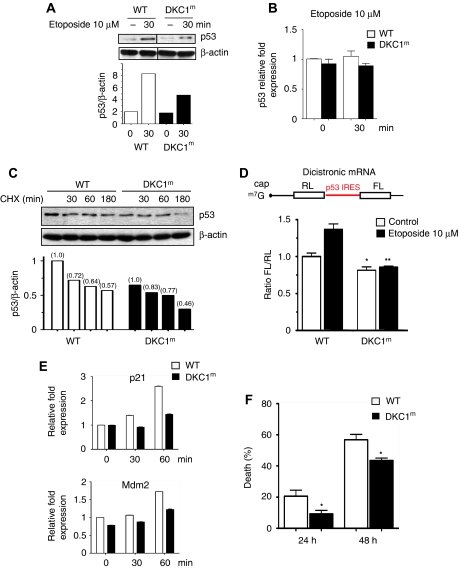
Figure 4.
p53 IRES-mediated translation is impaired in DKC1m cells after DNA damage. (A) Cells were cultured in the presence of etoposide and p53 protein levels were analysed at the indicated time by western blot analysis. Densitometry analysis of p53 normalized over β-actin levels is shown. (B) The p53 transcript levels in WT and DKC1m cells cultured in the presence or absence of etoposide were measured by Q–PCR. Mean±s.e.m. of p53 relative expression normalized over the levels of GAPDH mRNA from a representative experiment is shown. (C) Levels of p53 were measured in WT and DKC1m MEFs pre-cultured for 2 h with etoposide and subsequently exposed to CHX. Cells were harvested at the indicated time points. The graph shows the densitometric analysis of p53 over β-actin levels in a representative experiment. (D) WT and DKC1m MEFs were transduced with a dicistronic mRNA harbouring a p53 IRES element. Six hours after transfection, cells were cultured in the presence of 10 μM etoposide for 1 h and the FL/RL ratio was measured. Graph is mean±s.e.m. of FL/RL ratios measured in four independent experiments performed in duplicate *_P_=0.03 and **_P_=0.001. (E) WT and DKC1m MEFs were cultured in the presence of etoposide for the indicated time and p21 and Mdm2 mRNA levels were measured. Graphs show mean±s.e.m. of a representative experiment performed in triplicate. (F) WT and DKC1m cells were cultured in the absence or presence of 100 μM etoposide and cell death was measured by annexin V/PI co-staining after 24 and 48 h. Graph shows mean±s.e.m. of four experiments performed in duplicate; * are _P_=0.03 and _P_=0.01 for 24 and 48 h, respectively.
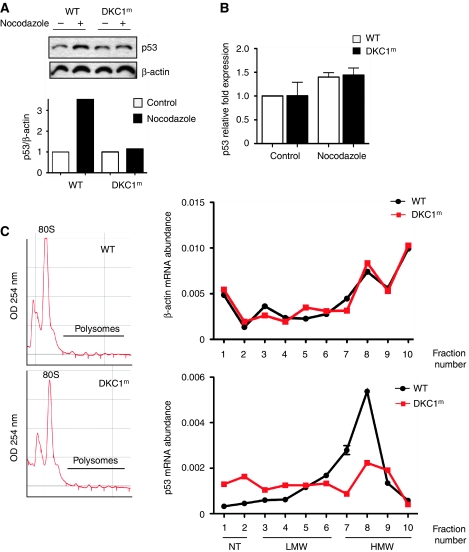
It has been shown that mitotic progression depends on precise translational control that is associated with a general reduction of cap-dependent translation and a concomitant increase of IRES-mediated translation (Pyronnet et al, 2000). We found that in cells arrested in the G2/M phase of the cell cycle, p53 protein levels were increased in WT, but not in DKC1m cells (Figure 5A). On the other hand, p53 transcript levels were comparable between WT and DKC1m cells (Figure 5B), and the percentage of cells in the G2/M phase was similar in both genotypes (Supplementary Figure S5).
Figure 5.
DKC1m cells show defects in p53 translational control during specific conditions that promote IRES-dependent translation. (A) Western blot analysis of p53 levels in WT and DKC1m cells treated with nocodazole for 18 h (top) and corresponding densitometric analysis normalized over β-actin (bottom). (B) Q–PCR analysis of p53 mRNA normalized over β-actin mRNA levels from total RNA prepared from cells in (A). Graph is mean±s.e.m. of a representative experiment performed in triplicate. (C) Polysomal fractions were prepared from WT and DKC1m MEFs after nocodazole treatment (see Materials and methods). Representative polysomal profiles of nocodazole-treated WT and DKC1m cells (left). The levels of β-actin (top) and p53 (bottom) mRNA were analysed in each polysomal fraction using Q–PCR analysis. The amount of p53 mRNA was normalized over the β-actin mRNA levels in three independent experiments performed in duplicate (right).
We took advantage of the ability to synchronize cells in a very controlled setting to perform polysome analysis. This enabled us to monitor p53 mRNA association with ribosomes at a moment when p53 protein levels are controlled through an IRES-dependent mechanism. We observed that the amount of p53 mRNA associated with fractions containing heavy polysomes is markedly reduced in DKC1m as compared with control cells (Figure 5C). Conversely, we did not find any significant differences in the levels of β-actin mRNA associated with the polysomes between the two genotypes (Figure 5C). Altogether, these results identify a function for DKC1 in the control of p53 IRES-mediated translation and uncover a novel regulatory circuitry that contributes, at least in part, to the accumulation of p53 during specific stress conditions such as mitosis, DNA damage, and in response to oncogenic insults.
p53 pro-apoptotic function is impaired in vivo in DKC1m mice
We next sought to investigate the consequences of deregulation of p53 IRES-mediated translation in a physiological setting in vivo in DKC1m mice. To this end, we analysed p53 expression and its pro-apoptotic activity in γ-irradiated thymocytes from WT and DKC1m mice. Importantly, we found that γ-irradiation-dependent induction of p53 protein expression was drastically impaired in DKC1m thymocytes (Figure 6A), whereas no differences in total p53 transcript levels were noticeable (Figure 6B). The induction of the two major p53 target genes, p21 and Mdm2, was also reduced in DKC1m compared with WT cells (Figure 6C). To validate the consequences of these molecular defects, we assessed the levels of apoptosis in WT and DKC1m thymi 6 h after irradiation (Figure 6D). Importantly, the number of DKC1m apoptotic (TUNEL-positive) thymocytes was significantly reduced as compared with WT cells (Figure 6D). Altogether, these data indicate that defective p53 translational control, because of impairments in DKC1 activity, is associated with defects in p53 tumour suppressive function in vivo.
Figure 6.
p53 function is impaired in vivo in DKC1m mice. (A) Representative western blot of p53 and β-actin protein levels in lysates from freshly isolated WT and DKC1m thymocytes after γ-irradiation. Densitometric analysis of p53 over β-actin levels is shown (bottom). (B) Analysis of p53 and (C) p21 and Mdm2 transcripts levels 1 h post-γ-irradiation were measured by using Q-PCR analysis. Graphs are mean±s.e.m. of one representative experiment. *P<0.01 was calculated combining three independent experiments performed in triplicate. (D) Quantification of γ-irradiation-induced apoptosis in thymocytes isolated from WT and DKC1m mice 6 h post-irradiation. Representative images in which TUNEL staining has been performed are shown. Apoptotic cells are shown in green (TUNEL-positive) and nuclei are shown in blue (DAPI staining). Graph shows percentage of apoptotic cells in WT and DKC1m thymi. Four mice for each genotype were used. *_P_=0.027.
X-DC human patient lymphoblasts show impaired p53 IRES-dependent translation
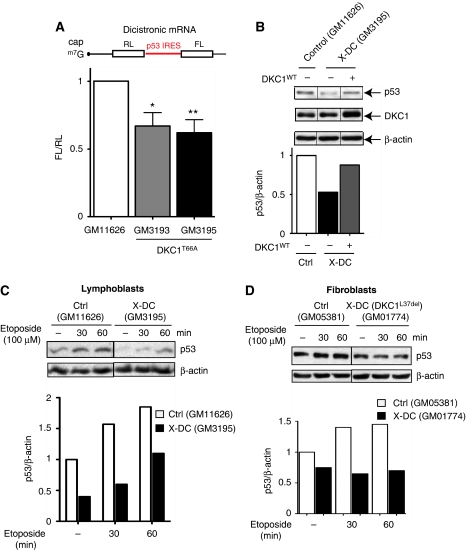
We next asked whether p53 IRES-mediated translation might be impaired in human patient cells that harbour point mutations in the DKC1 gene. We transfected control and X-DC lymphoblasts carrying a missense mutation leading to the amino-acid change T66A (DKC1T66A) (Knight et al, 1999) with the p53 dicistronic RNA reporter construct to monitor p53 IRES activity. We found that X-DC patient cell lines display impaired p53 IRES-mediated translation compared to controls (Figure 7A; Supplementary Figure S6). Accordingly, p53 protein levels were also reduced in the same X-DC patient cells and, most importantly, overexpression of an exogenous WT DKC1 (DKC1WT) completely restored p53 expression to levels of normal cells (Figure 7B). Furthermore, we found that induction of p53 expression was significantly hampered in X-DC lymphoblasts after etoposide treatment (Figure 7C). Similarly, we analysed early passage X-DC (GM01774) fibroblasts, which bear a distinct DKC1 mutation, a deletion of L37 (del37L referred to as DKC1L37del), and found that p53 levels were also decreased both at the steady state and after DNA damage (Figure 7D), which is consistent with our in vivo analysis in DKC1m mice. Taken together, these results suggest that impairments in DKC1 function also lead to defects in p53 IRES-dependent activity in X-DC human patient cells. Therefore, p53 IRES-mediated translation may represent one of the molecular underpinnings leading to increased cancer susceptibility in X-DC pathogenesis.
Figure 7.
p53 IRES-mediated translation is impaired in human cells derived from X-DC patients. (A) Normal control (Coriell # GM11626) and X-DC human lymphoblasts derived from two affected brothers carrying the missense mutation T66A (Coriell # GM3193 and GM3195) were electroporated with the dicistronic reporter mRNA harbouring a p53 IRES element. The FL/RL ratio was measured 6 h after transfection. Graph is mean±s.e.m. of FL/RL ratios measured in three independent experiments performed in duplicate (*_P_=0.015 and **_P_=0.005). (B) Normal control and X-DC lymphoblasts were electroporated with an empty or a DKC1 WT (DKC1WT) expression vector and p53, DKC1, and β-actin protein levels were measured 12 h post-transfection. Representative protein analysis in one of three experiments is shown. Graph shows densitometric analysis of p53 over β-actin levels. (C) Control and X-DC lymphoblasts were cultured in the absence or presence of etoposide and harvested at the indicated time points. The p53 and β-actin protein levels are shown. (D) Normal (GM05381) and X-DC (GM01774) fibroblasts carrying a deletion of L37 in DKC1 (referred to as DKC1L37del) were cultured in the absence or presence of etoposide. Cells were harvested at the indicated time points and p53 and β-actin protein levels were measured. Densitometric analysis of p53 over β-actin is shown (bottom).
Discussion
A growing number of human syndromes including Diamond Blackfan anaemia, Shwachman–Bodian–Diamond syndrome, and X-DC are characterized by defects in ribosome biogenesis and function and are associated with increased cancer susceptibility (Ruggero et al, 2003; Ganapathi and Shimamura, 2008; Alter et al, 2009). The molecular mechanisms that may account for increased cancer susceptibility as a consequence of impairments in ribosome activity remain poorly understood. In this study, we identified a translational switch from cap- to IRES-mediated translation in primary cells transduced with activated oncogenic RAS. This finding suggests that the synthesis of important proteins required for accurate OIS, the first cellular barrier against neoplastic transformation (Braig et al, 2005; Chen et al, 2005; Collado et al, 2005), may be regulated at the translational level. Importantly, we found that induction of OIS is impaired in DKC1m primary cells, which possess a specific defect in the IRES-mediated translation (Yoon et al, 2006). Our data further show that p53, a critical mediator of OIS, is regulated at the translational level through an IRES-dependent mechanism during this critical cellular event. Moreover, DKC1m cells show impairments in OIS that are accompanied by a significant reduction in p53 IRES-mediated translation and in the association of p53 mRNA with polysomes. The impairment in p53 expression present in DKC1m cells was sufficient to bypass senescence, leading to a dramatic increase in the clonogenic activity of the cells on overexpression of a single oncogene. IRES-mediated translation of p53 is also activated in response to cellular stress conditions (Halaby and Yang, 2007), and p53 IRES-mediated translation is impaired after DNA damage and during the mitotic transition in DKC1m cells. Furthermore, we show that this translational defect is associated with a severe impairment of p53 pro-apoptotic function in vivo. Importantly, human X-DC patient cells also show impairments in p53 IRES-mediated translation and display reduced p53 protein expression, which can be reverted back to normal levels by re-introduction of an exogenous form of WT DKC1, thus showing the importance of DKC1 in the regulation of p53 translational control. Therefore, modulation of p53 expression at the translation level may reflect a rapid, robust, and specific mechanism to counteract the earliest events during cellular transformation as well as during cellular stress conditions. Surprisingly, mouse cells harbouring a knock-in of a DKC1 human deletion, Dkc1Δ15, have been reported to show increased p53 expression levels (Gu et al, 2008). The discrepancy in these findings could be explained, at least in part, if DKC1 activity is impaired to a greater extent in Dkc1Δ15 cells. For example, upregulations in p53 expression could reflect an indirect stress response because of decreases in DKC1 activity below a certain threshold, as has been observed after nucleolar stress (Pestov et al, 2001; Olson, 2004).
Our results may, in part, explain what can be viewed as a paradox in X-DC pathogenesis. DKC1 has also been implicated in telomere maintenance, and X-DC patient cells show short telomeres (Mitchell et al, 1999; Ganapathi and Shimamura, 2008). Typically, as cells undergo telomere shortening, the p53 pathway is activated by different means to prevent the development of neoplastic lesions (Artandi and Attardi, 2005). In X-DC cells, a translational dysfunction in p53 expression may, on the contrary, promote cancer initiation even in the context of telomere shortening. The data presented in this study strongly suggest that deregulation of p53 translational control may contribute to increased cancer susceptibility in X-DC and possibly other diseases associated with ribosomal dysfunction.
Materials and methods
Transgenic mice
CMV-HCV-IREST mice were generated using a pCMV-Myc-RL-HCV IRES-FL construct. The HCV IRES element (Yoon et al, 2006) was subcloned into pCR 2.1 (Invitrogen), digested with EcoRI, and inserted into a pRF plasmid. The RL-HCV IRES-FL was amplified, digested using BglII and KpnI, and inserted into the pCMV-Myc expression vector. The resulting pCMV-Myc-RL-HCV IRES-FL was linearized using an Alw44I restriction enzyme and microinjected into mouse embryos, which were then implanted into female recipient mice. Founder lines were generated, genotyped, and crossed with WT mice to verify germline transmission. To confirm that there was no splicing of the transgenic HCV IREST dicistronic RNA, RT–PCR was performed to amplify a 1.9 kb fragment spanning from the sequence upstream of the Renilla coding region (P1) up to the Firefly coding region (P2). A second RT–PCR was carried out using primers on the Renilla (P3) and Firefly (P2) sequences to amplify a 700 bp fragment containing the HCV IRES element. The following primers were used: P1 5′-GCTAGCCACCATGACTTCGAAAG, P2 5′-GATGTTCACCTCGATATGTGC, and P3 5′-GTTTATTGAATCGGACCCAGG. In addition to the analysis at steady state conditions, the integrity of the endogenous transgenic HCV IREST reporter transcript was validated during OIS. To this end, an shRNAi expression vector targeting the RLuc portion was transfected along with a β-galactosidase plasmid (for transfection efficiency) into RAS-infected HCV IREST cells. Briefly, 100 000 RAS-infected HCV IREST cells were plated in 6 cm plates and transfected with a scramble or an shRNAi vector targeting the RLuc sequence (GenScript) by using Lipofectamice 2000 (Invitrogen) 4 days post-selection. Firefly-luciferase activity was determined using Dual-luciferase kit (Promega) using a single tube luminometer (Optocomp1, MGM Instruments).
Integrity of the exogenous dicistronic p53 IRES mRNA was also tested in MEFs after etoposide-induced DNA damage and RAS expression, as well as in human X-DC patient lymphoblasts. Total RNA was isolated, retro-transcribed, and cDNA was used to amplify a 680 bp (P3-P2). Furthermore, a 1.45 kb PCR fragment spanning the p53 IRES element was amplified by using the forward primer P4 5′-ATGATGACTCGAAAGTTTATGATCC-3′ along with the earlier described reverse primer P2.
Cell culture
MEFs were prepared from early generation WT, HCV IREST, and DKC1m male mice as described (Ruggero et al, 2003). Cells were isolated at embryonic 13.5 d.p.c. and cultured in high glucose Dulbecco Modified Eagle Media supplemented with 20% foetal bovine serum (FBS) and 100 U/ml of penicillin/streptomycin. Human control and X-DC cell lines were obtained from Coriell Cell Repositories (Coriell Institution for Medical Research, Camden, NJ). Control (GM11626) and X-DC human lymphoblasts derived from two affected brothers carrying the missense mutation T66A (GM3193 and GM3195) were maintained in RPMI supplemented with 20% FBS and 100 U/ml of penicillin/streptomycin. Similarly, human control (GM05381) and X-DC (GM01774) carrying a L37deletion (del37L) were obtained from Coriell Cell Repositories and maintained in MEM Eagle-Earle's Balance Salt Solution supplemented with non-essential amino acids, 20% FBS, and 100 U/ml of penicillin/streptomycin. All chemicals used in this study were purchased from Sigma-Aldrich unless otherwise stated.
Determination of senescence
Determination of RAS-induced senescence was carried out as described (Serrano et al, 1997) with some modifications. Briefly, early passage (p2) WT and DKC1m MEFs were subjected to multiple rounds of infection using control or RAS high-titer viral supernatants. Infected cells were selected for 3 to 4 days in the presence of 2 μg/ml of puromycin. After selection, cells were plated at 40 × 103 cells/well in six-well plates (NUNC). Cellular senescence was assayed after 6 and 8 days using a Senescence Detection kit (Calbiochem) after the manufacturer's instructions. Finally, the number of senescent SA-βgal-positive cells was assessed using a Nikon TE2000E inverted microscope.
Proliferation
Cell proliferation was determined using crystal violet staining. In brief, cells were plated at 20 × 103 cells/well in six-well plates and stained with a 0.1% crystal violet solution. Cells were rinsed with water to remove excess dye and air dried overnight at room temperature. Finally, crystal violet was solubilized using a 10% acetic acid solution for 10 min at room temperature and the 590 nm absorbance was measured using a fluorimeter (Safire). For colony forming assays, cells were plated at 20 × 103 cells per 6 cm plate and cultured for 3 weeks. Cells were fixed using 70% ice-cold methanol, air dried, and the number of colonies was determined after staining with a 0.5% crystal violet solution.
Protein analysis
Protein analysis was performed as described (Yoon et al, 2006) according to standard protocols. Proteins were analysed by SDS–PAGE and transferred to PVDF membranes (Millipore). Antibodies and dilutions used in this study were anti-mouse p53 dilution 1:400 (CM5 Novocastra), anti-ARF 1:200 (Santa Cruz Biotechnologies), anti-RAS 1:1000 (Cell Signaling), anti-DKC1 1:200 (Santa Cruz Biotechnologies), and anti-β-actin 1:5000 (Sigma). Anti-mouse and anti-rabbit horseradish peroxidase-conjugated secondary antibodies were purchased from Amersham and used at dilution 1:10 000. Secondary anti-rat horseradish peroxidase-conjugated antibody was purchased from Cell Signaling and used at 1:5000. Enhanced chemiluminescent solution was purchased from Pierce.
mRNA quantification by Q-PCR
Total RNA was purified using an RNAeasy kit (Qiagen) according to the manufacturer's instructions, and 1 μg was used for retro-transcription using the Superscript III—First-Strand cDNA Synthesis System (Invitrogen). Real-time Q-PCR was performed using SYBr green master mix (Applied Biosystem) and reactions were run on a real-time PCR system 7300 (Applied Biosystem). The following primers were used in this study: p53 Forward 5′-TGGAAGACTCCAGTGGGAAC-3′ and Reverse 5′-TCTTCTGTACGGCGGTCTCT-3′, p21 Forward 5′-TCCACAGCGATATCCAGACA-3′ and Reverse 5′-GGCACACTTTGCTCCTGTG-3′, GADD45 Forward 5′-AGAGCAGAAGACCGAAAGGA-3′ and Reverse 5′-CGTAATGGTGCGCTGACTC-3′, Mdm2 Forward 5′-TTAGTGGCTGTAAGTCAGCAAGA-3′ and Reverse 5′-CCTTCAGATCACTCCCACCT-3′, p16 Forward 5′-GGGTTTTCTTGGTGAAGTTCG-3′ and Reverse 5′-TTGCCCATCATCATCACCT-3′, p15 Forward 5′-GGCTGGATGTGTGTGACG-3′ and Reverse 5′-GCAGATACCTCGCAATGTCA-3′, 18S rRNA Forward 5′-AAACGGCTACCACATCCAAG-3′ and Reverse 5′-TACAGGGCCTCGAAAGAGTC-3′, β-actin Forward 5′-CTAAGGCCAACCGTGAAAAG-3′ and Reverse 5′-ACCAGAGGCATACAGGGACA-3′.
p53 IRES dicistronic reporter assay
The p53 IRES element was subcloned into the pRF dicistronic vector as described elsewhere (Ray et al, 2006; Chaudhuri et al, 2007). Measurement of p53 IRES-mediated translation was carried out as previously reported with some modifications (Yoon et al, 2006). Briefly, the pR-p53 IRES-F dicistronic plasmid was linearized using BamHI, and in vitro transcription of capped RNA was carried out using the mMessage mMachine T7 (Ambion) according to the manufacturer's instructions. Cells were plated at 100 × 103/well in six-well plates and transfected with 2 μg/well of dicistronic reporter capped RNA using Transmessenger (Qiagen). Finally, cells were harvested 6–7 h later and the activities of Renilla and Firefly luciferases were measured using the Dual-luciferase kit (Promega) using a single tube luminometer (Optocomp1, MGM Instruments). Transfections of human control and X-DC lymphoblasts were carried out using an Amaxa biosystem nucleofector device (Lonza). Briefly, four million cells were electroporated with 4 μg of dicistronic reporter mRNA using the nucleofector kit R (Program U-23) (Lonza). Luciferease activity was measured 6 h after transfection as previously described.
Isolation and analysis of polysomal mRNA during OIS and from nocodazole-treated MEFs
Polysomal mRNA was prepared as described (Yoon et al, 2006) with some modifications. In brief, for the analysis of polysomal mRNA during OIS, empty vector- and RAS-infected WT and DKC1m early passage MEFs were seeded in 15 cm tissue culture plates and harvested 6 days post-selection. For analysis of polysomal mRNA after nocodazole treatment, early passage MEFs were seeded in 15 cm tissue culture plates and cultured in the presence of 125 ng/ml nocodazole for 18 h. Cells were pre-incubated with 10 μg/ml of CHX before harvesting and lysed in ice-cold buffer A (10 mM Tris–HCl pH 7.4, 140 mM NaCl, 1.5 mM MgCl2, 0.25% NP-40, 0.5% Triton X-100 supplemented with 40 U/ml RNase inhibitor (Promega), 150 μg/ml CHX, and 20 mM dithiothreitol) for 40 min on ice. Lysates were centrifuged for 10 min at 10 000 r.p.m. at 4°C and supernatants with equal amounts of RNA was loaded onto 10–60% sucrose gradient in buffer B (25 mM Tris–HCl, 25 mM NaCl, 1.5 mM MgCl2). Sucrose gradients were centrifuged using an SW41Ti rotor (Beckman) at 36 000 r.p.m. for 3 h at 4°C. Polyribosome fractions were collected using an ISCO gradient fraction collector machine. RNA in each fraction was prepared using Trizol (Invitrogen), purified using Purelink RNA mini kit (Invitrogen), and DNAase treated with Turbo DNase (Ambion) before retro-transcription. Unfractionated and fractionated RNA samples were normalized by the 18S rRNA as earlier described (Koritzinsky et al, 2006).
Quantification of cell death
Apoptosis was determined by phosphatidylserine externalization in the presence of propidium iodide as described (Bellodi et al, 2009) with some modifications. Briefly, 60 × 103 cells/well were plated in six-well plates (NUNC) and cultured in the presence or absence of 100 μM etoposide for 24 and 48 h. Cells were harvested and stained with allophycocyanin-conjugated annexin V (BD Pharmigen) and propidium iodide (Sigma) and analysed using an FACSCalibur system (Becton Dickinson).
Determination of apoptosis in mouse thymocytes
WT and DKC1m mice were placed inside a cesium small animals irradiator and exposed to a 5 Gy dose of ionizing radiation. Mice were killed 6 h post-irradiation, thymi were dissociated in phosphate-buffered saline (PBS) supplemented with 3% FBS (PBS–FBS) and filtered through a 40 μm nylon mesh. Red blood cells were lysed by incubation in a solution of 10 mM KHCO3, 150 mM NH4Cl, and 0.1 mM EDTA (pH 8.0) for 2 min on ice. Thymocytes were then washed in PBS–FBS and pelleted at 1200 r.p.m. for 5 min and cytospun at 300 r.p.m. for 5 min using a cytocentrifuge (Shandon). Slides were dried for 30 min at room temperature and stained with the TACS 2 TdT-Fluor in situ Apoptosis Detection kit (Trevigen) following the manufacturer's instructions. Nuclei were counterstained for 10 min with DAPI (Invitrogen, Molecular Probes) and slides were analysed using a Nikon Eclipse TI-E motorized inverted microscope. Images were acquired using a Coolsnap HQ2 camera.
Statistical analysis
Statistical analysis of the data was performed using an unpaired Student's two-tailed _t_-test (Prism 5.0, Graphpad software Inc.). _P_-values <0.05 were considered statistically significant.
Supplementary Material
Supplementary Figures S1–S6
Review Process File
Acknowledgments
We are indebted to Maria Barna for critical discussion and editing the paper; Olivia F Siegel for editing the paper; and Olya Krasnykh for technical assistance. This work was supported by the NIH through R01 HL085572 and 3R01HL085572-05S1.
Footnotes
The authors declare that they have no conflict of interest.
References
- Alter BP, Giri N, Savage SA, Rosenberg PS (2009) Cancer in dyskeratosis congenita. Blood 113: 6549–6557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artandi SE, Attardi LD (2005) Pathways connecting telomeres and p53 in senescence, apoptosis, and cancer. Biochem Biophys Res Commun 331: 881–890 [DOI] [PubMed] [Google Scholar]
- Bellodi C, Lidonnici MR, Hamilton A, Helgason GV, Soliera AR, Ronchetti M, Galavotti S, Young KW, Selmi T, Yacobi R, Van Etten RA, Donato N, Hunter A, Dinsdale D, Tirro E, Vigneri P, Nicotera P, Dyer MJ, Holyoake T, Salomoni P et al. (2009) Targeting autophagy potentiates tyrosine kinase inhibitor-induced cell death in Philadelphia chromosome-positive cells, including primary CML stem cells. J Clin Invest 119: 1109–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braig M, Lee S, Loddenkemper C, Rudolph C, Peters AH, Schlegelberger B, Stein H, Dorken B, Jenuwein T, Schmitt CA (2005) Oncogene-induced senescence as an initial barrier in lymphoma development. Nature 436: 660–665 [DOI] [PubMed] [Google Scholar]
- Chaudhuri S, Vyas K, Kapasi P, Komar AA, Dinman JD, Barik S, Mazumder B (2007) Human ribosomal protein L13a is dispensable for canonical ribosome function but indispensable for efficient rRNA methylation. RNA 13: 2224–2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Trotman LC, Shaffer D, Lin HK, Dotan ZA, Niki M, Koutcher JA, Scher HI, Ludwig T, Gerald W, Cordon-Cardo C, Pandolfi PP (2005) Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature 436: 725–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado M, Gil J, Efeyan A, Guerra C, Schuhmacher AJ, Barradas M, Benguria A, Zaballos A, Flores JM, Barbacid M, Beach D, Serrano M (2005) Tumour biology: senescence in premalignant tumours. Nature 436: 642. [DOI] [PubMed] [Google Scholar]
- Efeyan A, Murga M, Martinez-Pastor B, Ortega-Molina A, Soria R, Collado M, Fernandez-Capetillo O, Serrano M (2009) Limited role of murine ATM in oncogene-induced senescence and p53-dependent tumor suppression. PLoS One 4: e5475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganapathi KA, Shimamura A (2008) Ribosomal dysfunction and inherited marrow failure. Br J Haematol 141: 376–387 [DOI] [PubMed] [Google Scholar]
- Gu BW, Bessler M, Mason PJ (2008) A pathogenic dyskerin mutation impairs proliferation and activates a DNA damage response independent of telomere length in mice. Proc Natl Acad Sci USA 105: 10173–10178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halaby MJ, Yang DQ (2007) p53 translational control: a new facet of p53 regulation and its implication for tumorigenesis and cancer therapeutics. Gene 395: 1–7 [DOI] [PubMed] [Google Scholar]
- Holcik M, Sonenberg N (2005) Translational control in stress and apoptosis. Nat Rev Mol Cell Biol 6: 318–327 [DOI] [PubMed] [Google Scholar]
- Kirwan M, Dokal I (2008) Dyskeratosis congenita: a genetic disorder of many faces. Clin Genet 73: 103–112 [DOI] [PubMed] [Google Scholar]
- Knight SW, Heiss NS, Vulliamy TJ, Greschner S, Stavrides G, Pai GS, Lestringant G, Varma N, Mason PJ, Dokal I, Poustka A (1999) X-linked dyskeratosis congenita is predominantly caused by missense mutations in the DKC1 gene. Am J Hum Genet 65: 50–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koritzinsky M, Magagnin MG, van den Beucken T, Seigneuric R, Savelkouls K, Dostie J, Pyronnet S, Kaufman RJ, Weppler SA, Voncken JW, Lambin P, Koumenis C, Sonenberg N, Wouters BG (2006) Gene expression during acute and prolonged hypoxia is regulated by distinct mechanisms of translational control. EMBO J 25: 1114–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin AW, Barradas M, Stone JC, van Aelst L, Serrano M, Lowe SW (1998) Premature senescence involving p53 and p16 is activated in response to constitutive MEK/MAPK mitogenic signaling. Genes Dev 12: 3008–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin AW, Lowe SW (2001) Oncogenic ras activates the ARF-p53 pathway to suppress epithelial cell transformation. Proc Natl Acad Sci USA 98: 5025–5030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier UT (2005) The many facets of H/ACA ribonucleoproteins. Chromosoma 114: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell JR, Wood E, Collins K (1999) A telomerase component is defective in the human disease dyskeratosis congenita. Nature 402: 551–555 [DOI] [PubMed] [Google Scholar]
- Ofir-Rosenfeld Y, Boggs K, Michael D, Kastan MB, Oren M (2008) Mdm2 regulates p53 mRNA translation through inhibitory interactions with ribosomal protein L26. Mol Cell 32: 180–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson MO (2004) Sensing cellular stress: another new function for the nucleolus? Sci STKE 2004: pe10. [DOI] [PubMed] [Google Scholar]
- Palmero I, Pantoja C, Serrano M (1998) p19ARF links the tumour suppressor p53 to Ras. Nature 395: 125–126 [DOI] [PubMed] [Google Scholar]
- Pestov DG, Strezoska Z, Lau LF (2001) Evidence of p53-dependent cross-talk between ribosome biogenesis and the cell cycle: effects of nucleolar protein Bop1 on G(1)/S transition. Mol Cell Biol 21: 4246–4255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyronnet S, Pradayrol L, Sonenberg N (2000) A cell cycle-dependent internal ribosome entry site. Mol Cell 5: 607–616 [DOI] [PubMed] [Google Scholar]
- Ray PS, Grover R, Das S (2006) Two internal ribosome entry sites mediate the translation of p53 isoforms. EMBO Rep 7: 404–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggero D, Grisendi S, Piazza F, Rego E, Mari F, Rao PH, Cordon-Cardo C, Pandolfi PP (2003) Dyskeratosis congenita and cancer in mice deficient in ribosomal RNA modification. Science 299: 259–262 [DOI] [PubMed] [Google Scholar]
- Ruggero D, Pandolfi PP (2003) Does the ribosome translate cancer? Nat Rev Cancer 3: 179–192 [DOI] [PubMed] [Google Scholar]
- Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW (1997) Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88: 593–602 [DOI] [PubMed] [Google Scholar]
- Takagi M, Absalon MJ, McLure KG, Kastan MB (2005) Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell 123: 49–63 [DOI] [PubMed] [Google Scholar]
- Yang DQ, Halaby MJ, Zhang Y (2006) The identification of an internal ribosomal entry site in the 5′-untranslated region of p53 mRNA provides a novel mechanism for the regulation of its translation following DNA damage. Oncogene 25: 4613–4619 [DOI] [PubMed] [Google Scholar]
- Yoon A, Peng G, Brandenburger Y, Zollo O, Xu W, Rego E, Ruggero D (2006) Impaired control of IRES-mediated translation in X-linked dyskeratosis congenita. Science 312: 902–906 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1–S6
Review Process File