Influenza Hemagglutinin and Neuraminidase Membrane Glycoproteins (original) (raw)
Abstract
Considerable progress has been made toward understanding the structural basis of the interaction of the two major surface glycoproteins of influenza A virus with their common ligand/substrate: carbohydrate chains terminating in sialic acid. The specificity of virus attachment to target cells is mediated by hemagglutinin, which acquires characteristic changes in its receptor-binding site to switch its host from avian species to humans. Anti-influenza drugs mimic the natural sialic acid substrate of the virus neuraminidase enzyme but utilize the much tighter binding of the drugs for efficacy. Resistance to one of the two main antiviral drugs is differentially acquired by the two distinct subsets of neuraminidase as a consequence of structural differences in the enzyme active site between the two phylogenetic groups.
Keywords: Viral Immunology, Viral Protein, Virus Assembly, Virus Entry, Virus Structure, Virus
Introduction
The two glycoproteins of the influenza virus membrane, hemagglutinin (HA)3 and neuraminidase (NA), both recognize sialic acid (1–3). Initiation of virus infection involves multiple HAs binding to sialic acids on carbohydrate side chains of cell-surface glycoproteins and glycolipids (4–6). Following virus replication, the receptor-destroying enzyme, NA, removes its substrate, sialic acid, from infected cell surfaces so that newly made viruses are released to infect more cells (7, 8). Both activities are the targets of antibodies that block infection (9–11), and as a result of immune pressure, the antigenic properties of the glycoproteins vary during a pandemic era, when repeated infections occur (e.g. Ref. 12). Such antigenic changes indicate the requirement for vaccine update. Antigenic differences are also used to classify influenza type A viruses into 16 HA (H1–H16) and 9 NA (N1–N9) subtypes (13). Phylogenetically, there are two groups of HAs: group 1 contains H1, H2, H5, H6, H8, H9, H11, H12, H13, and H16, and group 2 contains H3, H4, H7, H10, H14, and H15 (14, 15). NAs also form two groups: group 1 contains N1, N4, N5, and N8, and group 2 contains N2, N3, N6, N7, and N9 (16). Viruses of all subtypes are found in avian species, predominantly waterfowl, and viruses with numerous combinations of HA and NA subtypes have been isolated from them. In humans, the known pandemics in 1918, 1977, and 2009, in 1957, and in 1968 were caused by the H1N1, H2N2, and H3N2 viruses, respectively (17). For the 1918, 1957, and 1968 pandemics, evidence suggests that the HAs were derived from avian viruses, and the N1 and N2 NAs of 1918 and 1957 had a similar derivation (18, 19). In 1968, the N2 NA of the H3N2 virus was derived from the 1967 H2N2 virus (20). In 2009, both H1 HA and N1 NA appear to have been derived from a porcine source (21). The origin of the 1977 H1N1 virus is unknown (22, 23).
The focus of this minireview is sialic acid receptor binding by HAs of human, avian, and porcine viruses and antiviral drug complexes with N1 and N2 NAs and a drug-resistant mutant of N1 NA that has circulated worldwide in seasonal H1N1 viruses (24).
In addition to their role in sialic acid receptor binding, HAs are also membrane fusion glycoproteins (25). Receptor-bound viruses are taken into endosomes, and upon acidification, HAs are activated to fuse the virus and endosomal membranes. Activation involves extensive changes in HA conformation, and members of the two phylogenetic groups of HA are characterized by group-specific structural features at sites where these changes occur (Fig. 1) (26, 27) and by differences in response to compounds that inhibit the conformational changes and membrane fusion activity (28). The two phylogenetic groups of NA are also distinguished by group-specific differences in structure, prominently, in this case, in a region adjacent to the enzyme active site (see Fig. 3) (16).
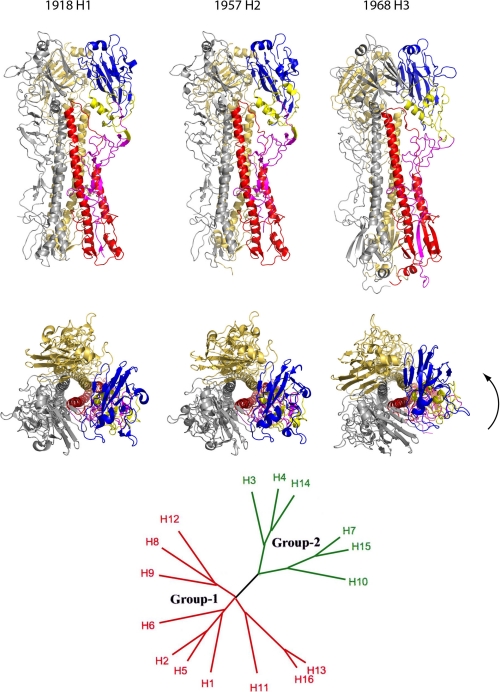
FIGURE 1.
Crystal structures and phylogenetic organization of pandemic HAs. The upper and middle panels show two orthogonal views of the H1, H2, and H3 HAs in ribbon representation. Two of the monomers from each trimer are in gold and silver, whereas the subunits that make up the third monomer are colored as follows: blue, receptor binding; yellow, vestigial esterase; and magenta and red, fusion subdomains (25). The lower panel shows a phylogenetic tree containing the 16 subtypes of HA that fall into two distinct groups. As well as local variations in structure, there are significant differences in rigid body orientation of subdomains between HAs in the groups. The arrow indicates rotation of the membrane-distal subdomains of group 2 H3 HA relative to those of group 1 H1 and H2 HAs.
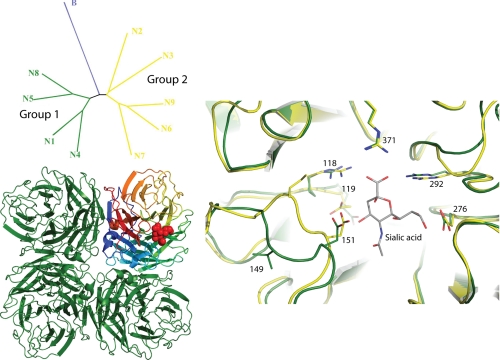
FIGURE 3.
Phylogenetic organization and crystal structures of NA. The upper left panel shows a phylogenetic tree of the nine NA subtypes of influenza A together with NA from influenza B. The influenza A NAs fall into two distinct groups. The lower left panel shows a ribbon representation of an NA tetramer viewed along the 4-fold axis. Three of the monomers are colored green, whereas for the fourth monomer, each of the six blades that make up the structure is separately colored (80). The right panel shows a detailed view of the NA active site in an overlap between a group 1 structure (in green) and a group 2 structure (in yellow), with some key side chains shown in ball-and-stick representation. Sialic acid has been docked into the overlapped structures.
Hemagglutinin
HA is a trimer of identical subunits, each of which contains two polypeptides that result from proteolytic cleavage of a single precursor (25). Cleavage of the precursor is essential for activation of membrane fusion potential and hence infectivity (29, 30). For HAs of most subtypes, the site of cleavage is a single arginine residue, and cleavage occurs extracellularly by an as yet unidentified enzyme (31). Some members of the H5 and H7 subtypes have acquired, however, multiple basic residues at the site of cleavage (32), which are recognized by an intracellular subtilisin-like enzyme (33). In these cases, cleavage is efficient, virus infectivity is high, and the viruses are highly pathogenic (29, 30, 34). The avian H5 influenza, which continues to spread throughout the world, excluding the Americas, is caused by such viruses. In all cases, enzymatic cleavage generates the N terminus of the “fusion peptide,” a conserved uncharged region of HA (25) that plays an essential but undefined role in membrane fusion.
Each HA monomer contains a receptor-binding site at its membrane-distal tip, which has, at its base, a number of conserved amino acids, Tyr-98, Trp-153, His-183, and Tyr-195, and at its edges, three conserved elements of secondary structure, the 130- and 220-loops and the 190-α-helix (Fig. 2) (25, 35). Sialic acid is bound similarly in all HAs examined by hydrophobic interactions and by hydrogen bonds with the 130- and 220-loops and conserved amino acids in the base of the site. In human H3 HA, for example, hydrogen bonds are formed between the carboxylate group of Sia-1 and Ser-136 and the main chain amide of Asn-137; between the acetamido nitrogen and main chain carbonyl of Gly-135; between the 8-OH group and the OH of Tyr-98; and between the 9-OH and His-183, Glu-190, and Ser-228. Additionally, the methyl group of the acetamido substituent is in van der Waals contact with the six-member ring of Trp-153 (36–38).
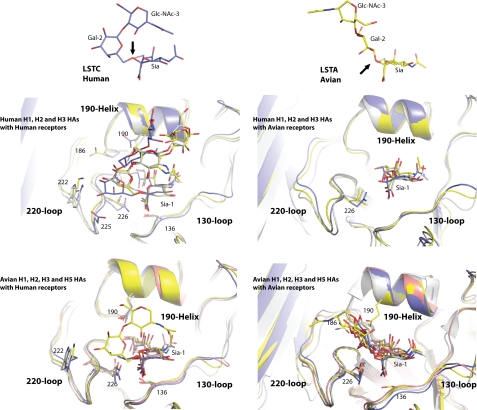
FIGURE 2.
Crystal structures of complexes between HA and receptor analogs. The upper panels show sialic acid linked to Gal-2 and GlcNAc-3 from the α2,6-linked human receptor analog LSTc (38) (left; carbons colored blue) and the α2,3-linked avian receptor analog LSTa (right; carbons colored yellow). The black arrows indicate the glycosidic oxygen in both cases. The middle and lower panels show overlaps of the receptor-binding domains of HAs from different species and subtypes in complex with the human receptor (left) and avian receptor (right). The three secondary structure elements of the site, the 130- and 220-loops and the 190-helix, are labeled. The HA and carbon atoms of the ligand are colored blue for H1, yellow for H2, gray for H3, and salmon for H5.
Receptor binding specificity and affinity have been estimated by a variety of methods, including the use of erythrocytes from different species (39) and specifically re-sialylated erythrocytes (40) in hemagglutination tests, solid-phase microplate assays (41), ligand microarray procedures (42, 43), surface plasmon resonance (5), and NMR (4). The main conclusion from these studies is that receptor binding is species-specific (44). Avian and equine viruses prefer to bind to sialic acid in α2,3-linkage to galactose, human viruses prefer α2,6-linked sialic acid, and swine viruses appear to bind sialic acid in both linkages (6, 45–48). These specificities may reflect the relative abundance of sialic acid in the different linkages to galactose on tissues at the sites of infection: human and swine respiratory epithelia and cells of avian enteric tracts (49, 50). Labeling the respiratory tract with specific lectins that bind to cells displaying either α2,6-linked sialic acid, Sambucus nigra lectin, or α2,3-linked sialic acid, Maackia amurensis lectin, supports the concept of anatomical differences in distribution (51). However, in cultures of differentiated human airway epithelia, ciliated cells that have sialic acid in α2,3-linkage together with non-ciliated cells that have sialic acid in α2,6-linkage can be identified using the same labeling procedures (52).
While sharing a general preference for sialic acid in α2,3-linkage to Gal-2, viruses from different avian species discriminate between alternative linkages from Gal-2 to GlcNAc-3 and in addition recognize modifications of GlcNAc-3. For example, the highly pathogenic H5N1 viruses isolated from chickens in Hong Kong in 1997 prefer the Gal-2β1,4-GlcNAc-3 linkage and sulfated GlcNAc-3. On the other hand, H5N1 and viruses of other subtypes from ducks appear to prefer the Gal-2β1,3-GalNAc-3 linkage (53). Correlation of such fine specificities, determined using chemically defined ligands (54) and the wide range of natural sialosides employed on glycan microarrays (42, 43), with species and tissue tropism, provides the detail required for assessing the role of HA-sialic acid recognition in virus pathogenesis.
Estimates of comparative affinities for sialylated ligands have been obtained by assays of competitive inhibition of binding using hemagglutination (55) and by solid-phase assays (56). Direct NMR estimates of affinity gave dissociation constants for human H3 HA of 2.1 mm for α2,6-sialyllactose and 3.2 mm for α2,3-sialyllactose (4). These estimates of low affinity imply that the tight binding of viruses to cells during infection is mediated by the simultaneous interaction of a number of HAs.
The structural basis of the receptor binding specificity of pandemic viruses and their possible avian and swine precursors has been deduced by correlating sequence changes in HAs with their different receptor binding properties (15, 46, 47, 56–65) and from x-ray crystallographic analyses of complexes formed by soaking HA crystals in defined ligands.
During antigenic variation, many amino acid sequence changes occur near the receptor-binding site. Some of these influence receptor binding affinity and specificity, presumably in some cases by steric hindrance with receptor association. This is the case, for example, when sequence changes introduce new sites for glycosylation of HA near the receptor-binding site (61, 66). Other changes influence receptor binding by specific interactions with the sugars that are linked to sialic acid. There are simple sequence changes of major significance associated with a change in receptor preference between pandemic viruses and their potential avian precursors. The changes observed are different in H1 HA compared with H2 and H3 HAs. In H1 HA, mutations at residue 138 (45) and residues 190 and 225 (56, 62, 67) were deduced to be important, with the substitution E190D found in viruses proposed to be evolutionary intermediates in the transfer of viruses from avian species to humans (56). In both H2 and H3 HAs, Q226L and G228S are the major differences between avian and human viruses, with Q226L observed in viruses isolated early in the pandemic (56). The primary role of the Q226L substitution is consistent with the major influence that it has on the structure of the receptor-binding site of mutant HA (57).
The structures of the receptor-binding sites of HA-ligand complexes determined by x-ray crystallography, in which the sialylpentasaccharide α2,6-linked LSTc was used as a human receptor analog and α2,3-linked LSTa was used as an avian receptor analog, are shown in Fig. 2. In the complexes formed by all three human HAs with LSTc (38, 68, 69), the α2,6-linkage between Sia-1 and Gal-2 adopts a _cis_-conformation in which the glycosidic oxygen faces out of the site (Fig. 2). The Gal-2 ring is oriented face on and together with C-6 presents a nonpolar surface toward the base of the site. The bound oligosaccharide forms a folded-back structure and exits toward the right side of the site (Fig. 2, upper left panel).
In avian H1, H2, H3, and H5 complexes with LSTa (35, 68–70), the first three sugars of the avian receptor form a more extended chain in which Gal-2 is projected upwards, and the oligosaccharide exits the site over the 220-loop, roughly opposite the direction taken by the folded human receptors (Fig. 2, lower right panel). The α2,3-linkage is in a _trans_-conformation, which exposes the glycosidic oxygen toward Gln-226 at the base of the site, and the Gal-2 ring is oriented edge on (Fig. 2, lower right panel). The _trans_-conformation allows the formation of additional hydrogen bonds between the amine and carbonyl groups of Gln-226 and the 4-OH of Gal-2 and the glycosidic linkage oxygen. These interactions do not occur with human receptors, but this binding motif is common to all avian HAs that have been examined.
In the human and swine H1 HA-human receptor complexes (68, 69), the oligosaccharide is not as folded back as in the H2 and H3 complexes. Hydrogen bonds are formed between Lys-222 and the 2-OH and 3-OH of Gal-2 and between Asp-225 (in humans) and the 3-OH of Gal-2. Asp-190 forms a hydrogen bond with GlcNAc-3. Gln-226 is positioned ∼1 Å lower in the site than in the avian receptor complex, allowing the hydrophobic C-6 of the glycosidic linkage to face into the site. This lower positioning of Gln-226 in the human receptor complex and in unliganded H1 HA appears to be a specific feature of H1 HA as a result of the conformation of the 130-loop. In the genetically closely related H5 HA, this feature of the 130-loop is not observed, and the human receptor is not bound. It should be noted, however, that mutations near the H5 HA receptor-binding site can increase the affinity for the human receptor (64, 71), and predictably, making the human-specific amino acid substitutions Q226L and G228S also leads to human receptor preference (63).
In both H2 and H3 HAs, the polar to hydrophobic substitution Q226L accommodates C-6 and the hydrophobic face of the Gal-2 ring. In both cases, also the human receptor is slightly more folded back over Sia-1 than in the human and swine H1 HA complexes (38, 68, 69). The G228S substitution, which also occurs in human H2 and H3 HAs by comparison with avian HAs, results in Ser-228 forming a hydrogen bond with the 9-OH of sialic acid (38, 69). This interaction substitutes for the one formed, through a water molecule, between the 9-OH and Gly-228 in human H1 HA (69) and in avian viruses (35, 70).
The electron density for the human receptor in avian H1 HA (69) and avian H3 HA (70) complexes is weak, indicating low affinity. By contrast, in the avian H2 HA-human receptor complex, there is well defined electron density for Sia-1, Gal-2, and GlcNAc-3 (69). In this case, effective binding of the human receptor appears to be due to interactions made by Asn-186 (Pro in H1 HA and Ser in H3 HA) and Gln-226, through a water molecule, with the 4-OH group of Gal-2. The receptor is bound in the preferred _cis_-conformation and folded back, much the same as in the human H2 HA and H3 HA complexes. This last observation indicates that the folded conformation of the human receptor observed in the H2 and H3 complexes is not a direct consequence of the Q226L and G228S mutations.
Further information on the basis of human HA specificity for the α2,6-glycosidic linkage is obtained by comparing complexes formed between human HAs and the avian receptor analog. For human H1 HAs such as swine H1 HA (68), electron density for the avian receptor is weak. This is because the E190D mutation in swine and human HAs disrupts the position of Gln-226 required for its interactions with the glycosidic oxygen and the 4-OH group of Gal-2. For the human H2 and H3 HAs (38, 69), Leu-226 creates a hydrophobic environment that is incompatible with the orientation of the Sia-1–Gal-2 glycosidic oxygen of the α2,3-linkage. There is a possible advantage for all three human HAs of this low preference for the avian receptor. Mucins in the human respiratory tract, which are rich in α2,3-linked sialosides (50), could block access, of viruses that bound them tightly, to receptors on cell surfaces.
Neuraminidase
NA is a mushroom-shaped tetramer of identical subunits, with the head of the mushroom suspended from the virus membrane on a thin ∼60-Å-long stalk, a length that is variable between virus strains. Each of the subunits that form the head of the mushroom is made up of a six-bladed propeller-like structure, the blades of which are formed by four antiparallel strands of β-structure (72, 73). The enzyme active site, containing a number of conserved charged amino acid residues, is located at roughly the center of each subunit (73, 74).
X-ray crystallographic analysis of NA-ligand complexes indicates that sialic acid is recognized in a different way by NA compared with HA (73, 75, 76). In NA complexes, the carboxylate group is equatorial, whereas in HA complexes, it is axial, pointing into the site. In further distinction, in NA, the carboxylate forms salt bridges with three conserved arginine residues, Arg-118, Arg-292, and Arg-371 (Fig. 3). In this orientation, the 2-OH of sialic acid is pointing out of the site. Superposition of the structures of NAs from phylogenetic groups 1 and 2 indicates that the positions of the active-site residues are very similar (16). However, there are notable differences between the group 1 and group 2 structures in the conformation of a loop, the 150-loop, adjacent to the active site. In group 1, the position of Val-149 in this loop is ∼7 Å distant from the group 2 equivalent residue, Ile-149. Moreover, the hydrophobic side chain of residue 149 is pointing away from the active site in group 1 but toward it in group 2. There are also significant differences in the positions of the conserved acidic residues Asp-151 and Glu-119 between the two groups (Fig. 3). The main consequence of these differences is that there is a large cavity adjacent to the active site in group 1 but not group 2 NAs, which is accessible from the active site. The possibility of exploiting this cavity to develop antiviral compounds has been considered (16, 77, 78).
During virus replication, NA removes sialic acid from cellular glycoproteins and glycolipids and from both of the virus glycoproteins. As a result, newly assembled viruses are prevented from binding to the infected cell surface and from aggregating with each other through HA-sialic acid interactions. Instead, they are released from the cell to infect new cells and spread the infection. Antibodies against NA block this receptor-destroying activity (10, 11) and, as a consequence, limit the infection (79). This effect appears to be significant because N1 and N2 NAs were observed to vary during the H3N2 pandemic to a similar extent as HAs (80). Amino acid sequence changes in monoclonal antibody-selected variant NAs show that the sites of antibody binding are on surface loops surrounding the enzyme active site (80), where many of the sequence changes that occurred during the pandemic are also located.
The NA active site is also the target of the anti-influenza drugs Relenza (zanamivir) and Tamiflu (oseltamivir), which act against both group 1 and group 2 enzymes and against influenza B NA (78). There have been numerous experiments designed to identify mutations that lead to drug resistance (81). There has also been extensive surveillance for drug-resistant viruses in circulation since the drugs were introduced (82). Studies in Japan, where there has been relatively high usage of anti-NA drugs, have identified drug-resistant mutants in 0.3% of H3N2 virus isolates and 3.0% of H1N1 viruses. The numbers of drug-resistant isolates varied from year to year between 2003 and 2007, but there was no consistently increased frequency in any strain or subtype, suggesting only low levels of transmission, if any (83).
However, despite little antiviral drug usage worldwide, in the winter of 2007–2008, Tamiflu-resistant H1N1 viruses accounted for the vast majority of influenza isolates, in a season when H1N1 viruses dominated (24). The reason for this occurrence is unknown, but it strikingly demonstrated the potential viability of drug-resistant viruses. It happened in the year before the H1N1 pandemic of 2009, in which, worldwide to date, 267 H1N1 Tamiflu-resistant isolates have been made. Fortunately, in this case, there is no evidence of human-to-human transmission of resistant viruses.
The drug-resistant N1 NA of 2008 contained the mutation H274Y. Its enzymatic properties were analyzed to show that the mutation reduced the binding of Tamiflu by a factor of 265 but had only an ∼2-fold effect on sensitivity to Relenza (84). In addition, the same mutation was without effect on group 2 NAs. Binding of Tamiflu to NA requires a conformational change in the side chain of Glu-276 so that its carboxyl group is oriented away from the hydrophobic pentyloxy substituent of Tamiflu, which enables hydrophobic contact with the C-β methylene of Glu-276. By contrast, binding of Relenza, like sialic acid, involves hydrogen bond formation between the carboxyl group of Glu-276 and the 8-OH and 9-OH groups of the sialic acid glycerol substituent. X-ray crystallographic analyses of the H274Y N1 NA-Tamiflu complex and the H274Y N1 NA-Relenza complex show that substitution of His-274 with the bulkier tyrosine residue pushes the carboxyl group of Glu-276 toward the Tamiflu-binding site (Fig. 4). In this position, the ionizable Glu-276 disrupts the otherwise hydrophobic site and perturbs the binding of Tamiflu such that its C-9 and C-91 move ∼2.5 Å from the positions they would occupy in wild-type N1 NA. By contrast, the structure of the H274Y N1 NA-Relenza complex shows that Relenza is accommodated in the active site of mutant NA by a small movement in the Glu-276 side chain and retains the hydrogen bonds made by wild-type NA. The lack of effect of the H274Y substitution on group 2 NAs results from the substituted Tyr-274 being able to adopt a different rotamer conformation because of an adjacent smaller residue, Thr-252 in group 2 NA rather than Tyr-252 in group 1 NA (Fig. 4).
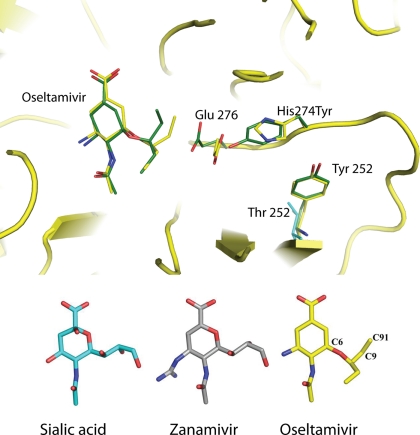
FIGURE 4.
Inhibitor binding to the active site of NA. The upper panel shows a C-α trace for wild-type N1 (in yellow), with bound oseltamivir and selected side chains colored yellow for the wild-type complex and green for the mutant complex. Thr-252 of group 2 NA is light blue. In the lower panel, the structures of sialic acid, zanamivir, and oseltamivir are shown in blue, gray, and yellow, respectively, with selected carbon atoms associated with the hydrophobic moiety at C-6 of oseltamivir numbered.
The N1 NA mutation H274Y was also dominant among drug-resistant mutant viruses selected in vitro; several other mutations were also identified, and the structures of the mutant NAs were analyzed (78, 85, 86). The results of these drug selection experiments included the important finding that the majority of mutations occurred not in NA but in HA (81, 87). The amino acid substitutions T155A, V223I, R229I, K222T, S186F, and S165N (which introduced a new carbohydrate side chain) in the HAs of drug-resistant H1 viruses were at sites in or near the sialic acid receptor-binding site. These mutations apparently decreased the affinity of the mutant HAs for sialic acid, and as a consequence, the newly made mutant viruses were less dependent on NA activity to release them from infected cells. In some instances such as the S165N mutation, the mutant viruses were dependent on anti-NA drugs for infectivity, an indication that NA can act in the early stages of infection and that viable viruses must contain HAs of sufficient affinity to balance NA activity. Interdependence of HA affinity and NA activity has been concluded from numerous genetic studies (88–92) particularly involving co-variation of virus NA stalk length and HA affinity for the receptor, with HA decreases in affinity often resulting from extra glycosylation of HA near the receptor-binding site.
The 2009 H1N1 pandemic exemplifies the unpredictability of human influenza and has emphasized the importance of the virus membrane glycoproteins in our response to new viruses. Both glycoproteins are important immunogens in anti-influenza vaccines, and the NA active site is the target of the available anti-influenza drugs.
The importance of immune recognition of HA and NA is evidenced by the extent of amino acid sequence variation, with time, during a pandemic period. This is much greater than for other influenza virus proteins despite the fact that the RNA-associated nucleoprotein, for example, is a very powerful immunogen. The regions of the glycoproteins that are recognized by antibodies that block virus infection are on their upper surfaces, in positions where binding of antibodies could prevent receptor binding in the case of HA (93) or the enzyme activity of NA (94). Details of the binding to HA or NA of specific monoclonal antibodies have been determined by x-ray crystallography to show the likely way in which they function and, in the case of HA, the relative efficiencies of virus infectivity neutralization that result from binding to different positions on the molecule.
As a result of antibody-mediated selection of antigenic variants during pandemics, antibodies produced following infection are virus strain-specific. This is largely the case also for vaccine-induced antibodies, hence the need for frequent, almost yearly updates of the viruses used to prepare vaccines, chosen on the basis of the results of international surveillance for antigenically distinguishable new viruses. An ideal vaccine would induce immune responses that would cross-neutralize either all viruses in a subtype or, better, all influenza viruses. A number of cross-reactive antibodies against HA that block virus infection have been prepared (95–97). Complexes that some of them form with HA have been analyzed by x-ray crystallography, and they are seen to bind relatively near to the region of HA that associates with the virus membrane (96, 97). Their binding is also reported to prevent the low pH-induced conformational changes in HA required for membrane fusion, and this may be the way that they influence virus infection. As an alternative, they may function to prevent virus assembly at the membranes of infected cells, at the time in infection when anti-NA antibodies have their effect (11). They would, however, have the additional attribute to anti-NA antibodies of being cross-reactive. If such antibodies can be induced by vaccination, they could be very valuable. As therapeutic antibodies, they could also join the anti-NA drugs in combination therapy. For treatment of infections with highly pathogenic viruses such as the H5N1 avian virus, they could be very valuable in this role.
The unexplained worldwide spread of Tamiflu-resistant H1N1 viruses in 2008 is a strong stimulus to the development of other anti-influenza drugs and therapies that could be used, together with Tamiflu, Relenza, or both drugs, like anti-human immunodeficiency virus drug cocktails, to combat the risk of the development and spread of drug-resistant influenza viruses (98). Their availability would add confidence to the tactic, in many pandemic plans, of stockpiling the anti-NA drugs.
Acknowledgments
We are grateful to Philip Walker for preparation of figures. We thank our many colleagues, past and present, for valuable discussions.
*
This is the first article in the Thematic Minireview Series on Influenza Virus. This minireview will be reprinted in the 2010 Minireview Compendium, which will be available in January, 2011.
3
The abbreviations used are:
HA
hemagglutinin
NA
neuraminidase
LST
lacto-series tetrasaccharide.
REFERENCES
- 1.Klenk E., Faillard H., Lempfrid H. (1955) Hoppe-Seyler's Z. Physiol. Chem. 301, 235–246 [PubMed] [Google Scholar]
- 2.Gottschalk A. (1957) Biochim. Biophys. Acta 23, 645–646 [DOI] [PubMed] [Google Scholar]
- 3.Gottschalk A. (1959) in The Viruses (Burnet F. M., Stanley W. M. eds) Vol. 3, pp. 51–61, Academic Press, New York [Google Scholar]
- 4.Sauter N. K., Bednarski M. D., Wurzburg B. A., Hanson J. E., Whitesides G. M., Skehel J. J., Wiley D. C. (1989) Biochemistry 28, 8388–8396 [DOI] [PubMed] [Google Scholar]
- 5.Takemoto D. K., Skehel J. J., Wiley D. C. (1996) Virology 217, 452–458 [DOI] [PubMed] [Google Scholar]
- 6.Gambaryan A. S., Tuzikov A. B., Piskarev V. E., Yamnikova S. S., Lvov D. K., Robertson J. S., Bovin N. V., Matrosovich M. N. (1997) Virology 232, 345–350 [DOI] [PubMed] [Google Scholar]
- 7.Palese P., Tobita K., Ueda M., Compans R. W. (1974) Virology 61, 397–410 [DOI] [PubMed] [Google Scholar]
- 8.Liu C., Eichelberger M. C., Compans R. W., Air G. M. (1995) J. Virol. 69, 1099–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirst G. K. (1942) J. Exp. Med. 75, 49–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Webster R. G., Laver W. G. (1967) J. Immunol. 99, 49–55 [PubMed] [Google Scholar]
- 11.Seto J. T., Rott R. (1966) Virology 30, 731–737 [DOI] [PubMed] [Google Scholar]
- 12.Bean W. J., Schell M., Katz J., Kawaoka Y., Naeve C., Gorman O., Webster R. G. (1992) J. Virol. 66, 1129–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.W. H. O. (1980) Bull. W.H.O. 58, 585–5916969132 [Google Scholar]
- 14.Air G. M. (1981) Proc. Natl. Acad. Sci. U.S.A. 78, 7639–7643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nobusawa E., Aoyama T., Kato H., Suzuki Y., Tateno Y., Nakajima K. (1991) Virology 182, 475–485 [DOI] [PubMed] [Google Scholar]
- 16.Russell R. J., Haire L. F., Stevens D. J., Collins P. J., Lin Y. P., Blackburn G. M., Hay A. J., Gamblin S. J., Skehel J. J. (2006) Nature 443, 45–49 [DOI] [PubMed] [Google Scholar]
- 17.Wright P. F., Neumann G., Kawaoka Y. (2006) in Fields Virology (Knipe D. M., Howley P. M. eds) 5th Ed., pp. 1691–1740, Lippincott Williams & Wilkins, Philadelphia [Google Scholar]
- 18.Reid A. H., Fanning T. G., Hultin J. V., Taubenberger J. K. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 1651–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawaoka Y., Krauss S., Webster R. G. (1989) J. Virol. 63, 4603–4608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coleman M. T., Dowdle W. R., Pereira H. G., Schild G. C., Chang W. K. (1968) Lancet 2, 1384–1386 [DOI] [PubMed] [Google Scholar]
- 21.Garten R. J., Davis C. T., Russell C. A., Shu B., Lindstrom S., Balish A., Sessions W. M., Xu X., Skepner E., Deyde V., Okomo-Adhiambo M., Gubareva L., Barnes J., Smith C. B., Emery S. L., Hillman M. J., Rivailler P., Smagala J., de Graaf M., Burke D. F., Fouchier R. A., Pappas C., Alpuche-Aranda C. M., López-Gatell H., Olivera H., López I., Myers C. A., Faix D., Blair P. J., Yu C., Keene K. M., Dotson P. D., Jr., Boxrud D., Sambol A. R., Abid S. H., St George K., Bannerman T., Moore A. L., Stringer D. J., Blevins P., Demmler-Harrison G. J., Ginsberg M., Kriner P., Waterman S., Smole S., Guevara H. F., Belongia E. A., Clark P. A., Beatrice S. T., Donis R., Katz J., Finelli L., Bridges C. B., Shaw M., Jernigan D. B., Uyeki T. M., Smith D. J., Klimov A. I., Cox N. J. (2009) Science 325, 197–20119465683 [Google Scholar]
- 22.Kendal A. P., Noble G. R., Skehel J. J., Dowdle W. R. (1978) Virology 89, 632–636 [DOI] [PubMed] [Google Scholar]
- 23.Nakajima K., Desselberger U., Palese P. (1978) Nature 274, 334–339 [DOI] [PubMed] [Google Scholar]
- 24.Meijer A., Lackenby A., Hungnes O., Lina B., van-der-Werf S., Schweiger B., Opp M., Paget J., van-de-Kassteele J., Hay A., Zambon M. (2009) Emerg. Infect. Dis. 15, 552–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skehel J. J., Wiley D. C. (2000) Annu. Rev. Biochem. 69, 531–569 [DOI] [PubMed] [Google Scholar]
- 26.Ha Y., Stevens D. J., Skehel J. J., Wiley D. C. (2002) EMBO J. 21, 865–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Russell R. J., Gamblin S. J., Haire L. F., Stevens D. J., Xiao B., Ha Y., Skehel J. J. (2004) Virology 325, 287–296 [DOI] [PubMed] [Google Scholar]
- 28.Russell R. J., Kerry P. S., Stevens D. J., Steinhauer D. A., Martin S. R., Gamblin S. J., Skehel J. J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 17736–17741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garten W., Klenk H. D. (1999) Trends Microbiol. 7, 99–100 [DOI] [PubMed] [Google Scholar]
- 30.Steinhauer D. A. (1999) Virology 258, 1–20 [DOI] [PubMed] [Google Scholar]
- 31.Böttcher E., Matrosovich T., Beyerle M., Klenk H. D., Garten W., Matrosovich M. (2006) J. Virol. 80, 9896–9898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perdue M. L., García M., Senne D., Fraire M. (1997) Virus Res. 49, 173–186 [DOI] [PubMed] [Google Scholar]
- 33.Stieneke-Gröber A., Vey M., Angliker H., Shaw E., Thomas G., Roberts C., Klenk H. D., Garten W. (1992) EMBO J. 11, 2407–2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klenk H. D., Rott R. (1988) Adv. Virus Res. 34, 247–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ha Y., Stevens D. J., Skehel J. J., Wiley D. C. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 11181–11186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weis W., Brown J. H., Cusack S., Paulson J. C., Skehel J. J., Wiley D. C. (1988) Nature 333, 426–431 [DOI] [PubMed] [Google Scholar]
- 37.Sauter N. K., Hanson J. E., Glick G. D., Brown J. H., Crowther R. L., Park S. J., Skehel J. J., Wiley D. C. (1992) Biochemistry 31, 9609–9621 [DOI] [PubMed] [Google Scholar]
- 38.Eisen M. B., Sabesan S., Skehel J. J., Wiley D. C. (1997) Virology 232, 19–31 [DOI] [PubMed] [Google Scholar]
- 39.Ito T., Suzuki Y., Mitnaul L., Vines A., Kida H., Kawaoka Y. (1997) Virology 227, 493–499 [DOI] [PubMed] [Google Scholar]
- 40.Carroll S. M., Higa H. H., Paulson J. C. (1981) J. Biol. Chem. 256, 8357–8363 [PubMed] [Google Scholar]
- 41.Gambaryan A. S., Matrosovich M. N. (1992) J. Virol. Methods 39, 111–123 [DOI] [PubMed] [Google Scholar]
- 42.Blixt O., Head S., Mondala T., Scanlan C., Huflejt M. E., Alvarez R., Bryan M. C., Fazio F., Calarese D., Stevens J., Razi N., Stevens D. J., Skehel J. J., van Die I., Burton D. R., Wilson I. A., Cummings R., Bovin N., Wong C. H., Paulson J. C. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 17033–17038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stevens J., Blixt O., Paulson J. C., Wilson I. A. (2006) Nat. Rev. Microbiol. 4, 857–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rogers G. N., Paulson J. C. (1983) Virology 127, 361–373 [DOI] [PubMed] [Google Scholar]
- 45.Rogers G. N., D'Souza B. L. (1989) Virology 173, 317–322 [DOI] [PubMed] [Google Scholar]
- 46.Connor R. J., Kawaoka Y., Webster R. G., Paulson J. C. (1994) Virology 205, 17–23 [DOI] [PubMed] [Google Scholar]
- 47.Matrosovich M. N., Gambaryan A. S., Teneberg S., Piskarev V. E., Yamnikova S. S., Lvov D. K., Robertson J. S., Karlsson K. A. (1997) Virology 233, 224–234 [DOI] [PubMed] [Google Scholar]
- 48.Ito T., Couceiro J. N., Kelm S., Baum L. G., Krauss S., Castrucci M. R., Donatelli I., Kida H., Paulson J. C., Webster R. G., Kawaoka Y. (1998) J. Virol. 72, 7367–7373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baum L. G., Paulson J. C. (1990) Acta Histochem. Suppl. 40, 35–38 [PubMed] [Google Scholar]
- 50.Couceiro J. N., Paulson J. C., Baum L. G. (1993) Virus Res. 29, 155–165 [DOI] [PubMed] [Google Scholar]
- 51.Shinya K., Ebina M., Yamada S., Ono M., Kasai N., Kawaoka Y. (2006) Nature 440, 435–436 [DOI] [PubMed] [Google Scholar]
- 52.Matrosovich M. N., Matrosovich T. Y., Gray T., Roberts N. A., Klenk H. D. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 4620–4624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gambaryan A., Yamnikova S., Lvov D., Tuzikov A., Chinarev A., Pazynina G., Webster R., Matrosovich M., Bovin N. (2005) Virology 334, 276–283 [DOI] [PubMed] [Google Scholar]
- 54.Gambaryan A. S., Tuzikov A. B., Pazynina G. V., Webster R. G., Matrosovich M. N., Bovin N. V. (2004) Virology 326, 310–316 [DOI] [PubMed] [Google Scholar]
- 55.Pritchett T. J., Paulson J. C. (1989) J. Biol. Chem. 264, 9850–9858 [PubMed] [Google Scholar]
- 56.Matrosovich M., Tuzikov A., Bovin N., Gambaryan A., Klimov A., Castrucci M. R., Donatelli I., Kawaoka Y. (2000) J. Virol. 74, 8502–8512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rogers G. N., Paulson J. C., Daniels R. S., Skehel J. J., Wilson I. A., Wiley D. C. (1983) Nature 304, 76–78 [DOI] [PubMed] [Google Scholar]
- 58.Daniels P. S., Jeffries S., Yates P., Schild G. C., Rogers G. N., Paulson J. C., Wharton S. A., Douglas A. R., Skehel J. J., Wiley D. C. (1987) EMBO J. 6, 1459–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hardy C. T., Young S. A., Webster R. G., Naeve C. W., Owens R. J. (1995) Virology 211, 302–306 [DOI] [PubMed] [Google Scholar]
- 60.Gambaryan A. S., Robertson J. S., Matrosovich M. N. (1999) Virology 258, 232–239 [DOI] [PubMed] [Google Scholar]
- 61.Gambaryan A. S., Marinina V. P., Tuzikov A. B., Bovin N. V., Rudneva I. A., Sinitsyn B. V., Shilov A. A., Matrosovich M. N. (1998) Virology 247, 170–177 [DOI] [PubMed] [Google Scholar]
- 62.Stevens J., Blixt O., Glaser L., Taubenberger J. K., Palese P., Paulson J. C., Wilson I. A. (2006) J. Mol. Biol. 355, 1143–1155 [DOI] [PubMed] [Google Scholar]
- 63.Stevens J., Blixt O., Tumpey T. M., Taubenberger J. K., Paulson J. C., Wilson I. A. (2006) Science 312, 404–410 [DOI] [PubMed] [Google Scholar]
- 64.Stevens J., Blixt O., Chen L. M., Donis R. O., Paulson J. C., Wilson I. A. (2008) J. Mol. Biol. 381, 1382–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xu R., McBride R., Paulson J. C., Basler C. F., Wilson I. A. (2010) J. Virol. 84, 1715–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gao Y., Zhang Y., Shinya K., Deng G., Jiang Y., Li Z., Guan Y., Tian G., Li Y., Shi J., Liu L., Zeng X., Bu Z., Xia X., Kawaoka Y., Chen H. (2009) PLoS Pathog. 5, e1000709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tumpey T. M., Maines T. R., Van Hoeven N., Glaser L., Solórzano A., Pappas C., Cox N. J., Swayne D. E., Palese P., Katz J. M., García-Sastre A. (2007) Science 315, 655–659 [DOI] [PubMed] [Google Scholar]
- 68.Gamblin S. J., Haire L. F., Russell R. J., Stevens D. J., Xiao B., Ha Y., Vasisht N., Steinhauer D. A., Daniels R. S., Elliot A., Wiley D. C., Skehel J. J. (2004) Science 303, 1838–1842 [DOI] [PubMed] [Google Scholar]
- 69.Liu J., Stevens D. J., Haire L. F., Walker P. A., Coombs P. J., Russell R. J., Gamblin S. J., Skehel J. J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 17175–17180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ha Y., Stevens D. J., Skehel J. J., Wiley D. C. (2003) Virology 309, 209–218 [DOI] [PubMed] [Google Scholar]
- 71.Yamada S., Suzuki Y., Suzuki T., Le M. Q., Nidom C. A., Sakai-Tagawa Y., Muramoto Y., Ito M., Kiso M., Horimoto T., Shinya K., Sawada T., Kiso M., Usui T., Murata T., Lin Y., Hay A., Haire L. F., Stevens D. J., Russell R. J., Gamblin S. J., Skehel J. J., Kawaoka Y. (2006) Nature 444, 378–382 [DOI] [PubMed] [Google Scholar]
- 72.Varghese J. N., Laver W. G., Colman P. M. (1983) Nature 303, 35–40 [DOI] [PubMed] [Google Scholar]
- 73.Burmeister W. P., Ruigrok R. W., Cusack S. (1992) EMBO J. 11, 49–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Colman P. M., Varghese J. N., Laver W. G. (1983) Nature 303, 41–44 [DOI] [PubMed] [Google Scholar]
- 75.Varghese J. N., McKimm-Breschkin J. L., Caldwell J. B., Kortt A. A., Colman P. M. (1992) Proteins 14, 327–332 [DOI] [PubMed] [Google Scholar]
- 76.Varghese J. N., Epa V. C., Colman P. M. (1995) Protein Sci. 4, 1081–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.von Itzstein M. (2007) Nat. Rev. Drug Discov. 6, 967–974 [DOI] [PubMed] [Google Scholar]
- 78.Colman P. M. (2009) Annu. Rev. Biochem. 78, 95–118 [DOI] [PubMed] [Google Scholar]
- 79.Monto A. S., Kendal A. P. (1973) Lancet 1, 623–625 [DOI] [PubMed] [Google Scholar]
- 80.Colman P. M. (1989) The Influenza Viruses, Plenum Publishing Corp., New York [Google Scholar]
- 81.McKimm-Breschkin J. L. (2000) Antiviral Res. 47, 1–17 [DOI] [PubMed] [Google Scholar]
- 82.Monto A. S., McKimm-Breschkin J. L., Macken C., Hampson A. W., Hay A., Klimov A., Tashiro M., Webster R. G., Aymard M., Hayden F. G., Zambon M. (2006) Antimicrob. Agents Chemother. 50, 2395–2402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tashiro M., McKimm-Breschkin J. L., Saito T., Klimov A., Macken C., Zambon M., Hayden F. G. (2009) Antiviral Ther. 14, 751–761 [DOI] [PubMed] [Google Scholar]
- 84.Collins P. J., Haire L. F., Lin Y. P., Liu J., Russell R. J., Walker P. A., Skehel J. J., Martin S. R., Hay A. J., Gamblin S. J. (2008) Nature 453, 1258–1261 [DOI] [PubMed] [Google Scholar]
- 85.Varghese J. N., Smith P. W., Sollis S. L., Blick T. J., Sahasrabudhe A., McKimm-Breschkin J. L., Colman P. M. (1998) Structure 6, 735–746 [DOI] [PubMed] [Google Scholar]
- 86.Smith B. J., McKimm-Breshkin J. L., McDonald M., Fernley R. T., Varghese J. N., Colman P. M. (2002) J. Med. Chem. 45, 2207–2212 [DOI] [PubMed] [Google Scholar]
- 87.Blick T. J., Sahasrabudhe A., McDonald M., Owens I. J., Morley P. J., Fenton R. J., McKimm-Breschkin J. L. (1998) Virology 246, 95–103 [DOI] [PubMed] [Google Scholar]
- 88.Kaverin N. V., Gambaryan A. S., Bovin N. V., Rudneva I. A., Shilov A. A., Khodova O. M., Varich N. L., Sinitsin B. V., Makarova N. V., Kropotkina E. A. (1998) Virology 244, 315–321 [DOI] [PubMed] [Google Scholar]
- 89.Castrucci M. R., Kawaoka Y. (1993) J. Virol. 67, 759–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mitnaul L. J., Matrosovich M. N., Castrucci M. R., Tuzikov A. B., Bovin N. V., Kobasa D., Kawaoka Y. (2000) J. Virol. 74, 6015–6020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wagner R., Wolff T., Herwig A., Pleschka S., Klenk H. D. (2000) J. Virol. 74, 6316–6323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Baigent S. J., McCauley J. W. (2001) Virus Res. 79, 177–185 [DOI] [PubMed] [Google Scholar]
- 93.Knossow M., Skehel J. J. (2006) Immunology 119, 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Colman P. M. (1988) Adv. Immunol. 43, 99–132 [DOI] [PubMed] [Google Scholar]
- 95.Khurana S., Suguitan A. L., Jr., Rivera Y., Simmons C. P., Lanzavecchia A., Sallusto F., Manischewitz J., King L. R., Subbarao K., Golding H. (2009) PLoS Med. 6, e1000049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sui J., Hwang W. C., Perez S., Wei G., Aird D., Chen L. M., Santelli E., Stec B., Cadwell G., Ali M., Wan H., Murakami A., Yammanuru A., Han T., Cox N. J., Bankston L. A., Donis R. O., Liddington R. C., Marasco W. A. (2009) Nat. Struct. Mol. Biol. 16, 265–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ekiert D. C., Bhabha G., Elsliger M. A., Friesen R. H., Jongeneelen M., Throsby M., Goudsmit J., Wilson I. A. (2009) Science 324, 246–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hayden F. (2009) Clin. Infect. Dis. 48, Suppl. 1, S3–S13 [DOI] [PubMed] [Google Scholar]